Abstract
The crystal structure of Mycobacterium tuberculosis D-3-phosphoglycerate dehydrogenase has been solved with bound effector, L-serine, and substrate, hydroxypyruvic acid phosphate, at resolutions of 2.7 Å and 2.4 Å, respectively. The structures display the same extreme asymmetry of the subunits as seen in the apo-structure and provide insight into the mode of serine binding and closure of the active site. Mutagenesis studies confirm the identity of the main residues involved in serine binding and suggest that the poly-glycine stretch in the loop that contains the locus for the 160° rotation that leads to subunit asymmetry may have a larger role in folding than in catalysis. The lack of electron density for the cofactor, NADH, in any of the crystals examined led us to study binding by stopped flow kinetic analysis. The kinetic data suggest that productive NADH binding, that would support catalytic turnover, is dependent on the presence of substrate. This observation, along with the binding of substrate at the correct place in the active site, but in an unproductive conformation, suggests a possible mechanism where initial binding of substrate leads to enhanced interaction with cofactor accompanied by a rearrangement of catalytically critical residue side chains. Furthermore, comparison to the structure of a truncated form of human D-3-phosphoglycerate dehydrogenase with cofactor and a substrate analog, provides insight into the conformational changes that occur during catalysis.
D-3-Phosphoglycerate dehydrogenase (PGDH, EC 1.1.1.95) from Mycobacterium tuberculosis (M.tb), a tetramer of identical polypeptide chains (1), is responsible for the reversible NAD+/ NADH dependent interconversion of D-3-phosphoglyceric acid and hydroxypyruvic acid phosphate. Each polypeptide chain consists of four distinct domains. Three of these domains, the nucleotide binding domain, the substrate binding domain, and the regulatory domain, are analogous to those found in E.coli PGDH (2, 3) and contain the primary binding determinants for the ligand reflected in their names. The fourth domain, called the intervening domain, is not present in E.coli PGDH, but rather intervenes between the substrate binding domain and the regulatory domain. This domain is also present in PGDH from B. subtilis, M. jannaschii, human and rat. Organisms whose PGDH lack this intervening domain include H. influenza and the eukaryotes S. cerevisiae, Leishmania and Neurospora.
The presence of the intervening domain changes the tertiary organization of this enzyme in comparison to E.coli PGDH. The crystal structure of M.tb PGDH apoprotein (1) showed that instead of the tetramer being composed of subunits with identical or similar structure, extreme asymmetry is observed in the orientation of the domains from one subunit to another. That is, in two of the subunits, the intervening and regulatory domains are rotated approximately 160° relative to the other two subunits. This results in the presence of two different conformations among the molecules of the asymmetric unit and within the subunits of the tetramer. As a result of this asymmetrical configuration, the face of the intervening domain in one subunit that is exposed to solvent is facing toward the interior of the tetramer in the other subunit. The nucleotide binding and substrate binding domains assume the same general orientation to each other as observed for E.coli PGDH. These two domains are joined to the intervening and regulatory domains by a long loop where the rotation takes place resulting in the inversion of the intervening domain-regulatory domain unit. Despite this asymmetry, the regulatory domains from adjacent subunits contact each other forming an eight-stranded anti-parallel β sheet, as seen in the E.coli enzyme (2, 3), which lead to formation of the biological tetramer. In E.coli PGDH, the effector, L-serine, binds at the interface between regulatory domains, which are members of the ACT domain family (4). ACT domains function in the regulation of a variety of proteins in response to the binding of small molecules which are often amino acids but also as diverse as nickel ions and the B vitamin, thiamine.
In addition to the active site where substrate and cofactor would bind, and the effector binding site for L-serine, the structure of M.tb PGDH revealed a potential fourth ligand binding site, called the anion binding site (1). The presence of this putative site was revealed because tartrate, a component in the crystallization buffer, was found at this site interacting with the side chains of several basic residues from both the intervening and regulatory domains within a subunit. However, tartrate is most likely not the physiological ligand since it is found to be involved in dicarboxylate metabolism not present in M.tb. Recently, mutation studies on the residues of this site (5) were consistent with it being a second binding site for the substrate resulting in substrate inhibition of the enzyme.
M.tb PGDH has an IC50 for L-serine of approximately 30 µM while E. coli PGDH has an IC50 of approximately 3 µM (6). PGDH from B. subtilis (7) and C. glutamicum (8) have been reported to be much less sensitive to serine with IC50 values of ~0.6mM and ~10mM, respectively. In contrast, serine has no effect on the enzyme activity of rat liver (9) and chicken liver PGDH (10). Human PGDH shares 94% sequence identity with rat liver PGDH and also is not affected by serine (unpublished result).
Two other crystal structures of PGDH have been deposited in the protein data bank since the structure determination of M.tb PGDH. No additional information is available for these structures. One is from Pyrococcus horikoshii (PDBcode-1WWK) that displays the simplest domain organization consisting only of a nucleotide and a substrate binding domain and exists as a dimer. Very little information is available about the substrate or effector recognition of the P. horikoshii enzyme. The other is a truncated form of human PGDH. This enzyme normally contains the same four domains found in M. tb PGDH. However, this truncated form consists of the nucleotide binding domain and the substrate binding domain (PDBcode-2G76) but lacks the intervening and regulatory domains.
In this study, we report the crystal structures of binary complexes of M.tb PGDH with substrate, hydroxypyruvic acid phosphate (HPAP) and with inhibitor, L-serine. These structures will be helpful in understanding the mechanism of enzyme action as well as pinpointing the residues interacting with the effector molecule and subsequent changes involved in allosteric inhibition in this enzyme in mycobacterium.
Experimental Procedures
M.tb PGDH was purified using the procedure described earlier (1). HPAP was produced from its dimethylketal tri-cyclohexylammonium salt (Sigma) using the procedure provided by the supplier. Small amounts were converted and stored frozen at − 20 ° C. The active concentration of HPAP was determined prior to performing the assay by monitoring the amount of NADH that is oxidized by limiting amounts of HPAP under conditions where the equilibrium lies far in the direction of HPAP reduction. NADH and the NAD analog (3-acetyl pyridine adenine dinucleotide) were also purchased from Sigma.
Crystallization and Structure Determination
Crystallization trials of PGDH (10 mg/ml) with 5 mM HPAP and 5 mM NAD analog, 3-acetyl pyridine adenine dinucleotide, and PGDH with 5 mM NADH and 5 mM L-serine were carried out using matrices from Hampton Research (crystal screen I and II, PEG/Ion screen) and Emerald Biosystems (Wizard I and II). There were several conditions that resulted in crystal formation with (NH4)2SO4 as the common precipitant, but they diffracted poorly. However, the crystals obtained in 1M Na K tartrate, 0.1M MES, pH 6.5, diffracted to good resolution. The datasets were collected at 100 ° K using crystals cryoprotected in 25% propylene glycol in mother liquor at 14BMC, Advanced Photon Source, Argonne National Laboratory, Chicago. The datasets were processed and scaled using the HKL2000 suite (11). The space group was found to be P6522.
As all the datasets possessed cell dimensions isomorphous to the apoenzyme structure (1, TABLE I), the M.tb PGDH apostructure (PDB code-1YGY) was refined against these datasets using CNS (rigid body refinement and simulated annealing refinement at 4000 ° K) (12) to minimize the model biasness as well as improve the geometry. These refined models were submitted to Shake&wARP (13) to get a bias minimized map. Further, iterative model building was performed using the Xfit module of Xtalview (14) and refined using REFMAC (15). As the models reached good statistics (i.e. R factor and R free below 30%), water molecules were added based on peaks at the 3σ level in difference map (Fobs −Fcal). Both the structures have good geometry as analyzed by PROCHECK (16). Data collection and refinement statistics are provided in Table I.
TABLE I.
Data collection, refinement and geometry statistics
| PGDH-HPAP | PGDH-serine | |
|---|---|---|
| Data collection | ||
| Space group | P6522 | P6522 |
| Unit cell dimensions | a=b=165.6 Å c=218.3Å |
a=b=165.2 Å c=218.9 Å |
| Molecules per ASUa | 2 | 2 |
| Wavelength (Å) | 0.9 | 0.9 |
| Resolution range (Å) | 48 – 2.4 | 46.5 – 2.7 |
| Highest resolution bin (Å) | 2.49 – 2.4 | 2.8 – 2.7 |
| Observed reflections | 1495125 | 1011203 |
| Unique reflections | 69375 | 48518 |
| Completeness (%)c | 100 (100) | 99.2 (99.9) |
| Average redundancyc | 21.6 (22.3) | 20.8 (19.5) |
| I/σ(I)c | 26 (3) | 19.2 (2.8) |
| Rsym c,d | 0.038 (0.3) | 0.037 (0.42) |
| Refinement statistics (REFMAC) | ||
| R value (%)e | 20 | 22 |
| Free R value (5%) | 24 | 26 |
| No. of protein residues | 1052 | 1049 |
| No. of water molecules | 165 | 77 |
| r.m.s.d.b bond length, (Å) | 0.011 | 0.016 |
| r.m.s.d.b bond angles, (°) | 1.42 | 1.75 |
| Ramchandran plot (PROCHECK) | ||
| Most favored region (%) | 819 (90) | 775 (85.4) |
| Additional allowed regions (%) | 89 (9.8) | 130 (14.3) |
| Generously allowed regions (%) | 2 (0.2) | 2 (0.2) |
| Disallowed regions (%) | 0 | 0 |
ASU - Asymmetric unit
r.m.s.d. - root mean square deviation
Values in parenthesis for the highest resolution bin
Rsym = Σ|I − <I>| /Σ<I>, where I is the observed intensity, and <I> is the average intensity of multiple observations of symmetry-related reflections.
R = Σ ||Fobs|−|Fcalc||/ Σ |Fobs| where Fobs and Fcalc are the observed and calculated structure factors
Steady-State Kinetic Analysis
The physiologic direction of the reaction catalyzed by PGDH is in the direction of phosphoglyceric acid oxidation and NAD+ reduction. However, the equilibrium of the reaction lies far in the direction of NADH oxidation, in vitro (17). Therefore, the enzyme is routinely assayed in this direction by measuring the decrease in absorbance at 340 nm during the conversion of NADH to NAD+ in the presence of substrate, HPAP. All assays were conducted in 200 mM potassium phosphate, pH 7.5, 1 mM EDTA, 1 mM dithiothreitol, 1 mM KCl.
Native PGDH as well as all mutants used in this investigation displayed substrate inhibition (5). The kinetic parameters were determined using the equation for complete uncompetitive substrate inhibition (18),
| (Eq. 1) |
where Vmax is the maximum velocity, K is the Km, and Ki is the inhibition constant. Serine inhibition plots were fit to the Hill equation (19, 20),
| (Eq. 2) |
where I is the fractional inhibition, L is the concentration of ligand, n is the Hill coefficient, and I 0.5 is the inhibitor concentration at 1/2 maximal inhibition. An increase in the I 0.5 indicates a decreased sensitivity to serine and is referred to as such in the text. All mutants could be inhibited to greater than 95% with saturating levels of serine.
Stopped-flow Analysis
Pre-steady state kinetic analyses were performed using an Applied Photophysics Model SX-20 stopped-flow spectrometer. When NADH binds to PGDH, a fluorescence resonance energy transfer (FRET) occurs between protein tryptophans and the bound NADH. This can be monitored by excitation of the protein tryptophan at 295 nm and observing either the decrease in tryptophan fluorescence at 340 nm or the increase in NADH fluorescence at 420 nm when NADH binds to the protein. In either case, emission fluorescence was monitored using a 10 nm band pass filter of the appropriate wavelength. Enzyme, in the absence of substrate, was rapidly mixed with varying concentrations of NADH as indicated in the figures. When substrate is included, the enzyme was rapidly mixed with the designated concentrations of NADH and HPAP. The enzyme subunit concentration used was 2 µM. A total of 10,000 points were collected in each trace and 5–18 individual traces were averaged at each set of conditions. The reaction and reagents were kept at a constant 25° C using a circulating water bath.
The binding transients were analyzed using software provided by the manufacturer or with Kaleidograph (Synergy Corp.) to exponential functions defined as
| (Eq. 3) |
where Y is the fluorescence intensity at time t, kobs i is the observed rate of the ith process with an amplitude of Ai and C is an offset value (21). Data that produce linear plots with a non-zero slope plus an apparent concentration independent component conform to a two step process that includes binding followed by a conformational change (21),
where
| (Eq. 4) |
and
| (Eq. 5) |
At high concentrations of [S], k2 + k−2 ≈ the maximum rate of the second step. Thus, kon for ligand binding is represented by k1, which is the slope of the kobs vs. ligand concentration plots. The rate of ligand dissociation, or koff, is represented by k−1 and equals the y intercept of the first step minus the maximum rate of the second step (i.e. k−1 = (k−1 + k2 + k−2) − (k2 + k−2) ).
Site-Specific Mutagenesis
The double stranded plasmid containing the M. tb serA1 gene coding for PGDH was isolated using a Column -Pure™ Plasmid Miniprep Kit from Lamda Biotech. Site-specific mutagenesis was performed by standard PCR methods (22). PCR mutagenesis was carried out in two different ways, depending on the proximity of the mutation site to a unique restriction site. In the case of the Y461A, D463A, and N 481A mutations, where the mutation sites were a considerable distance from usable restriction sites, four oligonucleotide primers were used for each mutation (see supplementary material). Primers A and B were made complementary to each DNA strand at unique restriction sites that flanked the area of intended mutation and contained the restriction site sequence. Primers C and D were made complementary to each DNA strand at the site of the intended mutation and contained the mutant sequence. Amplification of the DNA template was carried out with primers A and C and with primers B and D in separate tubes using the 2x Taq Plus Master Mix, Red from Lamda Biotech. Twenty-five amplification cycles were performed with a denaturation temperature of 94 °C, an annealing temperature of 50 °C, and an extension temperature of 72 °C. A second amplification was carried out using the products of the first amplifications combined with primer A and B. The resulting PCR products were purified with a Column-Pure™ PCR Clean-up Kit from Lamda Biotech. For the Gly316–318 mutants, where a usable restriction site was close to the mutation site, only two oligonucleotide primers were used (see supplementary material). One primer, Primer E, contained one restricition site as well as the mutations. The other primer, primer F, contained the other restriction site. A single PCR run was performed with the appropriate template and primers E and F. All other procedures were as described above.
The final PCR product and the plasmid were both digested with enzymes (EcoRI and HindIII) for the flanking restriction sites and the digestion products were separated on agarose gels. The plasmid DNA, with the native DNA sequence removed, and the PCR product were recovered from the gel with a Column-Pure™ DNA Gel Recovery Kit from Lamda Biotech. The PCR product containing the mutation was ligated into the expression vector and the sequence of the entire coding region of the protein was determined to be correct by sequencing on an Applied Biosystems 3730 DNA Sequencer. Mutant proteins were purified in the same manner as native enzyme (1). Protein purity was assessed on SDS-PAGE gels and all mutants were present as predominant single bands of the appropriate size.
Gel Filtration Chromatography
Gel filtration was performed at ambient temperature on a 1.6 × 115 cm column of Sephacryl S-300 (Sigma) in 200 mM potassium phosphate buffer, pH 7.5, at a flow rate of 1 ml/min. Fractions were collected every 2 minutes and the protein elution volume was determined from the fraction volumes. The void volume of the column was determined with blue dextran.
Results
The crystal structures of binary complexes of M.tb PGDH with substrate, HPAP, and allosteric inhibitor, L-serine, have been solved at resolutions of 2.4 Å and 2.7 Å, respectively. Both of these structures contain bound tartrate at the intervening domain and have two molecules in the asymmetric unit that forms a tetramer with its symmetry related molecule. The secondary structure folds are similar to the structure of the apoprotein described earlier (1). The data collection, refinement, and geometry statistics are presented in Table 1.
Architecture of the Active site
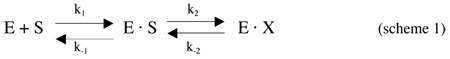
Each subunit has its own distinct active site with the distance between active sites in the dimer being 32 Å. In M.tb PGDH, the active site is lined by the loop regions Arg51-Asp57, Ala73-Val80, Pro279-Thr285, Ala231-Asp238, and Asp257-Thr265 (Figure 1). In addition, Ile374-Phe384 border the active site of side chain A (not shown). These residues are 76% conserved in E.coli PGDH (23) and 88% in human PGDH (24). The side chains of the basic residues of these loop regions are predominantly oriented in the active site for binding the negatively charged substrates of this enzyme. In addition to this site being formed by the cleft between the nucleotide and substrate binding domains, a loop region from the adjacent subunit (residues Leu125′-Lys131′) also contributes to its formation. This loop contains the conserved Trp residue (Trp130′) that supports one side of the hydrophobic pocket containing the catalytic dyad formed by the hydrogen bonding of Glu262-OE1 with His280-ND1 (2.7 Å) in M.tb PGDH. In E.coli PGDH, the residues in this loop (Asn140-Gly145) are dominated by Ala and Gly and presumably provide flexibility to this region. Mutagenesis studies in E.coli PGDH suggest this region to be important for the cooperativity of serine inhibition of catalytic activity (25, 26). In M.tb PGDH, this region is dominated by polar residues like Lys and Ser. Interestingly, in human PGDH, which is not inhibited by L-serine, this region is occupied by bulky side chains such as Lys, Phe and Met, suggesting it may be less flexible.
Figure 1. M.tb PGDH active site.
The figure shows the loops bordering the active site of M. tb PGDH in chain B in purple. The active site is formed by both chains. The nucleotide and substrate binding domain of chain B are shown in dark green. The intervening and regulatory domains are not shown for clarity. The nucleotide binding domain of chain A is in gold. The substrate, HPAP, is shown in space filling representation bound at the active site (carbon atoms are white, oxygen atoms are red, and phosphorus is cyan).
Substrate binding
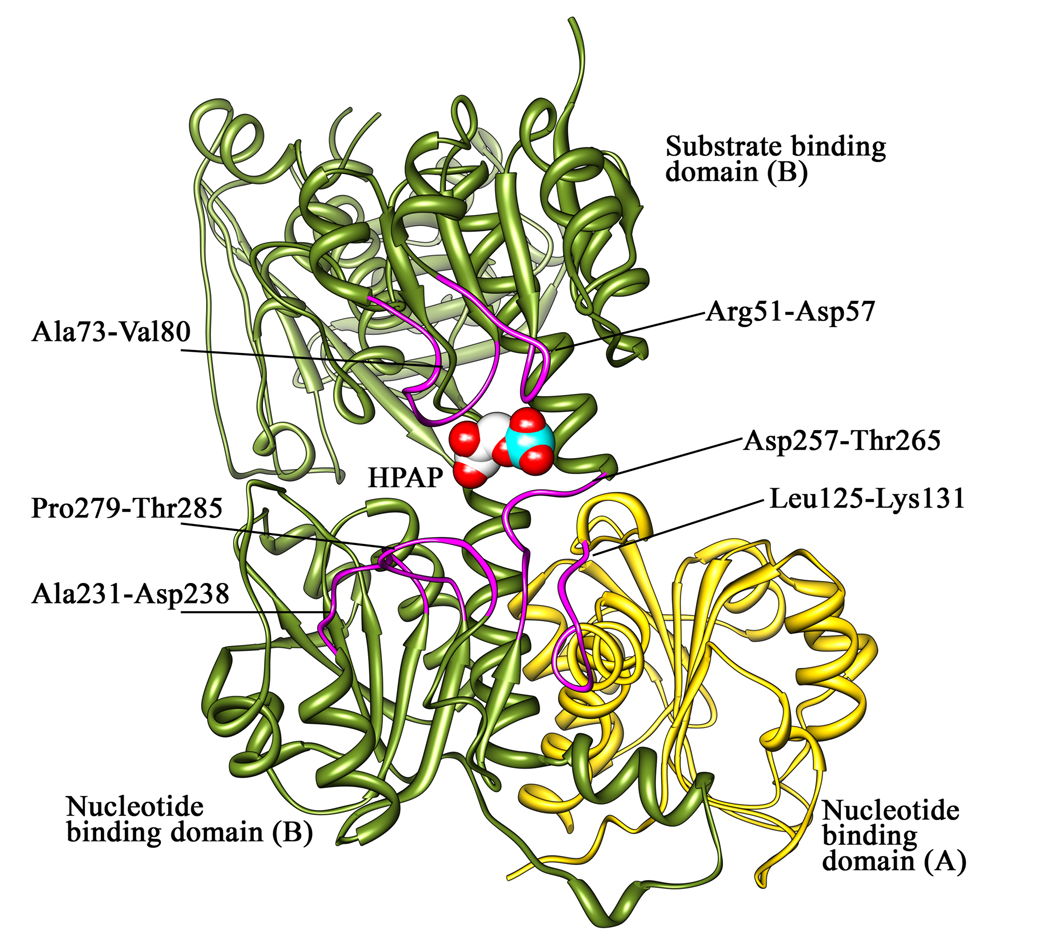
In the HPAP bound M.tb PGDH structure, the density for HPAP is visible only in chain B (Figure 1 and Figure 2). In this structure, the phosphate group of HPAP forms hydrogen bonds with Arg51-NE (3.5Å) and Gln289-OE1 (2.6 Å) from one subunit, and Arg132′-NH1 from the adjacent subunit (2.8 Å). The carboxyl group of the substrate is hydrogen bonded to Asn99-ND2 (3.16 Å) and the keto group of HPAP interacts with the main chain amide of Gly76 (3.08 Å). On comparing this HPAP binding with malate binding in the human PGDH structure, the HPAP is oriented differently, with the keto group pointing away from the catalytic His280. Also not seen is the expected interaction of the substrate C1-carboxyl group with an arginine residue as seen in the structure of human PGDH (Arg235) and many other dehydrogenases (27). Arg233 is a conserved residue in M.tb PGDH that corresponds to Arg235 in human PGDH, but in M.tb PGDH, the Arg233 side chain is facing away from the active site. In fact, in chain A, the side chain of Arg233 forms a salt bridge with Glu379 whereas in chain B it appears to be flexible, resulting in reduced density for the side chain. Thus, in the absence of cofactor, HPAP is not properly oriented for the enzyme to carry out catalysis in this structure. It may be possible that the presence of the cofactor plays a key role in the proper positioning of the substrate for hydride transfer to His280 by stabilizing the formation of a salt bridge between Arg233 and the carboxyl group of the substrate.
Figure 2. Interaction of HPAP at the M. tb PGDH active site.
This figure shows all the residues involved in the HPAP interaction in chain B of the substrate bound M.tb PGDH structure. Also shown are the catalytic dyad His280:Glu262 and Arg233 in relation to HPAP at the active site. The Shake &wARP unbiased 2Fo−Fc electron density map for HPAP (light blue) is contoured at 1σ level. HPAP is shown within the electron density. Oxygen atoms are shown in red, nitrogen atoms in blue, and phosphorus atoms in cyan.
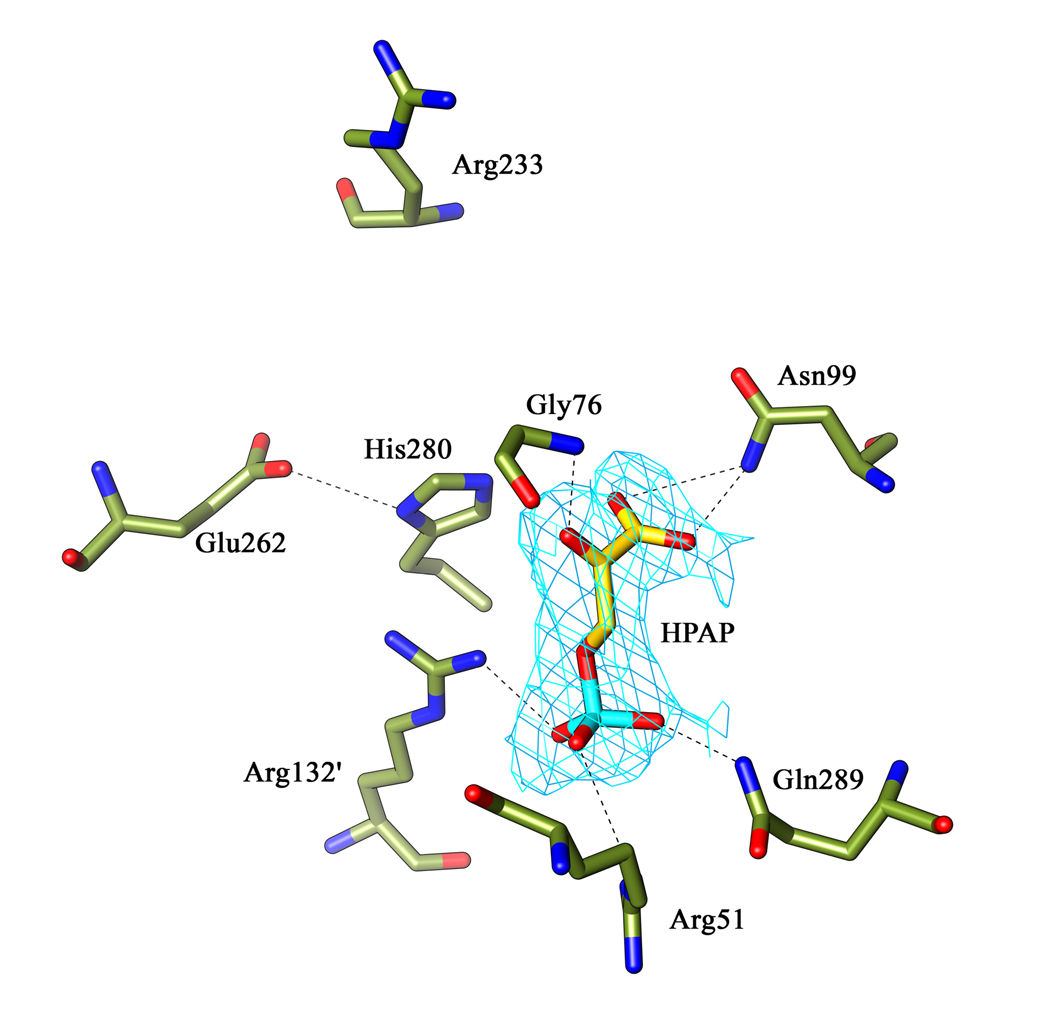
In the human PGDH structure, malate and NAD+ are bound at the active site. Superimposing the M.tb PGDH nucleotide binding and substrate binding domains with the same domains of substrate bound human PGDH (Figure 3) shows that the M.tb structure has a more open active site cleft that can be characterized by a 15° rotation of the substrate binding domain. Even with a more open cleft, the HPAP in M.tb PGDH occupies a position similar to malate in the human PGDH structure, suggesting that the binding of both cofactor and substrate is needed for the active site to close and productive contacts to be made with HPAP.
Figure 3. Model for the active site closure of M.tb PGDH upon substrate binding.
A ribbon diagram of the M.tb. apoenzyme (blue) is superimposed on a modeled ribbon diagram of M.tb. PGDH (gold) based on the malate and NAD bound human PGDH structure (pdb code 2G76). NADH (green) and HPAP (gray) are shown in ball and stick form. As seen in the figure, there is a rotation of approximately 15° (arrow) of the substrate binding domain in response to substrate and cofactor binding. The intervening domain and the regulatory domain are not shown for the sake of clarity.
The Cofactor Binding Site
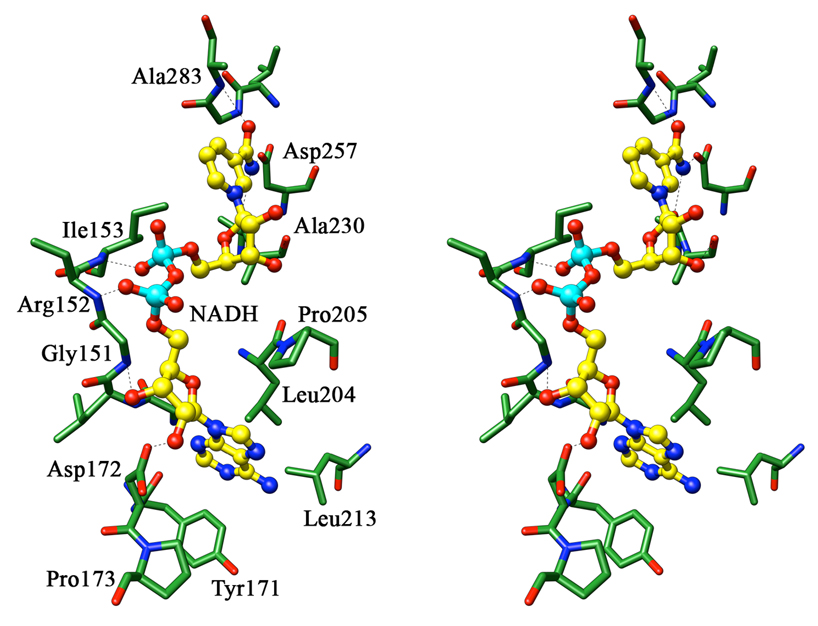
All dehydrogenases that bind NAD/NADH as a cofactor contain the Rossmann fold motif consisting of a GXGXXG…D sequence. Although the crystals were formed in the presence of NADH or its oxidized analog, 3-acetyl pyridine adenine dinucleotide, the density for the cofactor is not visible in any of the M.tb PGDH structures. However, NAD+/NADH can be easily modeled into the M.tb PGDH active site (Figure 4) by superimposing the nucleotide binding domain of either E.coli or human PGDH onto the M.tb PGDH structure as the residues interacting with the cofactor are conserved. The OD1 and OD2 atoms of the conserved Asp residue in the Rossmann fold (Asp172) should form hydrogen bonds with the O2 and O3 atoms of the ribose sugar of the adenosine moiety. The adenine ring of the cofactor is placed in a hydrophobic pocket made of Pro173 and 205, Val148, Tyr171 and Leu204 and 213. The two phosphate atoms connecting the nicotinamide and adenosine parts interact with the main chain amides of Arg152 and Ile153. The multiple Gly residues of the Rossmann fold prevent any spatial interference as the cofactor binds. The N7 atom of the nicotinamide ring forms hydrogen bonds with Asp257-OD1 as well as the main chain oxygen of Ala231.
Figure 4. The cofactor binding site of chain B of M. tb PGDH with cofactor modeled into the site.
A stereo diagram of the interactions of cofactor modeled into the cofactor binding site of chain B of M. tb PGDH is shown. The cofactor is shown in ball and stick representation with carbon atoms in yellow, nitrogen atoms in blue, oxygen atoms in red, and phosphorus atoms in cyan. The protein is shown in green. All hydrogen bonds, represented as dashed lines, are within the distance of 3.5 Å. In this figure, the side chain of Arg152 has been truncated in order to prepare this stereo representation.
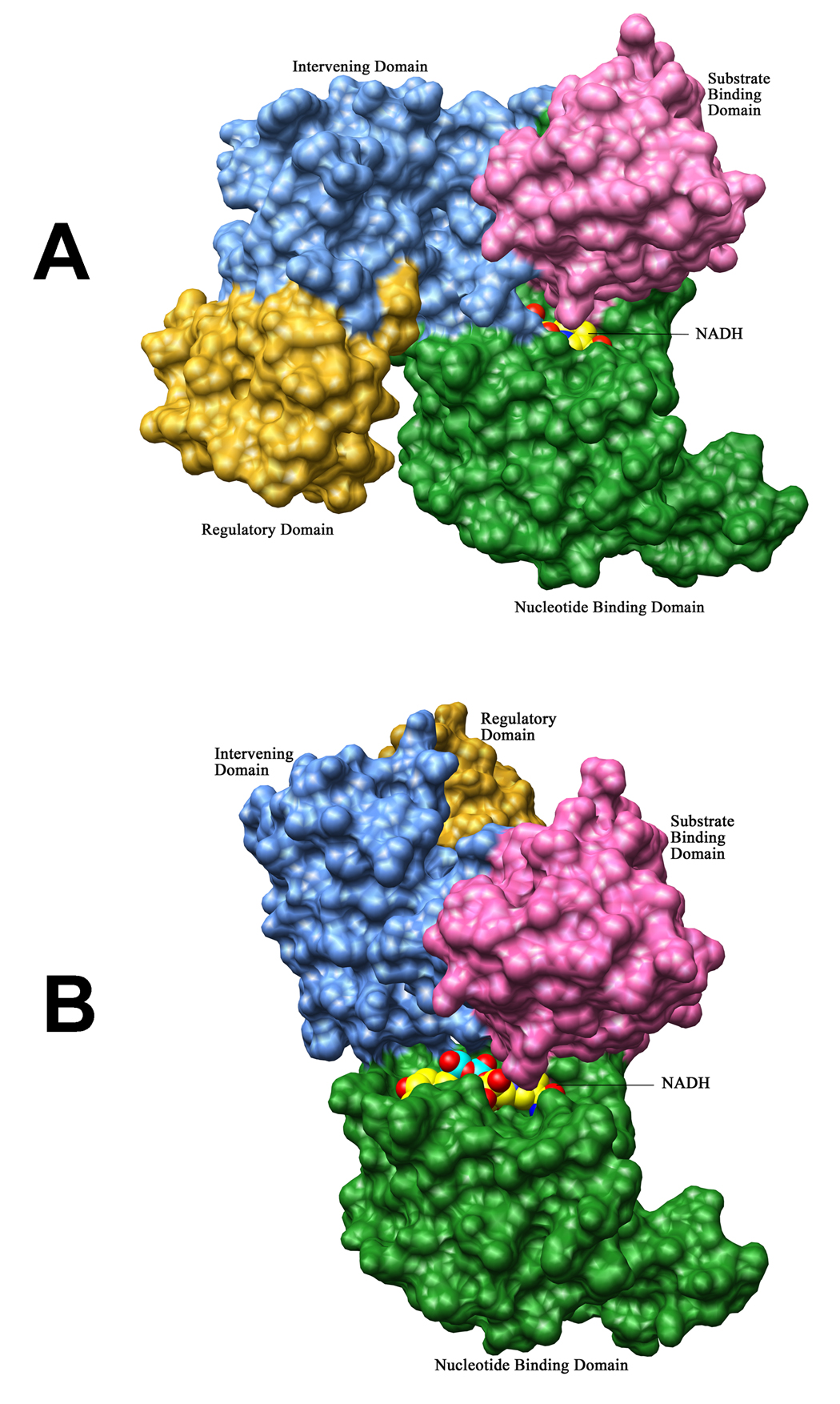
In M.tb PGDH, the cofactor binding sites are less accessible than they are in both the E. coli and human PGDH structures. Since the two subunits display significantly different domain orientations, the environment of the NADH binding sites in each is also significantly different. The binding of NADH is modeled in Figures 5A and B. In chain A (Figure 5A), the site is covered by the intervening domain, which results in the formation of a channel leading from the solvent to the binding site. Apparently, either the cofactor has to enter through the channel or the intervening domain has to change conformation to further expose the binding site. In either case, there is sufficient space to allow NADH to bind to the site. In chain B (Figure 5B) of all M.tb PGDH structures, the intervening domain is rotated away from the cofactor binding site, but the site appears to be blocked by the long flexible loop (Leu203-Ile213) present between strand β8 and helix α11 (Figure 6). However, this is due to the close proximity of the neighboring symmetry related molecule. In solution, it is possible that this loop can move to an orientation similar to chain A, providing sufficient exposure of the binding site to cofactor. This is modeled in Figure 5B.
Figure 5. Modeling of NADH binding to the cofactor binding sites in Chain A and Chain B.
Molecular surface representations of M.tb PGDH chains showing the two different cofactor binding sites. In both chains, the four domains are represented with different colors: nucleotide binding domain (green), substrate binding domain (pink), intervening domain (blue) and the regulatory domain (yellow). NADH has been modeled in both figures in space filling representation (carbon atoms are in yellow, oxygen atoms are in red, nitrogen atoms are in blue, and phosphorus atoms are in aquamarine). The nucleotide (green) and substrate binding (pink) domains are orientated in the same way in both figures. (A) In chain A, the cofactor binding site resides in a channel which is formed by the intervening domain (blue) and nucleotide binding domain (green). (B) In chain B, the loop (see Figure 4), which blocks the cofactor binding site in these structures, has been placed similar to chain A. As seen in this figure, this site is now totally exposed to the solvent as the intervening domain (blue) is oriented away from the binding site.
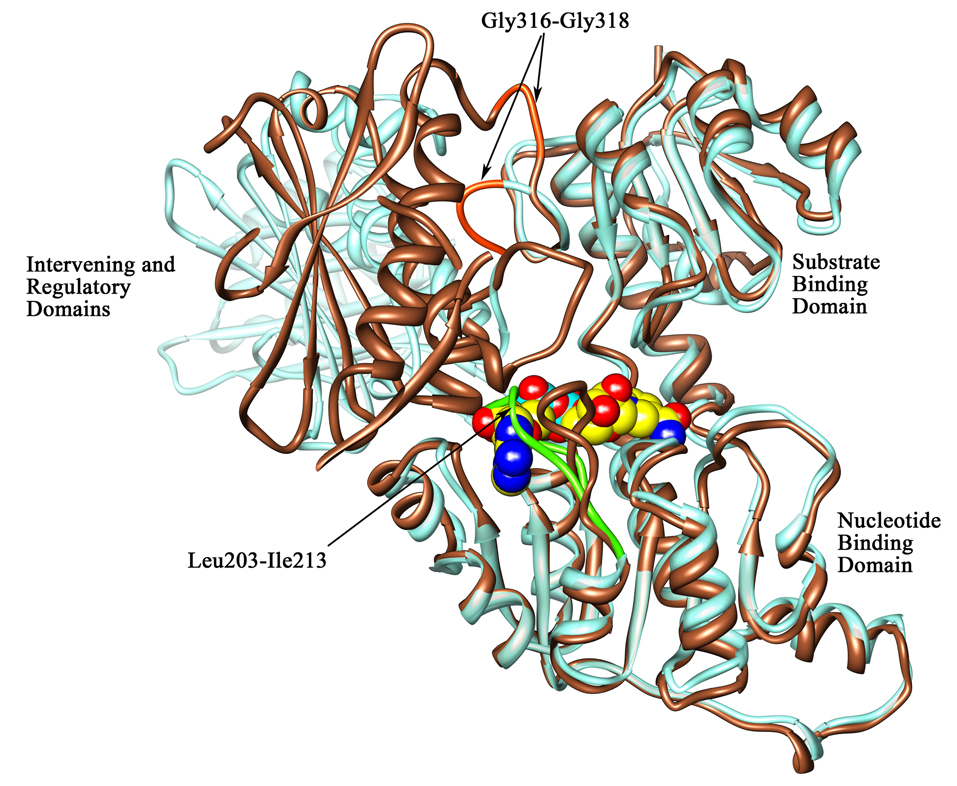
Figure 6. Orientation of the loop region at the cofactor binding site and the strand connecting the substrate binding domain to the intervening domain.
A ribbon diagram representation of the superimposition of chain A (in brown) on chain B (in aquamarine) showing the orientation of the loop region (residues 203–213 designated with arrows) at the cofactor binding site in the M.tb PGDH structures and the glycine containing strand (residues 316–318) connecting the substrate binding and intervening domains. The NADH molecule is represented in space filling representation (carbon atoms are in yellow, oxygen atoms are in red, nitrogen atoms are in blue, and phosphorus atoms are in aquamarine) and has been modeled into this site based on the E.coli PGDH structure. This figure depicts the loop movement near the cofactor binding site in both chains. As shown with an arrow, the loop region in chain B (green) falls over the cofactor binding site, thus blocking NADH from binding at this site in these crystal structures. Also apparent in this figure is the change in the domain orientation of the intervening domain-regulatory domain unit. A portion of the regulatory domain in chain A has been clipped in the front for clarity. The location of Gly316–Gly318 is shown in orange in the loop connecting the substrate binding and intervening domains.
Cofactor Binding
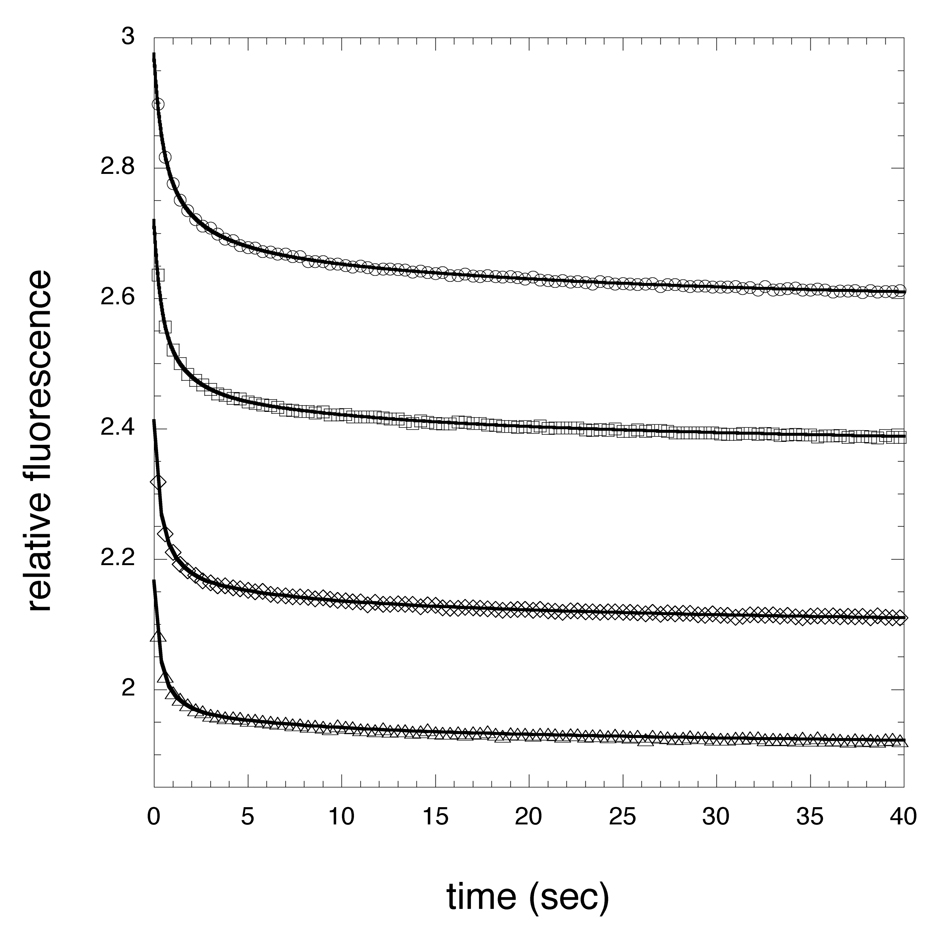
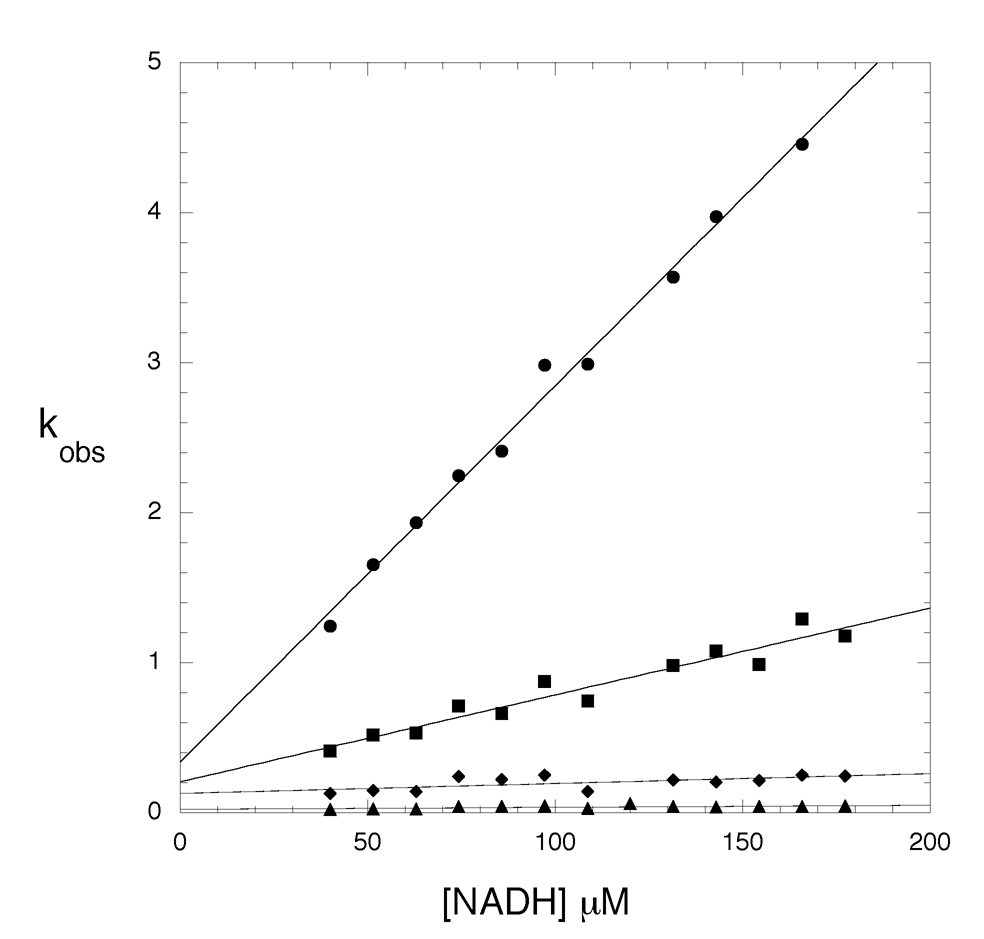
The lack of electron density for the cofactor in those crystals produced with high NADH concentrations prompted an investigation into the ability of M.tb PGDH to bind NADH in solution in the absence of substrate or products. Stopped-flow analysis of NADH binding to M.tb PGDH revealed that binding can be observed by monitoring the fluorescence resonance energy transfer (FRET) from protein tryptophans to NADH when the tryptophans are excited at 295 nm. The binding transients (Figure 7) fit best to four exponentials and a plot of kobs versus NADH concentration (Figure 8) revealed two NADH concentration dependent events and two NADH concentration independent events. The concentration dependent events occur within the first 1–2 sec of the transient. The concentration independent events occur more slowly as can be seen from the gradual decrease of the curves in the 5–40 sec range. The best interpretation of these data is that NADH is binding to two sets of sites within the tetramer followed by a conformational change for each binding event. The NADH concentration dependent plots for the binding step indicate on-rates in 200 mM potassium phosphate buffer, pH 7.5, of 0.025 and 0.006 µM −1 sec −1 and off-rates of 0.34 and 0.21 sec −1, respectively. Given that the kcat for this enzyme is approximately 2500 sec −1 per tetramer, the on-rates for NADH binding are too slow, by a very large degree, to be on the reaction path during continuous turnover of substrate. There was no NADH pre-bound to the enzyme since the absorbance due to NADH at 340 nm did not change when only HPAP was added to the enzyme preparation.
Figure 7. Pre-steady-state transients from the binding of NADH to M. tb PGDH.
The figure shows representative pre-steady state transients produced by the binding of NADH to the enzyme. The relative change in fluorescence of tryptophan upon binding of NADH is depicted by the symbols. The sample was excited at 295 nm and the fluorescence at 340 nm was recorded after rapid mixing of enzyme and NADH. Four representative transients are shown for 86 µM (○), 109 µM (□), 143 µM (◇), and 167 µM (△) NADH. The enzyme concentration was 2 µM in subunits. Only every 100th point is shown for clarity. The solid lines are the fits to the data using equation 3 for 4 exponentials. The binding exponentials are essentially complete within 2 sec. The remaining two exponentials represent a slow conformational change over the 2–40 sec range shown.
Figure 8. Plot of kobs vs NADH concentration.
The binding of NADH to M. tb. PGDH was investigated with pre-steady state transient kinetic analysis. The observed rates obtained from fitting the pre-steady state transients to an equation for four exponentials are plotted against the NADH concentration. Two concentration dependent (●, ■) and two concentration independent (◆, ▲) curves are fit to a straight line.
The very slow NADH binding indicates that either substrate binds first or there is a requirement for an enzyme/product binary complex for efficient NADH binding, similar to the mechanism for dihydrofolate reductase (28). Therefore, additional stopped flow studies were conducted in the presence of product and substrate. The inclusion of saturating levels of phosphoglyceric acid, which is the product of the reaction when assayed in the reverse direction, as is routinely done, did not significantly enhance the rate of NADH binding. It was observed that the enzyme fluorescence increases upon binding of HPAP. However, the transient is essentially complete within the dead time of the instrument, indicating rapid binding of HPAP. When HPAP and NADH are rapidly mixed with the enzyme, all that is observed is the steady state turnover, indicating that binding of both takes place within the dead time of the instrument.
Since the crystals were grown in conditions of very high ionic strength, 1 M NaK tartrate and 100 mM potassium phosphate, the effect of ionic strength on NADH binding was also investigated. These studies were conducted by increasing either phosphate buffer concentration or Na K tartrate concentration. They demonstrate that as ionic strength increases, the on-rates for NADH binding decrease, extrapolating to zero at 400 mM potassium phosphate and at 1 M Na K tartrate. Since the ionic strengths of potassium phosphate buffer at pH 7.5 (100 mM = 14.3 mS/cm) and Na K tartrate (100mM = 15.6 mS/cm) are similar on a molar basis, phosphate is more effective in reducing the binding of NADH. The lower concentration of phosphate required for a comparable decrease in rates suggests that the binding is decreased by competition with those determinants on the protein which specifically interact with the phosphate groups of NADH.
Hinge Regions
In M.tb PGDH, the nucleotide binding domain and substrate binding domain are linked to the intervening domain and regulatory domain by a long loop which is the locus of the 160° rotation between the domains for tetramer formation. The point of transition for this rotation is occupied by three consecutive glycine residues, Gly316-Gly318 (Figure 6). In E.coli PGDH, glycine residues are also found between domains that act as hinge regions and play a role in the cooperativity of serine inhibition. These are Gly294–Gly295, found between the nucleotide binding domain and the substrate binding domain (29), and Gly336–Gly337, found between the substrate binding domain and the regulatory domain (30). Thus, these hinge regions are present at both ends of a long helix that connects the catalytic site to the regulatory site. M.tb PGDH also contains a long helix connecting the substrate binding domain to the intervening domain. Gly282-Ala283 are found at one end in the loop region between the nucleotide binding domain and the substrate binding domain and Gly316–Gly318 are at the other end in a long loop connecting to the intervening domain. There is only a very short loop present between the regulatory domain and the intervening domain that is devoid of any Gly or Ala residues, indicating potentially less flexibility in this region. This arrangement may indicate that the intervening domain and regulatory domain may act together as an ensemble for allosteric inhibition. In the M. tb structure, the Gly316–318 locus would most closely correspond to the Gly336–337 locus in the E. coli structure since they both lead from the nucleotide binding domain-substrate binding domain pair to the rest of the molecule where the serine binding site is found.
Mutants of the glycine hinge between the substrate binding domain and the regulatory domain (Gly336, Gly337) in E. coli PGDH demonstrated that this region functioned in transmitting the effect of serine binding to the active site (29). In the E. coli enzyme, the glycine residues were converted to more bulky valine residues in order to restrict the range of the phi and psi angles. In order to test if the comparable region in M.tb PGDH played a similar role, the glycine residues between the substrate binding domain and the intervening domain-regulatory domain pair (Gly316–318), were mutated to valine residues, either singly or together (Table 2). For the single mutations, there was no significant difference in the parameters except for G318V where the sensitivity to serine was decreased about 5 fold and the cooperativity increased slightly. This is similar to what was seen in the E. coli enzyme for G336V (29). However, unlike the E. coli enzyme, mutation of a combination of 2 of these glycines in the same polypeptide did not have the effect of further intensifying the decrease in serine sensitivity over that for G318V or significantly lowering the Hill coefficient. On the contrary, the double mutations containing G318V increased serine sensitivity relative to G318V alone. This was particularly evident with the G316, 318V mutant. An unexpected result for G316, 317V and G316, 318V was an increase in the Km for substrate and a decrease in the Ki for substrate inhibition. In addition, the kcat for G317, 318V decreased about 4 fold. The triple mutation of this enzyme resulted in no detectable protein expression.
Table 2.
Kinetic Parameters of the Glycine Hinge Mutations
| Enzyme | Km (µM) | kcat(sec−1) | Ki(µM) | kcat/Km(M−1 sec−1) | Serine I 0.5b (µM) | Hill Coefficient |
|---|---|---|---|---|---|---|
| Native | 170 ± 50 | 2461 ± 281 | 950 ± 120 | 1.4 × 107 | 36 ± 6 | 1.8 ± 0.1 |
| G316V | 114 ± 4 | 1111 ± 30 | 2231 ± 190 | 1.0 × 107 | 23 ± 4 | 1.6 ± 0.1 |
| G317V | 165 ± 9 | 2111 ± 119 | 1110 ± 148 | 1.3 × 107 | 23 ± 3 | 1.5 ± 0.1 |
| G318V | 203 ± 23 | 2349 ± 177 | 1925 ± 437 | 1.2 × 107 | 166 ± 22 | 2.5 ± 0.1 |
| G316, 317V | 610 ± 95 | 2805 ± 310 | 380 ± 45 | 0.5 × 107 | 75 ± 17 | 1.7 ± 0.1 |
| G317, 318V | 220 ± 43 | 605 ± 27 | 1620 ± 402 | 0.3 × 107 | 53 ± 12 | 1.9 ± 0.1 |
| G316, 318V | 470 ± 43 | 2827 ± 197 | 220 ± 35 | 0.6 × 107 | 18 ± 5 | 1.9 ± 0.1 |
| G316,317, 318Va |
No detectable protein produced
All mutants are able to be inhibited by serine to < 95%.
Mode of Serine binding
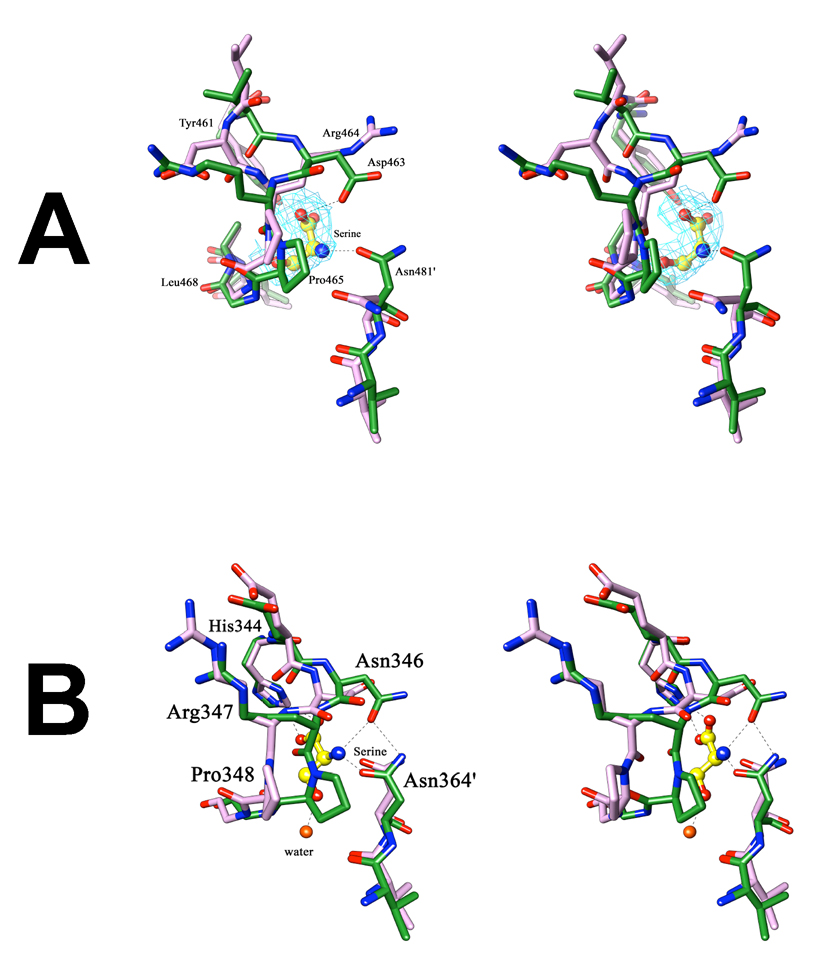
L-Serine binds at the interface of the ACT or regulatory domains, forming hydrogen bonds to both domains in a manner analogous to that seen in E.coli PGDH. Even though this domain shares the βαββαβ secondary structure with its E.coli counterpart, it lacks in sequence identity (30%) and the residues interacting with serine are also not well conserved. The serine bound M.tb PGDH structure shows full occupation of serine at both the subunits of the asymmetric unit. The L-serine carboxyl group forms a hydrogen bond (Figure 9A) with the hydroxyl group of Tyr461 (2.5 Å) and Asp463-OD2 (2.7 Å). The amino group forms a hydrogen bond with Asn481′ (3.0 Å) from the adjacent subunit. There is no water molecule present in the regulatory site for bond formation with the serine side chain hydroxyl group as seen in E.coli PGDH (Figure 9B), indicating the site to be more compact. Instead, the serine hydroxyl group makes direct contact with the main chain amide of Leu468.
Figure 9. Stereoview of the changes occurring at the allosteric site upon serine binding.
(A) In M.tb PGDH, superimposition of the regulatory domain of chain A of the apo-enzyme (purple) and the serine bound form (green) shows a flip of the side chains of Asp463 and Arg464, present on the loop region lining the allosteric site. Also seen is the change in orientation of Asn481′ from the adjacent subunit, which covers the allosteric site upon serine binding. Serine is depicted in yellow ball and stick. The Shake &wARP unbiased 2Fo−Fc electron density map of serine is shown in light blue (1σ level contour). (B) In E.coli PGDH, superimposition of the regulatory domain of chain A of the apo-form (purple) with the serine inhibited form (green) shows a transition of the loop region at Pro348, without side-chain flipping. In this case, transition of the Pro348 covers the allosteric site upon serine binding. Serine is shown at this site in the form of ball and stick (yellow). In both figures, the hydrogen bond interactions of serine with the protein are shown as dashed lines, oxygen atoms are shown in red, and nitrogen atoms in blue.
The functional roles of the residues that provide the primary interaction with serine (Tyr461, Asp463, and Asn481) were investigated by converting them to alanine residues (Table 3). All three mutations produced a very large reduction in the sensitivity of the enzymes to serine. In addition, N481A resulted in a loss of cooperativity in the serine inhibition of catalytic activity. In order to determine that the loss of cooperativity was not due to dissociation of subunits for N481A, both native and N481A M. tb PGDH were chromatographed on a column of Sephacryl S-300. They were first run separately and then as a mixture. Both proteins eluted with the same elution volume of 102 ml, while blue dextran, which is a measure of the void volume of the column, eluted at 70 ml. Since N481A co-elutes with native M. tb PGDH from a Sephacryl S-300 gel filtration column, the loss of cooperativity cannot be explained by a change in the quaternary structure of the enzyme from tetramer to dimer or monomer.
Table 3.
Kinetic Parameters of the Serine Binding Residue Mutations
| Enzyme | Km (µM) | kcat (sec−1) | Kia (µM) | kcat/Km ( M−1 sec−1) | Serine I 0.5b (µM) | Hill Coefficient |
|---|---|---|---|---|---|---|
| Native | 170 ± 50 | 2461 ± 281 | 950 ± 120 | 1.4 × 107 | 36 ± 6 | 1.8 ± 0.1 |
| Y461A | 270 ± 60 | 2754 ± 300 | 420 ± 65 | 1.0 × 107 | 6102 ± 106 | 1.7 ± 0.1 |
| D463A | 140 ± 10 | 720 ± 89 | 1090 ± 165 | 0.5 × 107 | 1886 ± 41 | 1.4 ± 0.1 |
| N481A | 120 ± 15 | 603 ± 77 | 2320 ± 337 | 0.5 × 107 | 2313 ± 366 | 1.1 ± 0.1 |
Ki refers to substrate inhibition.
All mutants are able to be inhibited by serine to < 95%.
No major conformational changes or large domain movements are seen when the serine bound structure is compared with the substrate bound structure or the apo-structure. However there are significant local changes in the orientations of the residues where serine binds (Figure 9A). The side chains of Asp463 and Arg464 present on the loop region flip their positions when serine binds in the regulatory domain. This flipping is necessary for hydrogen bond formation between the effector molecule and the side chain of Asp463. This flip is not seen in the E.coli allosteric site (Figure 9B). Rather, there is a transition of the loop region near Pro348 which covers the site in response to serine binding. Because of the flipping transition in M.tb PGDH, considerable movement in the neighboring residues (460–462 and 465–466) is also seen. A surface representation (not shown) of regulatory domains with bound serine shows that the path to the binding site is closed upon serine binding.
The orientation of the side chain of Asn481′ from the adjacent subunit also changes. When serine binds, it moves to face Asp463, forming a hydrogen bond with the serine amino group (2.9 Å). These interactions further strengthen the regulatory domain interface. In the absence of serine, Asn481′-ND2 is hydrogen bond interaction with Asp503-OD2 of the same chain (~3.4 Å). There is also a change in the orientation of the side chain of His460-NE2 which forms hydrogen bonds with Asp490-OD1 (3.1 Å) and OD2 (3.4 Å) when serine binds. In response to this change, the orientation of the phenolic group of Tyr521 also changes by 90°. There is also change in the orientation of Glu489 which, in the absence of serine, is hydrogen bonded to Arg464 but faces toward the anion binding site when serine binds.
Discussion
The crystal structures of M.tb PGDH have been solved with bound effector, L-serine, and bound substrate, HPAP. The ligand bound structures display the same asymmetry between subunits noted in the apo-enzyme structure (1), consisting of a domain rotation of 160° in one of the subunits in the asymmetric unit relative to the other. However, these ligand bound structures provide new insights into the conformational events that occur as a result of ligand binding. At the present time very little is known about the catalytic and regulatory mechanism of M. tb PGDH. It might be tempting to assume that these mechanisms will be essentially the same for all PGDH enzymes since they catalyze the same reaction and are regulated by the same small metabolite. However, this investigation has revealed that the interaction of M. tb PGDH with its effector and substrates is decidedly different than that determined for E. coli PGDH, which is the only other PGDH that has been studied in detail. The structural studies present initial evidence for a reversed order of substrate binding compared to E. coli PGDH that leads to a plausible explanation of the conformational changes that take place upon successive binding of substrate and cofactor. This is based on kinetic analysis that shows that cofactor binding is not productive in the absence of substrate and that substrate binds to the enzyme at the active site in a catalytically unproductive manner in the absence of cofactor. Thus, the substrates act synergistically in conforming the active site for catalysis. Furthermore, the structure of the serine binding site in M. tb PGDH is different than that in E. coli PGDH, both in regard to the identity and manner of amino acid residue interactions with L-serine and also in regard to the conformational changes that take place as a result of serine binding. Also, a poly-glycine hinge region is found in M. tb PGDH that is analogous to one found in E. coli PGDH. This poly-glycine hinge has been shown by mutagenesis in E. coli PGDH to be functional in the transmission of the signal from serine binding to the active site. Similar mutations to the poly-glycine hinge in M. tb PGDH demonstrates that it does not share the same function with the E.coli enzyme.
The respective catalytic characteristics of PGDH from M. tb and E. coli show significant differences. M.tb PGDH displays significant substrate inhibition while the E.coli enzyme does not under comparable conditions. This appears to be mediated by a second site, termed the anion binding site (5), formed largely by the intervening domain which is present in M. tb PGDH but not in E. coli PGDH. In E. coli PGDH, NADH binds very tightly and without cooperativity with a Kd of less than 0.5 µM (31). In M.tb PGDH, NADH displays homotropic positive cooperativity with a Km of approximately 50 µM (6). These differences in interaction with NADH may reflect the significantly different environments of the respective NADH binding sites. The positive cooperativity is likely due to the asymmetry of the tetramer. Furthermore, the observance of two sets of sites for NADH binding in the stopped-flow experiments indicates two different environments for NADH binding.
Noteworthy features of these structures are the lack of density for the cofactor at its binding sites and that substrate is bound in a seemingly non-productive manner insofar as the keto group is not pointed toward the catalytic His280 and the expected interaction of the substrate C1-carboxyl group with an arginine residue is not present. This arrangement, and the required order of binding of substrate before cofactor, suggests an interpretation where cofactor binding serves to re-orient the substrate for catalytic turnover. Re-orientation of the substrate would position its α-carboxyl adjacent to the ring nitrogen of His280 and cause the Arg233 side chain to rotate and make bonding contact with the carboxyl group of the substrate. Comparison of the nucleotide and substrate binding domain positions in M. tb PGDH with the structure of a truncated human PGDH with bound cofactor and a substrate analog, also demonstrates that a rotation of these domains relative to each other of approximately 15° takes place upon productive binding. Thus, the re-orientation of the substrate relative to the catalytic residues of the enzyme, and the closure of the active site, may occur in a concerted manner upon cofactor binding. Based on the substrate analog malate, bound to the human PGDH structure, a closed conformation of the active site cleft can be modeled for M.tb PGDH to elucidate the mechanism of action (Figure 3). Interestingly, all three catalytically important residues His280, Glu262 and Arg233 are present on different loop regions of the nucleotide binding domain. His280 is present on the Pro279-Thr285 loop connecting the nucleotide binding domain to the substrate binding domain, Glu262 is present on the loop between β10 and α13 and Arg233 is present on the loop between β9 and α12 near the nicotinamide ring at the entrance of the active site cleft (see Figure 1). The rest of the cofactor lies at the base of the cleft. If the presence of HPAP at the active site promotes the binding of NADH, then NADH in turn would promote the orientation of Arg233 toward the HPAP where its C1-carboxyl group would interact with the guanidium group of Arg233 closing the binding site cleft. The interaction of His280 with Asp262 properly positions the imidazole ring for proton transfer. The generation of a partial positive charge on C2 of HPAP will allow the transfer of hydride ion from NADH to the substrate resulting in the formation of NAD+ and phosphoglycerate.
Analysis of the cofactor binding sites demonstrates that in chains B and D of the tetramer, the loop consisting of residues 203–213 of the nucleotide binding domain partially occupies the space where NADH would bind. It would need to move out of the way in order to allow NADH to occupy the site. Since the loop placement is in close proximity to crystal lattice contacts from the symmetry related molecule, it is possible that the loop will move to a position in solution similar to that in chain A and allow NADH to bind more readily. This is, in fact, supported by the kinetic observation that NADH binds to two sets of sites. The extreme asymmetry between adjacent subunits in the asymmetric unit produces two distinct environments for the region around the cofactor site that appear to be reflected in the kinetic observations. In chains A and C, there is sufficient room for NADH to bind, but because of the proximity of the intervening domain, the cofactor must approach the binding site through a channel formed by the intervening and cofactor binding domains. In this regard, the channel opening appears to be large enough to allow the cofactor to enter (Figure 5).
The stopped flow NADH binding studies demonstrate that although NADH binds to M.tb PGDH in the absence of substrate, the on-rates are too slow to be operative during catalytic turnover. The concentration of NADH would have to be at least 65 mM in order to produce a rate equal to the kcat for the reaction. On the other hand, in the presence of substrate, the on-rate for NADH binding is not measurable since it occurs within the dead-time of the instrument and the only thing that can be observed is the steady state conversion of NADH to NAD+. Thus, the rate of binding of NADH is enhanced considerably when substrate is present. Since these conditions cannot be met in the crystal preparations without turnover occurring, and since the binding that does occur in the absence of substrate is sensitive to ionic strength, it is not surprising that crystals with bound NADH could not be obtained for this enzyme. This is in contrast to E. coli PGDH where NADH binds first in an ordered addition and with high affinity (32).
The binding of NADH within a channel is also consistent with what is observed with the enzyme in solution. E.coli PGDH is normally purified with a 5′-AMP sepharose affinity column. However, M.tb PGDH does not bind to this column (1). Even though binding of NADH to M.tb PGDH in the absence of other components is slow, the Kd for the binding (app. 15–30 µM) is sufficient to expect binding to occur to the 5′-AMP moiety of the affinity column and cause the enzyme to be retained. The observation that the enzyme does not bind to the affinity column could either be that the Kd for binding to 5′-AMP is increased even further or it could also be easily explained by the existence of the channel where the enzyme surface would encounter the sepharose beads before the 5′-AMP group could enter far enough to make productive contact to bind.
Another possibility may be that the tartrate bound at the anion binding site may stabilize the structure in a form that does not bind cofactor. However, crystals grown in the absence of tartrate, although they diffract poorly, do not show the distinct electron density for the cofactor and also have the same space group and cell dimensions as the crystals that contain tartrate. This suggests a similar structure in these crystals and argues against tartrate binding being responsible for the exclusion of NADH.
The presence of the intervening domain in M.tb PGDH produces a strikingly different structure from that of E.coli PGDH. All of the subunits in E. coli PGDH have similar overall conformations, while there are two extremely different conformations in the subunits of M.tb PGDH. The asymmetry observed in quaternary structure between the two subunits of the asymmetric unit is centered on a 160° rotation about a loop connecting the substrate binding domain to the intervening domain. This loop contains the triple glycine sequence Gly316–318. In this respect, the intervening domain and the regulatory or ACT domain can potentially be viewed as moving together as a single rigid body. In contrast, in E. coli PGDH, it is the substrate binding domain and the regulatory domain that appear to move together as a single unit. Comparison of the structures of E. coli PGDH, with (2) and without (3) bound serine, demonstrated that there is a coordinated rotation of the regulatory and substrate binding domain relative to the nucleotide binding domain of approximately 15°. Subsequently, the crystal structures of Gly336V and Gly337V mutations demonstrated that they could exert an effect on this rotation and these mutants defined a minimal limit on the degree of rotation leading to inhibition of enzyme activity (33).
The relative insensitivity to the placement of bulky side chains at the three glycine residues in this connecting loop could suggest that a large degree of rotation at this locus is not required for catalytic activity or serine inhibition. The loop may function mainly in the formation of the asymmetric subunits during folding and oligomerization of the subunits. This is consistent with the observation that the triple glycine mutation failed to produce significant levels of protein and the fact that there is no rotation observed at this locus when serine binds the enzyme. Clearly, the enzyme is not insensitive to changes in this region, but the effect is not the same as that seen for the E. coli enzyme in the poly-Gly hinge region between its regulatory domain and substrate binding domain. A particularly intriguing observation of these mutations is that the relatively modest decrease in serine sensitivity seen with the G318V mutation could be reversed with the addition of the G316V mutation (Gly316, 318V). In the crystal structure, this loop is exposed to solvent and does not appear to make any major contact with adjacent residues. However, the addition of a side chain at residue 316 could introduce enough bulk to force interaction with the nearby residues at positions 86–91, inducing a conformational change in the connecting loop that might compensate for the effect of the single mutation at G318. In this respect, the crystal structures of the enzymes containing these loop mutations may be revealing.
Despite strikingly different quaternary structures, E.coli and M.tb PGDH possess serine binding sites at the interface of adjacent ACT domains (1, 2, 34). Like E.coli PGDH (2), the serine forms hydrogen bonds across the interface to both domains. The M.tb site is more compact than the E.coli site in that the serine side chain hydroxyl makes direct contact with a main chain amide instead of being mediated by a water molecule as in the E. coli enzyme. Also, while the E. coli enzyme utilizes a His and two Asn residues to contact the α-amino (Asn346 and Asn364′) and α-carboxyl groups (His344) of serine, the M.tb enzyme utilizes a Tyr, an Asp, and an Asn residue with a different arrangement (Figure 9). In M.tb PGDH, two residues, Tyr461 and Asp463, interact with the carboxyl group while Asp463 and Asn481′ interact with the amino group. Once serine binds, both enzymes close the binding site to solvent through localized conformational changes. In E. coli PGDH, this is accompanied by the rotation of a short segment of polypeptide to bring the face of proline 348 over the serine ligand, completing the closing of the site. In M.tb PGDH, the rotation of the polypeptide chain causes a near 180° translocation of the side chains of Asp463 and Arg464 and the accompanying change in the polypeptide chain closes off the serine site. Neither the E.coli (3) nor the M.tb active sites appear to change appreciably in response to serine binding. This suggests that the overall effect of serine binding may be a more subtle alteration of the dynamics of the enzyme rather than on any specifically favored conformation.
Mutation of Y461, D463, and N481 to alanine residues greatly decreased serine inhibition, thus supporting their role as the main serine binding components. The decrease of the Hill coefficient to approximately 1 with N481A suggests that hydrogen bond interaction linking adjacent ACT domains contributes to the cooperativity of serine inhibition, since Asn 481 is the sole binding determinant spanning the ACT domain subunit interface when serine binds to Tyr461 and Asp463 of the other subunit. In addition, Y461A increases substrate inhibition while D463A and N481A decrease substrate inhibition. This effect may be transmitted through Arg 501 which extends from the ACT domain into the anion binding site and is one of the main binding residues for substrate whose binding at this site leads to substrate inhibition.
The structures reported here and the insights gained from them will be of considerable assistance in understanding the mode of action of the ACT domains in regulation not only for these PGDH enzymes but for other less closely related ACT domain containing enzymes and transcription factors.
Supplementary Material
The sequences of the oligonucleotide primers used for PCR mutagenesis are provided. This material is available free of charge via the Internet at http://pubs.acs.org.
Acknowledgments
We thank the scientists of the BioCARS beamline (14BM-C) at the Advanced Photon Source (APS) of the Argonne National Laboratory (ANL), Chicago, for their help in data collection. We also thank Shawei Chen and Xiao Lan Xu for excellent technical assistance.
Abbreviations
- HPAP
hydroxypyruvic acid phosphate
- ACT
Aspartate Kinase-Chorismate mutase-Tyr A
References
- 1.Dey S, Grant GA, Sacchettini JC. Crystal structure of Mycobacterium tuberculosis D-3- phosphoglycerate dehydrogenase: Extreme asymmetry in a tetramer of identical subunits. J. Biol. Chem. 2005;280:14892–14899. doi: 10.1074/jbc.M414489200. [DOI] [PubMed] [Google Scholar]
- 2.Schuller DJ, Grant GA, Banaszak LJ. The allosteric ligand site in the Vmax-type cooperative enzyme phosphoglycerate dehydrogenase. Nat. Struct. Biol. 1995;2:69–76. doi: 10.1038/nsb0195-69. [DOI] [PubMed] [Google Scholar]
- 3.Thompson JR, Bell JK, Bratt J, Grant GA, Banaszak LJ. Vmax regulation through domain and subunit changes. The active form of phosphoglycerate dehydrogenase. Biochemistry. 2005;44:5763–5773. doi: 10.1021/bi047944b. [DOI] [PubMed] [Google Scholar]
- 4.Grant GA. The ACT domain: A small molecule binding domain and its role as a common regulatory element. J. Biol. Chem. 2006;281:33825–33829. doi: 10.1074/jbc.R600024200. [DOI] [PubMed] [Google Scholar]
- 5.Burton RL, Chen S, Xu XL, Grant GA. A novel mechanism for substrate inhibition in Mycobacterium tuberculosis D-3-phosphoglycerate dehydrogenase. J. Biol. Chem. 2007;282:31517–31524. doi: 10.1074/jbc.M704032200. [DOI] [PubMed] [Google Scholar]
- 6.Dey S, Hu Z, Xu XL, Sacchettini JC, Grant GA. D-3-Phosphoglycerate dehydrogenase from Mycobacterium tuberculosis is a link between the Escherichia coli and mammalian enzymes. J. Biol. Chem. 2005;280:14884–14891. doi: 10.1074/jbc.M414488200. [DOI] [PubMed] [Google Scholar]
- 7.Saski R, Pizer LI. Regulatory properties of purified 3-phosphoglycerate dehydrogenase from Bacillus subtilis. Eur. J. Biochem. / FEBS. 1975;51(2):415–427. doi: 10.1111/j.1432-1033.1975.tb03941.x. [DOI] [PubMed] [Google Scholar]
- 8.Peters-Wendisch P, Netzer R, Eggeling L, Sahm H. 3-Phosphoglycerate dehydrogenase from Corynebacterium glutamicum: the C-terminal domain is not essential for activity but is required for inhibition by L-serine. Applied microbiol. biotech. 2002;60:437–441. doi: 10.1007/s00253-002-1161-y. [DOI] [PubMed] [Google Scholar]
- 9.Achouri Y, Rider MH, Schaftingen EV, Robbi M. Cloning, sequencing and expression of rat liver 3-phosphoglycerate dehydrogenase. Biochem. J. 1997;323:365–370. doi: 10.1042/bj3230365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Walsh DA, Sallach HJ. Purification and properties of chicken liver D-3-phosphoglycerate dehydrogenase. Biochemistry. 1965;4:1076–1085. doi: 10.1021/bi00882a015. [DOI] [PubMed] [Google Scholar]
- 11.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Meth. Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 12.Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. Sect. D: Biol. Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 13.Reddy V, Swanson SM, Segelke B, Kantardjieff KA, Sacchettini JC, Rupp B. Effective electron-density map improvement and structure validation on a Linux multi-CPU web cluster: The TB Structural Genomics Consortium Bias Removal Web Service. Acta Crystallogr. Sect. D: Biol. Crystallogr. 2003;59:2200–2210. doi: 10.1107/s0907444903020316. [DOI] [PubMed] [Google Scholar]
- 14.McRee DE. XtalView/Xfit--A versatile program for manipulating atomic coordinates and electron density. J. Struct. Biol. 1999;125:156–165. doi: 10.1006/jsbi.1999.4094. [DOI] [PubMed] [Google Scholar]
- 15.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. Sect. D: Biol. Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 16.Laskowski RA, MacArthur MW, Moss DS, Thornton JM. PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 1993;26:283–291. [Google Scholar]
- 17.Pizer LI, Sugimoto E. 3-Phosphoglycerate Dehydrogenase (Escherichia coli) Meth. Enzymol. 1989;178:325–331. [Google Scholar]
- 18.Cleland W. Substrate Inhibition. Meth. Enzymol. 1979;63:500–513. [PubMed] [Google Scholar]
- 19.Bell JE, Bell ET. Proteins and Enzymes. Prentice-Hall, Inc; 1988. pp. 465–470. [Google Scholar]
- 20.Grant GA. Methods for analyzing cooperativity in Phosphoglycerate Dehydrogenase. Meth. Enzymol. 2004;380:106–131. doi: 10.1016/S0076-6879(04)80005-3. [DOI] [PubMed] [Google Scholar]
- 21.Johnson KA. Transient-state kinetic analysis of enzyme reaction pathways. The Enzymes XX. 1992:1–61. [Google Scholar]
- 22.Cormack B. Mutagenesis by the polymerase chain reaction. In: Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K, editors. Current Protocols in Molecular Biology. New York: John Wiley and Sons; 1991. pp. 8.5.1–8.5.9. [Google Scholar]
- 23.Tobey KL, Grant GA. The Nucleotide Sequence of the serA Gene of Escherichia coli and the Amino Acid Sequence of the Encoded Protein, D-3-Phosphoglycerate Dehydrogenase. J. Biol. Chem. 1986;261:12179–12183. [PubMed] [Google Scholar]
- 24.Cho HM, Jun DY, Bae MA, Ahn JD, Kim YH. Nucleotide sequence and differential expression of the human 3-phosphoglycerate dehydrogenase gene. Gene. 2000;245:193–201. doi: 10.1016/s0378-1119(00)00009-3. [DOI] [PubMed] [Google Scholar]
- 25.Grant GA, Hu Z, Xu XL. Identification of amino acid residues contributing to the mechanism of cooperativity in Escherichia coli D-3-phosphoglycerate dehydrogenase. Biochemistry. 2005;44:16844–16852. doi: 10.1021/bi051681j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bell JK, Grant GA, Banaszak LJ. Multiconformational states in Phosphoglycerate dehydrogenase. Biochemistry. 2004;43:3450–3458. doi: 10.1021/bi035462e. [DOI] [PubMed] [Google Scholar]
- 27.Lamzin VS, Dauter Z, Popov VO, Harutyunyan EH, Wilson KS. High resolution structures of holo and apo formate dehydrogenase. J. Mol. Biol. 1994;236:759–785. doi: 10.1006/jmbi.1994.1188. [DOI] [PubMed] [Google Scholar]
- 28.Fierke CA, Johnson KA, Benkovic SJ. Construction and evaluation of the kinetic scheme associated with dihydrofolate reductase from Escherichia coli. Biochemistry. 1987;26:4085–4092. doi: 10.1021/bi00387a052. [DOI] [PubMed] [Google Scholar]
- 29.Grant GA, Hu Z, Xu Xiao Lan. Amino acid residues mutations Uncouple cooperative effects in Escherichia coli D-3-phosphoglycerate Dehydrogenase. J. Biol. Chem. 2001;276:17844–17850. doi: 10.1074/jbc.M009957200. [DOI] [PubMed] [Google Scholar]
- 30.Grant GA, Xu XL, Hu Z. Role of an interdomain Gly-Gly sequence at the regulatory-substrate domain interface in the regulation of Escherichia coli D-3-phosphoglycerate dehydrogenase. Biochemistry. 2000;39:7316–7319. doi: 10.1021/bi000218y. [DOI] [PubMed] [Google Scholar]
- 31.Sugimoto E, Pizer LI. The mechanism of end product inhibition of serine biosynthesis. II. Optical studies of phosphoglycerate dehydrogenase. J. Biol. Chem. 1968;243:2090–2098. [PubMed] [Google Scholar]
- 32.Dubrow R, Pizer LI. Transient kinetic and deuterium isotope effect studies on the catalytic mechanism of phosphoglycerate dehydrogenase. J. Biol. Chem. 1977;252:1539–1551. [PubMed] [Google Scholar]
- 33.Dey S, Hu Z, Xu XL, Sacchettini JC, Grant GA. The effect of hinge mutations on effector binding and domain rotation in Escherichia coli D-3-phosphoglycerate dehydrogenase. J. Biol. Chem. 2007;282:18418–18426. doi: 10.1074/jbc.M701174200. [DOI] [PubMed] [Google Scholar]
- 34.Al-Rabiee R, Zhanh Y, Grant GA. The mechanism of velocity modulated allosteric regulation in D-3-phosphoglycerate dehydrogenase: Site-directed mutagenesis of effector binding site residues. J. Biol. Chem. 1996;271:23235–23238. doi: 10.1074/jbc.271.38.23235. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The sequences of the oligonucleotide primers used for PCR mutagenesis are provided. This material is available free of charge via the Internet at http://pubs.acs.org.