Abstract
The DNA methylation capacity and some other properties of the related temperate Bacillus subtilis phages Z, SPR, SP beta, phi 3T, and rho 11 are compared. With phage mutants affected in their methylation potential, we show that phage-coded methyltransferase genes are interchangeable among the phages studied. DNA/DNA hybridization experiments indicate that phage methyltransferase genes are structurally related, whereas no such relationship is observed to a bacterial gene, specifying a methyltransferase with the same specificity.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bron S., Hörz W. Purification and properties of the Bsu endonuclease. Methods Enzymol. 1980;65(1):112–132. doi: 10.1016/s0076-6879(80)65017-4. [DOI] [PubMed] [Google Scholar]
- Bron S., Murray K. Restriction and modification in B. subtilis. Nucleotide sequence recognised by restriction endonuclease R. Bsu R from strain R. Mol Gen Genet. 1975 Dec 30;143(1):25–33. doi: 10.1007/BF00269417. [DOI] [PubMed] [Google Scholar]
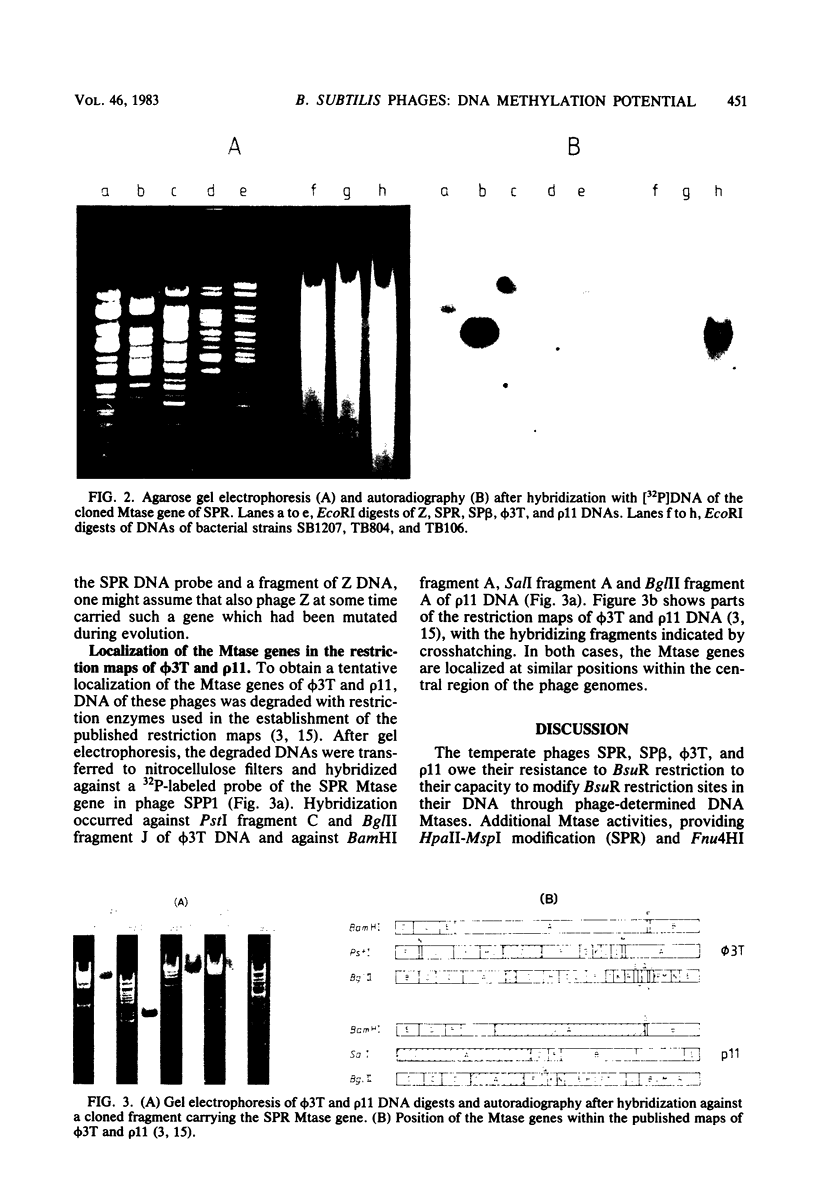
- Cregg J. M., Ito J. A physical map of the genome of temperate phage phi 3T. Gene. 1979 Jul;6(3):199–219. doi: 10.1016/0378-1119(79)90058-1. [DOI] [PubMed] [Google Scholar]
- Cregg J. M., Nguyen A. H., Ito J. DNA modification induced during infection of Bacillus subtilis by phage phi 3T. Gene. 1980 Dec;12(1-2):17–24. doi: 10.1016/0378-1119(80)90011-6. [DOI] [PubMed] [Google Scholar]
- Dean D. H., Orrego J. C., Hutchison K. W., Halvorson H. O. New temperate bacteriophage for Bacillus subtilis, rho 11. J Virol. 1976 Nov;20(2):509–519. doi: 10.1128/jvi.20.2.509-519.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fink P. S., Korman R. Z., Odebralski J. M., Zahler S. A. Bacillus subtilis bacteriophages SP beta c1 is a deletion mutant of SP beta. Mol Gen Genet. 1981;182(3):514–515. doi: 10.1007/BF00293946. [DOI] [PubMed] [Google Scholar]
- Gunthert U., Schweiger M., Stupp M., Doerfler W. DNA methylation in adenovirus, adenovirus-transformed cells, and host cells. Proc Natl Acad Sci U S A. 1976 Nov;73(11):3923–3927. doi: 10.1073/pnas.73.11.3923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Günthert U., Freund M., Trautner T. A. Restriction and modification in Bacillus subtilis: two DNA methyltransferases with BsuRI specificity. I. Purification and physical properties. J Biol Chem. 1981 Sep 10;256(17):9340–9345. [PubMed] [Google Scholar]
- Günthert U., Jentsch S., Freund M. Restriction and modification in Bacillus subtilis: two DNA methyltransferases with BsuRI specificity. II. Catalytic properties, substrate specificity, and mode of action. J Biol Chem. 1981 Sep 10;256(17):9346–9351. [PubMed] [Google Scholar]
- Hemphill H. E., Gage I., Zahler S. A., Korman R. Z. Prophage-mediated production of a bacteriocinlike substance by SP beta lysogens of Bacillus subtilis. Can J Microbiol. 1980 Nov;26(11):1328–1333. doi: 10.1139/m80-220. [DOI] [PubMed] [Google Scholar]
- Ikawa S., Shibata T., Ando T., Saito H. Genetic studies on site-specific endodeoxyribonucleases in Bacillus subtilis: multiple modification and restriction systems in transformants of Bacillus subtilis 168. Mol Gen Genet. 1980 Feb;177(3):359–368. doi: 10.1007/BF00271474. [DOI] [PubMed] [Google Scholar]
- Ito J., Roberts R. J. Unusual base sequence arrangement in phage phi 29 DNA. Gene. 1979 Jan;5(1):1–7. doi: 10.1016/0378-1119(79)90088-x. [DOI] [PubMed] [Google Scholar]
- Jentsch S., Günthert U., Trautner T. A. DNA methyltransferases affecting the sequence 5'CCGG. Nucleic Acids Res. 1981 Jun 25;9(12):2753–2759. doi: 10.1093/nar/9.12.2753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizukami T., Kawamura F., Takahashi H., Saito H. A physical map of the genome of the Bacillus subtilis temperate phage rho 11. Gene. 1980 Oct;11(1-2):157–162. doi: 10.1016/0378-1119(80)90095-5. [DOI] [PubMed] [Google Scholar]
- Noyer-Weidner M., Pawlek B., Jentsch S., Günthert U., Trautner T. A. Restriction and modification in Bacillus subtilis: gene coding for a BsuR-specific modification methyltransferase in the temperate bacteriophage phi 3T. J Virol. 1981 Jun;38(3):1077–1080. doi: 10.1128/jvi.38.3.1077-1080.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palefski S., Hemphill H. E., Kolenbrander P. E., Whiteley H. R. Dominance relationships in mixedly infected Bacillus subtilis. J Virol. 1972 Apr;9(4):594–601. doi: 10.1128/jvi.9.4.594-601.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratcliff S. W., Luh J., Ganesan A. T., Behrens B., Thompson R., Montenegro M. A., Morelli G., Trautner T. A. The genome of Bacillus subtilis phage SPP1: the arrangement of restriction endonuclease generated fragments. Mol Gen Genet. 1979 Jan 10;168(2):165–172. doi: 10.1007/BF00431442. [DOI] [PubMed] [Google Scholar]
- Reeve J. N., Amann E., Tailor R., Günthert U., Scholz K., Trautner T. A. Unusual behaviour of SPO1 DNA with respect to restriction and modification enzymes recognizing the sequence 5'-G-G-C-C. Mol Gen Genet. 1980 Apr;178(1):229–231. doi: 10.1007/BF00267234. [DOI] [PubMed] [Google Scholar]
- Rigby P. W., Dieckmann M., Rhodes C., Berg P. Labeling deoxyribonucleic acid to high specific activity in vitro by nick translation with DNA polymerase I. J Mol Biol. 1977 Jun 15;113(1):237–251. doi: 10.1016/0022-2836(77)90052-3. [DOI] [PubMed] [Google Scholar]
- Rottländer E., Trautner T. A. Genetic and transfection studies with B, subtilis phage SP 50. I. Phage mutants with restricted growth on B. subtilis strain 168. Mol Gen Genet. 1970;108(1):47–60. doi: 10.1007/BF00343184. [DOI] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Stroynowski I. T. Distribution of bacteriophage phi 3T homologous deoxyribonucleic acid sequences in Bacillus subtilis 168, related bacteriophages, and other Bacillus species. J Bacteriol. 1981 Oct;148(1):91–100. doi: 10.1128/jb.148.1.91-100.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stroynowski I. T. Integration of the bacteriophage phi 3T-coded thymidylate synthetase gene into the Bacillus subtilis chromosome. J Bacteriol. 1981 Oct;148(1):101–108. doi: 10.1128/jb.148.1.101-108.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trautner T. A., Pawlek B., Bron S., Anagnostopoulos C. Restriction and modification in B. subtilis. Biological aspects. Mol Gen Genet. 1974;131(3):181–191. doi: 10.1007/BF00267958. [DOI] [PubMed] [Google Scholar]
- Trautner T. A., Pawlek B., Günthert U., Canosi U., Jentsch S., Freund M. Restriction and modification in Bacillus subtilis: identification of a gene in the temperate phage SP beta coding for a BsuR specific modification methyltransferase. Mol Gen Genet. 1980;180(2):361–367. doi: 10.1007/BF00425849. [DOI] [PubMed] [Google Scholar]
- Vogelstein B., Gillespie D. Preparative and analytical purification of DNA from agarose. Proc Natl Acad Sci U S A. 1979 Feb;76(2):615–619. doi: 10.1073/pnas.76.2.615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahler S. A., Korman R. Z., Rosenthal R., Hemphill H. E. Bacillus subtilis bacteriophage SPbeta: localization of the prophage attachment site, and specialized transduction. J Bacteriol. 1977 Jan;129(1):556–558. doi: 10.1128/jb.129.1.556-558.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]