Abstract
Adaptor protein complexes (APs) function as vesicle coat components in different membrane traffic pathways; however, there are a number of pathways for which there is still no candidate coat. To find novel coat components related to AP complexes, we have searched the expressed sequence tag database and have identified, cloned, and sequenced a new member of each of the four AP subunit families. We have shown by a combination of coimmunoprecipitation and yeast two-hybrid analysis that these four proteins (ε, β4, μ4, and ς4) are components of a novel adaptor-like heterotetrameric complex, which we are calling AP-4. Immunofluorescence reveals that AP-4 is localized to ∼10–20 discrete dots in the perinuclear region of the cell. This pattern is disrupted by treating the cells with brefeldin A, indicating that, like other coat proteins, the association of AP-4 with membranes is regulated by the small GTPase ARF. Immunogold electron microscopy indicates that AP-4 is associated with nonclathrin-coated vesicles in the region of the trans-Golgi network. The μ4 subunit of the complex specifically interacts with a tyrosine-based sorting signal, indicating that, like the other three AP complexes, AP-4 is involved in the recognition and sorting of cargo proteins with tyrosine-based motifs. AP-4 is of relatively low abundance, but it is expressed ubiquitously, suggesting that it participates in a specialized trafficking pathway but one that is required in all cell types.
INTRODUCTION
Protein trafficking between organelles is performed by transport vesicles, which bud from a donor membrane and fuse with a target acceptor membrane. Vesicle budding has been shown to require the recruitment of soluble factors from the cytosol onto the membrane to form a coat. This coat may play two roles: 1) to select the cargo for inclusion in the transport vesicle and 2) to provide a scaffold for vesicle formation.
Clathrin-coated vesicles were the first coated transport vesicles to be identified. Their coats have been shown to consist of clathrin, which forms the vesicle scaffold, and adaptor protein complexes or APs, which choose the vesicle cargo by interacting with sorting signals on the cytoplasmic domains of selected transmembrane proteins. Clathrin-coated vesicles bud from the plasma membrane and the trans-Golgi network (TGN), where they concentrate proteins destined for the endocytic pathway. Although the clathrin is the same at both locations, different adaptor complexes are found; the AP-1 complex associates with the TGN and directs the transport of lysosomal enzymes to endosomes, whereas the AP-2 complex associates with the plasma membrane and directs the internalization of trafficking cell surface proteins.
Recently, another ubiquitously expressed adaptor-related complex, AP-3, was described (Simpson et al., 1996, 1997; Dell’Angelica et al., 1997) and shown to be involved in the delivery of proteins to lysosomes and lysosome-related organelles, including the yeast vacuole (Cowles et al., 1997; Panek et al., 1997) and insect and mammalian pigment granules (Dell’Angelica et al., 1997; Simpson et al., 1997; Kantheti et al., 1998). Conflicting results have been obtained as to whether the AP-3 complex is associated with clathrin. In vitro experiments have shown that clathrin and AP-3 are able to interact with each other (Dell’Angelica et al., 1998); however, AP-3 is not enriched in purified clathrin-coated vesicle preparations (Simpson et al., 1996), and at least in yeast, genetic studies show that AP-3 and clathrin participate in different and nonoverlapping pathways (Panek et al., 1997; Vowels and Payne, 1998).
Two other distinct types of coated vesicles have been identified that act at early stages of the secretory pathway: COPI-coated vesicles, which bud from membranes of the Golgi stack and intermediate compartment, and COPII-coated vesicles, which bud from the endoplasmic reticulum (ER). Although distinct from the adaptor complexes, four of the subunits of COPI coats show a limited degree of homology to AP subunits (Lowe and Kreis, 1998).
A common feature of all of the coats described so far is their ability to be recruited from the cytosol onto the membrane in a GTP-dependent manner. Thus, the recruitment of AP-1, AP-3, and COPI is greatly enhanced by the addition of GTPγS and blocked by the addition of the fungal metabolite brefeldin A, which inactivates the guanine nucleotide exchange factor for the small GTPase ADP ribosylation factor (ARF) (Donaldson et al., 1992; Robinson and Kreis, 1992; Peyroche et al., 1996; Simpson et al., 1996). Although AP-2 at the plasma membrane is insensitive to brefeldin A, in the presence of GTPγS or constitutively active ARF1 it is mistargeted onto late endosomes, and this recruitment is sensitive to brefeldin A (Seaman et al., 1993; West et al., 1997). COPII recruitment onto the ER requires another small GTPase, Sar1p (Barlowe et al., 1994).
The three AP complexes, AP-1, AP-2, and AP-3, share a number of additional common features. They are all heterotetramers composed of two large subunits of ∼100–130 kDa, a medium subunit of ∼50 kDa, and a small subunit of ∼20 kDa. Thus, AP-1 consists of the large subunits γ and β1, the medium subunit μ1, and the small subunit ς1, whereas the AP-2 complex consists of α, β2, μ2, and ς2 subunits, and the AP-3 complex consists of δ, β3, μ3, and ς3 subunits. The corresponding subunits in the three complexes generally share between 20 and 40% amino acid identity as well as a number of conserved motifs. The three complexes also all bind to similar types of sorting signals on the cytoplasmic domains of transmembrane proteins: tyrosine-based sorting signals, which have been shown to interact with the μ subunits of the three AP complexes (Ohno et al., 1995, 1998; Owen and Evans, 1998), and dileucine signals, which may interact with their β subunits (Greenberg et al., 1998).
There are still a number of vesicular transport pathways for which no coat has been identified. These include trafficking from the endosome to the lysosome, recycling from the endosome to the TGN (Luzio and Banting, 1993), and trafficking of basolaterally targeted plasma membrane proteins in polarized epithelial cells (Matter and Mellman, 1994). Interestingly, proteins that take these pathways have all been shown to make use of tyrosine and/or dileucine sorting signals. A recent report has suggested that the AP-1 complex may play a role in the movement of proteins from endosomes to the basolateral membrane (Futter et al., 1998); however, there may be additional adaptor-related complexes involved in other trafficking events.
In this study we set out to identify novel adaptor-related complexes that could play a role in sorting steps for which there is, at present, no known coat. We have cloned and sequenced cDNAs encoding four novel proteins, one belonging to each of the four families of AP subunits, and have shown that these four proteins interact with each other in a complex that we are calling AP-4. We have localized the complex at both the light and the electron microscope level, we have investigated the effects of brefeldin A on its distribution, and we have looked at the ability of the μ subunit of the complex to interact with tyrosine-based sorting signals.
MATERIALS AND METHODS
Cloning and Sequencing
The expressed sequence tag (EST) database was searched for proteins with homology to the γ/α/δ, β, μ, and ς families of AP subunits. Clones were then obtained from the IMAGE Consortium, sequenced, and expressed as recombinant fusion proteins for the production of antibodies. Most molecular biology techniques were performed as described by Sambrook et al. (1989).
The ε subunit was first identified as a human testis EST (IMAGE Consortium clone ID 1031294). Sequencing of this clone indicated that both the 5′ and 3′ ends of the cDNA were missing (the clone encodes amino acids 35–455 of the full-length ε subunit). To obtain the 5′ and 3′ ends of ε, we screened a human heart cDNA library (Clontech Laboratories, Palo Alto, CA) with a PCR fragment of the EST clone encoding amino acids 36–128. This screen yielded clones that covered amino acids 1–737 of the full-length ε. A second library screen was performed with a restriction fragment encoding amino acids 621–737 of ε to obtain the 3′ sequence. This screen yielded a single clone that extended over amino acids 453-1079 of full-length ε but did not include an obvious stop codon. The 3′ sequence of this clone was used to search the EST database again, and a single mouse EST entry (IMAGE Consortium clone ID 581775) was identified that was 90% identical to the human library clone and extended further in the 3′ direction. A third library screen was performed using a PCR fragment of the mouse EST corresponding to the 3′-coding sequence, and two clones were obtained that included the rest of the coding sequence of human ε (amino acids 1080–1108).
Similar strategies were used to clone β4, μ4, and ς4. β4 was first identified as a human brain EST (IMAGE Consortium clone ID 51694). Sequencing of this clone indicated that the 5′ end of the cDNA was missing (the clone encodes amino acids 387–739 of the full-length β4). To obtain the 5′ end of β4, we screened a human brain cDNA library (Clontech Laboratories) with a restriction fragment of the EST corresponding to amino acids 388–547 of the full-length β4. This screen yielded a clone that extended from amino acid 1 to amino acid 443 and was used to search the EST database. This search identified human (IMAGE Consortium clone ID 547064) and mouse (IMAGE Consortium clone ID 312360) ESTs. Sequencing of the ESTs revealed that both contained full-length β4-adaptin corresponding to amino acids 1–739, but the human EST had a deletion of amino acids 416–451.
μ4 was first identified as a human brain EST (IMAGE Consortium clone ID 48136). Sequencing of this clone (in collaboration with A. Whitney, University of Geneva, Geneva, Switzerland) indicated that the 5′ end of the cDNA was missing (the clone encodes amino acids 49–453 of the full-length μ4). To obtain the full-length μ4, we screened a human brain cDNA library with a restriction fragment corresponding to amino acids 49–202. This screen yielded a clone that extended the full length of μ4, corresponding to 453 amino acids.
ς4 was first identified as ESTs from human placenta (IMAGE Consortium clone IDs 259587 and 259562). Sequencing indicated that these clones encoded full-length ς4 corresponding to 144 amino acids.
Sequencing of all clones was performed by John Lester (University of Cambridge, Cambridgeshire, UK) on an automated ABI sequencer using oligonucleotide primers to “walk out” along the DNA. The entire coding sequence was read in both directions.
Sequences of related AP subunits were compared using the SIP program to calculate percent identities and make diagon plots (Staden, 1990). To construct a phylogenetic tree, we compared the sequences using the Clustal method with the PAM250 residue weight table, part of the MegAlign package (DNASTAR, Madison, WI).
Northern Blotting
Human multiple tissue Northern blots (Clontech Laboratories) were probed according to the manufacturer’s instructions using probes that had been labeled with 32P by random priming. The ε, β4, and μ4 probes were the same DNA sequence as those used for library screening, corresponding to amino acids 36–128, 388–547, and 49–202, respectively. The ς4 probe corresponded to a restriction fragment encoding the full-length protein.
Antibody Production
Fusion proteins of ε and β4 with glutathione S-transferase were constructed by the addition of appropriate restriction sites by PCR and cloning into pGEX-4T-1 (Pharmacia, Piscataway, NJ). Primers were designed to amplify amino acids 607–717 of full-length ε and amino acids 387–739 of full-length human β4 and to add SalI and EcoRI restriction sites for ε and SmaI and NotI restriction sites for β4. A fusion protein of μ4 with glutathione S-transferase was constructed by the restriction digestion of μ4 in Bluescript with BamHI (fragment encodes amino acids 49–202 of μ4).
The PCR products and restriction fragment were ligated into pGEX-4T-1 and transformed into MC1061 cells, and expression of the fusion protein was induced. The ε fusion protein was partially soluble and was purified using glutathione-Sepharose affinity chromatography (Pharmacia). The β4 and μ4 fusion proteins were found to be insoluble and so were purified from inclusion body preparations as described previously (Page and Robinson, 1995). In each case the antigens were injected into at least two rabbits. The immunization protocol and the affinity purification of the resulting antisera have been described previously (Page and Robinson, 1995). After affinity purification, the antisera were tested on Western blots of whole–pig brain cytosol, as well as by immunoprecipitation followed by Western blotting.
The anti-ε antisera raised in four different rabbits all recognized a predominant protein band of 130 kDa by Western blotting. The anti-β4 antisera were not able to detect any protein bands on a Western blot of brain cytosol, but the antisera raised in two different rabbits both recognized the same protein band of 85 kDa by immunoprecipitation followed by Western blotting. The anti-μ4 antisera were able to detect a protein band of 50 kDa, but only under reducing conditions. Unfortunately, attempts to raise an antiserum against ς4 were not successful.
Epitope Tagging
An epitope-tagged version of β4 was constructed by the insertion of a 22-amino acid sequence (ELEPPAPESPMALLADPAPAAD), derived from the hinge domain (amino acids 706–727) of the αA subunit of the AP-2 complex. Previous studies have shown that this sequence is encoded by an alternatively spliced exon that is normally only expressed in neurons, and we have raised both rabbit and mouse antisera against it (rabbit anti-A706–727 and mouse anti-A706–727) (Seaman et al., 1993; Ball et al., 1995). The epitope was constructed by designing two complementary primers that when annealed gave PvuII and EcoRI restriction site overhangs. The double-stranded oligonucleotide was ligated to two β4 PCR products: one encoding amino acids 1–579, with KpnI and PvuII restriction sites at the 5′ and 3′ ends, and the other encoding amino acids 580–739, with EcoRI and NotI sites at the 5′ and 3′ ends. This construct was cloned into the KpnI and NotI sites of ΔpMEP4 (Reaves and Banting, 1994; Girotti and Banting, 1996), downstream from an inducible metallothionein IIA promoter. Expression of the epitope-tagged β4 was induced by the addition of 5 mM ZnCl2 to the culture medium for 15 h.
Transfection of Rat 1 Cells
Rat 1 fibroblasts maintained in DME supplemented with 10% fetal calf serum (DME-FCS) were transfected with the epitope-tagged β4 in ΔpMEP4 using Lipofectamine reagent (Life Technologies, Gaithersburg, MD). Cells were grown in 6-cm dishes until 70% confluent and rinsed in serum-free Optimem medium. Fifteen microliters (3 μg) of DNA were added to 185 μl of Optimem 1 (Life Technologies), and 20 μl of Lipofectamine was separately added to 180 μl of Optimem. The two were then mixed and incubated for 25 min at room temperature. The DNA–Lipofectamine mix was then added to the cells in 1.6 ml of DME-FCS and incubated for 5 h, after which the cells were rinsed and the medium was replaced with DME-FCS.
The cells were trypsinized the following day, split into two 15-cm dishes, and left for another day before the addition of Hygromycin B (Boehringer Mannheim, Indianapolis, IN) at a concentration of 0.2 mg/ml. After 2 weeks, control untransfected Rat 1 cells had all died, and by immunofluoresence >90% of transfected Rat 1 cells stained positive with rabbit anti-A706–727.
Yeast Two-Hybrid Systems
The “Matchmaker” two-hybrid system was obtained from Clontech Laboratories, and all procedures were performed according to the manufacturer’s instructions. PCR was used to generate appropriate sites to clone the full-length β4, μ4, and ς4 and amino acids 80–737 of ε into the yeast expression vectors pGBT9 and pGAD424. Insertion into the vector pGBT9 results in the fusion of the protein with the GAL4 DNA-binding domain, whereas insertion into the vector pGAD424 results in the fusion of the protein with the GAL4 transcriptional activation domain. Interactions between proteins result in the production of β-galactosidase activity.
The yeast strain Y187 was cotransformed with the plasmids using a polyethylene glycol–lithium acetate protocol and was grown on selection plates lacking leucine and tryptophan to select for colonies containing both plasmids. The colonies were then transferred onto filter paper and permeabilized by freezing in liquid nitrogen and thawing at room temperature. The filters were then placed on another piece of filter paper that had been presoaked in a solution containing 0.33 mg/ml 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside and were incubated at 30°C for 15 h to test for β-galactosidase activity by the production of a blue reaction product. β-Galactosidase activity was also measured using a liquid culture assay as described previously (Page and Robinson, 1995).
The yeast two-hybrid system was also used to investigate interactions between AP μ subunits and the cytoplasmic domain of the lysosomal membrane protein CD63. Constructs encoding full-length μ1, μ2, and μ3 and partial μ4, in the two-hybrid transcriptional activation domain vector pVP16, were kindly provided by Banting (Stephens and Banting, 1998). Wild-type (KSIRSGYEVM) and mutant (KSIRSGAEVM) versions of the cytoplasmic tail of CD63 were amplified by PCR and subcloned into the two-hybrid DNA-binding domain vector pBTM116. The yeast strain L40, maintained as described by Stephens and Banting (1998), was cotransformed with two plasmids using a polyethylene glycol–lithium acetate procedure and plated onto selective medium lacking leucine and tryptophan to select for colonies containing both plasmids. After 3–4 d, colonies were inoculated into liquid cultures to perform quantitative growth assays. Liquid cultures were set up by inoculating 0.15 OD600 units of cells into 2 ml of selective medium lacking leucine, tryptophan, and histidine and were assayed for growth after 0–148 h of incubation at 30°C by measurement of OD600. Each time point was assayed in triplicate.
Immunoprecipitations and Western Blotting
Immunoprecipitations were performed either on pig brain cytosol, prepared in PBS as described previously (Seaman et al., 1993), or on Rat 1 cells under nondenaturing conditions. The Rat 1 cells were washed in PBS, drained of excess buffer, and then incubated with immunoprecipitation buffer (150 mM NaCl, 1% Nonidet P-40, and 50 mM Tris-HCl, pH 8) for 10 min at 4°C with rocking. The lysed cells were harvested with a cell scraper and clarified by centrifugation for 20 min at 14,000 rpm.
All samples were precleared by the addition of 100 μl of 50% protein A-Sepharose (Pharmacia) for each milliliter of cytosol or cell lysate. To each of the 200-μl aliquots was added 20 μl of affinity-purified antibodies, and the samples were incubated for 2 h at room temperature. The antibodies used were those against ε, β4, and μ4 and rabbit anti-A706–727. After the antibody incubation step, 70 μl of 50% protein A-Sepharose was added, and the samples were incubated for an additional hour at room temperature, after which the Sepharose was collected by centrifugation and washed five times with PBS containing 0.1% Nonidet P-40. The samples were then boiled in sample buffer, run on SDS polyacrylamide gels, and subjected to Western blotting. Blots were probed with various antibodies followed by 125I-labeled protein A, as described previously (Robinson and Pearse, 1986).
Immunofluorescence
Rat 1 cells stably expressing the epitope-tagged version of human β4 were grown on multiwell test slides and fixed either with 3% paraformaldehyde followed by 0.1% saponin or with methanol and acetone, as described previously (Robinson, 1987). For some experiments the cells were treated with 10 μg/ml brefeldin A (Sigma Chemical, St. Louis, MO) before fixation. The cells were then labeled with rabbit anti-A706–727 either alone or together with a mouse monoclonal antibody. The monoclonal antibodies used included anti-transferrin receptor (Chemicon International, Temecula, CA), anti-mannosidase II (a gift from Graham Warren, ICRF, London, UK), anti-TGN38 (a gift from George Banting, University of Bristol, Bristol, UK), anti-lgp120 (GM10) (Grimaldi et al., 1987), and anti-clathrin (X22) (Brodsky, 1985). Alternatively, the cells were labeled with anti-ε together with mouse anti-A706–727. The secondary antibodies used were fluorescein-conjugated donkey anti-rabbit IgG and Cy3-conjugated goat anti-mouse IgG (when mouse anti-A706–727 was used) or fluorescein-conjugated sheep anti-mouse IgG and Cy3-conjugated goat anti-rabbit IgG (when rabbit anti-A706–727 or anti-ε was used). All secondary antibodies were supplied by Amersham (Arlington Heights, IL).
Electron Microscopy
For immunogold localization of AP-4, Rat 1 cells expressing the epitope-tagged β4 and nontransfected Rat 1 control cells were trypsinized and fixed for 1 h with 4% paraformaldehyde in 0.1 M sodium cacodylate, pH 7.2, at room temperature, pelleted, and embedded in gelatin. The cells were then prepared for ultrastructural immunocytochemistry essentially as outlined by Griffiths (1993). Briefly, the embedded cell pellets were infused with 1.7 M sucrose and 15% polyvinylpyrrolidone in PBS overnight at 4°C and then frozen on aluminum stubs in liquid nitrogen. Frozen ultrathin sections were cut using a Reichert Ultracut S Ultramicrotome equipped with an FCS cryochamber attachment (Leica, Milton Keynes, UK). Sections were collected and labeled with rabbit anti-A706–727 using the protein A-gold technique (Slot and Geuze, 1983). The sections were then contrasted by embedding them in freshly prepared 1.8% methyl cellulose and 0.3% uranyl acetate (Tokuyasu, 1978), allowed to air dry, and observed in a transmission electron microscope (CM100; Philips Electronic Instruments, Mahwah, NJ).
RESULTS
Identification of Novel Proteins Related to AP Subunits
To identify subunits of novel AP complexes, we searched the EST database for mammalian sequences with 15–40% identity to sequences of subunits in the AP-1, AP-2, and AP-3 complexes. Sequences that were much more closely related to subunits of one of the complexes than to those of the others were ruled out, because these were likely to be novel isoforms of subunits of a known complex rather than subunits of a novel complex. Using these criteria, we identified, cloned, and sequenced a number of ESTs. In some cases the ESTs did not encode a full-length protein, so library screens were performed to obtain the rest of the sequence. Figure 1A shows the sequences of four candidate novel AP subunits, ε, β4, μ4, and ς4, whereas Figure 1B shows diagon plots of these sequences against those of the corresponding subunits in the AP-1, AP-2, and AP-3 complexes to illustrate their relationship. Figure 1C shows the positioning and sequences of various conserved motifs.
Figure 1.
Sequences of the novel adaptor subunit–related proteins and comparison with their counterparts in AP-1, AP-2, and AP-3. (A) cDNAs encoding the four proteins were originally identified as ESTs encoding homologues of the known adaptor subunits. The complete sequences of the human ε, mouse β4, and human μ4 subunits were obtained by library screening, whereas a full-length human ς4 clone was available as an EST. These sequence data are available from GenBank/EMBL/DDBJ under accession numbers AF155156 (ε), AF155157 (β4), AF155158 (μ4), and AF155159 (ς4). μ4 has been identified previously as μ-adaptin–related protein 2 (μARP2 [Wang and Kilimann, 1997]). (B) The sequences of ε, β4, μ4, and ς4 were compared with the sequences of their counterparts in the AP-1 (left), AP-2 (middle), and AP-3 (right) complexes using the SIP program (Staden, 1990), which was also used to calculate the percent amino acid identity. The novel proteins are no more like any one subunit than the others, indicating that they are novel homologues of the known subunits rather than isoforms. (C) The four members of the γ/α/δ/ε, β, and μ families are shown diagrammatically, together with the positions of some of the conserved motifs: the KRIGYL (K) and WIIGEY (W) motifs in γ, α, δ, and ε; the KKLVYLY (K) and WIIGEY (W) motifs in the β family; and the YELLDE (Y) motif in the μ family.
ε has a predicted size of 127 kDa and is ∼20% identical to the γ, α, and δ subunits. This homology is restricted to the first ∼600 amino acids, as is also found when γ, α, and δ are compared with each other (Robinson, 1990; Simpson et al., 1997). Although the homology between the various members of the γ/α/δ family is relatively weak, some conserved stretches of sequence have been found, including a KRIGYL motif (Figure 1C, position K) at approximately amino acid 100 and a WIIGEY motif (Figure 1C, position W) at approximately amino acid 500 (Robinson, 1989, 1990; Simpson et al., 1997), both of which are also present in ε (Figure 1C, KRVGYL at amino acids 113–118 and WVLGEY at amino acids 514–519). γ, α, and δ also all have a three-domain structure consisting of a conserved N-terminal domain of ∼600 amino acids, a hydrophilic, proline- and glycine-rich domain of ∼100–150 amino acids that is thought to act as a hinge, and a C-terminal “ear” domain of ∼100–300 amino acids. ε shares the conserved N-terminal domain, but its putative hinge domain, comprising amino acids 600–750, is only moderately hydrophilic and enriched in proline and glycine residues. However, by every other criterion ε is clearly a member of the same family as γ, α, and δ, and is equally related to all three of them, whereas δ is only ∼16% identical to either γ or α, which are 26% identical to each other (Simpson et al., 1997).
β4 has a predicted size of 83 kDa and is also ∼20% identical to its counterparts in AP-1, AP-2, and AP-3, with the homology again restricted to the ∼600 amino acid N-terminal domain. Like members of the γ/α/δ family, members of the β family have also all been shown to have a WIIGEY motif (Figure 1C, position W) at approximately amino acid 500 (interestingly, this motif is also present in the COPI subunit β-COP [Duden et al., 1991]) as well as a KKLVYLY motif (Figure 1C, position K) near the N terminus, probably related to the KRIGYL motif in the γ/α/δ family (Kirchhausen et al., 1989; Ponnambalam et al., 1990; Newman et al., 1995; Simpson et al., 1997). β4 shares both of these motifs (Figure 1C, KKLVYLY at amino acids 65–71 and WLLGVH at amino acids 441–446). However, β4 is smaller than β1, β2, or β3 and appears to be missing most of its C-terminal hinge and/or ear domain. This is not unprecedented, however, because one of the members of the β family in the budding yeast Saccharomyces cerevisiae, Apl1p, is also truncated at its C-terminal end, with a predicted size of 80 kDa.
μ4 has a predicted size of 50 kDa and is ∼25% identical to its counterparts in the other three complexes. Again, it shares the motifs that have been found in other μ family members, including the YELLDE motif (Figure 1C, position Y) at amino acids 110–115. Like other μ subunits, it also has weak homology with the ς subunits and with δ-COP over its N-terminal 150 amino acids. This subunit has been independently cloned and sequenced by Wang and Kilimann (1997). ς4 has a predicted size of 17 kDa and shows the greatest homology of any of the novel sequences to its counterparts in the other three complexes, being ∼30–40% identical to ς1, ς2, and ς3.
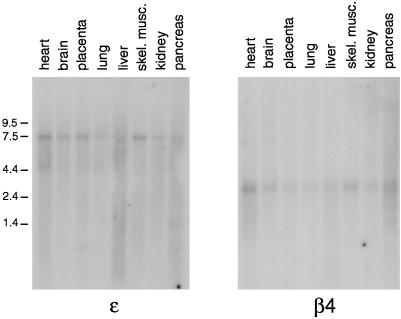
Multiple tissue Northern blots were probed to determine the expression patterns of the four novel proteins. Figure 2 shows blots probed for ε and β4 and illustrates that both are expressed ubiquitously, although at low levels, and that they show similar distribution patterns. Similar results were obtained for μ4 and ς4 (our unpublished results) (see also Wang and Kilimann, 1997; Dell’Angelica et al., 1999a).
Figure 2.
Expression patterns of ε and β4. Multiple tissue human Northern blots were probed with cDNAs specific for the two novel adaptor subunit–related proteins. Both genes appear to be expressed ubiquitously, but labeling is relatively weak, suggesting that the mRNAs are of low abundance. skel. musc., skeletal muscle.
ε, β4, μ4, and ς4 Form a Complex
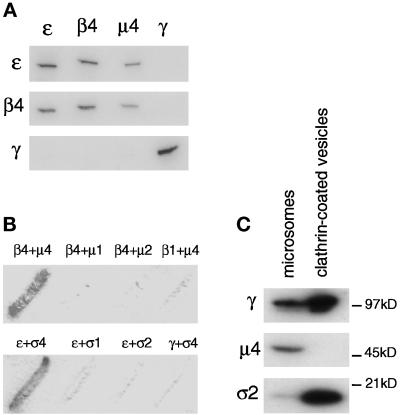
To characterize the four novel adaptor subunit homologues further and to determine whether they associate with each other to form a fourth AP complex, we expressed the fusion proteins and injected them into rabbits for antibody production. The fusion proteins were not full-length but were designed to include the portions of the proteins that are the most divergent and that have been successfully used to raise antibodies against related subunits of the other three AP complexes. The antibodies were affinity purified and used for immunoprecipitation and Western-blotting experiments. Figure 3A shows the results of one such experiment. Pig brain cytosol was immunoprecipitated under nondenaturing conditions with anti-ε, anti-β4, and anti-μ4. As a control, cytosol was also immunoprecipitated with an antibody against the γ subunit of the AP-1 complex. The immunoprecipitates were subjected to SDS-PAGE, blotted onto nitrocellulose, and probed with anti-ε, anti-β4, and anti-γ. The results show that anti-ε, anti-β4, and anti-μ4 all bring down both ε and β4. However, none bring down γ, nor does anti-γ bring down ε or β4, as expected if they form distinct complexes. It was not possible to probe blots with the μ4 antibody because the protein band was masked by the presence of immunoglobulin heavy chain. In addition, attempts to raise a specific antibody against ς4 were unsuccessful.
Figure 3.
The four proteins are subunits of a novel adaptor-related complex not associated with clathrin. (A) Pig brain cytosol was immunoprecipitated under nondenaturing conditions using affinity-purified antibodies raised against ε, β4, and μ4 and against the γ subunit of the AP-1 complex. The samples were separated by SDS-PAGE and blotted. The appropriate region of the gel was cut out and probed with ε, β4, and γ antibodies. The results show that the novel proteins, ε, β4 and μ4, all coimmunoprecipitate but do not associate with the γ subunit of the AP-1 complex. (B) Interactions between the four novel proteins were investigated using the yeast two-hybrid system. Yeast cells were transformed with pairs of constructs containing ε, β4, μ4, or ς4 fused with either the transcriptional activation domain (in pGAD424) or the DNA-binding domain (in pGBT9) of the GAL4 promoter. As controls, various combinations of constructs containing subunits of AP-1 and AP-2 complexes were transformed with either ε, β4, μ4, or ς4. The β + μ filters were incubated with substrate for 24 h, whereas the ε + ς filters were incubated with substrate for 8 h. Results show that the combinations of β4 and μ4 and of ε and ς4 interact with each other in a specific manner to produce β-galactosidase activity. No interaction can be detected in alternative pairings with subunits in AP-1 and AP-2 complexes. (C) AP-4 is not enriched in purified clathrin-coated vesicles. Equal protein loadings of clathrin-coated vesicles purified from pig brain and a crude microsomal fraction from a previous stage in the preparation were subjected to SDS-PAGE followed by Western blotting. The blot shows that the γ subunit of AP-1 and the ς2 subunit of AP-2 are strongly enriched in the clathrin-coated vesicles but the μ4 subunit of AP-4 is not.
The interactions between the four proteins were also investigated using the yeast two-hybrid system. Fusion proteins of ε, β4, μ4, and ς4 were made with both the GAL4 transcriptional activation domain (in pGAD424) and the GAL4 DNA-binding domain (in pGBT9). Pairs of constructs were then coexpressed in yeast, and interactions were detected by the presence of β-galactosidase activity. As negative controls, constructs were expressed either on their own, with an empty vector, or with subunits of the AP-1 and AP-2 complexes, with which they should not interact. None of the constructs on their own or with an empty vector produced any β-galactosidase activity (our unpublished results). However, as shown in Figure 3B, β4 specifically interacted with μ4 but not with μ1 or μ2, nor did μ4 interact with β1. In addition, there was a very strong and specific interaction between ε and ς4, but ε did not interact with ς1 or ς2, and ς4 did not interact with the γ subunit of the AP-1 complex (Figure 3B).
Together, the native immunoprecipitation experiments and yeast two-hybrid studies indicate that we have identified a novel adaptor-related complex. Like the conventional adaptor complexes AP-1 and AP-2 and the adaptor-related complex AP-3, it consists of a large γ/α/δ–related subunit (ε), a β subunit (β4), a μ subunit (μ4), and a ς subunit (ς4). We propose that this complex be called AP-4.
AP-1 and AP-2 are both associated with clathrin-coated vesicles, whereas there is some controversy as to whether AP-3 is clathrin associated or not. To determine whether AP-4 is associated with clathrin-coated vesicles, equal protein loadings of clathrin-coated vesicles purified from pig brain and crude microsomal membranes from a previous stage in the preparation were subjected to SDS-PAGE, and Western blots were probed with antibodies specific for the γ subunit of the AP-1 complex, the μ4 subunit of the AP-4 complex, and the ς2 subunit of the AP-2 complex. Both γ and ς2 were highly enriched in the clathrin-coated vesicle sample. However, the antiserum against μ4 was unable to detect a band in the clathrin-coated vesicle sample, although it labeled a band of the appropriate size in the microsome sample (Figure 3C). Thus, unlike AP-1 and AP-2, AP-4 does not appear to be associated with clathrin-coated vesicles.
Localization of the AP-4 Complex
In our first attempts to localize the AP-4 complex, we performed immunofluorescence experiments using our antibodies against the ε, β4, and μ4 subunits. The three antibodies all gave different patterns (our unpublished results), even though the results shown in Figure 3A indicate that the three proteins are part of the same complex. Because all three antibodies label more than one band on whole-cell Western blots (our unpublished results), it is likely that the different immunofluorescence patterns are a result of the antibodies cross-reacting with other proteins. To determine the localization of AP-4 with little or no background, we inserted an epitope tag into the β4 subunit. The epitope that we chose was a 22-amino acid sequence from the hinge domain of one of the isoforms of the α subunit of the AP-2 complex, which is only expressed in neuronal cells because of alternative splicing (Ball et al., 1995). We have used antibodies against this epitope previously for immunofluorescence and immunoelectron microscopy and know that they are highly specific (Seaman et al., 1993; Ball et al., 1995; West et al., 1997). In addition, because the epitope is derived from the hinge domain of one of the AP large subunits, we reasoned that we could transplant it into the comparable domain of another large subunit without interfering with protein folding or function.
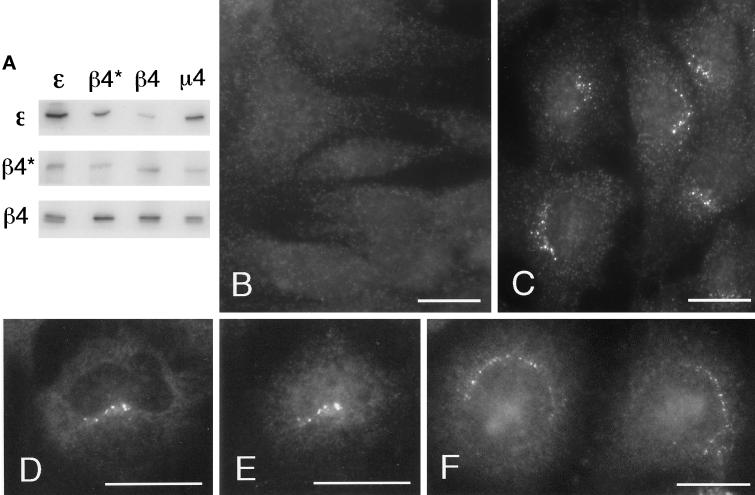
Rat 1 cells were stably transfected with the epitope-tagged β4 construct in the vector ΔpMEP4, which has a metallothionine promoter, and expression was induced by the addition of zinc chloride to the culture medium for 15 h. To ensure that the epitope-tagged β4 was correctly incorporated into AP-4 complexes, native immunoprecipitations were performed. Figure 4A shows an experiment in which cytosol from cells expressing the tagged β4 was immunoprecipitated with anti-ε, an antibody against the tag (anti-β4*), anti-β4, and anti-μ4. Western blots were then probed with anti-ε, anti-β4*, and anti-β4. The anti-ε, anti-β4, and anti-μ4 antibodies were all able to bring down the tagged β4 as well as ε and endogenous β4, and the antibody against the tag brought down ε as well as tagged β4. In the blot labeled with anti-β4, two bands can be seen in the anti-ε, anti-β4, and anti-μ4 immunoprecipitates. The more slowly migrating band is the only band visible in the anti-β4* immunoprecipitate and thus presumably corresponds to the tagged β4, whereas the lower band corresponds to endogenous β4. The upper band is somewhat more intense than the lower band in the anti-ε and anti-μ4 immunoprecipitates, indicating that at least one-half of the AP-4 complexes in the transfected cells contain the tagged β4.
Figure 4.
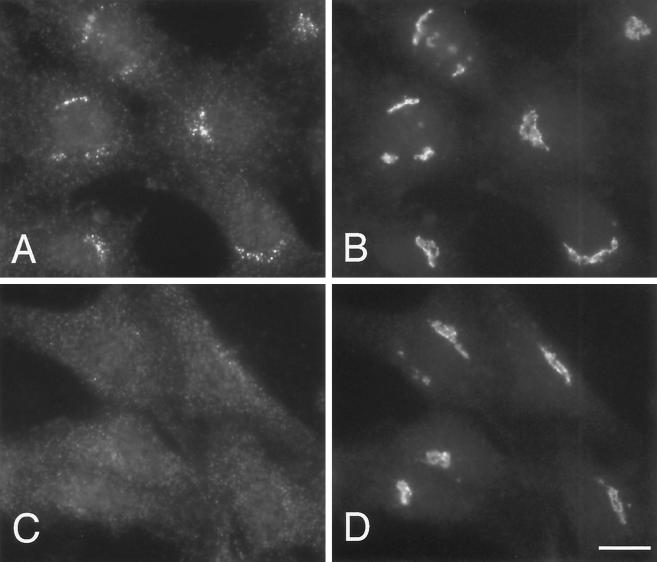
Expression and localization of epitope-tagged β4. (A) β4 tagged with an epitope derived from a neuronal-specific variant of the α subunit of AP-2 was stably transfected into Rat 1 cells. Cytosol from these cells was immunoprecipitated under nondenaturing conditions using affinity-purified antibodies raised against ε, β4, and μ4 and an antibody against the epitope tag that only recognizes the transfected β4 (β4*). The samples were subjected to SDS-PAGE and blotted, and the appropriate region of the blot was cut out and probed with antibodies against ε, tagged β4 (β4*), and total β4. The blot shows that antibodies against ε, β4, and μ4 all bring down tagged β4 as well as endogenous β4, whereas the antibody against the tag brings down ε as well as tagged β4. (B and C) Both control nontransfected cells (B) and cells stably transfected with tagged β4 (C) were labeled for immunofluorescence with a rabbit antibody against the epitope tag. The tagged β4 is localized to a fine pattern of discrete dots in the perinuclear region. This pattern is not seen in the nontransfected cells. (D and E) Stably transfected Rat 1 cells were double labeled with a rabbit antibody against the AP-4 ε subunit (D) and a mouse antibody against the epitope tag (E). Although there is more background than with the rabbit antibody against the tag, the same fine pattern of dots is seen with both antibodies. (F) Nontransfected HeLa cells were labeled with rabbit anti-ε. Again, a fine perinuclear pattern of dots is visible. Bars, 20 μm.
By immunofluorescence, the antibody against the epitope tag labeled a discrete pattern of ∼10–20 (15.7 ± 3.7; SEM) dots in the perinuclear region of transfected cells (Figure 4C). There was also a somewhat grainy background fluorescence, but this was seen in nontransfected as well as transfected cells and thus is presumably nonspecific (Figure 4B). As a further control to ensure that the labeled dots contain AP-4 complexes and not just the tagged β4 subunit, transfected cells were double labeled with rabbit anti-ε (Figure 4D) and a mouse antibody against the tag (Figure 4E). Although both antibodies gave more background than did the rabbit antibody against the tag, they both label perinuclear dots that can be seen to coincide with each other. In addition, when nontransfected HeLa cells were labeled with rabbit anti-ε, a similar pattern was seen (Figure 4F).
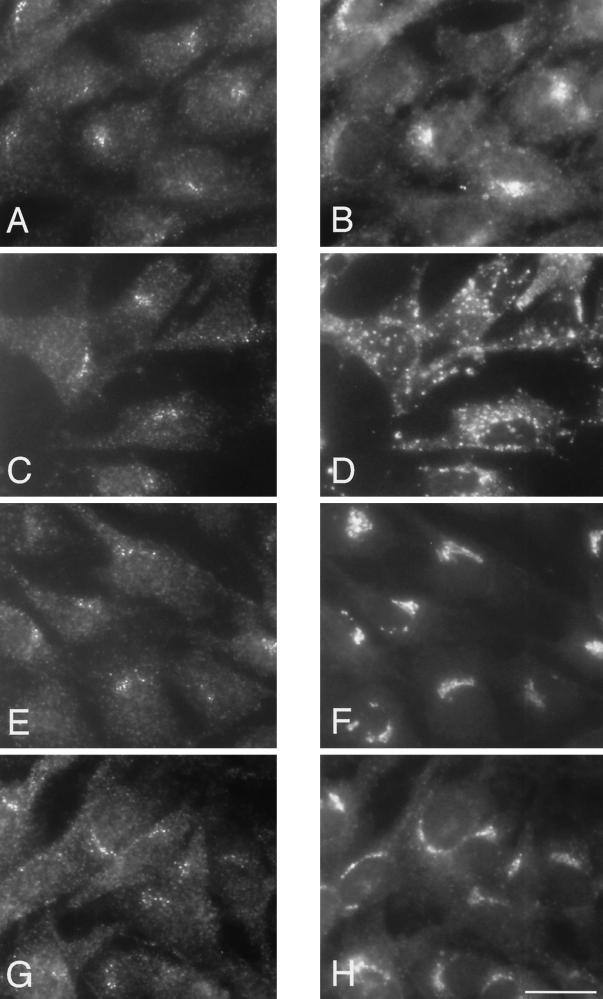
The perinuclear distribution of the epitope-tagged β4 and the endogenous ε suggests that the AP-4 complex is localized in the Golgi region of the cell. Because many organelles are found in this location, double-labeling immunofluorescence was performed to compare the distribution of AP-4 with that of various marker proteins. For these experiments, the stably transfected Rat 1 cells were labeled with the rabbit antibody against the epitope tag because it gives the best signal-to-background ratio. Figure 5 shows cells that were double labeled with the rabbit antibody against the tagged β4 (Figure 5, A, C, E, and G) and mouse monoclonal antibodies against the transferrin receptor, a marker for the early and/or recycling endosomal compartment (Figure 5B), lgp120, a marker for late endosomes and lysosomes (Figure 5D), mannosidase II, a marker for the Golgi stack (Figure 5F), and TGN38, a marker for the TGN (Figure 5H). Although most of these antibodies give a perinuclear-labeling pattern, the patterns are distinct from that seen with the antibody against the epitope-tagged β4. However, the AP-4–positive dots are most closely associated with the Golgi stack and the TGN, suggesting that they may correspond to vesicles budding from one or both of these compartments.
Figure 5.
Immunofluorescent double labeling of the tagged β4 subunit of the AP-4 complex with markers for the endocytic and exocytic pathways. Stably transfected Rat 1 cells were double labeled with an antibody against the epitope tag (A, C, E, and G) together with antibodies against the transferrin receptor (B), lgp120 (D), mannosidase II (F), and TGN38 (H). Although all the antibodies give perinuclear labeling, the patterns are distinct from that seen with the antibody against the epitope-tagged β4. However, both mannosidase II and TGN38 are in the same location as the tagged β4, suggesting that AP-4 may be associated with one or both of these compartments. Bar, 20 μm.
The other three coats that are associated with the Golgi stack and TGN, AP-1, AP-3, and COPI, are all sensitive to the drug brefeldin A. To determine whether the same is true for AP-4, Rat 1 cells transfected with the epitope-tagged β4 were treated either with or without 10 μg/ml brefeldin A for 2 min. The cells were then double labeled with antibodies specific for the tagged β4 and for the Golgi marker mannosidase II. Figure 6, A and B, shows that in the absence of brefeldin A, β4 and mannosidase II are both found in the perinuclear region of the cell (see also Figure 5, E and F). However, after the cells were incubated with the drug for 2 min, the β4-staining pattern had completely redistributed to the cytoplasm (Figure 6C). In contrast, the distribution of mannosidase II was unchanged at this time point (Figure 6D), although longer incubations caused it to redistribute to the ER (our unpublished results). The rapid response of AP-4 to brefeldin A suggests that, like the other three Golgi-associated coats, the membrane localization of AP-4 is dependent on the small GTPase ARF.
Figure 6.
Effect of brefeldin A on the distribution of AP-4. Rat 1 cells stably expressing epitope-tagged β4 were treated with (C and D) or without (A and B) 10 μg/ml brefeldin A for 2 min at 37°C, fixed, and double labeled with antibodies against tagged β4 (A and C) and the Golgi membrane protein mannosidase II (B and D). A complete redistribution of AP-4 can be seen, whereas the mannosidase II labeling is unaffected at this time point. Bar, 20 μm.
Electron Microscopy Localization of AP4
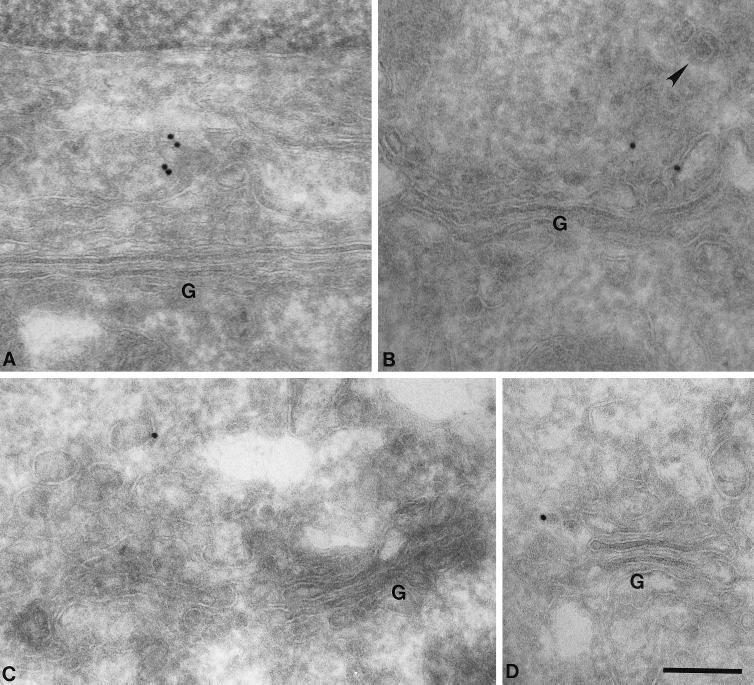
To determine the localization of AP-4 more precisely, immunogold electron microscopy (EM) was performed on Rat 1 cells stably transfected with epitope-tagged β4. Gold particles were very hard to find, consistent with the sparse-labeling pattern seen at the light microscope level (see Figures 4–6). Indeed, we calculate that if each fluorescent dot that we see in the light microscope represents a single coated bud or vesicle, in a typical cell 15 μm in diameter, containing 15 AP-4–coated buds or vesicles, and cut into 50-nm sections for electron microscopy, only ∼5% of all the sections of that cell are likely to contain AP-4–positive membranes. In addition, not all of the AP-4–positive membranes will be labeled, because even when we perform immunogold EM using a well-characterized antibody against clathrin, at least one-half of the structures that are clearly clathrin-coated vesicles remain unlabeled (Simpson et al., 1996). Because this degree of labeling is difficult to distinguish from background, it was necessary to look at a large number of cells and to compare transfected cells with nontransfected controls. Sections were cut from both sets of cells, and 10 grids of each were prepared and labeled with the rabbit antibody against the epitope tag followed by protein A coupled to 15-nm colloidal gold. The grids were then scrambled to ensure that scoring would be completely objective. Because the immunofluorescence labeling indicates that AP-4 is close to or coincident with the Golgi complex, Golgi stacks were found and scored as either positive or negative (to score positive, a Golgi stack had to have a membrane-associated gold particle within 500 nm). Positive Golgi stacks were also photographed. Out of 1109 Golgi stacks found in the transfected cells, 40 (3.6%) scored positive, whereas out of 677 Golgi stacks found in the control cells, 11 (1.6%) scored positive. Thus, the transfected cells had 2.25 times more Golgi-associated labeling than did the control cells. In addition, 11 of the positive Golgi complexes in the transfected cells had more than one gold particle associated with them, compared with only 2 of the positive Golgi complexes in the control cells.
Because at least one-half of the Golgi-associated labeling seen in the transfected cells is likely to be specific, it was possible to draw some conclusions about the distribution of AP-4. Figure 7 shows four examples of positive Golgi complexes from transfected cells. The labeling can be seen to be associated with tubulovesicular membranes near the Golgi stack, but not with the stack itself. The appearance of these membranes and the close proximity of clathrin-coated vesicles (Figure 7B, arrowhead) indicate that AP-4 is associated with the TGN. However, in agreement with the Western-blotting data shown in Figure 3, AP-4 does not appear to be clathrin associated.
Figure 7.
EM localization of AP-4. Stably transfected Rat 1 cells expressing epitope-tagged β4 were labeled with an antibody against the epitope tag followed by protein A coupled to 15-nm colloidal gold. Although the labeling was very low density, consistent with the immunofluorescent results shown in Figures 4–6, transfected cells had more than twofold higher labeling of membranes in the vicinity of the Golgi (G) stack than did nontransfected control cells. (A–D) Four examples of labeled membranes are shown. The label is associated with tubulovesicular membranes, which are likely to correspond to the TGN, but not with the Golgi stack itself. Clathrin-coated buds are frequently seen in the same general area (arrowhead in B), but the AP-4 does not appear to be clathrin associated. Bar, 200 nm.
Binding of μ4 to a YXXØ Sequence
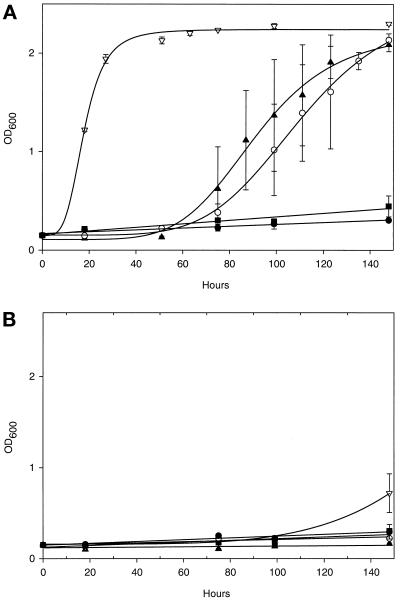
The μ subunits of AP-1, AP-2, and AP-3 have all been shown to interact specifically with tyrosine-based sorting signals with the consensus sequence YXXØ, in which Ø is a bulky hydrophobic residue (Ohno et al., 1995, 1998). However, the three μ subunits differ in their relative abilities to interact with different YXXØ sequences, and it has been postulated that these differences contribute to the different localizations of different YXXØ-containing proteins (Ohno et al., 1998). To determine whether μ4 also has the ability to interact with such sequences, μ1, μ2, μ3, and μ4 were all cloned into the two-hybrid transcriptional activation domain vector pVP16, and part of the cytoplasmic domain of the lysosomal membrane protein CD63, containing the sorting signal YEVM, was cloned into the two-hybrid DNA-binding domain vector pBTM116. As a control, the same sequence from CD63 but with the tyrosine mutated to an alanine was also cloned into pBTM116. Pairs of constructs were then transformed into yeast cells, and the ability of the various μ subunits to interact with the YEVM sorting signal was assayed by the ability of cells coexpressing them to grow in medium lacking histidine. Figure 8A shows that μ3 interacts most strongly with the YEVM sorting signal, followed by μ2 and μ4, whereas μ1 shows little or no interaction. None of the μ subunits interacted significantly with the AEVM control (Figure 8B). Thus, although μ4 appears to bind relatively weakly to tyrosine-based sorting signals, its binding is specific in that the tyrosine residue is absolutely required.
Figure 8.
The AP-4 μ4 subunit interacts with a YXXØ motif in a tyrosine-dependent manner. Yeast cells were cotransformed with a vector containing LexA fused with the cytoplasmic domain of wild-type CD63 (KSIRSGYEVM) (A) or a tyrosine-to-alanine mutant (KSIRSGAEVM) (B) and a vector containing VP16 fused to μ1 (●), μ2 (▴), μ3A (▿), or μ4 (○) or a VP16-only control (▪). Cultures were set up containing 0.15 OD600 units of cells in 2 ml of selective medium lacking histidine and grown at 30°C with shaking. OD600 readings were taken between 0 and 148 h to monitor growth in the absence of histidine, which only occurs if the two fusion proteins interact. Each time point represents the mean ± SEM of three separate cultures. (A) μ3A, μ2, and μ4 all interact with the wild-type tail, as measured by the ability of the cells coexpressing the two fusion proteins to grow. (B) This interaction is specific, because mutating the tyrosine to an alanine abolishes growth.
DISCUSSION
In a search to identify novel coat components, we have found four adaptor subunit homologues that specifically associate with each other to form a new adaptor-related protein complex, which we are calling AP-4. These subunits were discovered by searching through human and mouse cDNA sequences in the EST database. On the basis of the level of homology with subunits of the AP-1, AP-2, and AP-3 complexes, it was possible to predict that these proteins would be novel homologues rather than isoforms of the known AP subunits. Thus, whereas the various isoforms of AP subunits that have so far been described are from 60 to 90% identical (Robinson, 1989; Simpson et al., 1997; Takatsu et al., 1998), the four novel proteins that we identified are from 17 to 43% identical to their counterparts in the AP-1, AP-2, and AP-3 complexes, which is the level of homology that we see when we compare subunits from two different complexes.
We used two approaches to demonstrate that the subunits are all components of the same complex: immunoprecipitation and yeast two-hybrid analysis. By immunoprecipitation we were able to show that the ε, β4, and μ4 subunits were associated with each other, but we were unable to investigate ς4 in this way because we have not yet succeeded in raising ς4-specific antibodies. However, using the yeast two-hybrid system, we were able to demonstrate a very strong and specific interaction between ς4 and ε, consistent with our previous studies using this system to investigate interactions between subunits of the AP-1 and AP-2 complexes in which we showed specific interactions between ς1 and γ and between ς2 and α (Page and Robinson, 1995). In our previous studies on AP-1 and AP-2, we were also able to detect interactions between the β and μ subunits and between the β and γ/α subunits (Page and Robinson, 1995). Similarly, we have now shown that β4 and μ4 interact, but so far we have been unable to detect any interactions between β4 and ε (our unpublished results). It is possible that interactions between the large subunits of AP complexes are more difficult to reconstitute using the yeast two-hybrid system, because we have also been unable to detect any interactions between the β3 and δ subunits of the AP-3 complex, although β3 interacts with μ3 and δ interacts with ς3 (Lui and Robinson, unpublished observations).
The μ4 subunit has been independently cloned by Wang and Kilimann and named μARP2 (μARP1 was cloned from chicken and is probably its μ4 orthologue) (Wang and Kilimann, 1997). In addition, while our manuscript was in preparation, Dell’Angelica and coworkers published the sequences of the β4 and ς4 subunits of the AP-4 complex and showed by coimmunoprecipitation that these subunits were associated with each other and with μ4, although they did not have a full-length ε clone (Dell’Angelica et al., 1999a) (both β4 and ς4 are now available as full-length EST clones). They also performed immunofluorescence studies in HeLa cells, using an antibody raised against the C-terminal domain of β4, and showed that it gave perinuclear labeling that was sensitive to brefeldin A, as we have found as well. However, we believe that the labeling pattern that we see is different from theirs. Our antibodies against both tagged β4 and endogenous ε label a very fine pattern of discrete dots, whereas their perinuclear labeling is much more “chunky,” even in the same cell type. Indeed, we saw similar patterns in our initial immunofluorescence experiments using antibodies that we had raised against β4 and ς4, and the “ς4” pattern was even brefeldin A sensitive, but we subsequently found that in both cases the antibodies were cross-reacting with other proteins. In contrast, our ε antibody, although it gave a relatively high cytosolic background, labeled the same pattern of dots that we saw when we epitope-tagged β4 and localized it with a highly specific antibody. Because so many organelles are clustered in the pericentriolar region of the cell, we feel that it is important to be cautious when interpreting immunofluorescence labeling that gives this sort of pattern, especially when the protein is of very low abundance, as appears to be the case for AP-4 (see below).
What is the evidence that AP-4 is a coat component? First, it is highly sensitive to brefeldin A, indicating that like other coats, its association with membranes is regulated by a GTP-binding protein, probably ARF. Second, it has a punctate distribution by immunofluorescence, indicating that it is concentrated in discrete loci, most likely corresponding to buds and vesicles. Similarly, by immunogold EM, much of the AP-4 labeling is seen on budding profiles. In addition, both our studies and those of Stephens and Banting (1998) indicate that the μ4 subunit of the AP-4 complex can interact with the cytoplasmic tails of proteins with tyrosine-based sorting signals, consistent with a role in cargo selection. Thus, Stephens and Banting (1998) showed that μ4 binds to the cytoplasmic tail of the lysosomal membrane protein lgp120, whereas in the present study we showed an interaction with the cytoplasmic tail of CD63, which is tyrosine dependent because mutation of the tyrosine to an alanine abolishes this interaction. However, in general μ4 appears to interact with such signals relatively weakly when compared with μ2 and μ3 (Stephens and Banting, 1998).
If AP-4 is indeed a coat component, one would predict that another component of the coat would be a scaffolding protein such as clathrin. However, so far we have no evidence that AP-4 is clathrin associated. We cannot detect AP-4 subunits in samples of purified clathrin-coated vesicles by Western blotting, and there is no obvious colocalization of AP-4 and clathrin by immunofluorescence (our unpublished results), although there are difficulties in distinguishing the two patterns at this level. Immunogold EM also indicates that AP-4 is not clathrin associated. In addition, none of the AP-4 subunits contains the consensus clathrin-binding motif as defined by Dell’Angelica et al. (1998) (L[LI] [DEN][LF][DE]). Thus, AP-4 may require an as yet unidentified structural protein to facilitate vesicle budding.
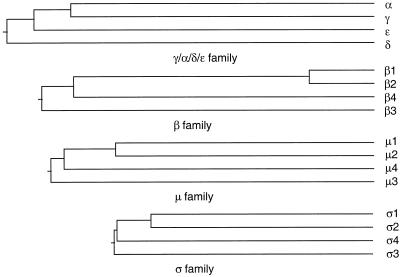
What is the evolutionary relationship between AP-4 and the other AP complexes? We have constructed a phylogenetic tree by aligning the sequences of the various subunits from each of the four AP complexes using the Clustal method. Figure 9 demonstrates that for all four subunit families, the AP-3 subunit appears to have diverged from its progenitor first, followed by the AP-4 subunit and then by the AP-1 and AP-2 subunits. Further insights into evolutionary relationships can be obtained by looking for homologues of the AP subunits in lower eukaryotes. In the budding yeast S. cerevisiae, in which the entire genome is known, genes have been identified that encode subunits sufficient to form three complete AP complexes, and there is strong genetic and biochemical evidence that yeast has AP-1 and AP-3 complexes that are functionally equivalent to AP-1 and AP-3 in mammals (Phan et al., 1994; Rad et al., 1995; Stepp et al., 1995; Cowles et al., 1997; Panek et al., 1997). It has been proposed that yeast also has an AP-2 complex, because the remaining AP subunits in yeast are more homologous to mammalian AP-2 subunits than to the subunits of any of the other AP complexes (Odorizzi et al., 1998), including AP-4. However, there is no evidence that yeast AP-2 is functionally equivalent to mammalian AP-2. Thus, it is possible that yeast “AP-2” is in fact neither AP-2 nor AP-4 and that at the time that the ancestors of fungi and animals diverged from each other, there were only two AP complexes, AP-3 and an AP-1–like complex that subsequently gave rise to the other AP complexes. Alternatively, the AP-4 complex may have been lost from some organisms. This is suggested by the observation that in the nematode worm Caenorhabditis elegans, in which the entire genome is also now known, there again appear to be no AP-4 subunits, although the homology between worm and mammalian AP-1, AP-2, and AP-3 subunits is extremely high. Is there any evidence of AP-4 in an organism other than a vertebrate? In searching through the database, we have found an EST from rice (accession number C27315) encoding a member of the ς family that is more homologous to ς4 in mammals (38% identity) than to any of the other ς subunits (28, 28, and 20% for ς1, ς2, and ς3, respectively). This suggests that some plants, at least, may have an AP-4 complex, even though it may have been lost from some animals such as the nematode worm.
Figure 9.
The phylogenetic relationships of subunits of the four AP complexes. A progressive alignment of the amino acid sequences of the four members of the γ/α/δ/ε, β, μ, and ς families was performed using the Clustal method. In every case, the AP-3 subunit appears to have branched off first, followed by the AP-4 subunit and then the AP-1 and AP-2 subunits.
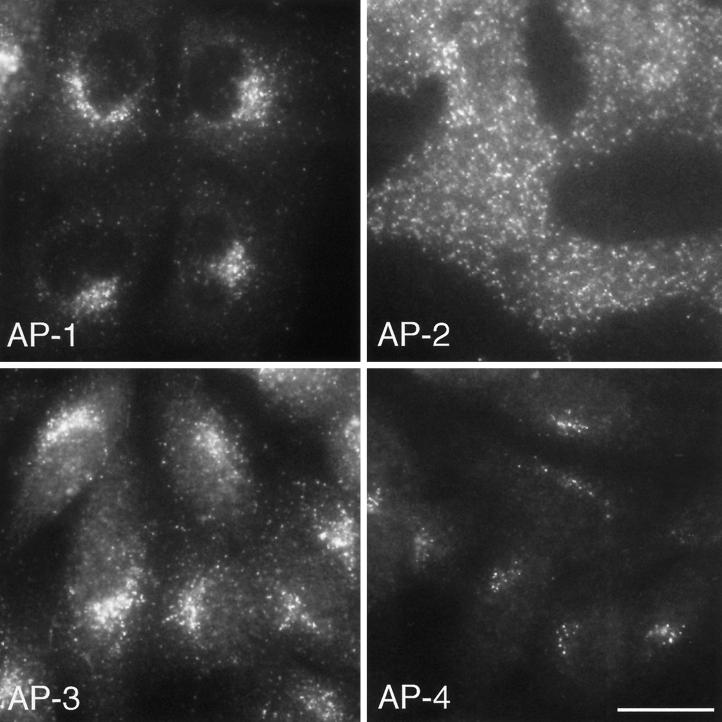
Although the AP-4 subunits are ubiquitously expressed, there are very few hits for them in the EST database when compared with their counterparts in the other three AP complexes. Indeed, as of April, 1999, there was only a single hit for ε in the human EST database. There are more hits for the other subunits in the human EST database (37 for β4, 19 for μ4, and 4 for ς4), but these are still relatively infrequent when compared with that for the other AP complexes. In addition, it is possible that the expression level of the ε subunit may be the most important in determining the amount of complex that gets made, because in the mocha mouse, which has a null mutation in the AP-3 δ subunit (the counterpart of ε in the AP-3 complex), none of the other AP-3 subunits are detectable by Western blotting, indicating that in the absence of δ they are unstable and get degraded (Kantheti et al., 1998). In comparison with the single human hit for ε, there are over 50 hits each for its homologues γ, α, and δ, suggesting that AP-4 may be as much as two orders of magnitude less abundant than AP-1, AP-2, or AP-3. Similarly, by immunofluorescence, AP-4 labeling is extremely sparse, with only ∼10–20 discrete dots per cell. We suspect that each dot may correspond to a single coated bud or vesicle, because we have used the same antibody to localize AP-2 and get a similar signal for each dot, where the number of dots seen by immunofluorescence correlates well with the number of plasma membrane–associated clathrin-coated pits and vesicles seen by electron microscopy (Ball et al., 1995). Thus, whereas there are ∼1000 or more AP-2–coated buds and vesicles per cell at steady state, there may be only 10–20 AP-4–coated buds and vesicles, indicating that the AP-4 pathway is not very extensively used (Figure 10).
Figure 10.
Immunofluorescence labeling comparing the distribution of the four AP complexes in Rat 1 cells. AP-4 is most closely associated with the perinuclear region of the cell, followed by AP-1 and AP-3, whereas AP-2 shows no perinuclear localization because it is associated with the plasma membrane. All four complexes have a punctate pattern, consistent with their association with buds and vesicles; however, there are many fewer dots seen with anti-AP-4 than with antibodies against the other three complexes. Bar, 20 μm.
What might the AP-4 pathway be? The localization of AP-4 indicates that it acts at the level of the TGN, whereas the ability of the AP-4 μ4 subunit to interact with the cytoplasmic domains of two lysosomal membrane proteins suggests that it may help direct such proteins from the TGN to lysosomes, although we cannot rule out an involvement in another pathway or pathways. AP-1 and AP-3 have also been implicated in the TGN-to-lysosome pathway, and it is possible that all four of the AP complexes act together to facilitate sorting to lysosomes, and possibly to other compartments as well. This is consistent with the phenotype of AP-3–deficient cells in which the steady-state distribution of lysosomal membrane proteins is still primarily lysosomal, even though some of the proteins get there less efficiently, escaping more frequently to the plasma membrane and then being reinternalized via the AP-2 pathway (Le Borgne et al., 1998; Dell’Angelica et al., 1999b) (Peden and Robinson, unpublished observations). A comparison of the distribution of the four AP complexes reveals that AP-4 is the most closely juxtaposed to the nucleus (Figure 10), whereas much of the AP-1 labeling and even more of the AP-3 labeling are found on more peripheral membranes, some of which are thought to be endosomal (Dell’Angelica et al., 1997; Simpson et al., 1997; Futter et al., 1998), as well as in the perinuclear Golgi region. In contrast, the AP-2 complex shows no perinuclear localization but is associated with the plasma membrane (Figure 10). Thus, a newly synthesized membrane protein that has just passed through the Golgi stack may encounter AP-4 first. If it fails to be recognized by AP-4, it may have “another chance” when it encounters AP-1, AP-3, and/or AP-2, possibly in that order. The differences in the steady-state distributions and trafficking pathways of membrane proteins bearing similar types of sorting signals probably reflect, at least in part, their relative abilities to interact with different AP complexes in the various compartments through which they travel.
Although AP-4 is expressed ubiquitously in small amounts, it is possible that there may be a particular type of cell in which it plays a crucial role and in which it may be much more abundant. So far we have looked at a number of different cell lines by Western blotting, including polarized epithelial cells, melanocytes, neuroblastoma cells, keratinocytes, B cells, and erythroblastoid cells, and we have not found any appreciable differences in the expression levels of AP-4 subunits, but there are many other types of cells in tissues. Techniques such as in situ hybridization or immunolocalization on whole-animal sections may help to identify a highly expressing cell type, if it exists. It is also possible that AP-4 is essential for the sorting of a very specific type of cargo. A combinatorial library screen of the sort performed by Ohno et al. (1998) may provide clues about sorting signals preferentially recognized by μ4. Are there any more AP complexes in mammals? So far, we have not found any candidate subunits in the EST database, but again, they may be only expressed in specialized types of cells that are not represented in the database. The completion of the human genome-sequencing project should reveal whether there are any more AP complexes or whether all of them have now been identified. The challenge for the future will be to find out precisely how all the AP complexes function and how they contribute to protein sorting in the context of the whole cell.
ACKNOWLEDGMENTS
We thank Andy Whitney, Ira Mellman, and Thomas Kreis for communicating their μ4- and β4-sequencing results and for helpful discussions; George Banting for the μ subunit two-hybrid clones; Paul Luzio, John Kilmartin, and members of the Robinson lab for reading and commenting on the manuscript; and members of the Cambridge and London membrane traffic community for their constructive suggestions. This work was supported by grants from the Human Frontier Science Program, the Wellcome Trust, and the Medical Research Council.
REFERENCES
- Ball CL, Hunt SP, Robinson MS. Expression and localization of α-adaptin isoforms. J Cell Sci. 1995;108:2865–2875. doi: 10.1242/jcs.108.8.2865. [DOI] [PubMed] [Google Scholar]
- Barlowe C, Orci L, Yeung T, Hosobuchi M, Hamamoto S, Salama N, Rexach MF, Ravazzola M, Amherdt M, Schekman R. COPII: a membrane coat formed by sec proteins that drive vesicle budding from the endoplasmic reticulum. Cell. 1994;77:895–907. doi: 10.1016/0092-8674(94)90138-4. [DOI] [PubMed] [Google Scholar]
- Brodsky FM. Clathrin structure characterized with monoclonal antibodies. I. Analysis of multiple antigenic sites. J Cell Biol. 1985;101:2047–2054. doi: 10.1083/jcb.101.6.2047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowles CR, Odorizzi G, Payne GS, Emr SD. The AP-3 adaptor complex is essential for cargo-selective transport to the yeast vacuole. Cell. 1997;91:109–118. doi: 10.1016/s0092-8674(01)80013-1. [DOI] [PubMed] [Google Scholar]
- Dell’Angelica EC, Klumperman J, Stoorvogel W, Bonifacino JS. Association of the AP-3 complex with clathrin. Science. 1998;280:431–434. doi: 10.1126/science.280.5362.431. [DOI] [PubMed] [Google Scholar]
- Dell’Angelica EC, Mullins C, Bonifacino JS. AP-4, a novel protein complex related to clathrin adaptors. J Biol Chem. 1999a;274:7278–7285. doi: 10.1074/jbc.274.11.7278. [DOI] [PubMed] [Google Scholar]
- Dell’Angelica EC, Ohno H, Ooi CE, Rabinovich E, Roche KW, Bonifacino JS. An adaptor-like protein complex with ubiquitous expression. EMBO J. 1997;16:917–928. doi: 10.1093/emboj/16.5.917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dell’Angelica EC, Shotelersuk V, Aguilar RC, Gahl WA, Bonifacino JS. Altered trafficking of lysosomal proteins in Hermansky-Pudlak syndrome due to mutations in the beta 3A subunit of the AP-3 adaptor. Mol Cell. 1999b;3:11–21. doi: 10.1016/s1097-2765(00)80170-7. [DOI] [PubMed] [Google Scholar]
- Donaldson JG, Finazzi D, Klausner RD. Brefeldin A inhibits Golgi membrane-catalyzed exchange of guanine nucleotide onto ARF protein. Nature. 1992;360:350–352. doi: 10.1038/360350a0. [DOI] [PubMed] [Google Scholar]
- Duden R, Griffiths G, Frank R, Argos P, Kreis TE. β-COP, a 100 kDa protein associated with nonclathrin-coated vesicles and the Golgi complex, shows homology to β-adaptin. Cell. 1991;64:649–665. doi: 10.1016/0092-8674(91)90248-w. [DOI] [PubMed] [Google Scholar]
- Futter CE, Gibson A, Allchin EH, Maxwell S, Ruddock LJ, Odorizzi G, Domingo D, Trowbridge IS, Hopkins CR. J. Cell Biol. 141:611–623. 1998. In polarized MDBK cells basolateral vesicles arise from clathrin-gamma-adaptin-coated domains on endosomal tubules. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girotti M, Banting G. TGN38-green fluorescent protein hybrid proteins expressed in stably transfected eukaryotic cells provide a tool for the real-time, in vivo study of membrane traffic pathways and suggest a possible role for rat TGN38. J Cell Sci. 1996;109:2915–2925. doi: 10.1242/jcs.109.12.2915. [DOI] [PubMed] [Google Scholar]
- Greenberg M, DeTulleo L, Rapoport I, Skowronski J, Kirchhausen T. A dileucine motif in HIV-1 Nef is essential for sorting into clathrin-coated pits and for down-regulation of CD4. Curr Biol. 1998;8:1239–1242. doi: 10.1016/s0960-9822(07)00518-0. [DOI] [PubMed] [Google Scholar]
- Griffiths G. Fine Structure Immunocytochemistry. Berlin: Springer-Verlag; 1993. [Google Scholar]
- Grimaldi KA, Hutton JC, Siddle K. Production and characterization of monoclonal antibodies to insulin secretory granule membranes. Biochem J. 1987;245:557–566. doi: 10.1042/bj2450557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kantheti P, et al. Mutation in AP-3 in the mocha mouse links endosomal transport to storage deficiency in platelets, melanosomes, and synaptic vesicles. Neuron. 1998;21:111–122. doi: 10.1016/s0896-6273(00)80519-x. [DOI] [PubMed] [Google Scholar]
- Kirchhausen T, Nathanson KL, Matsui W, Vaisberg A, Chow EP, Burne C, Keen JH, Davis AE. Structural and functional division into two domains of the large (100–100 kDa) chains of the clathrin-associated protein complex AP-2. Proc Natl Acad Sci USA. 1989;86:2612–2616. doi: 10.1073/pnas.86.8.2612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Borgne R, Alconda A, Bauer U, Hoflack B. The mammalian AP-3 adaptor-like complex mediates the intracellular transport of lysosomal membrane glycoproteins. J Biol Chem. 1998;273:29451–29461. doi: 10.1074/jbc.273.45.29451. [DOI] [PubMed] [Google Scholar]
- Lowe M, Kreis TE. Regulation of membrane traffic in animal cells by COPI. Biochim Biophys Acta. 1998;1404:53–66. doi: 10.1016/s0167-4889(98)00046-9. [DOI] [PubMed] [Google Scholar]
- Luzio JP, Banting G. Eukaryotic membrane traffic: retrieval and retention mechanisms to achieve organelle residence. Trends Biochem Sci. 1993;18:395–398. doi: 10.1016/0968-0004(93)90097-7. [DOI] [PubMed] [Google Scholar]
- Matter K, Mellman I. Mechanisms of cell polarity: sorting and transport in epithelial cells. Curr Opin Cell Biol. 1994;6:545–554. doi: 10.1016/0955-0674(94)90075-2. [DOI] [PubMed] [Google Scholar]
- Newman LS, McKeever MO, Hirotaka OJ, Darnell RB. β-NAP, a cerebellar degeneration antigen, is a neuron-specific vesicle coat protein. Cell. 1995;82:773–783. doi: 10.1016/0092-8674(95)90474-3. [DOI] [PubMed] [Google Scholar]
- Odorizzi G, Cowles CR, Emr SD. The AP-3 complex: a coat of many colors. Trends Cell Biol. 1998;8:282–288. doi: 10.1016/s0962-8924(98)01295-1. [DOI] [PubMed] [Google Scholar]
- Ohno H, Aguilar RC, Yeh D, Taura D, Saito T, Bonifacino JS. The medium subunits of adaptor complexes recognize distinct but overlapping sets of tyrosine-based sorting signals. J Biol Chem. 1998;273:25915–25921. doi: 10.1074/jbc.273.40.25915. [DOI] [PubMed] [Google Scholar]
- Ohno H, Stewart J, Fournier MC, Bosshart H, Rhee I, Miyatake S, Saito T, Gallusser A, Kirchhausen T, Bonifacino JS. Interaction of tyrosine-based sorting signals with clathrin-associated proteins. Science. 1995;269:1872–1875. doi: 10.1126/science.7569928. [DOI] [PubMed] [Google Scholar]
- Owen DJ, Evans PR. A structural explanation for the recognition of tyrosine-based endocytic signals. Science. 1998;282:1327–1332. doi: 10.1126/science.282.5392.1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page LJ, Robinson MS. Targeting signals and subunit interactions in coated vesicle adaptor complexes. J Cell Biol. 1995;131:619–630. doi: 10.1083/jcb.131.3.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panek HR, Stepp JD, Engle HM, Marks KM, Tan P, Lemmon SK, Robinson LC. Suppressors of YCK-encoded yeast casein kinase 1 define the four subunits of a novel clathrin AP-like complex. EMBO J. 1997;16:4194–4204. doi: 10.1093/emboj/16.14.4194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peyroche A, Paris S, Jackson CL. Nucleotide exchange on ARF mediated by yeast Gea1 protein. Nature. 1996;384:479–481. doi: 10.1038/384479a0. [DOI] [PubMed] [Google Scholar]
- Phan HL, Finlay JA, Chu DS, Tan P, Kirchhausen T, Payne GS. The S. cerevisiae APS1 gene encodes a homologue of the small subunit of the mammalian clathrin AP-1 complex: evidence for functional interaction with clathrin at the Golgi complex. EMBO J. 1994;13:1706–1717. doi: 10.1002/j.1460-2075.1994.tb06435.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponnambalam S, Robinson MS, Jackson AP, Peiperl L, Parham P. Conservation and diversity in families of coated vesicle adaptins. J Biol Chem. 1990;265:4814–4820. [PubMed] [Google Scholar]
- Rad MR, Phan HL, Kirchrath L, Tan PK, Kirchhausen T, Hollenberg CP, Payne GS. Saccharomyces cerevisiae Apl2p, a homologue of the mammalian clathrin AP beta subunit, plays a role in clathrin-dependent Golgi functions. J Cell Sci. 1995;108:1605–1615. doi: 10.1242/jcs.108.4.1605. [DOI] [PubMed] [Google Scholar]
- Reaves B, Banting G. Overexpression of TGN38/41 leads to mislocalisation of gamma-adaptin. FEBS Lett. 1994;351:448–456. doi: 10.1016/0014-5793(94)00813-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MS. 100-kDa coated vesicle proteins: molecular heterogeneity and intracellular distribution studied with monoclonal antibodies. J Cell Biol. 1987;104:887–895. doi: 10.1083/jcb.104.4.887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MS. Cloning of cDNAs encoding two related 100-kDa coated vesicle proteins (α-adaptins) J Cell Biol. 1989;108:833–842. doi: 10.1083/jcb.108.3.833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MS. Cloning and expression of g-adaptin, a component of clathrin-coated vesicles associated with the Golgi apparatus. J Cell Biol. 1990;111:2319–2326. doi: 10.1083/jcb.111.6.2319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MS, Kreis TE. Recruitment of coat proteins onto Golgi membranes in intact and permeabilized cells: effects of brefeldin A and G protein activators. Cell. 1992;69:129–138. doi: 10.1016/0092-8674(92)90124-u. [DOI] [PubMed] [Google Scholar]
- Robinson MS, Pearse BMF. Immunofluorescent localization of 100K coated vesicle proteins. J Cell Biol. 1986;102:48–54. doi: 10.1083/jcb.102.1.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning, a Laboratory Manual. New York: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Seaman MNJ, Ball CL, Robinson MS. Targeting and mistargeting of plasma membrane adaptors in vitro. J Cell Biol. 1993;123:1093–1105. doi: 10.1083/jcb.123.5.1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson F, Bright NA, West MA, Newman LS, Darnell RB, Robinson MS. A novel adaptor-related protein complex. J Cell Biol. 1996;133:749–760. doi: 10.1083/jcb.133.4.749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson F, Peden AA, Christopoulou L, Robinson MS. Characterization of the adaptor-related protein complex, AP-3. J Cell Biol. 1997;137:835–845. doi: 10.1083/jcb.137.4.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slot JW, Geuze HJ. The use of protein A-colloidal gold (PAG) complexes as immunolabels in ultrathin frozen sections. In: Cuello AC, editor. Immunohistochemistry. Chichester, UK: John Wiley & Sons; 1983. pp. 323–346. [Google Scholar]
- Staden R. Searching for patterns in protein and nucleic acid sequences. Methods Enzymol. 1990;183:193–211. doi: 10.1016/0076-6879(90)83014-z. [DOI] [PubMed] [Google Scholar]
- Stephens DJ, Banting G. Specificity of interactions between adaptor-complex medium chains and the tyrosine-based sorting motifs of TGN38 and lgp120. Biochem J. 1998;335:567–572. doi: 10.1042/bj3350567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepp JD, Pellicena-Palle A, Hamilton S, Kirchhausen T, Lemmon S. A late Golgi sorting function for Saccharomyces cerevisiae Apm1p, but not for Apm2p, a second yeast clathrin AP medium chain-related protein. Mol Biol Cell. 1995;6:41–58. doi: 10.1091/mbc.6.1.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takatsu H, Sakurai M, Shin F, Murakami K, Nakayama K. Identification and characterization of novel clathrin adaptor-related proteins. J Biol Chem. 1998;38:24693–24700. doi: 10.1074/jbc.273.38.24693. [DOI] [PubMed] [Google Scholar]
- Tokuyasu KT. A study of positive staining of ultrathin frozen sections. J Ultrastruct Res. 1978;63:287–307. doi: 10.1016/s0022-5320(78)80053-7. [DOI] [PubMed] [Google Scholar]
- Vowels JJ, Payne GS. A dileucine-like sorting signal directs transport into an AP-3-dependent, clathrin-independent pathway to the yeast vacuole. EMBO J. 1998;17:2482–2493. doi: 10.1093/emboj/17.9.2482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Kilimann MW. Identification of two new mu-adaptin-related proteins, mu-ARP1 and mu-ARP2. FEBS Lett. 1997;402:57–61. doi: 10.1016/s0014-5793(96)01500-1. [DOI] [PubMed] [Google Scholar]
- West MA, Bright NA, Robinson MS. The role of ADP-ribosylation factor and phospholipase D in adaptor recruitment. J Cell Biol. 1997;138:1239–1254. doi: 10.1083/jcb.138.6.1239. [DOI] [PMC free article] [PubMed] [Google Scholar]