Abstract
Asymmetric delivery and distribution of macromolecules are essential for cell polarity and for cellular functions such as differentiation, division, and signaling. Injury of podocytes, which are polarized epithelial cells, changes the dynamics of the actin meshwork, resulting in foot process retraction and proteinuria. Although the spatiotemporal control of specific protein–protein interactions is crucial for the establishment of cell polarity, the mechanisms controlling polarity-dependent differentiation and division are incompletely understood. In this study, yeast two-hybrid screens were performed using a podocyte cDNA library and the polarity protein PATJ as bait. The protein KIBRA was identified as an interaction partner of PATJ and was localized to podocytes, tubular structures, and collecting ducts. The last four amino acids of KIBRA mediated binding to the eighth PDZ domain of PATJ. In addition, KIBRA directly bound to synaptopodin, an essential organizer of the podocyte cytoskeleton. Stable knockdown of KIBRA in immortalized podocytes impaired directed cell migration, suggesting that KIBRA modulates the motility of podocytes by linking polarity proteins and cytoskeleton-associated protein complexes.
Cell polarity regulates important processes, such as asymmetric cell division, cellular morphology, intracellular signaling, and cell migration. So far, it is known that specific protein–protein interactions are important for these processes, but the detailed molecular mechanisms that control cell polarity are poorly understood. Podocytes are highly polarized epithelial cells that play a key role in the maintenance of the size-selective filtration barrier of the kidney.1 They consist of a cell body with primary and highly branched secondary foot processes, leading to a complex “neuron-like” cell architecture. The interdigitating secondary foot processes mediate the adhesion to the glomerular basement membrane and form the slit diaphragm, unique cell–cell contacts that serve as a final filtration barrier.1
Injury of podocytes leads to dynamic changes of the actin cytoskeleton, resulting in foot process retraction (foot process effacement) and proteinuria. Obviously, a regulated cell polarity is essential for podocyte function. Nevertheless, the expression, function, and cross-talk of polarity regulators and the molecular links between polarity proteins and downstream effects such as signaling, differentiation, and directional migration of podocytes are unknown.
During the past decade, two polarity complexes have been described, the aPKC-PAR3-PAR6 (aPKC for atypical protein kinase C; PAR for partitioning defective) and the Pals1-PATJ-Crb complex (Pals1 for protein-associated with Lin7–1; PATJ for Pals1-associated tight junction protein; and Crb3 for Crumbs3).2–4 It has been shown that these complexes are part of an evolutionarily conserved system that regulates apicobasal polarity, tight junction formation, signaling, and directional migration of eukaryotic cells. All core components of the complexes carry multiple protein–protein interaction modules, suggesting that they are part of multiprotein complexes.2–4
Interestingly, PATJ (also called INADL or CIPP) was first identified as a highly abundant protein in brain and kidney.5–7 In addition to the N-terminal L27 domain, PATJ contains ten PDZ (for PSD95/discs large/zonula occludens 1 [ZO-1]) domains, suggesting that PATJ acts as a scaffolding protein that is able to bind to many cellular partners through these domains.3,6 Previously, it was shown that PATJ associates with neuronal proteins and channels (e.g., neurexin, neuroligin, members of the Kir-family, ASIC3, 5HT2A), indicating that it recruits receptors and structural proteins at synaptic sites.8,9 A recent study reported that PATJ interacts with the tuberous sclerosis complex protein 1 and 2 (TSC1/2) and thereby links the mammalian target of rapamycin (mTOR) signaling pathway to the Pals1-PATJ-Crb3 cell polarity complex, supporting the idea that cell polarity proteins are also involved in signaling pathways.10
In addition, PATJ directly binds to the tight junction proteins ZO-3 and claudin 1 via the sixth and eighth PDZ domains, emphasizing that PATJ plays an important role in the maintenance of cell polarity and tight junction establishment of epithelial cells.11–14 Recently, it was shown that PATJ is part of a huge Rich/Amot complex by interacting with members of the Amot protein family.12,15 This family plays a role in the regulation of cell–cell junctions and cell motility.16 In this context, it is interesting that the knockdown of PATJ in epithelial cells results in an impaired migration of MDCK II cells, suggesting that PATJ regulates not only cell polarity and tight junction establishment but also the directional migration of epithelial cells.
In this study, we performed a yeast two-hybrid (Y2H) screen, using a cDNA library from immortalized podocytes and PATJ as bait. We found KIBRA (for kidney brain), a protein with high expression in the brain and kidney that probably plays a central role in human memory, as interaction partner of PATJ.17–21
Our investigations revealed that in the kidney, KIBRA is expressed in glomerular podocytes, in some tubules, and in the collecting duct. In addition to the PATJ interaction, we found that KIBRA binds to synaptopodin, an essential protein of podocytes. Furthermore, a knockdown of the KIBRA expression resulted in an impairment of podocyte directional migration.
RESULTS
KIBRA Directly Interacts with Cell Polarity Protein PATJ
During the past decade, it was shown that the Pals1-PATJ-Crb3 and aPKC-PAR3-PAR6 cell polarity complexes are especially important in diverse epithelia of various tissues; therefore, we first tested whether these core proteins of the apical polarity complexes are expressed in immortalized cultured podocytes.22 We found mRNA expression of PATJ; Pals1; both Crumbs3 isoforms (Crb3a/b); and aPKCζ, PAR3, and three PAR6 isoforms in podocyte cDNA library PCR reactions (Figure 1A). This expression pattern might be a first hint that the cell polarity of podocytes could be regulated in a similar manner as already shown for many other cell types.
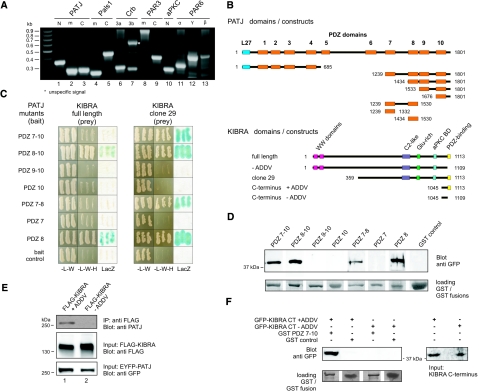
Figure 1.
KIBRA directly interacts with PDZ8 domain of cell polarity protein PATJ. (A) Expression of apical cell polarity proteins in a human podocyte cell line. A PCR analysis of a cDNA library derived from the human differentiated podocytes (AB 8 cells) demonstrates the expression of apical cell polarity proteins in podocytes. cDNA for PATJ (lanes 1 through 3), PAls1 (lanes 4 through 5), Crumbs isoforms Crb3a ad Crb3b (lanes 6 through 7), PAR3 (lanes 8 through 9), aPKCζ (lane 10), and three PAR6 isoforms (αβγ, lanes 11 through 13) were detected (details in Supplementary Table 2). (B) Domain architecture of PATJ, KIBRA, and used deletion mutants. Full-length PATJ (1801 aa) consists of one N-terminal L27 (blue) and 10 PDZ domains (orange). KIBRA (1113 aa) comprises two N-terminal WW domains (pink), a C2-like domain (magenta), a glutamate-rich region (green), an aPKC-binding domain (light blue), and a C-terminal PDZ-binding motif (yellow). An N-terminal deletion mutant of KIBRA (clone 29 [aa 359 through 1113]) was isolated in a Y2H screen, using the last four PDZ domains of PATJ as bait. (C) Y2H assays using different PATJ mutants and prey plasmids (clone 29/full-length KIBRA) to map the binding site. (D) KIBRA binds to PATJ PDZ 8 in a GST pull-down assay. GFP-tagged KIBRA C-terminus (aa 1045 through 1113) were incubated with equal amounts of various GST-PATJ fusion proteins. GST was used as a control. (E and F) Co-IP experiments (E) and GST pull-down assays (F) revealed that the last four aa of KIBRA (ADDV) mediate the interaction with PATJ.
For further examinations, we focused on the multi-PDZ protein PATJ and performed a Y2H screen, using a podocyte cDNA library and PATJ as bait. One of the isolated yeast clones encoded an N-terminal deletion of KIBRA.23 Mapping studies using a set of PATJ deletion mutants in the Y2H system (Figure 1, B and C) and GST pull-down assays revealed that PDZ8 mediates the PATJ–KIBRA interaction (Figure 1D).
The identified KIBRA clone (clone 29) of the Y2H screen lacks the two N-terminal WW domains (amino acids [aa] 1 through 39 and 54 through 86) but contains a putative calcium-sensitive C2-like domain (aa 726 through 787), a glutamate-rich region (aa 845 through 868), and the aPKCζ-binding domain (aa 953 through 996; Figure 1B, bottom). The C-terminal four aa of KIBRA (ADDV, single-letter code) contain a putative class III PDZ-binding site.24 To find out whether the KIBRA-PATJ interaction is mediated by this motif, we performed co-immunoprecipitation (Co-IP) assays with lysates from HEK293T cells expressing FLAG-tagged full-length KIBRA with and without the ADDV motif and EYFP-tagged full-length PATJ, respectively. Only KIBRA containing this motif was able to precipitate EYFP-tagged PATJ in these assays (Figure 1E). In addition, lysates from HEK293T expressing the GFP-tagged C-terminus of KIBRA with and without the ADDV motif (aa 1045 through 1113/1109) were incubated with recombinant GST PDZ7–10 fusion proteins. Again, only KIBRA deletion mutants that contained the ADDV motif interacted with PATJ (Figure 1F).
KIBRA Is Expressed in Various Renal Tissues
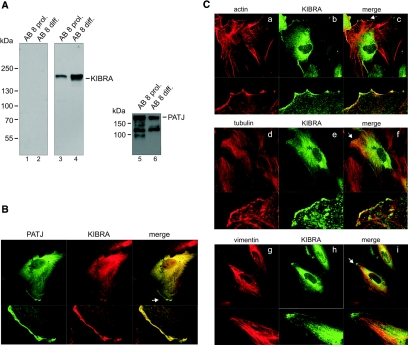
KIBRA and PATJ both are expressed in immortalized podocytes (Figure 2A). Furthermore, immunofluorescence analysis revealed that KIBRA and PATJ co-localize throughout the cytoplasm in a predominant perinuclear pattern. Minor fractions of both proteins were also found at the lamellipodia leading edge, similar to the recently described PATJ distribution in migrating MDCK II cells (Figure 2B).25 In addition, we found that KIBRA partially co-localizes with the actin and tubulin cytoskeleton at these leading edges (Figure 2C, a through f).
Figure 2.
Expression of KIBRA and PATJ in a human podocyte cell line. (A, left) Affinity-purified KIBRA antiserum (Supplemental Figure S2A) specifically recognize KIBRA in extracts from proliferating (lane 3) and differentiated human podocytes (lane 4). Preimmune serum shows no reactivity to KIBRA (lanes 1 and 2). (A, right) PATJ is also expressed in both proliferating (lane 5) and differentiated (lane 6) human podocytes. (B) Co-localization of myc-KIBRA and EYFP-PATJ. Both proteins display a cytosolic distribution with perinuclear enrichment in proliferating and differentiated human podocytes. Small amounts of both proteins are localized at the cell periphery (arrow). (C) Myc-KIBRA co-localizes at lamellipodia of the leading edge (arrows) with actin (a through c) and tubulin (d through f) but not with vimentin (g through i).
By contrast, vimentin, a marker for intermediate filaments, does not co-localize with KIBRA (Figure 2C, g through i). After treatment of the cells with tubulin or actin de-polymerizing agents (nocodazole and cytochalasin D, respectively), a fraction of KIBRA accumulates with retracted tubulin or actin (Supplemental Figure S1).
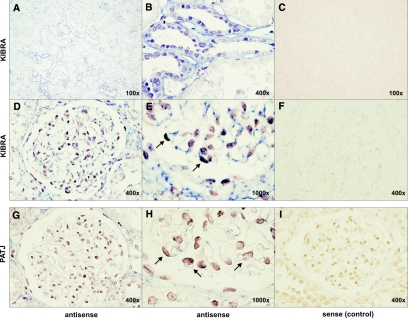
An in situ hybridization analysis of total human kidney revealed an mRNA expression of KIBRA in podocytes and tubular structures (Figure 3). These observations were confirmed by immunohistochemistry analyses on human kidney sections showing a predominant KIBRA expression in glomerular and tubular cells (Supplemental Figure S2B). Cells of the proximal tubules seemed to express only low levels of KIBRA. These data are supported by a Western blot analysis of IHKE-1 cells, an immortalized cell line of the human proximal tubule. Here, KIBRA shows only low expression levels when compared with lysates from total kidney or the podocyte cell line (Supplemental Figure S2C).26
Figure 3.
In situ hybridization analysis of KIBRA and PATJ mRNA expression. Representative pictures after incubation with specific antisense probes against KIBRA mRNA revealed the expression of KIBRA in the glomeruli, distal tubule, and collecting duct of the kidney (A, overview; B, tubule). Higher magnification of human glomeruli showed a KIBRA (D and E) and PATJ (G and H) expression in podocytes (arrows). (C, F, and I) Sense negative control.
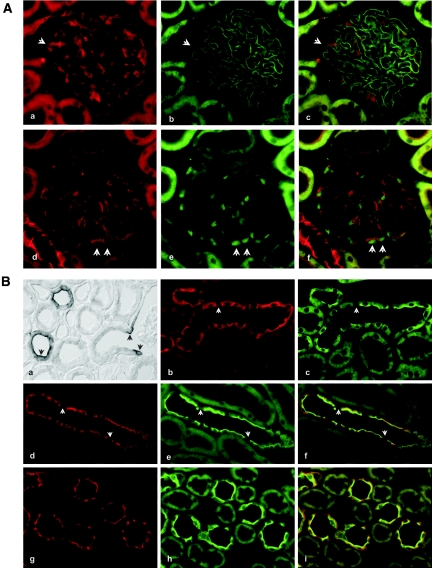
An additional analysis using rodent tissue confirmed this KIBRA expression pattern. In the adult rat, KIBRA staining of the glomerular tuft was restricted to the podocytes. These cells were identified by double staining of the filtration slit using anti–ZO-1 antibodies (Figure 4A, a through c) and of their nuclei with anti–Wilms Tumor Antigen-1 (WT-1) (Figure 4A, d through f). In the glomeruli, all podocytes seemed to be KIBRA positive, albeit at a relatively low signal intensity compared with collecting duct staining.
Figure 4.
KIBRA expression in rodent renal tissue. (A) KIBRA immunostaining of the glomerular tuft. (a through c) Double staining of glomeruli for KIBRA (a) and ZO-1 (b) is displayed. Only the podocytes are immunostained, which is indicated by signals in epimembranous and membranous location; ZO-1 shows the slit membrane. The position of one representative podocyte is indicated by arrowheads. (c) Merged image. (d through f) Double staining of glomeruli for KIBRA (d) and podocyte marker protein WT-1 (e). All WT-1–positive podocytes are also KIBRA positive; two representative, double-stained podocytes are marked by arrowheads. (f) Merged image. (B) KIBRA immunostaining of the renal cortex and medulla. KIBRA immunoperoxidase staining shows significantly labeled cells at the “bottleneck” region near the urinary pole of the glomerular parietal epithelium (a, arrowheads). Note that immunoreactive profiles of cortical collecting ducts (CCD) show strong epithelial staining with luminally enhanced signal. (b and c) Double staining for KIBRA (b) and aquaporin (AQP-2; c) of a connecting tubule profile. Signals overlap at the luminal cell pole, whereas only KIBRA staining extends to the lateral cell borders and the intracellular/basolateral aspect. Intercalated cells (arrowheads) are negative for either staining. (d through i) Double staining for KIBRA (d and g) and AQP-2 (e and h) of a CCD profile (d through f) and medullary collecting duct profiles (MCD; g through i). As in CNT, principal cells show overlap at the luminal cell pole, whereas only KIBRA staining extends to the lateral cell borders and the intracellular/basolateral aspect in CCD and MCD principal cells. Intercalated cells (arrowheads in d through f) are negative. (f and i) Merged images.
Parietal cells of the “bottleneck” region of Bowman's capsule and parietal epithelium at the transition to the proximal tubule were regularly positive with two to four cells stained on each side in transverse sections through the glomerulus (Figure 4B). Equally strong staining was seen in principal cells of the entire connecting tubule (Figure 4B, b and c) and collecting duct (Figure 4B, d through i). Apparently, the intercalated cells were negative throughout. The KIBRA signal was strong at the luminal cell membrane, equally significant at the lateral cell membrane, and weaker and diffuse intracellularly and at the basal cell aspects of the principal cells. Double staining with anti–aquaporin-2 antibodies revealed co-localization with KIBRA chiefly at the luminal cell aspect. There were no major differences between cortical collecting duct and medullar collecting duct principal cell staining patterns (Figure 4B, d through i).
KIBRA Binds Directly to the Slit Diaphragm Protein Synaptopodin
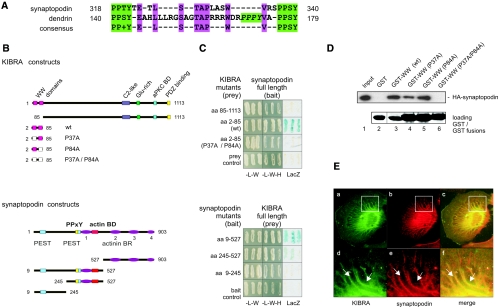
The PATJ–KIBRA interaction raises the question of whether KIBRA might act as a linker protein that mediates the binding of PATJ-containing complexes to proteins that specifically interact with the N-terminal WW domains of KIBRA. WW domains are protein–protein interaction modules that bind to PPxY aa motifs.27 This is interesting because KIBRA directly binds to the actin cytoskeleton-associated protein dendrin that was recently described as a slit diaphragm protein.28–30 Dendrin directly interacts with CD2AP and nephrin and shares several properties with synaptopodin that is involved in migration of podocytes and the development of kidney diseases.31,32 In addition, dendrin and synaptopodin are found in the dendrites of neurons and are associated with the cytoskeleton via binding to actin and α-actinin isoforms.31–34
An alignment of synaptopodin and dendrin showed no obvious homology between both proteins; however, the PPxY sites of human synaptopodin and dendrin elucidated a striking similarity between aa 140 through 179 of dendrin and aa 318 through 340 of synaptopodin (Figure 5A, green boxes). Both proteins contain two flanking PPxY sites with a serine or threonine at the third position and have additional identical aa (TAPxxxW) inside these flanking PPxY motifs (Figure 5A, magenta boxes). To test whether KIBRA binds not only to dendrin but also to synaptopodin, we performed Y2H assays with various synaptopodin deletion mutants and full-length KIBRA. Only deletion mutants (aa 9 through 527 and aa 245 through 527) that include the two PPxY sites (aa 318 through 340) interact with KIBRA (Figure 5, B and C, bottom). This interaction was further investigated by truncated KIBRA mutants that encode only the N-terminal two WW domains (aa 2 through 85). We applied the wild-type WW domains as well as point mutants for single (P37A or P84A) or both WW domains (P37A/P84A) that should abrogate the binding to PPxY motifs.23 The binding assays revealed that the wild-type WW domains of KIBRA bind to synaptopodin, whereas the double mutant (P37A/P84A) did not. A KIBRA mutant lacking the WW domains (aa 85 through 1113) and an empty bait or prey vectors were used as controls (Figure 5C). These results were confirmed by GST pull-down experiments (Figure 5D). For these, recombinant HA-tagged synaptopodin (903 aa) was incubated with GST or GST KIBRA-WW fusion proteins. Again, the wild-type KIBRA WW domains bind to HA-synaptopodin, whereas the WW P37A/P84A double mutant failed. Fusion proteins that composed only a single mutated WW domain were still able to interact with synaptopodin, suggesting that one KIBRA WW domain is sufficient to recognize the PPxY motifs of synaptopodin. The interaction of KIBRA and synaptopodin was further supported by our findings that both proteins display a high degree of co-localization in podocytes (Figure 5E).
Figure 5.
KIBRA binds directly to synaptopodin via the WW domains. (A) Synaptopodin and dendrin carry two independent PPxY motifs with an S/T at the third position. The regions between the flanking PPxY motifs contain additional identical aa (magenta). Dendrin comprises one further PPxY motif (green italic letters). (B) Schematic structure of KIBRA and synaptopodin deletion constructs used for the binding assays. The N-terminal region of KIBRA contains two WW domains (pink/white boxes; aa 7 through 39 and aa 54 through 84). The substitution of a conserved proline into alanine (P37A and/or P84A) abrogates binding of the KIBRA WW domains. The deletion mutant lacking the WW domains (aa 85 through 1113) was used as negative control. The long variant of synaptopodin (903 aa) that is expressed in podocytes contains two internal PPxY sites (yellow), two PEST sequences (light blue), an actin-binding domain (red), and four α-actinin–binding regions (actinin BR, purple). (C) Y2H assay with bait constructs expressing different synaptopodin deletion mutants and prey plasmids encoding KIBRA, illustrating a direct interaction between KIBRA and synaptopodin. (control: empty bait/prey plasmid). (D) KIBRA WW domains bind to synaptopodin. HA-tagged synaptopodin was incubated with equal amounts of GST-fusion proteins that comprise KIBRA wild-type or mutated WW domains as shown in B. (E) Co-localization of KIBRA and synaptopodin in podocytes. Differentiated AB 8 cells coexpressing HA-synaptopodin and myc-KIBRA display a partial co-localization of both proteins.
A Knockdown of KIBRA Disturbs Directional Cell Migration
Previous studies reported that synaptopodin regulates the bundling activity of the actin cross-linking protein α-actinin in an isoform-specific manner.32 Furthermore, synaptopodin regulates the integrity of the cytoskeleton and the cell motility of podocytes, because gene silencing of synaptopodin leads to defects in directional podocyte migration.31 In addition, a recently published study showed that a PATJ knockdown (k/d) resulted in an impaired migration of epithelial cells, suggesting that PATJ regulates not only tight junction formation but also the mobility of migrating cells. This leads to the intriguing hypothesis that KIBRA may take part in podocyte migration, because KIBRA directly binds to both of these motility regulating proteins.
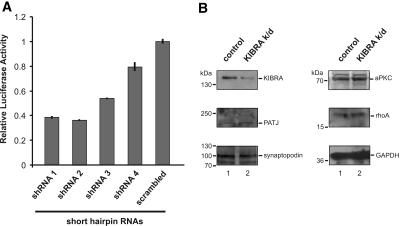
We generated KIBRA k/d podocytes to investigate a possible involvement of KIBRA in cell migration. For testing the efficiency of various short hairpin RNA (shRNA), human KIBRA cDNA was cloned into the bicistronic luciferase vector (psiCHECK2; Promega, Madison, WI) encoding a fusion protein with Renilla reniformis luciferase. Within this system, R. reniformis luciferase activity is a quantitative parameter of RNA degradation mediated by co-transfected shRNA. Coexpressed firefly (Photinus pyralis) luciferase served as control to normalize for transfection efficiency, expression level, and cell number. ShRNA 1 and 2 resulted in at least 60% k/d of the reporter luciferase (Figure 6A). The expression levels of KIBRA-interacting proteins PATJ, synaptopodin, and aPKCζ and the cytoskeleton-regulating protein rhoA were not changed in KIBRA k/d cells (Figure 6B).
Figure 6.
KIBRA k/d in podocytes. (A) For determination of the KIBRA k/d efficiencies of different shRNA, luciferase assays using the psiCHECK2 (Promega) system were performed. shRNA 1 and 2 resulted in approximately 60 to 65% k/d of the reporter luciferase (Supplemental Figure S3). (B) The expression level for PATJ, synaptopodin, aPKCζ, and rhoA is not changed in KIBRA k/d podocytes (lane 2) compared with control cell lines (empty vector–transduced cells, lane 1), emphasizing that the migration impairment is caused by the decreased endogenous KIBRA expression.
shRNA 1 and 2 were subcloned into a lentiviral vector to k/d KIBRA expression selectively in human podocytes (Supplemental Figure S3). Relying on a highly efficient retroviral gene transfer method, we obtained a pool of polyclonal cells harboring integration of the transgene at various positions in the genome. Expression of shRNA was monitored by simultaneous coexpression of GFP from the same construct (Supplemental Figure S3).
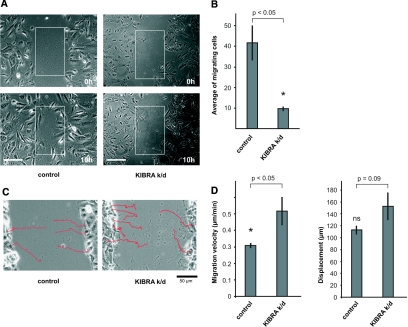
Next, we performed wound-healing assays clearly showing the incapability of KIBRA k/d podocytes to close a wound via directed migration within 10 h. By contrast, control cells achieved wound healing within the same time (Figure 7, A and B [n = 3; 41.6 ± 8.4 in control cells versus 9.6 ± 1.0 in KIBRA k/d cells; P = 0.04, t test], and Supplemental Movies). To investigate further whether KIBRA k/d podocytes are impaired in the migration process itself or alternatively in polarized migration (toward a wound), we analyzed trajectories of single cells, migration velocity, and the distance covered within the experimental period. Control cells migrate in a persistent way into the wounded area and rarely make major changes in the direction of movement. Figure 7C, left, displays original paths of migrating control cells (magnification ×20). In contrast, KIBRA k/d cells migrate ineffectively with frequent turns (Figure 7C, right; magnification ×20). Surprising, our data show KIBRA k/d podocytes to migrate significantly faster than control cells (Figure 7D, migration velocity [0.3 ± 0.01 μm/min in control cells versus 0.5 ± 0.09 μm/min in KIBRA k/d cells; P = 0.01, t test; means of 21 (control) or 17 (KIBRA k/d) cells are given]). Nevertheless, podocytes from both cell lines cover approximately the same distance within the same period (10 h; Figure 7D, displacement). This apparent discrepancy can be accounted for by the frequent occurrence of changes in the direction of movement of KIBRA k/d cells. These data point to KIBRA's being required for efficient directed migration.
Figure 7.
KIBRA regulated cell migration of podocytes. (A) A wound was scraped into confluent cell culture monolayers of control and KIBRA k/d podocytes (top, 0 h). In contrast to control cells, KIBRA k/d podocytes are incapable of closing a wound within 10 h via directed migration (bottom, 10 h). Square fields of identical size were superimposed to count cells that had migrated into the wound area. Bar = 50 μm. (B) The number of cells that had migrated into equal areas within 10 h was plotted (square fields in A). KIBRA k/d podocytes are severely impaired in migrating into the denuded area. Experiments were performed in triplicate. (C) Typical original trajectories of control and KIBRA k/d cells are superimposed on the starting images of wound-healing assays. Migration of control podocytes is characterized by a high degree of persistence, and cells rarely change the direction of migration (left). In contrast, KIBRA k/d podocyte migration is ineffective and direction of movement changes frequently (right). (D) Statistical evaluation of migration velocity and displacement. KIBRA k/d podocytes move significantly faster (P < 0.05; P = 0.01) than control cells. Despite the higher velocity of KIBRA k/d podocytes, their displacement (Ø 152.8 ± 22.9 μm) is not significantly different (P = 0.09) from that of control cells (Ø 112.8 ± 7.0 μm). This can be accounted for by the frequent turns in the direction of movement of KIBRA k/d cells. Means of 21 (control) or 17 (KIBRA k/d) cells are given.
DISCUSSION
It is still an open question of which mechanism links cell polarity to the diverse cellular programs such as cell differentiation and motility in podocytes. Here we used the cell polarity and scaffold protein PATJ to screen a podocyte library for interacting partners. We identified KIBRA as a new interacting partner of PATJ and described the expression and localization of KIBRA in the kidney. KIBRA directly interacts with the eighth PDZ domain of PATJ. The C-terminal ADDV motif of KIBRA is similar to that of the previously characterized PATJ interacting protein claudin 1 (KDYV), emphasizing that the eighth PDZ domain favors class III PDZ-binding motifs.11 In addition, we elucidated that the N-terminal WW domains of KIBRA directly interact with synaptopodin, a podocyte protein that plays a role as cytoskeletal organizer and that is also associated with synaptic plasticity in neurons.34,35 In the kidney, we found KIBRA expression in glomeruli, tubules, and collecting duct. Furthermore, we described for the first time that KIBRA is important for directional migration.
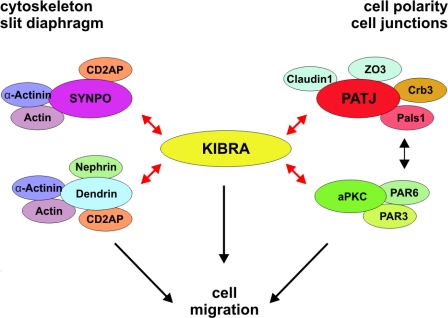
Initially, KIBRA was identified as an interacting partner of dendrin, a protein that controls synaptic plasticity and that has been recently identified as a slit diaphragm–associated protein.23,29 Interestingly, dendrin binds to nephrin and CD2AP, two other essential components of the slit diaphragm.30 Notably, protein kinase aPKC that is part of the aPKC-PAR3-PAR6 complex directly binds to and phosphorylates KIBRA.36 The C-terminus of KIBRA, which carries the aPKCζ binding as well as the PATJ binding site, might be involved in the assembly or the maintenance of both apical polarity complexes (see model Figure 8).37
Figure 8.
Model of the cellular function of KIBRA. KIBRA directly interacts with dendrin, synaptopodin (SYNPO), and PATJ and aPKC (red arrows). Thus, KIBRA could serve as a linker molecule between polarity proteins and components of the cytoskeleton (actin, α-actinin, CD2AP, synaptopodin, and dendrin), thereby regulating cell motility of podocytes.
The N-terminal part of KIBRA contains two WW domains that mediate the binding of KIBRA to dendrin and synaptopodin. It remains to be shown whether the PPxY motifs of both proteins compete or cooperate in binding to KIBRA; however, both KIBRA interactors, synaptopodin as well as dendrin, bind to actin and α-actinin isoforms, thereby acting as regulators of the actin-based cytoskeleton.30–32
In addition to the putative indirect association to the actin cytoskeleton, it has been demonstrated that KIBRA directly binds to dynein light chain 1 (DLC1) and sorting nexin 4 (SNX4).38,39 The DLC1-SNX4-KIBRA complex associates with the minus-end directed microtubule motor dynein complex.38 These studies support our observation that KIBRA at least indirectly associates with the microtubule network; therefore, KIBRA might act as a bridge between actin- and/or microtubule-associated networks and cell polarity complexes.
In this context, it is worth mentioning that in podocytes, MAGI-1 also interacts with dendrin and synaptopodin.28,40,41 MAGI-1 belongs to the MAGUK (for membrane associated guanylate kinase) family and has a guanylate kinase domain and two WW domains that are flanked by one N- and five C-terminal PDZ domains. Similar to KIBRA, MAGI-1 associates with synaptopodin and dendrin via the WW domains. In addition, the PDZ2 and PDZ3 domains of MAGI-1 bind to the C-terminus of the slit diaphragm protein nephrin, suggesting that MAGI-1 provides a molecular link between the slit diaphragm and cytoskeletal-associated proteins40; however, because of the different domain architecture of KIBRA and MAGI-1, it is likely that they have only partial analogous functions in podocytes.
Interestingly, in brain, KIBRA represents a component of the postsynaptic density (data not shown). Thus, our data support the hypothesis that the motility of podocyte foot processes and the flexibility of synaptic contacts of neurons could be regulated by an analogous set of molecules, including proteins such as KIBRA, synaptopodin, dendrin, actin, and α-actinin.
Both foot processes and dendrites are long F-actin–rich, cellular extensions, suggesting a role for KIBRA in the continuous regeneration and plasticity of both cell types. Remarkably, several recently published studies reported that the memory performance that depends on synaptic plasticity correlates with different KIBRA alleles.17–20,38 Hence, the functional data provided may also provide a first hint of how synaptic plasticity could be regulated in neurons and how KIBRA might be involved in learning and memory processes on a molecular level.
Foot processes of podocytes are highly flexible and dynamic structures that play a key role in withstanding the continuous filtration pressure. In addition, it is assumed that foot process retraction is a migration event caused by podocyte injury.42 Key aspects of migratory processes are rearrangements of the actin and microtubule cytoskeleton and polarization of the cell along the front-rear axis. We propose that a possible physiologic function of KIBRA is to channel actin/microtubule cytoskeleton dynamics into a polarized way. Interestingly, KIBRA k/d does not influence expression levels of interaction partners PATJ, synaptopodin, aPKCζ, and associated protein rhoA, indicating that the observed phenotype of KIBRA k/d cells is directly based on KIBRA gene silencing and not on indirect effects. Here we observed that KIBRA k/d leads to an increased velocity without affecting the displacement of cells. That is, directed migration of KIBRA k/d cells is less efficient than that of control cells. Our data suggest that the higher migration velocity displayed by KIBRA k/d cells is due to increased dynamics of lamellipodia. Any rearrangement of the cytoskeleton resulting in protrusions or retraction of lamellipodia leads to a shift of the cell center (i.e., cells may reach high migration velocities without truly covering a distance [“running on the spot”]). Thus, the inefficient migration of KIBRA k/d podocytes results in the impairment of closing a wound via directed movement.
Our data provide evidence that KIBRA is part of multiprotein complexes that probably link cell polarity components with cytoskeleton networks. Future studies are required to elucidate the detailed composition of these complexes and their spatially and temporally controlled interactions in the different cell types and how they regulate efficient directional migration.
CONCISE METHODS
All procedures performed were in accordance with the ethics commission guidelines of the University of Münster Ethic Commission.
Plasmid Constructs
To generate human PATJ cDNA (accession no. NM_176877.2) mutants by PCR, we used the pEYFP-PATJ construct as a template (provided by Dr. Ben Margolis, Division of Nephrology, Department of Internal Medicine, The University of Michigan Health System, Ann Arbor, MI). KIBRA cDNA truncation mutants have been described previously or were amplified from pSV42-myc-KIBRA (Supplemental Table 1). Full-length human KIBRA (accession no. NM_15238.1) cDNA or fragments encoding deletion mutants were cloned into modified pcDNA6 (details available from T.B.) or pEGFP-C2 (Clontech, Heidelberg, Germany) expression plasmids, respectively. The corresponding PCR products were subcloned into yeast GAL4 DNA-binding or GAL4-activation domain vectors (pDB-Leu, pEX-AD502, and pDEST32/22 from Invitrogen (Karlsruhe, Germany) or pAS2–1 and pACT2 from Clontech, respectively) or into a modified pGEX-KG expression construct. Vectors for the expression of HA-tagged synaptopodin or synaptopodin bait fragments for yeast co-transformation assays were described previously.33 Details of constructs (Supplemental Table 1) and primers (Supplemental Tables 2 and 3) are also available from T.W. or H.P.
Cell Culture and Transient Transfection
Human immortalized podocytes (AB 8 cells) were cultivated as described previously.22 In brief, cells were grown in standard RPMI 1640 medium containing 10% FCS and supplements either at the permissive temperature of 33°C (in 5% CO2) to promote cell propagation or at the nonpermissive temperature of 37°C (in 5% CO2) to allow the terminal differentiation. HEK293T cells were cultivated and transfected as described previously.23 Transient transfection of human podocytes was performed using the nucleofector technology (Amaxa, Cologne, Germany) according to the manufacturer's instructions.
Yeast Two-Hybrid Screen and Yeast Co-transformation Assays
S. cerevisiae MaV203 yeast cells containing the bait plasmid pDB-Leu encoding the last four PDZ of murine PATJ (provided by Dr. Eric Lingueglia, Institute of Pharmacology, Valbonne, France) were transformed with a cDNA library derived from a human podocyte cell line (differentiated AB 8) in pEXP-AD502 (Invitrogen) according to the manufacturer's instructions. For co-transformation assays, yeast cells were simultaneously transformed with bait and prey plasmids encoding various deletion mutants of human KIBRA, human and murine PATJ, and human synaptopodin (Supplemental Table 1). Interaction of bait and prey molecules were tested by growth of yeast cells on selective medium containing 75 mM 3-amino 1,2,4,-triazole but lacking leucine, tryptophan, and histidine and by LacZ-assays with filter-immobilized cells.
Generation of Polyclonal Antibodies against Human KIBRA
Polyclonal KIBRA antibodies were raised in rabbits against a purified recombinant KIBRA fragment (aa 661 to 796) fused to GST (Eurogentec, Cologne, Germany). The obtained antisera were affinity-purified using a GST-KIBRA661 to 796 affinity column and were tested by Western blot analysis (Supplemental Figure S2A).
Extract Preparation and Western Blotting
Cellular lysates were prepared by scraping cells into IP buffer (1% Triton-X 100, 20 mM Tris-HCl [pH 7.5], 25 mM NaCl, 50 mM NaF, 15 mM Na4P2O7, and 1.5 mM EDTA) containing protease inhibitor (Complete; Roche, Mannheim, Germany). Lysates were centrifuged at 10,000 × g for 30 min at 4°C. Supernatants were removed and stored at −80°C until further use. For Western blot analysis, samples were mixed with Laemmli buffer and boiled, and proteins were separated by 6 to 12% SDS-PAGE (Bio-Rad, Munich, Germany). After transfer of proteins onto a polyvinylidene fluoride membrane (Millipore, Schwalbach, Germany), reactive sites were blocked for 30 min at 37°C with 5% skim milk powder dissolved in TBS containing 0.05% Tween-20 (TBS-T). Primary antibodies were diluted in blocking buffer and incubated with membranes for 1 h at 37°C or overnight at 4°C. KIBRA and rabbit anti-PATJ antibodies were used at 1:500 dilutions. The other primary antibodies used in this work were as follows: Rabbit anti-GFP (Santa Cruz Biotechnology, Santa Cruz, CA), mouse anti–glyceraldehyde-3-phosphate dehydrogenase (Covance, Muenster, Germany), mouse anti-FLAG (Sigma-Aldrich, Munich, Germany), mouse anti-HA-tag (Roche), rabbit anti-aPKCζ (Sigma), rabbit anti-RhoA (Santa Cruz Biotechnology), mouse anti-synaptopodin (Progen, Heidelberg, Germany), mouse anti–β-tubulin (Sigma), and rabbit anti-V5 (Abcam, Heidelberg, Germany). After washing in TBS-T, the membranes were incubated with a secondary horseradish peroxidase–coupled antibody directed against the primary antibody (Dianova, Hamburg, Germany). Finally, the membranes were washed and developed using a chemiluminescence detection reagent (Roche).
Bacterial Protein Synthesis and In Vitro Binding Assays
Constructs for bacterial expression of PATJ and KIBRA fragment mutants are described in Supplemental Table 1. Synthesis of recombinant proteins in Escherichia coli strain BL21 was induced with 1 mM IPTG for 3 h at 30°C. Affinity purification of GST fusion proteins was performed according to the manufacturer's instructions (GE Healthcare, Munich, Germany). Purified proteins were stored at −80°C until further use. For GST-binding assays, equal amounts of GST fusion proteins (approximately 5 μg) were immobilized on GST-Sepharose beads (GE Healthcare) for 2 h at 4°C. The beads were washed five times with PBS and were subsequently incubated at 4°C with cell lysates containing recombinant FLAG-KIBRA, GFP-KIBRA fragments, or HA-tagged synaptopodin, respectively. Loaded beads were washed five times in PBS, and bound proteins were eluted in SDS sample buffer by boiling samples for 5 min at 95°C. Finally, probes were analyzed by SDS-PAGE and Western blot analysis as described already.
Co-immunoprecipitation
For Co-IP assays, single or co-transfected HEK293T cells were first lysed in IP buffer. Aliquots of lysates were incubated with anti-FLAG affinity gel (Sigma) overnight at 4°C on a rocking platform. In a next step, beads were washed five times in IP buffer. For elution of bound proteins, beads were resolved in sample buffer and boiled for 5 min at 95°C. Samples were subsequently subjected to SDS-PAGE and Western blot analysis as described already.
Staining of Cells and Tissues
For indirect immunofluorescence analysis on cultured podocytes, cells were grown on collagen-coated coverslips and fixed in 4% paraformaldehyde (PFA) supplemented with 4% sucrose in PBS at room temperature for 20 min. Samples were washed with PBS and then incubated for 10 min in 50 mM NH4Cl in PBS to quench reactive amino groups. After washing with PBS, cells were permeabilized in PBS containing 0.2% Triton-X 100 for 5 min and washed three times in PBS containing 0.2% Triton-X 100 and 0.2% gelatin (PBS-TG). Next, samples were blocked with 10% goat serum diluted in PBS-TG for 20 min at room temperature. Immunofluorescence staining was performed by incubating the coverslips for 1 h at room temperature with primary antibodies diluted in PBS-TG containing 2% goat serum. Primary antibodies were the polyclonal, affinity-purified anti-KIBRA serum, mouse anti–β-tubulin (Sigma), mouse anti-vimentin (Sigma), and phalloidin–Alexa 594 (Molecular Probes, Eugene, OR). For detection of overexpressed KIBRA and synaptopodin, the mAb against HA-tag (Roche) and myc-tag (Abcam, Santa Cruz Biotechnology) were used. Coverslips were washed after antibody incubation in PBS-TG and incubated 20 min at room temperature with fluorochrome-conjugated secondary antibodies (Molecular Probes) diluted in PBS-TG containing 2% goat serum. After washing in PBS, coverslips were rinsed in water and cells were mounted in Crystal Clear Mount Medium (Sigma). Images were obtained using a Leica photomicroscope attached to a Spot 2 slider digital camera. Immunofluorescence stainings of rat kidneys were performed as described previously.43 Adult male Sprague-Dawley rats (250 g body wt) were perfusion-fixed using 3% PFA/cacodylate. Tissues were prepared for histochemical analysis using paraffin or cryotechniques. Sections were incubated with specific antisera and fluorescence- or peroxidase-labeled secondary antibodies. For signal enhancement, an amplification kit (CSA; Dako, Glastrup, Denmark) was used. Specific antibodies comprised anti-KIBRA (see “Generation of Polyclonal Antibodies against Human KIBRA”), goat anti–aquaporin-2 (Santa Cruz Biotechnology), mouse anti–WT-1 (Dako), and mouse anti–ZO-1 (Zymed). Signals were evaluated using a Leica (Bensheim, Germany) fluorescence microscope equipped with a SPOT camera and Visitron (Puchheim, Germany) software. Human kidney slices were fixed in 4% PFA, embedded in paraffin, and cut into 4-μm-thick slices. Slices were deparaffinized in Xylol for 1 h, gradually hydrated through graded alcohols (100 to 50%), and washed in deionized water. After incubation in 3% H2O2 for 20 min, slices were rehydrated with PBS. Antigen unmasking was performed by incubation of the slices in 30% FCS and 3% BSA in PBS for 20 min at room temperature. Furthermore, slices were incubated for 3 min at 120°C in 0.01 M citrate/PBS. Thereafter, sections were incubated overnight in a humidified chamber at 4°C, with rabbit anti-KIBRA (1:400) or with KIBRA serum preincubated for 1 h at room temperature with 10 μg of blocking agent (Supplemental Figure S1A), respectively. Rabbit anti–WT-1 (1:400; Santa Cruz Biotechnology) was applied as positive control. Slices were washed extensively with PBS and incubated for 40 min with an anti-rabbit antibody included in the commercially available Vectastain ABC kit (Vector Laboratories, Burlingame, CA). Slices were washed with PBS, incubated with avidin-biotin for 40 min, and stained with diaminobenzidine. Sections were examined with a conventional light microscope (Zeiss, Cologne, Germany).
In Situ Hybridization
DNA probes comprising nucleotides (nt) 1402 through 1680 of human KIBRA cDNA and nt 5026 through 5407 of human PATJ cDNA were cloned in pGEM-T (Promega, Madison, WI). Recombinant RNA probes were transcribed in vitro from linearized pGEM-T using digoxygenin-labeled UTP and the DIG RNA Labeling Kit (Roche). In situ hybridization experiments on kidney sections were performed as described previously44 except that the probes were hybridized at 50°C for 16 h.
RNA Interference Experiments
shRNA were designed with publicly available prediction programs and are summarized in Supplemental Table 3. shRNA were cloned into the RNA expression vector pSuper (Oligoengine, Seattle, WA). To determine the efficiency of shRNA-mediated KIBRA k/d, we used psiCHECK2 (Promega) in which the coding sequence and the 3′ untranslated region of KIBRA were fused to the coding sequence of R. reniformis luciferase as an artificial 3′ untranslated region. In addition to R. reniformis luciferase, psiCHECK2 encodes for firefly luciferase as internal control. A total of 50 ng of the reporter plasmid and 50 ng of the respective pSuper shRNA constructs were co-transfected into HEK 293T cells in a 96-well format using Lipofectamine 2000 (Invitrogen). Luciferase activities were measured by a dual-luciferase reporter assay system (Promega) in a luminometer (Mithras LB940; Berthold Technologies, Pforzheim, Germany) 24 h after transfection. Transfections and measurements were performed in triplicate. Selected hairpins (shRNA 1 and 2) were then subcloned into pLVTHM for stable lentiviral expression in human podocyte cell lines. Empty vector–transduced podocytes served as control cells as described previously.45
Wound-Healing Assays
Migration of empty vector–transduced control cells and KIBRA k/d podocytes was assessed with wound-healing assays. Cells were grown in collagen-coated (50 μg/ml) tissue culture flasks to confluence. A wound was scraped into the cell layers, using a sterile 10-μl pipette tip. Then cells were washed with PBS and incubated with fresh medium for 1 h in the incubator to allow recovering of the cells. Thereafter, the cells were placed in a heating chamber (37°C) on the stage of an inverted microscope (Axiovert, Zeiss, Cologne, Germany). Constant CO2 contents for the length of the experiments were ensured by gas-tight culture flasks. Migration was monitored with a video camera for 10 h (Hamamatsu, Hersching, Germany) and controlled by HiPic software (Hamamatsu). Images were taken at 10-min intervals.
The number of cells that had migrated into same-sized square fields after 10 h was counted. Migration itself was quantified on a single-cell level. To this end, the outlines of individual cells were marked semiautomatically at each time step throughout the entire image stacks with Amira software (Mercury Computer Systems, Duesseldorf, Germany) as described previously.46,47 These segmentation data were used for further processing. Migration was quantified as the movement of the cell center. The x and y coordinates of the cell center were calculated as geometric means of equally weighted pixel positions within the cell outlines as function of time. We used two parameters to reveal the effect of KIBRA k/d on podocyte migration: Velocity and displacement. The velocity of migrating cells (μm/min) was calculated for each time interval by applying a three-point difference quotient. The displacement (μm) is the distance between the position of cells at the beginning and at the end of the experiment (after 10 h). Because the formation of lamellipodia leads to a shift of the cell center, it has to be mentioned that cells may reach high migration velocities without really covering a distance (“running on the spot”). Experiments were performed in triplicate. Representatively, original trajectories of control cells and KIBRA k/d podocytes are displayed in Figure 7C.
DISCLOSURES
None.
Acknowledgments
This study was supported by the Deutsche Forschungsgemeinschaft Pa 483/14-1 and IZKF Schw2/030/08 (A.S.).
We thank Nina Meyer, Katja Brinkmann, and Sabine Mally for excellent technical assistance and Kerstin Riskowsky for expert help in immunohistochemistry and imaging. We thank Drs. Hsiang-Hao Hsu and Roman Preston for critical reading of the manuscript and all members of our laboratory for helpful comments and discussions. We are grateful to Hermann Krähling for technical support during the migration assays. In addition, we thank Dr. Eric Lingueglia (Valbonne, France) for the mCIPP construct and Dr. Ben Margolis (Ann Arbor, MI) and Dr. Elior Peles (Rehovot, Israel) for human PATJ cDNA and PATJ antibody gifts.
Published online ahead of print. Publication date available at www.jasn.org.
K.D. and E.-M.S. contributed equally to this work, and T.W. and H.P. contributed equally to this work.
Supplemental information for this article is available online at http://www.jasn.org/.
REFERENCES
- 1.Pavenstadt H, Kriz W, Kretzler M: Cell biology of the glomerular podocyte. Physiol Rev 83: 253–307, 2003 [DOI] [PubMed] [Google Scholar]
- 2.Suzuki A, Ohno S: The PAR-aPKC system: Lessons in polarity. J Cell Sci 119: 979–987, 2006 [DOI] [PubMed] [Google Scholar]
- 3.Margolis B, Borg JP: Apicobasal polarity complexes. J Cell Sci 118: 5157–5159, 2005 [DOI] [PubMed] [Google Scholar]
- 4.Shin K, Fogg VC, Margolis B: Tight junctions and cell polarity. Annu Rev Cell Dev Biol 22: 207–235, 2006 [DOI] [PubMed] [Google Scholar]
- 5.Roh MH, Fan S, Liu CJ, Margolis B: The Crumbs3-Pals1 complex participates in the establishment of polarity in mammalian epithelial cells. J Cell Sci 116: 2895–2906, 2003 [DOI] [PubMed] [Google Scholar]
- 6.Roh MH, Makarova O, Liu CJ, Shin K, Lee S, Laurinec S, Goyal M, Wiggins R, Margolis B: The Maguk protein, Pals1, functions as an adapter, linking mammalian homologues of Crumbs and Discs Lost. J Cell Biol 157: 161–172, 2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kurschner C, Mermelstein PG, Holden WT, Surmeier DJ: CIPP, a novel multivalent PDZ domain protein, selectively interacts with Kir4.0 family members, NMDA receptor subunits, neurexins, and neuroligins. Mol Cell Neurosci 11: 161–172, 1998 [DOI] [PubMed] [Google Scholar]
- 8.Anzai N, Deval E, Schaefer L, Friend V, Lazdunski M, Lingueglia E: The multivalent PDZ domain-containing protein CIPP is a partner of acid-sensing ion channel 3 in sensory neurons. J Biol Chem 277: 16655–16661, 2002 [DOI] [PubMed] [Google Scholar]
- 9.Becamel C, Gavarini S, Chanrion B, Alonso G, Galeotti N, Dumuis A, Bockaert J, Marin P: The serotonin 5-HT2A and 5-HT2C receptors interact with specific sets of PDZ proteins. J Biol Chem 279: 20257–20266, 2004 [DOI] [PubMed] [Google Scholar]
- 10.Massey-Harroche D, Delgrossi MH, Lane-Guermonprez L, Arsanto JP, Borg JP, Billaud M, Le BA: Evidence for a molecular link between the tuberous sclerosis complex and the crumbs complex. Hum Mol Genet 16: 529–536, 2007 [DOI] [PubMed] [Google Scholar]
- 11.Roh MH, Liu CJ, Laurinec S, Margolis B: The carboxyl terminus of zona occludens-3 binds and recruits a mammalian homologue of discs lost to tight junctions. J Biol Chem 277: 27501–27509, 2002 [DOI] [PubMed] [Google Scholar]
- 12.Wells CD, Fawcett JP, Traweger A, Yamanaka Y, Goudreault M, Elder K, Kulkarni S, Gish G, Virag C, Lim C, Colwill K, Starostine A, Metalnikov P, Pawson T: A Rich1/Amot complex regulates the Cdc42 GTPase and apical-polarity proteins in epithelial cells. Cell 125: 535–548, 2006 [DOI] [PubMed] [Google Scholar]
- 13.Shin K, Straight S, Margolis B: PATJ regulates tight junction formation and polarity in mammalian epithelial cells. J Cell Biol 168: 705–711, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lemmers C, Medina E, Delgrossi MH, Michel D, Arsanto JP, Le BA: hINADl/PATJ, a homolog of discs lost, interacts with crumbs and localizes to tight junctions in human epithelial cells. J Biol Chem 277: 25408–25415, 2002 [DOI] [PubMed] [Google Scholar]
- 15.Sugihara-Mizuno Y, Adachi M, Kobayashi Y, Hamazaki Y, Nishimura M, Imai T, Furuse M, Tsukita S: Molecular characterization of angiomotin/JEAP family proteins: Interaction with MUPP1/Patj and their endogenous properties. Genes Cells 12: 473–486, 2007 [DOI] [PubMed] [Google Scholar]
- 16.Bratt A, Birot O, Sinha I, Veitonmaki N, Aase K, Ernkvist M, Holmgren L: Angiomotin regulates endothelial cell-cell junctions and cell motility. J Biol Chem 280: 34859–34869, 2005 [DOI] [PubMed] [Google Scholar]
- 17.Almeida OP, Schwab SG, Lautenschlager NT, Morar B, Greenop KR, Flicker L, Wildenauer D: KIBRA genetic polymorphism influences episodic memory in later life, but does not increase the risk of mild cognitive impairment. J Cell Mol Med January 11, 2008. [epub ahead of print] [DOI] [PMC free article] [PubMed]
- 18.Schaper K, Kolsch H, Popp J, Wagner M, Jessen F: KIBRA gene variants are associated with episodic memory in healthy elderly. Neurobiol Aging 29: 1123–1125, 2007 [DOI] [PubMed] [Google Scholar]
- 19.Rodriguez-Rodriguez E, Infante J, Llorca J, Mateo I, Sanchez-Quintana C, Garcia-Gorostiaga I, Sanchez-Juan P, Berciano J, Combarros O: Age-dependent association of KIBRA genetic variation and Alzheimer's disease risk. Neurobiol Aging August 16, 2007. [epub ahead of print] [DOI] [PubMed]
- 20.Papassotiropoulos A, Stephan DA, Huentelman MJ, Hoerndli FJ, Craig DW, Pearson JV, Huynh KD, Brunner F, Corneveaux J, Osborne D, Wollmer MA, Aerni A, Coluccia D, Hanggi J, Mondadori CR, Buchmann A, Reiman EM, Caselli RJ, Henke K, de Quervain DJ: Common Kibra alleles are associated with human memory performance. Science 314: 475–478, 2006 [DOI] [PubMed] [Google Scholar]
- 21.Lauriat TL, Dracheva S, Kremerskothen J, Duning K, Haroutunian V, Buxbaum JD, Hyde TM, Kleinman JE, McInnes LA: Characterization of KIAA0513, a novel signaling molecule that interacts with modulators of neuroplasticity, apoptosis, and the cytoskeleton. Brain Res 1121: 1–11, 2006 [DOI] [PubMed] [Google Scholar]
- 22.Saleem MA, O'Hare MJ, Reiser J, Coward RJ, Inward CD, Farren T, Xing CY, Ni L, Mathieson PW, Mundel P: A conditionally immortalized human podocyte cell line demonstrating nephrin and podocin expression. J Am Soc Nephrol 13: 630–638, 2002 [DOI] [PubMed] [Google Scholar]
- 23.Kremerskothen J, Plaas C, Buther K, Finger I, Veltel S, Matanis T, Liedtke T, Barnekow A: Characterization of KIBRA, a novel WW domain-containing protein. Biochem Biophys Res Commun 300: 862–867, 2003 [DOI] [PubMed] [Google Scholar]
- 24.Harris BZ, Lim WA: Mechanism and role of PDZ domains in signaling complex assembly. J Cell Sci 114: 3219–3231, 2001 [DOI] [PubMed] [Google Scholar]
- 25.Shin K, Wang Q, Margolis B: PATJ regulates directional migration of mammalian epithelial cells. EMBO Rep 8: 158–164, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tveito G, Hansteen IL, Dalen H, Haugen A: Immortalization of normal human kidney epithelial cells by nickel(II). Cancer Res 49: 1829–1835, 1989 [PubMed] [Google Scholar]
- 27.Einbond A, Sudol M: Towards prediction of cognate complexes between the WW domain and proline-rich ligands. FEBS Lett 384: 1–8, 1996 [DOI] [PubMed] [Google Scholar]
- 28.Kremerskothen J, Kindler S, Finger I, Veltel S, Barnekow A: Postsynaptic recruitment of Dendrin depends on both dendritic mRNA transport and synaptic anchoring. J Neurochem 96: 1659–1666, 2006 [DOI] [PubMed] [Google Scholar]
- 29.Patrakka J, Xiao Z, Nukui M, Takemoto M, He L, Oddsson A, Perisic L, Kaukinen A, Szigyarto CA, Uhlen M, Jalanko H, Betsholtz C, Tryggvason K: Expression and subcellular distribution of novel glomerulus-associated proteins dendrin, ehd3, sh2d4a, plekhh2, and 2310066E14Rik. J Am Soc Nephrol 18: 689–697, 2007 [DOI] [PubMed] [Google Scholar]
- 30.Asanuma K, Campbell KN, Kim K, Faul C, Mundel P: Nuclear relocation of the nephrin and CD2AP-binding protein dendrin promotes apoptosis of podocytes. Proc Natl Acad Sci U S A 104: 10134–10139, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Asanuma K, Yanagida-Asanuma E, Faul C, Tomino Y, Kim K, Mundel P: Synaptopodin orchestrates actin organization and cell motility via regulation of RhoA signalling. Nat Cell Biol 8: 485–491, 2006 [DOI] [PubMed] [Google Scholar]
- 32.Asanuma K, Kim K, Oh J, Giardino L, Chabanis S, Faul C, Reiser J, Mundel P: Synaptopodin regulates the actin-bundling activity of alpha-actinin in an isoform-specific manner. J Clin Invest 115: 1188–1198, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kremerskothen J, Plaas C, Kindler S, Frotscher M, Barnekow A: Synaptopodin, a molecule involved in the formation of the dendritic spine apparatus, is a dual actin/alpha-actinin binding protein. J Neurochem 92: 597–606, 2005 [DOI] [PubMed] [Google Scholar]
- 34.Mundel P, Heid HW, Mundel TM, Kruger M, Reiser J, Kriz W: Synaptopodin: An actin-associated protein in telencephalic dendrites and renal podocytes. J Cell Biol 139: 193–204, 1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Deller T, Mundel P, Frotscher M: Potential role of synaptopodin in spine motility by coupling actin to the spine apparatus. Hippocampus 10: 569–581, 2000 [DOI] [PubMed] [Google Scholar]
- 36.Buther K, Plaas C, Barnekow A, Kremerskothen J: KIBRA is a novel substrate for protein kinase Czeta. Biochem Biophys Res Commun 317: 703–707, 2004 [DOI] [PubMed] [Google Scholar]
- 37.Hurd TW, Gao L, Roh MH, Macara IG, Margolis B: Direct interaction of two polarity complexes implicated in epithelial tight junction assembly. Nat Cell Biol 5: 137–142, 2003 [DOI] [PubMed] [Google Scholar]
- 38.Traer CJ, Rutherford AC, Palmer KJ, Wassmer T, Oakley J, Attar N, Carlton JG, Kremerskothen J, Stephens DJ, Cullen PJ: SNX4 coordinates endosomal sorting of TfnR with dynein-mediated transport into the endocytic recycling compartment. Nat Cell Biol 9: 1370–1380, 2007 [DOI] [PubMed] [Google Scholar]
- 39.Rayala SK, den Hollander P, Manavathi B, Talukder AH, Song C, Peng S, Barnekow A, Kremerskothen J, Kumar R: Essential role of KIBRA in co-activator function of dynein light chain 1 in mammalian cells. J Biol Chem 281: 19092–19099, 2006 [DOI] [PubMed] [Google Scholar]
- 40.Patrie KM, Drescher AJ, Welihinda A, Mundel P, Margolis B: Interaction of two actin-binding proteins, synaptopodin and alpha-actinin-4, with the tight junction protein MAGI-1. J Biol Chem 277: 30183–30190, 2002 [DOI] [PubMed] [Google Scholar]
- 41.Hirabayashi S, Mori H, Kansaku A, Kurihara H, Sakai T, Shimizu F, Kawachi H, Hata Y: MAGI-1 is a component of the glomerular slit diaphragm that is tightly associated with nephrin. Lab Invest 85: 1528–1543, 2005 [DOI] [PubMed] [Google Scholar]
- 42.Reiser J, Oh J, Shirato I, Asanuma K, Hug A, Mundel TM, Honey K, Ishidoh K, Kominami E, Kreidberg JA, Tomino Y, Mundel P: Podocyte migration during nephrotic syndrome requires a coordinated interplay between cathepsin L and alpha3 integrin. J Biol Chem 279: 34827–34832, 2004 [DOI] [PubMed] [Google Scholar]
- 43.Theilig F, Kriz W, Jerichow T, Schrade P, Hahnel B, Willnow T, Le HM, Bachmann S: Abrogation of protein uptake through megalin-deficient proximal tubules does not safeguard against tubulointerstitial injury. J Am Soc Nephrol 18: 1824–1834, 2007 [DOI] [PubMed] [Google Scholar]
- 44.Schaefer L, Raslik I, Grone HJ, Schonherr E, Macakova K, Ugorcakova J, Budny S, Schaefer RM, Kresse H: Small proteoglycans in human diabetic nephropathy: Discrepancy between glomerular expression and protein accumulation of decorin, biglycan, lumican, and fibromodulin. FASEB J 15: 559–561, 2001 [DOI] [PubMed] [Google Scholar]
- 45.Schermer B, Ghenoiu C, Bartram M, Muller RU, Kotsis F, Hohne M, Kuhn W, Rapka M, Nitschke R, Zentgraf H, Fliegauf M, Omran H, Walz G, Benzing T: The von Hippel-Lindau tumor suppressor protein controls ciliogenesis by orienting microtubule growth. J Cell Biol 175: 547–554, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dreval V, Dieterich P, Stock C, Schwab A: The role of Ca2+ transport across the plasma membrane for cell migration. Cell Physiol Biochem 16: 119–126, 2005 [DOI] [PubMed] [Google Scholar]
- 47.Dieterich P, Klages R, Preuss R, Schwab A: Anomalous dynamics of cell migration. Proc Natl Acad Sci U S A 105: 459–463, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]