Abstract
Classically, infants with mutations in NPHS1, which encodes nephrin, present with nephrotic syndrome within the first 3 mo of life (congenital nephrotic syndrome of the Finnish-type), and children with mutations in NPHS2, which encodes podocin, present later with steroid-resistant nephrotic syndrome. Recently, however, NPHS2 mutations have been identified in children with congenital nephrotic syndrome. Whether NPHS1 mutations similarly account for some cases of childhood steroid-resistant nephrotic syndrome is unknown. In this study, 160 patients who belonged to 142 unrelated families and presented with nephrotic syndrome at least 3 mo after birth were screened for NPHS1 variants once mutations in NPHS2 had been excluded. Compound heterozygous NPHS1 mutations were identified in one familial case and nine sporadic cases. Mutations included protein-truncating nonsense and frameshift mutations, as well as splice-site and missense variants. Mutations were classified as “severe” or “mild” using prediction algorithms and functional assays. Most missense variants trafficked normally to the plasma membrane and maintained the ability to form nephrin homodimers and to heterodimerize with NEPH1, suggesting retained function. The presence of at least one “mild” mutation in these patients likely explains the later onset and milder course of disease. These results broaden the spectrum of renal disease related to nephrin mutations.
Idiopathic nephrotic syndrome (NS) represents a heterogeneous group of glomerular disorders occurring mainly in children and may be classified as steroid-sensitive (SSNS) or steroid-resistant (SRNS) on the basis of response to corticosteroid therapy. Gene discovery efforts in the past decade have led to an improved understanding of the hereditary basis of NS.1 Mutations in the NPHS1 gene, encoding the podocyte-expressed protein nephrin, lead to the congenital NS of the Finnish-type (CNF), which is inherited in an autosomal recessive manner. It affects approximately 1:10,000 newborns in Finland2 but has since been described in other populations.3–5 Nephrin is a single-pass transmembrane protein consisting of eight extracellular Ig-like modules, a fibronectin type III–like motif, and a cytosolic C-terminal tail. Homodimers of nephrin and heterodimers with the glomerular protein NEPH1 constitute the structural basis of the slit diaphragm.6,7 In addition to its structural function, nephrin is involved in podocyte signaling events.8
Mutations in the NPHS2 gene encoding podocin were, thereafter, described in patients presenting with autosomal recessive SRNS with onset typically between 3 mo and 5 yr of age.9 NPHS2 mutations account for 42% of familial and 10% of sporadic cases of SRNS.10 Proper assembly of nephrin and other slit diaphragm constituents and trafficking to the plasma membrane and to lipid rafts require interaction with podocin.11,12
Classically, mutations in the NPHS1 and NPHS2 genes have been distinguished by their implications in familial congenital (onset at birth to 3 mo) and in childhood-onset (later than 3 mo) cases, respectively. Hinkes et al.13 recently confirmed the findings of others that patients with NPHS1 mutations present with NS exclusively during the first 3 mo of life.3–5,14 They also identified NPHS2 mutations in 39% of children with congenital onset of NS,13 as described previously,15 therein broadening the spectrum of NPHS2-associated renal disease. We therefore sought to determine whether mutations in the nephrin gene may similarly account for a wider range of disease presentations to include childhood-onset SRNS. Our studies revealed that compound heterozygous mutations in the NPHS1 gene were responsible for SRNS in a cohort of patients in whom NPHS2 mutations had been excluded and who presented with noncongenital onset and a more protracted course of renal disease.
Mutation screening was performed in a cohort of familial and sporadic cases of SRNS, for which NPHS2 mutations had been excluded by sequencing the exonic regions and intronic junctions. Nephrin mutations were found in one family with two affected siblings, among 44 families with familial SRNS, using a combination of linkage analysis and NPHS1 sequencing (Table 1). Of 98 patients with sporadic SRNS, nine individuals were compound heterozygotes for NPHS1 mutations (Table 1). The mean age of onset of NS in these 11 patients was 3.0 yr (range 6 mo to 8 yr); hence, later than previously described for patients with NPHS1 mutations. The NS was resistant to corticosteroids in all cases, as well as to cyclosporine and cyclophosphamide, when additional treatments were attempted. Renal biopsy performed at the time of presentation revealed mesangioproliferative lesions in one patient, minimal-change disease in six patients, and FSGS in three patients (Table 1). Characteristic tubular lesions of CNF were not observed. No extrarenal involvement was reported. At the end of follow-up, only five patients had reached ESRD, at a mean age of 13.6 yr (range 6 to 25 yr), whereas six patients had normal serum creatinine (Table 1). Four patients successfully received a renal allograft, with no disease recurrence.
Table 1.
Clinical data of patients who had SRNS and in whom NPHS1 mutations were identifieda
| Patient | Gender | Age of Onset Pu (NS) (yr) | Biopsy | Therapy | Evolution | Mutation 1 Severe | Mutation 2 Mild |
|---|---|---|---|---|---|---|---|
| 1420 | F | 0.25 (3.00) | MCNS | CS, CP | Normal Cr at 14 yr | c.609–2A→C (M) | c.319G→A |
| p.A107T (P) | |||||||
| 446 | F | 0.80 (0.80) | MCNS | CS | ESRF at 9 yr | c.3720_3735del16 | c.1724C→A |
| p.L1240fs1286Xb (P) | p.P575Q (M) | ||||||
| 1075 | F | 0.50 (0.50) | FSGS | Unknown | ESRF at 13 yr | c.1379G→A | c.2928G→T |
| p.R460Qb (?) | p.R976S (?) | ||||||
| 841 | F | 3.80 (3.80) | FSGS | CS, CsA | Normal Cr at 6 yr | c.468C→G | c.2928G→T |
| p.Y156X (P) | p.R976S (M) | ||||||
| 466 | F | 0.25 (0.75) | MCNS | CS, CP | Normal Cr at 10 yr | c.2479C→T | c.2928G→T |
| p.R827X (M) | p.R976S (P) | ||||||
| 1167 | F | 3.10 (3.10) | FSGS | CS, CP | ESRF at 15 yr | c.516delC | c.2928G→T |
| p.T712fs175X (M) | p.R976S (P) | ||||||
| 693 | M | 8.00 (8.00) | MCNS | CS | Normal Cr at 16 yr | c.1134–1135delGC | c.286C→G |
| p.R379fs417X (M) | p.L96V (P) | ||||||
| 1407 | F | 5.00 (5.00) | MCNS | CS, CP | ESRF at 25 yr | c.516delC | c.2928G→T |
| p.T712fs175X (M) | p.R976S (P) | ||||||
| 634 | M | 3.00 (3.00) | MP | Unknown | ESRF at 6 yr | c.1491delC | c.2072–6C→G (P) |
| p.S494fs547X (M) | |||||||
| 771c | F | 2.20 (2.80) | MCNS | CS, CsA | Normal Cr at 9 yr | c.2495T→C | c.2928G→T |
| 1462c | M | 2.50 (2.50) | Not performed | No treatment | Normal Cr at 6 yr | p.L832P (P) | p.R976S (M) |
CP, cyclophosphamide; Cr, creatinine; CS, corticosteroids; CsA, cyclosporin A; ESRF, end-stage renal failure; MCNS, minimal-change glomerulonephritis; MP, mesangioproliferative glomerulonephritis; Pu, proteinuria.
This mutation may potentially be a “mild” one. Mode of transmission of mutant alleles are indicated in parentheses: M, maternal; P, paternal; ?, unknown.
Siblings with the same parents.
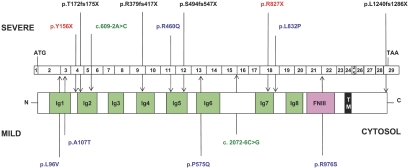
In total, 14 mutations, 12 of which are novel, were identified in 10 unrelated patients. These included six nonsense and frameshift, two splice-site, and six missense mutations, uniformly distributed throughout the NPHS1 gene (Figure 1). Segregation of mutations confirmed recessive inheritance. All nonsense and frameshift mutations are predicted to result in a truncated protein. In addition, the c.3720_3735delC (p.L1240fs1286X) mutation is unusual because it involves a deletion of the last seven nucleotides of the NPHS1 coding and of 9 bp of the 3′-untranslated regions. The resulting protein lacks the usual terminal valine residue and instead bears an additional 45 amino acids with no known sequence homology.
Figure 1.
Schematic diagram of the NPHS1 mutations detected in a cohort of patients with SRNS and their localization with respect to exons and protein functional domains. Nephrin consists of Ig-like domains, a fibronectin III (FNIII)-like domain, a transmembrane (TM) region, and a cytosolic C-terminal tail. Mutations included nonsense and frameshift, splice-site, and missense mutations and were classified as severe or mild on the basis of prediction algorithms and functional analyses.
Six missense mutations were identified in this cohort of patients, and all were ruled out as polymorphisms by sequencing at least 176 control chromosomes, and 352 control chromosomes for the most commonly detected p.R976S mutation. These missense changes were evaluated for pathogenicity using two widely used prediction methods: SIFT (Sorting Intolerant From Tolerant)16 and PolyPhen (Polymorphism Phenotyping).17 SIFT predicts whether an amino acid substitution will affect protein function on the basis of the degree to which the amino acid residue is conserved during evolution, thereafter designating changes as either tolerated or deleterious.16 Of the six missense mutations, five were classified as deleterious, with the exception of the p.R460Q mutation (Table 2). PolyPhen designates variants as either probably or possibly damaging or benign on the basis of sequence annotation, sequence alignment, and structural parameters.17 PolyPhen predicted only three of the missense changes to be damaging: p.P575A, p.L832P, and p.R976S mutations (Table 2).
Table 2.
Effects of NPHS1 mutations according to prediction algorithms and functional studies
| Nucleotide Alteration | Coding Sequence Alteration | Exon Involved | Score of Donor/Acceptor Site | Control Chromosomes | PolyPhen Score | SIFT Prediction | Cellular Localization | Nephrin Homodimerization | NEPH1 Heterodimerization |
|---|---|---|---|---|---|---|---|---|---|
| c.286C→G | p.L96V | 3 | 0/188 | 1.34 (benign) | Not tolerated | Plasma membrane | Intact | Intact | |
| c.319G→A | p.A107T | 3 | 0/188 | 1.50 (benign) | Not tolerated | Plasma membrane | Intact | Intact | |
| c.379G→A22 | p.R460Q | 11 | 0/190 | 1.40 (benign) | Tolerated | Plasma membrane | Intact | Intact | |
| c.1724C→A | p.P575Q | 13 | 0/176 | 1.80 (possibly damaging) | Not tolerated | Plasma membrane | Intact | Intact | |
| c. 2495T→C | p.L832P | 18 | 0/182 | 2.20 (probably damaging) | Not tolerated | Endoplasmic reticulum | |||
| c.2928G→T | p.R976S | 22 | Normal: 0.95 | 0/352 | 2.20 (probably damaging) | Not tolerated | Plasma membrane | ||
| Splice site (?) | Mutant: 0.79 | ||||||||
| c.609–2A→C | Splice site | IVS5 | Normal: 0.88 | ||||||
| Mutant: 0.00 | |||||||||
| c.2072–6C→G | Splice site | IVS15 | Normal: 0.88 | 0/186 | |||||
| Mutant: 0.26 | |||||||||
| c.468C→G22 | p.Y156X | 4 | |||||||
| c.516delC | p.T172fs175X | 4 | |||||||
| c.1134–1135delGC | p.R379fs417X | 9 | |||||||
| c.1491delC | p.S494fs547X | 12 | |||||||
| c.2479C→T | p.R827X | 18 | |||||||
| c. 3720–3735del16 | p.L1240fs1286X | 29 | 0/176 |
Two splice-site mutations were identified. Potential splice sites were additionally evaluated using the neural networks program Genie, and scores ranging from 0 to 1 (where 1 is indicative of the presence of an ideal splice site) were assigned and compared between wild-type and mutant sequences to determine the extent to which the mutation is predicted to alter a splice site.18 The c.609–2A→C mutation affects the invariant A of the splice acceptor site, resulting in a decrease in the score from 0.88 to 0.00 and hence would be expected to lead to absence of correct splicing (Table 2). Conversely, the c.2072–6C→G mutation only moderately decreases the score of the normal splice junction sequence from 0.88 to 0.26. Normally spliced nephrin transcripts are, therefore, likely to exist, although in reduced amounts (Table 2). In addition, the c.2928G→T (p.R976S) mutation, which leads to a probably damaging substitution in the fibronectin-like domain (according to the PolyPhen program)17 affects the first base of exon 22. The Genie program predicted a slight decrease in the splicing score from 0.96 to 0.79. Despite this modest score change, an in vitro exontrap system demonstrated an alteration in the splicing around exon 22 (Supplemental Figure S1). Unfortunately, verification in vivo was not possible given the absence of available RNA from these patients.
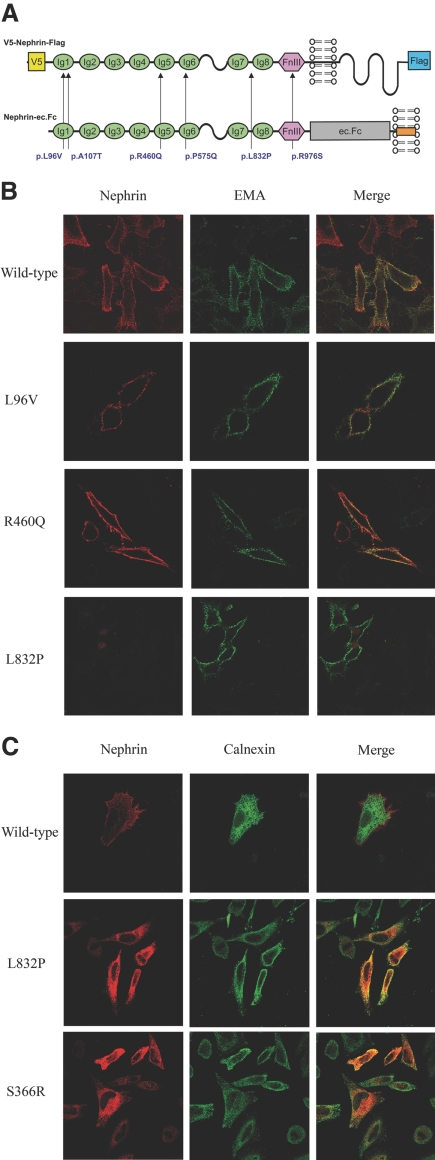
To determine the putative effects of missense mutations, we additionally performed functional studies. The intracellular trafficking of V5- and Flag-tagged missense nephrin variants (Figure 2A) was studied upon transient transfection in HeLa cells. Except for the p.L832P mutant, co-localization with fluorescently labeled epithelial membrane antigen, a plasma membrane marker, was observed for the p.L96V, p.A107T, p.P575Q, p.R460Q, and p.R976S mutants, just as with wild-type nephrin, under nonpermeabilized conditions (Figure 2B, Supplemental Figure S2). Furthermore, immunolabeling under permeabilized conditions revealed co-localization of the p.L832P variant with calnexin, similar to the p.S366R mutant,19 suggesting retention in the endoplasmic reticulum (Figure 2C).
Figure 2.
Effects of missense mutations on nephrin intracellular trafficking. (A) Schematic representation of full-length V5- and Flag-tagged and ec.Fc-tagged nephrin constructs and the locations of missense variants. (B) The p.L96V and p.R460Q mutants co-localized with epithelial membrane antigen (green) at the plasma membrane, similar to wild-type nephrin, when transiently transfected into HeLa cells. Nephrin was detected using an anti-V5 antibody (red), under nonpermeabilized conditions. Conversely, the p.L832P mutant failed to traffic to the membrane. (C) Similar to the p.S366R variant used as a positive control, the p.L832P mutant co-localized with calnexin (green) in transiently transfected HeLa cells, suggesting retention in the endoplasmic reticulum. Cells were permeabilized with saponin before immunolabeling. Magnification, ×1000.
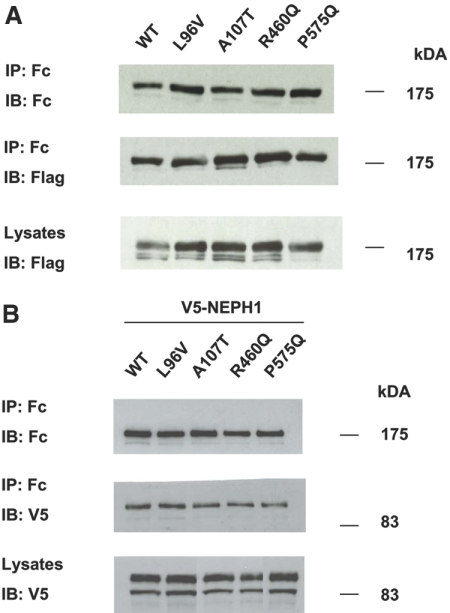
Thereafter, the effects on nephrin homodimerization and NEPH1 heterodimerization of the missense p.L96V, p.A107T, p.R460Q, and p.P575Q mutants, which affect the nephrin Ig domains, were tested. Upon co-transfection of mutagenized nephrin-ec.Fc and Flag-tagged nephrin constructs in HEK293 cells, co-immunoprecipitation experiments demonstrated maintenance of nephrin–nephrin interaction, despite the missense changes (Figure 3A). Similarly, variants heterodimerized with full-length V5-tagged NEPH1 in transiently transfected cells (Figure 3B). Our assays were not, however, sufficiently quantitative to assess whether the strengths of these interactions were altered.
Figure 3.
Effects of missense mutations on nephrin homodimerization and heterodimerization with NEPH1. (A) HEK 293T cells were transfected with wild-type or mutant full-length nephrin-ec.Fc and V5-nephrin-Flag constructs. Nephrin-ec.Fc was immunoprecipitated using Protein A-Sepharose beads. Immunoblotting using anti-Flag antibody revealed intact dimerization between the two nephrin constructs. These data are representative of three experiments. (B) Similarly, cells were transfected with wild-type or mutant full-length nephrin-ec.Fc and full-length V5-NEPH1 plasmid constructs. Protein A-Sepharose beads were used to immunoprecipitate nephrin. Immunoblotting using anti-V5 antibody revealed intact heterodimerization between missense nephrin mutants and NEPH1. These data represent five replicate experiments.
Our analysis of a cohort of patients presenting with familial and sporadic SRNS after 3 mo of life broadens the phenotypic spectrum of renal disease associated with mutations in the NPHS1 gene encoding nephrin. Previously, NS caused by nephrin mutations had been described exclusively in children presenting at the time of birth or within the first 3 mo of life. Our study identified patients bearing NPHS1 mutations in which the renal disease presented later in childhood, was resistant to steroids, and had a spectrum of renal histologic findings atypical for CNF,20 ranging from minimal-change disease to FSGS. These features are reminiscent of SRNS caused by NPHS2 mutations. A protracted course was also observed, averaging 13.6 yr from disease onset to end-stage renal failure in five of 11 patients.
We attempted to classify these mutations on the basis of prediction algorithms and functional studies as either “severe” or “mild.” Nephrin mutations were classified as severe when they result in truncated, aberrantly spliced, or nonfunctional proteins. This includes all nonsense and frameshifting, the c.609–2A→C splice, and the endoplasmic reticulum–retained p.L832P missense mutations. Greater than 75% of missense nephrin variants leading to CNF have previously been shown to be retained in the endoplasmic reticulum.21 Mutations were designated mild when partial function of nephrin was likely maintained. The five other missense mutations were shown to retain the abilities to traffic in the cell properly and to homo- and heterodimerize with NEPH1 and thus were classified as mild. The c.2072–6C→G splice mutant likely allows for some correct splicing and therefore is included as a mild mutation.
The mutation detection rate in familial cases was lower than might normally be expected, because most of these patients were offsprings of consanguineous relationships; thus, compound heterozygosity is less likely. Compound heterozygosity for at least one mild mutation in all cases may explain the lesser severity of disease in our cohort of patients. Indeed, less severe phenotypes of cystinosis, Pierson syndrome, and autosomal recessive polycystic kidney disease have been observed in patients bearing missense mutations in the CTNS,22 LAMB2,23 and PKHD124 genes, respectively, which maintained residual protein function. At present, however, it is difficult to be certain of the functional effects of these mutations, as demonstrated by patient 446, in whom two potentially mild mutations were detected. Additional work is required, but such genotype–phenotype correlations exemplify the importance of screening for NPHS1 mutations in patients with SRNS for disease prognostication.
Our method of classifying mutations as either severe or mild was based on applying the best available prediction algorithms and coupling these with established functional assays of known nephrin functions. Despite this added rigor, there were, however, discrepancies in the results of functional assays, of prediction algorithms, and in clinical correlation in the case of previously described mutations. For instance, although the p.R460Q mutant is predicted to be a tolerated variant and was shown to traffic normally in the cell and to homodimerize and to heterodimerize with NEPH1, it had previously been identified in cases of CNF,3,5,25 suggesting that the mutation may disrupt other structural or functional properties of nephrin. It is likewise possible that this mutation leads to a decrease in the ability to homo- or heterodimerize, a feature that we were unable to quantify with precision in our assays. Future studies will additionally need to address whether missense variants that traffic to the membrane affect nephrin phosphorylation, actin reorganization in the cytoskeleton of podocytes,26 or downstream signaling events involved in transcriptional regulation27 and apoptosis.28 Conversely, the protein-truncating p.L1240fs1286X mutation might be considered a mild mutation because it involves the very C-terminal end of the protein. This is reminiscent of the p.R1160X mutant, which results in a truncation of the nephrin protein downstream of the last amino acid residue of exon 27 and which has been shown to be expressed in the kidney.4 This mutation results in renal disease of lesser severity in 50% of female patients.4
Mutations in the NPHS2 gene and in exons 8 and 9 of the WT1 gene were excluded in our cohort of patients after sequencing the exonic and intronic splice junctional regions. We are, therefore, confident that the renal disease in these patients may be attributed to the pathogenic NPHS1 mutations we have identified, rather than to the triallelic inheritance of NPHS1 and NPHS2 mutations in a few rare cases, as had been reported by others.4,29 It is conceivable that variants in the promoter and intronic regions of the NPHS2 gene may have been missed, but no pathogenic mutations in the promoter region of the gene have previously been associated with SRNS; however, variants and haplotypes in the promoter region of the podocin gene have been associated with variations in the levels of proteinuria in patients with IgA nephropathy but not in FSGS.30 It will be of great interest in the future to explore the possibility of genetic modification as a basis for phenotypic variability in patients with familial forms of NS. These studies, however, will require larger numbers of patients and should involve genome-wide approaches.
Finally, our findings bear significant implications on the therapy of NS. As with patients bearing NPHS2 mutations,15 our limited cohort revealed that patients carrying similar NPHS1 mutations should be spared immunosuppressive therapies. Furthermore, therapies aimed at correcting mis-trafficking of misfolded mutant proteins in the cell may provide hope for retarding the progression of SRNS by preserving residual nephrin function.21,31
CONCISE METHODS
Patients
A total of 160 patients belonging to 142 families were included in this study. They originated from a large number of countries worldwide, but most are from Europe (mainly from various regions in France) and from North Africa. All patients presented between 3 mo and 18 yr of age with SRNS, defined as a lack of response to three bolus infusions of methylprednisolone, followed by 4 wk of treatment with prednisone. Among them, 62 patients belonging to 44 families (31 consanguineous) were classified as familial cases, defined previously10 either as families in which two or more children were affected or families in which one affected child was the product of a consanguineous relationship. Cases were defined as sporadic when only one child, in families with no history of consanguinity, was affected. Mean age at discovery (proteinuria and/or NS) was 67 ± 49 mo in AR SRNS (n = 57), whereas it was lower in sporadic cases (54 ± 39 mo; n = 98). Informed consent was obtained from patients or their parents. Experiments were performed in accordance with French ethical committee recommendations.
Mutation Analysis
Genomic DNA was extracted from peripheral blood by standard methods. Linkage analyses were carried out using four polymorphic microsatellite markers (D19S224, D19S225, D19S425, and D19S608), spanning a 0.5-Mb region containing the NPHS1 locus. Pedigrees were constructed with CYRILLIC 2.1 (Cherwell Scientific, Oxford, UK). Mutations in the NPHS2 gene encoding podocin were excluded in all cases by sequencing exonic and intronic junctional regions, as were mutations in exons 8 and 9 of the WT1 gene in phenotypically female patients. Mutation screening of NPHS1 was performed by direct sequencing of the 29 coding exons and the adjacent intronic junctions. All variants were screened in at least 88 unrelated controls using direct sequence analysis and/or the SNAPshot method.10 In silico analyses of missense mutations was carried out using the SIFT (http://blocks.fhcrc.org/sift/SIFT.html)16 and PolyPhen (http://genetics.bwh.harvard.edu/pph/)17 software. The evaluation of splice sites and potential splice sites was performed using the Genie program (http://www.fruitfly.org/seq_tools/splice.html).32
Plasmid Constructs and Antibodies
The generation of full-length Nephrin-Flag, Nephrin-ec.Fc, and V5-NEPH1 constructs has previously been described.7,24 For subcloning of the NPHS1-Flag cDNA into the V5 plasmid, a unique MluI restriction site was inserted in position 57 by site-directed mutagenesis (Quick-Change Kit; Stratagene, La Jolla, CA), digested with MluI and NotI and ligated into corresponding sites in the plasmid. Site-directed mutagenesis was similarly used to generate all missense variants, followed by verification by automated sequencing. Commercial antibodies used for this study are detailed in Supplemental Table S1. Affinity-purified rabbit anti-podocin antibodies were generated in the laboratory and have previously been described.33
Cellular Immunolocalization Studies
HeLa cells (7 × 105) were plated on poly-l-lysine–coated glass coverslips (Sigma, St. Louis, MO) and grown to 90% confluence in DMEM supplemented with 10% FBS. Cells were transiently transfected 24 h later with 0.5 μg of plasmid DNA, using Lipofectamine according to the manufacturer's instructions (Invitrogen, Carlsbad, CA), thereby achieving 80% transfection efficiency. After 48 h of incubation, HeLa cells were rinsed in PBS and incubated with rabbit anti-V5 (diluted 1:1300 in PBS) and/or mouse anti–epithelial membrane antigen (diluted 1:50) antibodies for 15 min at 4°C. Cells were fixed in 4% paraformaldehyde for 20 min and subsequently treated with 50 mM NH4Cl. Alternatively, cells were incubated for 1 h with mouse anti-V5 (diluted 1:200) or anti-calnexin (diluted 1:100) in PBS/BSA/saponin for permeabilization. Coverslips were then incubated with fluorescently labeled secondary antibodies for 1 h, before mounting using Fluoprep (Biomérieux, Lyon, France). Images were obtained using a Zeiss Pascal confocal laser scanning microscope (Carl Zeiss, Jena, Germany)
Homo- and Heterodimerization Studies
HEK293T cells grown to 60% confluence in DMEM supplemented with 10% FBS were transiently co-transfected using calcium phosphate with 5 μg of wild-type, mutant nephrin-Flag, or V5-NEPH1 and 5 μg of wild-type or mutant nephrin-ec.Fc plasmids. After incubation for 24 h, cells were washed with PBS and lysed in buffer containing 20 mM Tris-HCl (pH 7.5), 50 mM NaCl, 0.1 mM EDTA, 1% Triton X-100, and protease inhibitors (Complete Mini; Roche, Basel, Switzerland). After centrifugation at 15,000 × g for 15 min at 4°C and ultracentrifugation at 40,000 × g for 35 min at 4°C, cell lysates containing 1 mg of total proteins were incubated at 4°C for 1 h with 50 μl of Protein A-Sepharose beads (Sigma). The beads were washed extensively with lysis buffer, and bound proteins were split in two and resolved on a 6% SDS-PAGE. Western blot analysis was performed with anti-IgG (diluted 1:5000), anti-Flag (diluted 1:2000), or mouse anti-V5 (diluted 1:2000) antibodies followed by incubation with appropriate horseradish peroxidase–coupled secondary antibodies.
DISCLOSURES
C.A. and E.L.E. are recipients of a Renal Innovations Program grant from Genzyme Corp.
Acknowledgments
We acknowledge financial support for this work from the Association pour l'Information et la Recherche sur les Maladies Rénales Génétiques (AIRG), the Programme Hospitalier de Recherche Clinique (AOM02123), and the GIS-Institute des Maladies Rares (4MR02F). A.P. was the recipient of a PhD grant from the Ministère de l'Enseignement Supérieur et de la Recherche. E.L.E. was supported by a Ruth Kirchstein National Research Service Award (DK065409) from the National Institutes of Health, National Institute of Diabetes and Digestive and Kidney Diseases, and by the EuReGene Project, an integrated project (5085) of the 6th Framework Program of the European Commission.
Published online ahead of print. Publication date available at www.jasn.org.
A.P., F.N., and E.L.E. contributed equally to this work.
See related editorial, “Yet More Ways to Skin a Cat: Nephrin Mutations outside the Neonatal Period,” on pages 1837–1838.
Supplemental information for this article is available online at http://www.jasn.org/.
References
- 1.Tryggvason K, Patrakka J, Wartiovaara J: Hereditary proteinuria syndromes and mechanisms of proteinuria. N Engl J Med 354: 1387–1401, 2006 [DOI] [PubMed] [Google Scholar]
- 2.Kestila M, Lenkkeri U, Mannikko M, Lamerdin J, McCready P, Putaala H, Ruotsalainen V, Morita T, Nissinen M, Herva R, Kashtan CE, Peltonen L, Holmberg C, Olsen A, Tryggvason K: Positionally cloned gene for a novel glomerular protein—nephrin—is mutated in congenital nephrotic syndrome. Mol Cell 1: 575–582, 1988 [DOI] [PubMed] [Google Scholar]
- 3.Gigante M, Monno F, Roberto R, Laforgia N, Assael MB, Livolti S, Caringella A, La Manna A, Masella L, Iolascon A: Congenital nephrotic syndrome of the Finnish type in Italy: A molecular approach. J Nephrol 15: 696–702, 2002 [PubMed] [Google Scholar]
- 4.Koziell A, Grech V, Hussain S, Lee G, Lenkkeri U, Tryggvason K, Scambler P: Genotype/phenotype correlations of NPHS1 and NPHS2 mutations in nephrotic syndrome advocate a functional inter-relationship in glomerular filtration. Hum Mol Genet 11: 379–388, 2002 [DOI] [PubMed] [Google Scholar]
- 5.Sako M, Nakanishi K, Obana M, Yata N, Hoshii S, Takahashi S, Wada N, Takahashi Y, Kaku Y, Satomura K, Ikeda M, Honda M, Iijima K, Yoshikawa N: Analysis of NPHS1, NPHS2, ACTN4, and WT1 in Japanese patients with congenital nephrotic syndrome. Kidney Int 67: 1248–1255, 2005 [DOI] [PubMed] [Google Scholar]
- 6.Liu G, Kaw B, Kurfis J, Rahmanuddin S, Kanwar YS, Chugh SS: Neph1 and nephrin interaction in the slit diaphragm is an important determinant of glomerular permeability. J Clin Invest 112: 209–221, 2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gerke P, Huber TB, Sellin L, Benzing T, Walz G: Homodimerization and heterodimerization of the glomerular podocyte proteins nephrin and NEPH1. J Am Soc Nephrol 14: 918–926, 2003 [DOI] [PubMed] [Google Scholar]
- 8.Benzing T: Signaling at the slit diaphragm. J Am Soc Nephrol 15: 1382–1391, 2004 [DOI] [PubMed] [Google Scholar]
- 9.Boute N, Gribouval O, Roselli S, Benessy F, Lee H, Fuchshuber A, Dahan K, Gubler MC, Niaudet P, Antignac C: NPHS2, encoding the glomerular protein podocin, is mutated in autosomal recessive steroid-resistant nephrotic syndrome. Nat Genet 24: 349–354, 2000 [DOI] [PubMed] [Google Scholar]
- 10.Weber S, Gribouval O, Esquivel EL, Moriniere V, Tete MJ, Legendre C, Niaudet P, Antignac C: NPHS2 mutation analysis shows genetic heterogeneity of steroid-resistant nephrotic syndrome and low post-transplant recurrence. Kidney Int 66: 571–579, 2004 [DOI] [PubMed] [Google Scholar]
- 11.Schwarz K, Simons M, Reiser J, Saleem MA, Faul C, Kriz W, Shaw AS, Holzman LB, Mundel P: Podocin, a raft-associated component of the glomerular slit diaphragm, interacts with CD2AP and nephrin. J Clin Invest 108: 1621–1629, 2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huber TB, Simons M, Hartleben B, Sernetz L, Schmidts M, Gundlach E, Saleem MA, Walz G, Benzing T: Molecular basis of the functional podocin-nephrin complex: Mutations in the NPHS2 gene disrupt nephrin targeting to lipid raft microdomains. Hum Mol Genet 12: 3397–3405, 2003 [DOI] [PubMed] [Google Scholar]
- 13.Hinkes BG, Mucha B, Vlangos CN, Gbadegesin R, Liu J, Hasselbacher K, Hangan D, Ozaltin F, Zenker M, Hildebrandt F: Nephrotic syndrome in the first year of life: Two thirds of cases are caused by mutations in 4 genes (NPHS1, NPHS2, WT1, and LAMB2). Pediatrics 119: e907–e919, 2007 [DOI] [PubMed] [Google Scholar]
- 14.Lenkkeri U, Mannikko M, McCready P, Lamerdin J, Gribouval O, Niaudet PM, Antignac CK, Kashtan CE, Homberg C, Olsen A, Kestila M, Tryggvason K: Structure of the gene for congenital nephrotic syndrome of the finnish type (NPHS1) and characterization of mutations. Am J Hum Genet 64: 51–61, 1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ruf RG, Lichtenberger A, Karle SM, Haas JP, Anacleto FE, Schultheiss M, Zalewski I, Imm A, Ruf EM, Mucha B, Bagga A, Neuhaus T, Fuchshuber A, Bakkaloglu A, Hildebrandt F: Patients with mutations in NPHS2 (podocin) do not respond to standard steroid treatment of nephrotic syndrome. J Am Soc Nephrol 15: 722–732, 2004 [DOI] [PubMed] [Google Scholar]
- 16.Ng PC, Henikoff S: Predicting deleterious amino acid substitutions. Genome Res 11: 863–874, 2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sunyaev S, Ramensky V, Koch I, Lathe W 3rd, Kondrashov AS, Bork P: Prediction of deleterious human alleles. Hum Mol Genet 10: 591–597, 2001 [DOI] [PubMed] [Google Scholar]
- 18.Reese MG, Eeckman FH, Kulp D, Haussler D: Improved splice site detection in Genie. J Comput Biol 4: 311–323, 1997 [DOI] [PubMed] [Google Scholar]
- 19.Liu L, Done SC, Khoshnoodi J, Bertorello A, Wartiovaara J, Berggren PO, Tryggvason K: Defective nephrin trafficking caused by missense mutations in the NPHS1 gene: Insight into the mechanisms of congenital nephrotic syndrome. Hum Mol Genet 10: 2637–2644, 2001 [DOI] [PubMed] [Google Scholar]
- 20.Huttunen NP, Rapola J, Vilska J, Hallman N: Renal pathology in congenital nephrotic syndrome of Finnish type: A quantitative light microscopic study on 50 patients. Int J Pediatr Nephrol 1: 10–16, 1980 [PubMed] [Google Scholar]
- 21.Liu XL, Done SC, Yan K, Kilpelainen P, Pikkarainen T, Tryggvason K: Defective trafficking of nephrin missense mutants rescued by a chemical chaperone. J Am Soc Nephrol 15: 1731–1738, 2004 [DOI] [PubMed] [Google Scholar]
- 22.Kalatzis V, Nevo N, Cherqui S, Gasnier B, Antignac C: Molecular pathogenesis of cystinosis: Effect of CTNS mutations on the transport activity and subcellular localization of cystinosin. Hum Mol Genet 13: 1361–1371, 2004 [DOI] [PubMed] [Google Scholar]
- 23.Hasselbacher K, Wiggins RC, Matejas V, Hinkes BG, Mucha B, Hoskins BE, Ozaltin F, Nurnberg G, Becker C, Hangan D, Pohl M, Kuwertz-Broking E, Griebel M, Schumacher V, Royer-Pokora B, Bakkaloglu A, Nurnberg P, Zenker M, Hildebrandt F: Recessive missense mutations in LAMB2 expand the clinical spectrum of LAMB2-associated disorders. Kidney Int 70: 1008–1012, 2006 [DOI] [PubMed] [Google Scholar]
- 24.Furu L, Onuchic LF, Gharavi A, Hou X, Esquivel EL, Nagasawa Y, Bergmann C, Senderek J, Avner E, Zerres K, Germino GG, Guay-Woodford LM, Somlo S: Milder presentation of recessive polycystic kidney disease requires presence of amino acid substitution mutations. J Am Soc Nephrol 14: 2004–2014, 2003 [DOI] [PubMed] [Google Scholar]
- 25.Beltcheva O, Martin P, Lenkkeri U, Tryggvason K: Mutation spectrum in the nephrin gene (NPHS1) in congenital nephrotic syndrome. Hum Mutat 17: 368–373, 2001 [DOI] [PubMed] [Google Scholar]
- 26.Jones N, Blasutig IM, Eremina V, Ruston JM, Bladt F, Li H, Huang H, Larose L, Li SS, Takano T, Quaggin SE, Pawson T: Nck adaptor proteins link nephrin to the actin cytoskeleton of kidney podocytes. Nature 440: 818–823, 2006 [DOI] [PubMed] [Google Scholar]
- 27.Huber TB, Kottgen M, Schilling B, Walz G, Benzing T: Interaction with podocin facilitates nephrin signaling. J Biol Chem 276: 41543–41546, 2001 [DOI] [PubMed] [Google Scholar]
- 28.Huber TB, Hartleben B, Kim J, Schmidts M, Schermer B, Keil A, Egger L, Lecha RL, Borner C, Pavenstadt H, Shaw AS, Walz G, Benzing T: Nephrin and CD2AP associate with phosphoinositide 3-OH kinase and stimulate AKT-dependent signaling. Mol Cell Biol 23: 4917–4928, 2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schultheiss M, Ruf RG, Mucha BE, Wiggins R, Fuchshuber A, Lichtenberger A, Hildebrandt F: No evidence for genotype/phenotype correlation in NPHS1 and NPHS2 mutations. Pediatr Nephrol 19: 1340–1348, 2004 [DOI] [PubMed] [Google Scholar]
- 30.Di Duca M, Oleggini R, Sanna-Cherchi S, Pasquali L, Di Donato A, Parodi S, Bertelli R, Caridi G, Frasca G, Cerullo G, Amoroso A, Schena FP, Scolari F, Ghiggeri GM: Cis and trans regulatory elements in NPHS2 promoter: Implications in proteinuria and progression of renal diseases. Kidney Int 70: 1332–1341, 2006 [DOI] [PubMed] [Google Scholar]
- 31.Egan ME, Pearson M, Weiner SA, Rajendran V, Rubin D, Glockner-Pagel J, Canny S, Du K, Lukacs GL, Caplan MJ: Curcumin, a major constituent of turmeric, corrects cystic fibrosis defects. Science 304: 600–602, 2004 [DOI] [PubMed] [Google Scholar]
- 32.Brunak S, Engelbrecht J, Knudsen S: Prediction of human mRNA donor and acceptor sites from the DNA sequence. J Mol Biol 220: 49–65, 1991 [DOI] [PubMed] [Google Scholar]
- 33.Roselli S, Gribouval O, Boute N, Sich M, Benessy F, Attie T, Gubler MC, Antignac C: Podocin localizes in the kidney to the slit diaphragm area. Am J Pathol 160: 131–139, 2002 [DOI] [PMC free article] [PubMed] [Google Scholar]