Abstract
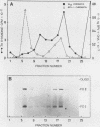
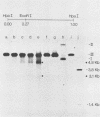
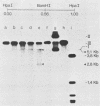
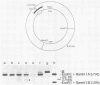
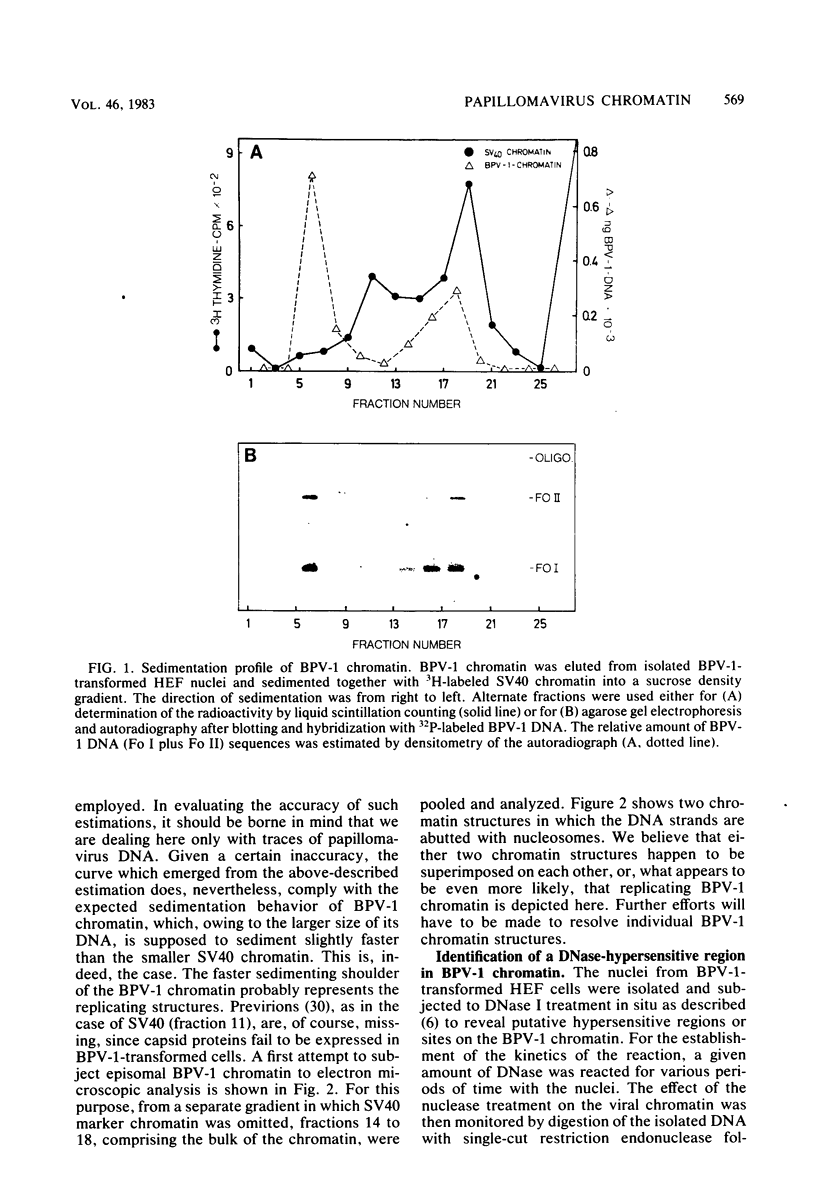
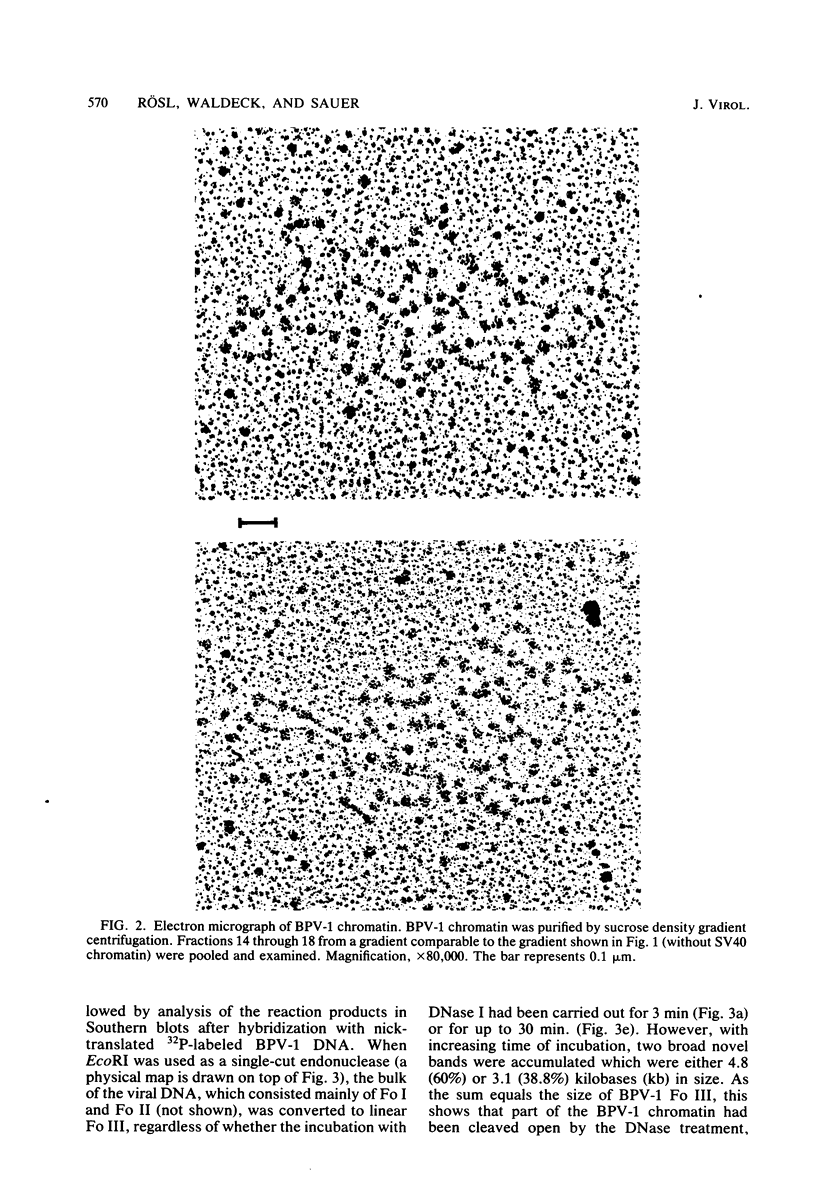
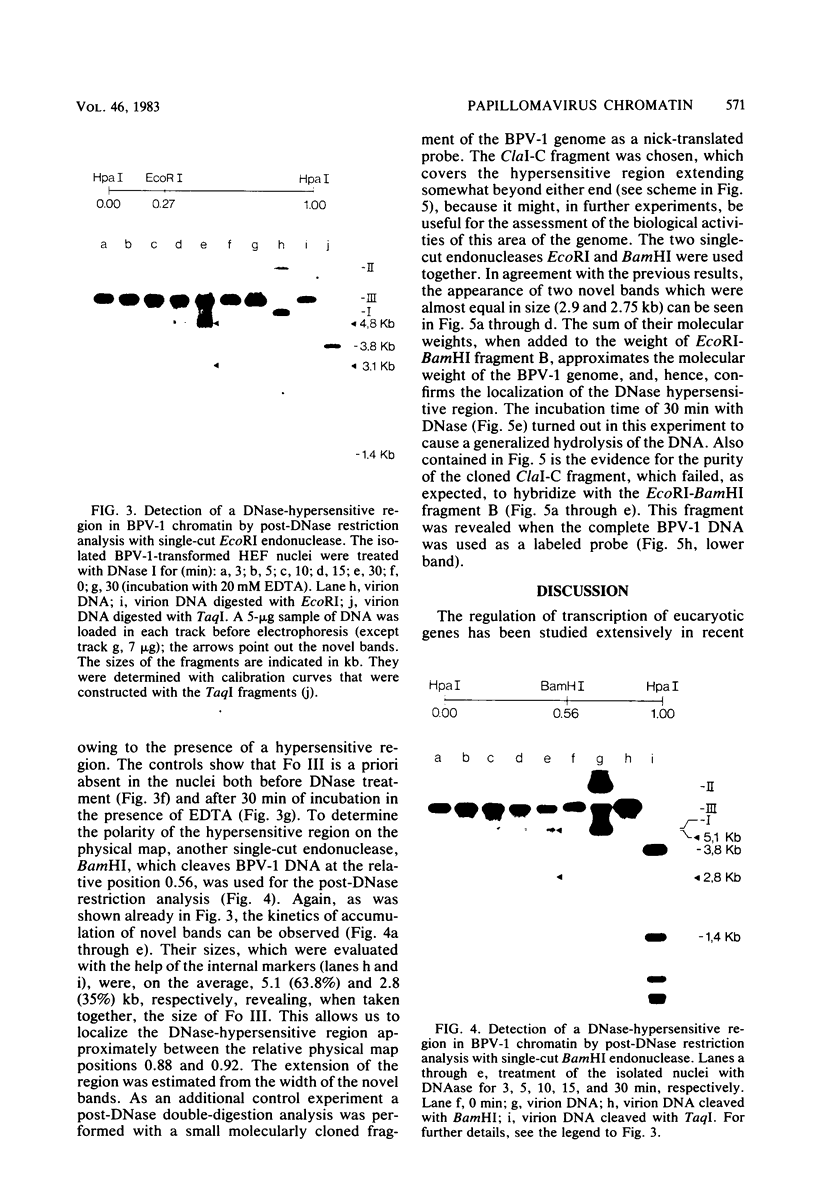
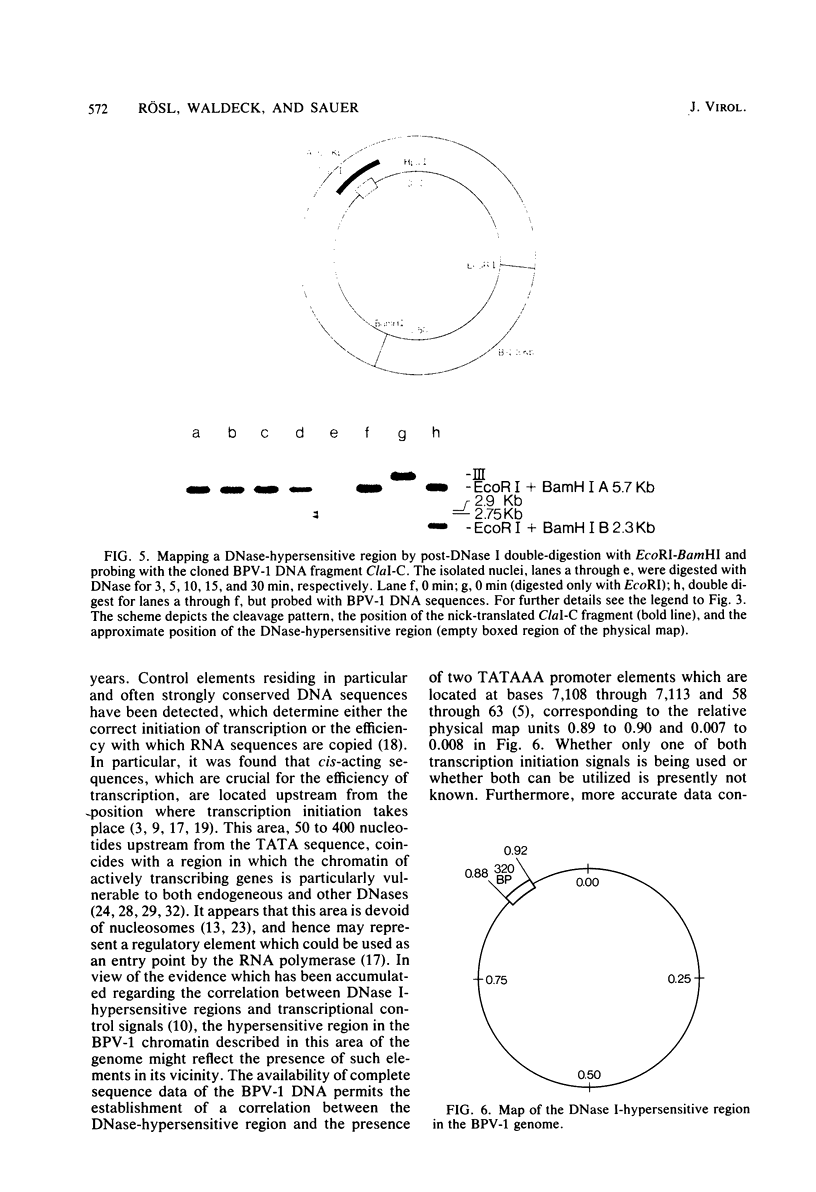
The investigation of papillomavirus chromatin has been hampered by the unavailability of a tissue culture system for vegetative growth of these viruses. We have used, therefore, bovine papillomavirus type 1-transformed hamster embryo fibroblasts containing 200 to 250 episomal genome equivalents per cell as a source of viral chromatin. The selectively isolated chromatin was shown to be slightly larger (80S) than the mature simian virus 40 chromatin, which was cosedimented in a sucrose density gradient. Both Fo I and Fo II were present in the bovine papillomavirus type 1 chromatin. A fast-sedimenting fraction, whose structure is still unknown, also contained oligomeric bovine papillomavirus type 1 DNA. By in situ DNase digestion of isolated nuclei and subsequent cleavage of the bovine papillomavirus type 1 DNA with various restriction endonucleases, a major DNase-hypersensitive region was detected in the chromatin. This region, comprising approximately 320 base pairs, is located between the relative physical map positions 0.88 and 0.92.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Amtmann E., Müller H., Sauer G. Equine connective tissue tumors contain unintegrated bovine papilloma virus DNA. J Virol. 1980 Sep;35(3):962–964. doi: 10.1128/jvi.35.3.962-964.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amtmann E., Sauer G. Bovine papilloma virus transcription: polyadenylated RNA species and assessment of the direction of transcription. J Virol. 1982 Jul;43(1):59–66. doi: 10.1128/jvi.43.1.59-66.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benoist C., Chambon P. In vivo sequence requirements of the SV40 early promotor region. Nature. 1981 Mar 26;290(5804):304–310. doi: 10.1038/290304a0. [DOI] [PubMed] [Google Scholar]
- Bolivar F., Backman K. Plasmids of Escherichia coli as cloning vectors. Methods Enzymol. 1979;68:245–267. doi: 10.1016/0076-6879(79)68018-7. [DOI] [PubMed] [Google Scholar]
- Bolivar F., Rodriguez R. L., Greene P. J., Betlach M. C., Heyneker H. L., Boyer H. W., Crosa J. H., Falkow S. Construction and characterization of new cloning vehicles. II. A multipurpose cloning system. Gene. 1977;2(2):95–113. [PubMed] [Google Scholar]
- Chen E. Y., Howley P. M., Levinson A. D., Seeburg P. H. The primary structure and genetic organization of the bovine papillomavirus type 1 genome. Nature. 1982 Oct 7;299(5883):529–534. doi: 10.1038/299529a0. [DOI] [PubMed] [Google Scholar]
- Cremisi C. The appearance of DNase I hypersensitive sites at the 5' end of the late SV40 genes is correlated with the transcriptional switch. Nucleic Acids Res. 1981 Nov 25;9(22):5949–5964. doi: 10.1093/nar/9.22.5949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danos O., Katinka M., Yaniv M. Human papillomavirus 1a complete DNA sequence: a novel type of genome organization among papovaviridae. EMBO J. 1982;1(2):231–236. doi: 10.1002/j.1460-2075.1982.tb01152.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denhardt D. T. A membrane-filter technique for the detection of complementary DNA. Biochem Biophys Res Commun. 1966 Jun 13;23(5):641–646. doi: 10.1016/0006-291x(66)90447-5. [DOI] [PubMed] [Google Scholar]
- Dierks P., van Ooyen A., Mantei N., Weissmann C. DNA sequences preceding the rabbit beta-globin gene are required for formation in mouse L cells of beta-globin RNA with the correct 5' terminus. Proc Natl Acad Sci U S A. 1981 Mar;78(3):1411–1415. doi: 10.1073/pnas.78.3.1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elgin S. C. DNAase I-hypersensitive sites of chromatin. Cell. 1981 Dec;27(3 Pt 2):413–415. doi: 10.1016/0092-8674(81)90381-0. [DOI] [PubMed] [Google Scholar]
- Heilman C. A., Engel L., Lowy D. R., Howley P. M. Virus-specific transcription in bovine papillomavirus-transformed mouse cells. Virology. 1982 May;119(1):22–34. doi: 10.1016/0042-6822(82)90061-7. [DOI] [PubMed] [Google Scholar]
- Herbomel P., Saragosti S., Blangy D., Yaniv M. Fine structure of the origin-proximal DNAase I-hypersensitive region in wild-type and EC mutant polyoma. Cell. 1981 Sep;25(3):651–658. doi: 10.1016/0092-8674(81)90172-0. [DOI] [PubMed] [Google Scholar]
- Jakobovits E. B., Bratosin S., Aloni Y. A nucleosome-free region in SV40 minichromosomes. Nature. 1980 May 22;285(5762):263–265. doi: 10.1038/285263a0. [DOI] [PubMed] [Google Scholar]
- Krieg P., Amtmann E., Jonas D., Fischer H., Zang K., Sauer G. Episomal simian virus 40 genomes in human brain tumors. Proc Natl Acad Sci U S A. 1981 Oct;78(10):6446–6450. doi: 10.1073/pnas.78.10.6446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lancaster W. D. Apparent lack of integration of bovine papillomavirus DNA in virus-induced equine and bovine tumor cells and virus-transformed mouse cells. Virology. 1981 Jan 30;108(2):251–255. doi: 10.1016/0042-6822(81)90433-5. [DOI] [PubMed] [Google Scholar]
- Law M. F., Lowy D. R., Dvoretzky I., Howley P. M. Mouse cells transformed by bovine papillomavirus contain only extrachromosomal viral DNA sequences. Proc Natl Acad Sci U S A. 1981 May;78(5):2727–2731. doi: 10.1073/pnas.78.5.2727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKnight S. L., Gavis E. R., Kingsbury R., Axel R. Analysis of transcriptional regulatory signals of the HSV thymidine kinase gene: identification of an upstream control region. Cell. 1981 Aug;25(2):385–398. doi: 10.1016/0092-8674(81)90057-x. [DOI] [PubMed] [Google Scholar]
- McKnight S. L., Kingsbury R. Transcriptional control signals of a eukaryotic protein-coding gene. Science. 1982 Jul 23;217(4557):316–324. doi: 10.1126/science.6283634. [DOI] [PubMed] [Google Scholar]
- Mellon P., Parker V., Gluzman Y., Maniatis T. Identification of DNA sequences required for transcription of the human alpha 1-globin gene in a new SV40 host-vector system. Cell. 1981 Dec;27(2 Pt 1):279–288. doi: 10.1016/0092-8674(81)90411-6. [DOI] [PubMed] [Google Scholar]
- Moar M. H., Campo M. S., Laird H. M., Jarrett W. F. Unintegrated viral DNA sequences in a hamster tumor induced by bovine papilloma virus. J Virol. 1981 Sep;39(3):945–949. doi: 10.1128/jvi.39.3.945-949.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfister H., Fink B., Thomas C. Extrachromosomal bovine papillomavirus type 1 DNA in hamster fibromas and fibrosarcomas. Virology. 1981 Dec;115(2):414–418. doi: 10.1016/0042-6822(81)90125-2. [DOI] [PubMed] [Google Scholar]
- Rigby P. W., Dieckmann M., Rhodes C., Berg P. Labeling deoxyribonucleic acid to high specific activity in vitro by nick translation with DNA polymerase I. J Mol Biol. 1977 Jun 15;113(1):237–251. doi: 10.1016/0022-2836(77)90052-3. [DOI] [PubMed] [Google Scholar]
- Saragosti S., Moyne G., Yaniv M. Absence of nucleosomes in a fraction of SV40 chromatin between the origin of replication and the region coding for the late leader RNA. Cell. 1980 May;20(1):65–73. doi: 10.1016/0092-8674(80)90235-4. [DOI] [PubMed] [Google Scholar]
- Scott W. A., Wigmore D. J. Sites in simian virus 40 chromatin which are preferentially cleaved by endonucleases. Cell. 1978 Dec;15(4):1511–1518. doi: 10.1016/0092-8674(78)90073-9. [DOI] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Su R. T., DePamphilis M. L. In vitro replication of simian virus 40 DNA in a nucleoprotein complex. Proc Natl Acad Sci U S A. 1976 Oct;73(10):3466–3470. doi: 10.1073/pnas.73.10.3466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tegtmeyer P., Macasaet F. Simian virus 40 deoxyribonucleic acid synthesis: analysis by gel electrophoresis. J Virol. 1972 Oct;10(4):599–604. doi: 10.1128/jvi.10.4.599-604.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varshavsky A. J., Sundin O., Bohn M. A stretch of "late" SV40 viral DNA about 400 bp long which includes the origin of replication is specifically exposed in SV40 minichromosomes. Cell. 1979 Feb;16(2):453–466. doi: 10.1016/0092-8674(79)90021-7. [DOI] [PubMed] [Google Scholar]
- Waldeck W., Föhring B., Chowdhury K., Gruss P., Sauer G. Origin of DNA replication in papovavirus chromatin is recognized by endogenous endonuclease. Proc Natl Acad Sci U S A. 1978 Dec;75(12):5964–5968. doi: 10.1073/pnas.75.12.5964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waldeck W., Spaeren U., Mastromei G., Eliasson R., Reichard P. Replication of polyoma DNA in nuclear extracts and nucleoprotein complexes. J Mol Biol. 1979 Dec 15;135(3):675–689. doi: 10.1016/0022-2836(79)90171-2. [DOI] [PubMed] [Google Scholar]
- Weintraub H., Groudine M. Chromosomal subunits in active genes have an altered conformation. Science. 1976 Sep 3;193(4256):848–856. doi: 10.1126/science.948749. [DOI] [PubMed] [Google Scholar]
- Weisbrod S. Active chromatin. Nature. 1982 May 27;297(5864):289–295. doi: 10.1038/297289a0. [DOI] [PubMed] [Google Scholar]
- Worcel A., Han S., Wong M. L. Assembly of newly replicated chromatin. Cell. 1978 Nov;15(3):969–977. doi: 10.1016/0092-8674(78)90280-5. [DOI] [PubMed] [Google Scholar]
- Zenke M., Sauer G. Spliced and unspliced virus specific RNA sequences are associated with purified simian virus 40 chromatin. Nucleic Acids Res. 1982 Aug 11;10(15):4543–4550. doi: 10.1093/nar/10.15.4543. [DOI] [PMC free article] [PubMed] [Google Scholar]