Abstract
OBJECTIVE—MicroRNAs are short, noncoding RNAs that regulate gene expression. We hypothesized that the phosphatidylinositol 3-kinase (PI 3-kinase) cascade known to be important in β-cell physiology could be regulated by microRNAs. Here, we focused on the pancreas-specific miR-375 as a potential regulator of its predicted target 3′-phosphoinositide–dependent protein kinase-1 (PDK1), and we analyzed its implication in the response of insulin-producing cells to elevation of glucose levels.
RESEARCH DESIGN AND METHODS—We used insulinoma-1E cells to analyze the effects of miR-375 on PDK1 protein level and downstream signaling using Western blotting, glucose-induced insulin gene expression using quantitative RT-PCR, and DNA synthesis by measuring thymidine incorporation. Moreover, we analyzed the effect of glucose on miR-375 expression in both INS-1E cells and primary rat islets. Finally, miR-375 expression in isolated islets was analyzed in diabetic Goto-Kakizaki (GK) rats.
RESULTS—We found that miR-375 directly targets PDK1 and reduces its protein level, resulting in decreased glucose-stimulatory action on insulin gene expression and DNA synthesis. Furthermore, glucose leads to a decrease in miR-375 precursor level and a concomitant increase in PDK1 protein. Importantly, regulation of miR-375 expression by glucose occurs in primary rat islets as well. Finally, miR-375 expression was found to be decreased in fed diabetic GK rat islets.
CONCLUSIONS—Our findings provide evidence for a role of a pancreatic-specific microRNA, miR-375, in the regulation of PDK1, a key molecule in PI 3-kinase signaling in pancreatic β-cells. The effects of glucose on miR-375 are compatible with the idea that miR-375 is involved in glucose regulation of insulin gene expression and β-cell growth.
Type 2 diabetes currently affects >170 million people worldwide (1). The disease is characterized by an inability of the functional β-cell mass to meet chronically increased metabolic demands for insulin, as occurring under variable states of insulin resistance. The normal β-cell population can adapt to a sustained stimulation by recruiting β-cells into a higher translational and insulin synthetic activity (2,3) and possibly by an expansion of its total cell number (4,5). The molecular mechanisms involved in this chronic adaptation of the β-cell population are not completely understood. Several reports have highlighted the importance of phosphatidylinositol 3-kinase (PI 3-kinase) signaling in β-cell physiology. For example, glucose stimulates the insulin gene promoter activity via a cascade involving PI 3-kinase. Indeed, glucose triggers phosphorylation of PDX-1 via the PI 3-kinase pathway, which induces nuclear translocation of PDX-1, and the latter then increases insulin gene transcription (6,7). Furthermore, glucose promotes β-cell survival through the PI 3-kinase/protein kinase B (PKB) cascade (8). Different actors of the PI 3-kinase cascade have been identified as critical control points in insulin signaling (9), among them 3′-phosphoinositide–dependent protein kinase-1 (PDK1). This kinase was initially recognized by its ability to phosphorylate in the presence of lipid products generated by PI 3-kinase, the activation loop of PKB on Thr-308 (10,11). The role of PDK1 in vivo has been addressed in different organisms by genetic deletion of PDK1 homologs. In short, knockout studies revealed a central role of PDK1 in regulation of cell growth and organ development (12,13). Importantly, PDK1 ablation in β-cells induces diabetes consecutively to a reduction in β-cell mass (14).
The discovery of microRNAs has opened an entirely new line of thoughts regarding the regulation of signaling by growth factors and hormones and its perturbations in situations associated to disease processes. MicroRNAs are 21- to 25-nucleotide–long noncoding RNA molecules first identified as regulators of the level of several proteins in Caenorhabditis elegans (15). Currently, hundreds of microRNAs have been cloned in mammalian species, i.e., 285 in rats, 442 in mice, and 533 in humans. They are collectively annotated and indexed in the microRNA registry (http://microrna.sanger.ac.uk). Remarkably, some microRNAs are evolutionarily conserved, heralding important biological functions (16,17). Another feature of several microRNAs is their time- and tissue-specific expression, which points to their role in development and organ function. Since their discovery in 1993 by Lee et al. (15), a growing list of pleiotropic effects of microRNAs has appeared. Indeed, microRNAs have been shown to regulate development in C. elegans (18) and metabolism in Drosophila (19). More recently, they have been implicated in mammals in adipocyte differentiation (20), lipid metabolism (21,22), heart (23) and brain (24) development, and β-cell physiology (25–27).
Here, we were interested in identifying microRNAs targeting molecules involved in insulin signaling in pancreatic β-cells. Using computational analysis, we found that miR-375, previously described as decreasing glucose-induced insulin secretion and characterized by pancreas-specific expression pattern (25), targets PDK1, a key player in the PI 3-kinase cascade. By gain- and loss-of-function experiments, we found that miR-375 regulates PDK1 protein level, resulting in modulation of glucose-stimulatory action on insulin gene expression and DNA synthesis. We showed that miR-375 interacts directly with the 3′ untranslated region (3′UTR) of PDK1 mRNA. Furthermore, exposure of either INS-1E cells or freshly isolated rat islets to glucose modulates endogenous miR-375 precursor levels, suggesting its involvement in regulation of glucose responsiveness of β-cells. Finally, miR-375 expression is found to be decreased in diabetic Goto-Kakizaki (GK) rats compared with Wistar rats.
RESEARCH DESIGN AND METHODS
Cell culture and transfections.
INS-1E β-cells were maintained in RPMI 1640 containing 11 mmol/l glucose supplemented with 10% (vol/vol) heat-inactivated FCS, 100 units/ml penicillin, 100 μg/ml streptomycin, 2 mmol/l l-glutamine, 10 mmol/l HEPES, 1 mmol/l sodium pyruvate, and 50 μmol/l β-mercaptoethanol in humidified 5% (vol/vol) CO2, 95% (vol/vol) air at 37°C and used between passages 50 and 75. LipofectAMINE 2000 transfection reagent (Invitrogen, Life Technologies) was used to transfect INS-1E cells. A total of 3 μg pmiR-375 or pNeg and the indicated amounts of 2′-O-methyl-miR-375 or 2′-O-methyl-GFP antisense oligonucleotides and 2 μl LipofectAMINE 2000 were used per well with each containing 5 × 105 cells (six-well plate).
Islet preparation.
Islets were isolated by collagenase digestion, elutriation, and manual handpicking from adult male Wistar rats (150–250 g; Janvier, Le Genest Saint Isle France). Animals were bred according to Belgian regulations of animal welfare and used in experiments that were approved by the local ethical committee. Islets were cultured in Ham's F10 nutrient mixture (Gibco, Invitrogen, Carlsbad, CA) supplemented with 0.5% (wt/vol) BSA (Cohn Analog; Sigma), 2 mmol/l glutamine, 100 units/ml penicillin, 0.1 mg/ml streptomycin, 2% (vol/vol) FCS (Hyclone), and the indicated glucose concentration.
Generation of DNA constructs.
Expression vector driving expression of miR-375 was prepared by introducing oligonucleotides corresponding to the murine precursor sequence of mir-375 into pcDNA6.2 (pmiR-375) (Invitrogen). The oligonucleotide sequences were as follows: sense, 5′-TGCTGCCCCGCGACGAGCCCC-TCGCACAAACCGGACCTGAGCGTTTTGTTCGTTCGGCTCGCGTGAGGC-3′; and antisense, 5′-CCTGGCCTCACGCGAGCCGAACGAACAAAACGCTCAGGTCCGGTTTGTG CGAGGGGCTCGTCGCGGGGC-3′. As negative control, we used pNeg driving the expression of an unrelated known microRNA precursor (Invitrogen). The rat PDK1 3′UTR target site was cloned using the following oligonucleotides: sense, 5′-ACCCAACCACACAAAGAACAAAA-3′; and antisense, 5′-TTTTGTTCTTTGTGTGGTTGGGT-3′ in the 3′UTR of the Renilla luciferase reporter vector, pmiR-Report luciferase (Ambion), as described previously (28). As a negative control response element, we used a mutated sequence by inserting the following oligonucleotides: sense, 5′-ACCCAACCACACCCCTCCTGGGG-3′; and antisense, 5′-CCCCAGGAGGGGTGTGGTT-GGGT-3′. Loss of function experiments were carried out using the following: 2′-O-methyl-375, UGCAUCACGCGAGCCGAACGAACAAAUAAGL, and 2′-O-methyl-eGFP, AAGGCAAGCUGACCCUGAAGUL.
Luciferase assays.
INS-1E cells were cultured in six-well plates and transfected with different reporter vectors (p-Luc-Empty, p-Luc 3′UTR PDK1 or p-Luc-3′UTR MUT PDK1) and cotransfected with pNeg or pmiR-375. Cells were assayed 48 h after transfection with the dual-luciferase reporter assay system (Promega, Madison, WI). Luciferase activity was normalized by β-galactosidase activity.
RNA RT-PCR and real-time PCR.
RNA from transfected β-cells and rat islets was isolated using TRIzol reagent (Invitrogen), and its quality was verified by Agilent Bioanalyzer (minimal cutoff RNA integrity number ≥8). From each transfected well, 1 μg total RNA was reverse-transcribed. For microRNAs precursor detection, RT was performed as described (29). MicroRNA precursor primer sequences are described by Jiang et al. (30). The following forward and reverse primers were used for amplification: PDK1, 5′-CCCACGTGATGGACTCAAAGA-3′ (reverse) and 5′-AAGGGTACGGGCCTCTCAAA-3′ (forward); Insulin-1, 5′-GTGCACCAACAGGGCCAT-3′ (reverse) and 5′-CAGAGACCATCAGCAAGCAGG-3′ (forward); U6, 5′-AACGCTTCACGAATTTGCGT-3′ (reverse) and 5′-CTCGCTTCGGCAGCACA-3′ (forward); and 36B4, 5′-ATGATCAGCCCGAAGGAGAAGG-3′ (reverse) and 5′-CCACGAAAATCTCCAGAGGCAC-3′ (forward).
Analysis of total cell extracts and Western blotting.
Two days after transfection, INS-1E cells were washed with ice-cold PBS and processed for protein isolation. For Western blotting, total proteins were separated by electrophoresis and transferred to polyvinylidene difluoride membranes (Immobilon-P; Millipore, Bedford, MA) followed by blotting. Immunodetection was performed using affinity-purified polyclonal antibodies to PDK1 and phospho–glycogen synthase kinase (GSK)3 (Cell Signaling, Beverly, MA) and to phospho-Thr-308 PKB, total PKB, or total GSK3 (Santa Cruz Biotechnology, Santa Cruz, CA). To assess the total protein amount, membranes were stripped and reprobed with antibody to β-tubulin (Sigma-Aldrich, St. Quentin-Fallavier, France).
Cell viability assay.
Cells (2 × 105 per well) seeded in 12-well plastic plates were transfected with pmiR-375 or pNeg and incubated at 37°C. After 48 h, cell viability was assessed by the ability of metabolically active cells to reduce tetrazolium salt to orange-colored formazan compounds. The absorbance of the samples was measured with a spectrophotometer reader (wavelength 450 nm). Data shown correspond to mean values from three independent experiments measured in sextuplicate.
Measurement of DNA synthesis using [methyl-3H]thymidine incorporation.
Cells were plated in six-well plates at a density of 5 × 105 cells per well. After reaching 60% confluence, they were transfected with pmiR-375 or pNeg. Twenty-four hours later, cells were starved in RPMI containing 0.5% FCS (vol/vol) for 24 h and then replaced in RPMI 10% (vol/vol) FCS. After 24 h, DNA synthesis was assayed by adding 1 μCi [methyl-3H]thymidine/well and by incubating the cells for another 2 h. Then cells were washed twice with PBS, fixed with 10% (vol/vol) trichloroacetic acid for 30 min, and solubilized by adding 300 μl 0.2 N NaOH to each well. Radioactivity, reflecting incorporation of [methyl-3H]thymidine into DNA, was measured by adding scintillation liquid and counting.
RESULTS
miR-375 regulates PDK1 protein level.
To identify potential targets of miR-375, we used computational algorithms designed to predict mRNA targets of microRNAs. One of the predicted targets for miR-375 is PDK1. The predicted miR-375 binding site in the 3′UTR of PDK1 mRNA (Fig. 1A) appears to be phylogenetically maintained. This would suggest that the function of miR-375 as potential regulator of PDK1 has been conserved in mice, rats, and humans. To investigate whether miR-375 affects PDK1, plasmids driving the expression of miR-375 precursor (pmiR-375) or control (pNeg) were transfected in INS-1E cells. pmiR-375 increased miR-375 precursor levels by ∼500-fold, as assayed by real-time PCR (Fig. 1B). Functional analysis shows that miR-375 precursor overexpression in INS-1E cells results in a reduction of ∼40% of PDK1 protein (Fig. 1C and D) without affecting its mRNA level (Fig. 1E), indicating that miR-375 acts as a translational repressor. To evaluate whether the predicted miR-375 target site in the 3′UTR of PDK1 mRNA was directly involved in miR-375–induced reduction in PDK1 protein, we cloned the putative 3′UTR target site downstream of a luciferase reporter gene and cotransfected this p-Luc-3′UTR PDK1 construct into INS-1E cells with pmiR-375 or pNeg. Luciferase activity of cells transfected with pmiR-375 and p-Luc-3′UTR PDK1 was decreased by ∼25% compared with cells cotransfected with control pNeg and p-Luc-3′UTR PDK1 (Fig. 1F). With negative control constructs p-Luc-Empty and p-Luc-3′UTR MUT PDK1, no reduced luciferase activity was observed when cells were cotransfected with pmiR-375 compared with pNeg. Taken together, our data argue for a direct interaction between miR-375 and PDK1 mRNA.
FIG. 1.
PDK1 is a target of miR-375. A: Scheme of the interaction between miR-375 and the 3′UTR of mouse, rat, and human PDK1. B: Quantification of miR-375 precursor levels. INS-1E cells were transfected with pNeg or pmiR-375 for 48 h. miR-375 precursor was then quantified by quantitative RT-PCR. Values for miR-375 precursor were normalized to U6 RNA. C: Analysis of PDK1 protein. INS-1E cells were transfected as above, and protein extracts were analyzed by Western blot using antibody to PDK1 or to β-tubulin. D: Relative quantification of PDK1 protein. Data represent three independent transfections done in triplicate, ±SE, with n = 3. *P < 0.05. E: Relative quantification of PDK1 mRNA levels. RNA extracts were used for quantitative RT-PCR analysis of PDK1 mRNA normalized to 36B4 mRNA. Data represent three independent transfections done in triplicate, ±SE, with n = 3. F: Study of the interaction between miR-375 and 3′UTR of PDK1 mRNA. INS-1E cells were cotransfected with one of the following pmiR-reporter luciferase vectors (Ambion): empty vector, vector containing the 3′UTR PDK-1 oligonucleotide predicted to interact with miR-375, or vector containing a mutated sequence. The cells were also transfected with either pmiR-375 or pNeg. Forty-eight hours after transfection, cells were assayed for luciferase and β-galactosidase activity; β-galactodidase was used as control to normalize for transfection efficiency. Data represent three independent transfections, each carried out in duplicate, ±SE, with n = 3. *P < 0.05.
miR-375 decreases insulin signaling downstream of PDK1.
The PI 3-kinase/PDK1/PKB signaling pathway is used by insulin in pancreatic β-cells to elicit several actions of the hormone. It is generally believed that after insulin stimulation of cells, PDK1 is recruited to the plasma membrane and phosphorylates PKB on Thr-308, which becomes activated and phosphorylates a series of substrates, including GSK3α/β. Downregulation of PDK1 in β-cells is expected to cause a decrease in insulin-induced signaling dependent on this particular enzyme. To study the effect of miR-375 on insulin signaling, we examined the phosphorylation state of molecules functioning downstream of PDK1. Immunoblot analysis showed that in response to insulin, PKB phosphorylation on Thr-308 is less abundant in cells overexpressing miR-375 precursor compared with control cells (Fig. 2A and B). Consistent with this latter observation, ectopic expression of miR-375 precursor also reduces insulin-induced phosphorylation of GSK3β (Fig. 2C and D).
FIG. 2.
Effect of miR-375 on PKB and GSK3 phosphorylation. A: Analysis of Thr-308-phosphorylation of PKB. INS-1E cells were transfected with pNeg or pmiR-375. Forty-eight hours later, cells were starved in Krebs-Ringer bicarbonate HEPES medium for 2 h and either were or were not stimulated with 100 nmol/l insulin for 5 min. Protein extracts were analyzed by Western blot using antibody to phospho308Thr PKB and total PKB. B: Quantification of Thr-308-phosphorylated PKB. C: Analysis of GSK3β phosphorylation. Protein extracts were analyzed by Western blot using antibody to phosphoGSK3β and total GSK3. D: Quantification of phosphorylated GSK3β. The data presented correspond to three independent experiments, each done in duplicate, ±SE, with n = 3. *P < 0.05.
miR-375 decreases glucose-induced insulin gene expression and DNA synthesis.
Because PI 3-kinase signaling has been implicated in glucose-induced upregulation of insulin gene expression and because we found that miR-375 decreases PDK1 protein, we examined the impact of miR-375 on insulin gene expression in response to glucose in INS-1E cells. We found that, as expected, high glucose concentration (20 mmol/l) induced insulin gene expression in control cells, but this effect was lost in cells overexpressing pre-miR-375 (Fig. 3A). To test whether PDK1 is involved in the glucose-induced increase in cell proliferative activity, we analyzed the impact of miR-375 on cellular [methyl-3H]thymidine incorporation. As illustrated in Fig. 3B, the stimulatory effect of glucose on DNA synthesis was reduced by ∼50% when miR-375 precursor was overexpressed.
FIG. 3.
Effect of miR-375 on glucose-enhanced insulin gene expression and cell proliferation. A: Quantification of insulin mRNA. INS-1E cells were transfected as above. Forty-eight hours later, cells were starved in RPMI 1640 with 0.5% (vol/vol) FCS containing 2 mmol/l glucose for 16 h and treated with 2 or 20 mmol/l glucose for 1 h. Insulin mRNA expression was analyzed by quantitative RT-PCR and normalized to 36B4 transcript. Data represent five independent experiments carried out in triplicate, ±SE, with n = 5. *P < 0.05, **P < 0.005. B: Measurement of [methyl-3H]thymidine incorporation. INS-1E cells were transfected as above. Twenty-four hours later, cells were starved in RPMI 1640 with 0.5% (vol/vol) FCS containing 2 mmol/l glucose for 16 h and treated with 2 or 20 mmol/l glucose for 24 h, and cell proliferation was assessed by measuring [methyl-3H]thymidine incorporation. Data represent three independent experiments done in triplicate, ±SE, with n = 3. *P < 0.05.
miR-375 attenuates cell viability and proliferation.
Because the PI 3-kinase/PKB cascade has been involved in cell survival and proliferation, we investigated the consequences of miR-375 expression on cell viability and proliferation. Thus, INS-1E cells were transfected with pmiR-375 or pNeg, and 48 h after transfection, cell number and viability were measured. We found that miR-375 overexpression reduced cell number by ∼25% (Fig. 4A) and cell viability by ∼20% (Fig. 4B). To further determine the effects of miR-375 on cell proliferation, DNA synthesis was measured using [methyl-3H]thymidine incorporation. As shown in Fig. 4C, transfection with miR-375 precursor inhibited INS-1E cell proliferative activity by 20% compared with control cells.
FIG. 4.
Effect of miR-375 on cell number, viability, and proliferation. A: Cell number counting. INS-1E cells were seeded in six-well plates (5 × 105/well) and transfected with pNeg or premiR-375. Cells were starved in RPMI 1640 containing 0.5% (vol/vol) FCS and 11 mmol/l glucose for 24 h and then replaced in RPMI 1640 10% (vol/vol) FCS and 11 mmol/l glucose for 24 h. Cells were counted using Coulter counter. B: Cell viability assay. INS-1E cells were seeded in 12-well plates (2 × 105/well) and transfected as above. Forty-eight hours later, cell viability was assessed as described in research design and methods. Data represent three independent transfections done in sextuplicate, ±SE, with n = 3. *P < 0.05. C: Measurement of [methyl-3H]thymidine incorporation. Cells were seeded in six-well plates (5 × 105/well) and treated as in A. Cell proliferation was assessed by measuring [methyl-3H]thymidine incorporation. Data represent three independent experiments, each run in triplicate, ±SE, with n = 3. *P < 0.05.
2′-O-methyl-miR-375 increases PDK1 protein level and glucose-stimulatory action on insulin mRNA and DNA synthesis.
Because miR-375 targets PDK1 and impairs glucose-stimulated insulin gene expression and cell proliferation, we investigated whether antisense oligonucleotides of miR-375 induce effects opposite to those seen after miR-375 overexpression. Using 2′-O-methyl-miR-375 antisense oligonucleotides, we found that blocking miR-375 augments PDK1 protein. This increase reaches ∼40% when cells are transfected with either 250 or 500 nmol/l 2′-O-methyl-miR-375 (Fig. 5A and B). Importantly, we found that 2′-O-methyl-miR-375–induced miR-375 depletion increases both basal and glucose-enhanced insulin mRNA (Fig. 5C). Finally, as shown in (Fig. 5D), 2′-O-methyl-miR-375 increases the glucose stimulatory action on [3H]thymidine incorporation compared with 2′-O-methyl-GFP.
FIG. 5.
Effect of 2′-O-methyl-miR-375 antisense oligonucleotides on PDK1, glucose-enhanced insulin mRNA, and cell proliferation. A: Analysis of PDK1 protein. INS-1E cells were transfected with indicated amounts of 2′-O-methyl-miR-375. After 48 h, protein extracts were analyzed by Western blot using antibody to PDK1 or to β-tubulin. B: Quantification of PDK1 protein. Data represent three independent transfections, ±SE, with n = 3. **P < 0.01. C: Quantification of insulin mRNA level. INS-1E cells were transfected with either 2′-O-methyl-GFP or 2′-O-methyl-miR-375 at 500 nmol/l. Twenty-four hours later, cells were starved in RPMI 1640 with 0.5% (vol/vol) FCS containing 2 mmol/l glucose for 16 h and treated with 2 or 20 mmol/l glucose for 1 h. RNA extracts were reverse-transcribed and analyzed by RT-PCR for the expression of insulin gene normalized to the 36B4 transcript level. Data represent three independent experiments done in triplicate, ±SE, with n = 3. *P < 0.05, **P < 0.01. D: Measurement of [methyl-3H]thymidine incorporation. INS-1E cells were transfected as described above. Twenty-four hours later, cells were starved in RPMI 1640 with 0.5% (vol/vol) FCS containing 2 mmol/l glucose for 16 h and treated with 2 or 20 mmol/l glucose for 24 h, and [methyl-3H]thymidine incorporation was measured. Data represent four independent experiments, done in triplicate, ±SE, with n = 4. *P < 0.05, ***P < 0.001.
Glucose specifically decreases miR-375 expression.
To investigate whether glucose-induced responses in INS-1E cells could be mediated by microRNAs, we analyzed the expression of a series of microRNAs in cells maintained for 4 days with different glucose concentrations. Using quantitative RT-PCR as described previously (29), we found that miR-375 precursor is negatively regulated by glucose. Indeed, 11 and 22 mmol/l glucose induce a decrease of ∼60 and 70%, respectively, in miR-375 precursor levels compared with 5.5 mmol/l glucose (Fig. 6A). To look at whether this glucose effect was limited to miR-375, we measured the level of three other microRNAs expressed in INS-1E cells. In contrast to miR-375, both miR-296 and miR-9 are positively regulated by prolonged glucose treatment. Indeed, glucose leads to a concentration-dependent increase in miR-296 precursor (Fig. 6B), whereas miR-9 precursor is robustly increased after cell exposure to 11 mmol/l glucose. At higher glucose concentration, miR-9 expression tends to decline but remains higher compared with low glucose (5.5 mmol/l) condition (Fig. 6C). Finally, miR-122, which presents a specific liver expression, is detected in INS-1E cells, but glucose does not modulate its expression (Fig. 6D).
FIG. 6.
Expression of microRNAs in glucose-stimulated INS-1E cells. INS-1E cells were cultured in six-well plates (5 × 105/well) and starved in RPMI 1640 with 0.5% (vol/vol) FCS containing 2 mmol/l glucose for 24 h and thereafter treated with 5.5, 11, or 22 mmol/l glucose for 4 days. RNA extracts were reverse-transcribed and analyzed by RT-PCR for the expression of miR-375 (A), miR-296 (B), miR-9 (C), and miR-122 (D) precursors. Expression levels of microRNA precursors were normalized to U6 transcript. Data represent three independent experiments each done in duplicate, ±SE, with n = 3. *P < 0.05, **P < 0.005. Results were expressed relative to the low glucose condition (5.5 mmol/l Glc).
The levels of miR-375 and PDK1 are inversely correlated in glucose-stimulated INS-1E cells.
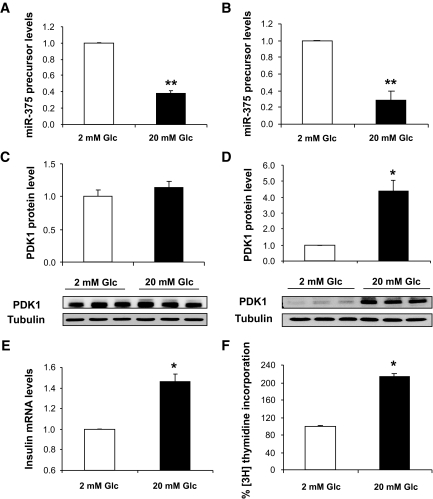
To further document the role of miR-375, we analyzed the expression of premiR-375 in INS-1E cells treated for 1 or 24 h with 2 or 20 mmol/l glucose. We found that glucose induces a robust decrease in miR-375 after 1 and 24 h (Fig. 7A and B). Furthermore, immunoblot analyses show that PDK1 protein level is slightly, but not significantly, increased within 1 h of glucose treatment (Fig. 7C). Remarkably, in INS-1E cells exposed for 24 h to 20 mmol/l glucose, PDK1 protein is increased (fourfold), and its level thus inversely correlates with that of miR-375 (Fig. 7D). Finally, decreased miR-375 without change in PDK1 level seen after 1 h of glucose treatment occurs with enhanced insulin gene expression (Fig. 7E). Decreased miR-375 and increased PDK1 levels are associated with enhanced DNA synthesis as reflected by increased thymidine incorporation seen within 24 h of glucose treatment (Fig. 7F).
FIG. 7.
Effect of glucose on endogenous miR-375 and PDK1 expression in INS-1E cells. INS-1E cells were cultured in six-well plates (5 × 105/well) and starved in RPMI 1640 with 0.5% (vol/vol) FCS containing 2 mmol/l glucose for 24 h and thereafter treated with 2 or 20 mmol/l glucose for 1 h (A) or 24 h (B). RNA extracts were analyzed for miR-375 precursor. Expression of miR-375 precursor was normalized to the U6 transcript level. Protein extracts from cells stimulated for 1 h (C) or 24 h (D) with 2 or 20 mmol/l glucose were analyzed by Western blot using antibody to PDK1 or to β-tubulin. INS-1E cells were starved in RPMI 1640 with 0.5% (vol/vol) FCS containing 2 mmol/l glucose for 16 h and treated with 2 or 20 mmol/l glucose for 1 h (E) or 24 h (F). Cell proliferation was assessed by measuring [methyl-3H]thymidine incorporation. Data represent three independent experiments done in triplicate, ±SE, with n = 3. *P < 0.05, **P < 0.005.
Glucose regulates miR-375 expression in freshly isolated rat pancreatic islets.
To characterize the glucose-mediated miR-375 regulation, we analyzed the expression level of miR-375 precursor in isolated rat islets that had been exposed to 5, 10, or 20 mmol/l glucose for 2- or 72-h periods, which represent conditions in which the acute and chronic influences, respectively, of glucose can be studied. After 2 h at stimulatory glucose concentrations (10 or 20 mmol/l), miR-375 expression is lower than in the basal 5 mmol/l condition (Fig. 8A). After 72 h, a lower expression level was measured at 5 mmol/l than at 10 and 20 mmol/l, whereas the level at 20 mmol/l tended to be lower than at 10 mmol/l (Fig. 8B).
FIG. 8.
Study of endogenous miR-375 expression in freshly isolated rat islets. Rat islets were isolated as described in research design and methods. Islets were maintained overnight in Ham's F10 containing 1.8 g/l glucose supplemented with 300 mg/l glutamine, 100 mg/l streptomycin, 75 mg/l penicillin, 0.5% (wt/vol) BSA, and 2% (vol/vol) FCS. Thereafter, islets were treated with either 5, 10, or 20 mmol/l glucose for 2 h (A) or 72 h (B). RNA extracts were reverse-transcribed and analyzed by quantitative RT-PCR for miR-375 precursor. Expression of miR-375 precursor was normalized to the U6 transcript level. Results are means ± SE (n = 3/condition). *P < 0.05, **P < 0.005. Freshly isolated islets from Wistar (n = 5) and GK rats (n = 6) were prepared as previously described. RNA extracts were analyzed by quantitative RT-PCR for pre-miR-375 (C), pre-miR-124a2 (D), and insulin mRNA (E). Results are means ± SE (n = 5 for Wistar and n = 6 for GK), *P < 0.05.
Reduced miR-375 expression in freshly isolated islets from diabetic GK rats.
To study miR-375 expression under in vivo conditions of hyperglycemia, we analyzed islets from diabetic GK rats. We found that miR-375 is downregulated in diabetic GK rats compared with control Wistar rats (Fig. 8C). Furthermore, in GK rats, this decrease in miR-375 expression is associated with a modest, albeit not significant, increase in insulin mRNA level (Fig. 8E). Finally, the expression of miR-124a2, which has been involved in insulin mRNA expression (27), is upregulated in GK rats (Fig. 8D).
DISCUSSION
During recent years, microRNAs have emerged as important regulators of cell fate and metabolism. Although several microRNAs have been implicated in a variety of disease processes (31), only a few have been linked to insulin signaling and diabetes (32). Using either a gain- or loss-of-function approach, we show here in a pancreatic β-cell line that miR-375, previously described to reduce glucose-induced insulin secretion by inhibiting Mtpn protein (25), is able to downregulate PDK1 protein by interfering directly with its mRNA. By targeting PDK1, a key player in the PI 3-kinase signaling cascade, miR-375 decreases insulin-induced phosphorylation of PKB and GSK3, both acting downstream of PDK1.
It is interesting to mention that pancreatic β-cell specific PDK1 knockout in mice results in a reduced number and size of β-cells, and in decreased islet density (14). These data and ours are intriguing in the context of a report showing that miR-375 knockout by morpholinos in zebrafish embryos leads to defects in the development of pancreatic islets (33).
Previous studies have shown that inhibition of the PI 3-kinase cascade dampens glucose-induced insulin gene expression (6). To document the biological role of miR-375 as an inhibitor of PI 3-kinase signaling downstream of PDK1, we analyzed the effect of miR-375 overexpression or depletion on glucose-induced insulin gene expression. Our findings that miR-375 controls insulin gene expression stimulated by glucose are consistent with the notion that the PI 3-kinase cascade is important for glucose stimulatory action on insulin gene expression. A chief observation of our study is that microRNA expression seems to be glucose sensitive. Importantly, the increased insulin gene expression seen in INS-1E cells exposed to high glucose is associated with reduced expression of miR-375 precursor. Our in silico analysis (data not shown) reveals that miR-375 is located in an intergenic region between cryba2 and Ccdc108 genes on the mouse chromosome 1. Comparison of genomic sequences (http://ecrbrowser.decode.org/) across humans, mice, and rats shows that a 5-kb genomic region downstream of the miR-375 precursor contains highly conserved elements. Moreover, according to a recent publication, several regions located downstream and upstream of the miR-375 gene interact with NeuroD1/BETA2 and PDX-1 (34). Taken together, these facts suggest that the gene encoding for miR-375 may behave as a locus controlled by its own promoter.
Although glucose decreased miR-375 expression within 1 h in INS-1E cells, a longer exposure time was needed to observe the downstream stimulatory effect on PDK1 protein expression. This suggests that miR-375 suppresses synthesis of PDK1 protein. Interestingly, prolonged glucose exposure of INS-1E cells (35) has been shown to increase insulin receptor substrate 2 gene expression, protein localization to the plasma membrane, and PKB phosphorylation. These observations together with ours indicate that in INS-1E cells, glucose induces a decrease in miR-375 followed by an increase in PDK1 protein and an activation of the PI 3-kinase cascade and, hence, β-cell proliferation.
Glucose was also found to regulate miR-375 expression in primary islet tissue. A 2-h incubation was used to measure levels in freshly isolated rat islets at glucose concentrations known to have exerted dose-dependent stimulations of their metabolic, secretory, and protein biosynthetic activities. We found miR-375 expression to be lower at 10 and 20 mmol/l than at 5 mmol/l glucose, which is similar to the effect seen in INS-1E cells. The 72-h culture condition compares expression in islets that have maintained this functional responsiveness (10 and 20 mmol/l glucose during culture) with that in islets having lost this responsiveness and showing increased susceptibility to apoptosis (5 mmol/l glucose during culture) (36–38). Expression of miR-375 is lower in islets showing viability consequences of prolonged glucose deprivation; it is unknown whether this reduction participates in the apoptotic pathway and can be interpreted in light of a report showing that miR-375 knockout by morpholinos in zebrafish leads to defects in islet development (33). When comparing functionally competent islets cultured at control (10 mmol/l) versus elevated (20 mmol/l) glucose concentrations, miR-375 levels appeared lower after prolonged glucose hyperactivation (20 mmol/l). Islets isolated from diabetic GK rats exhibited a lower miR-375 expression than islets from Wistar rats, which can be considered as further evidence for the suppressing effect of supraphysiological glucose concentrations. Such reduced miR-375 level is expected to be associated with increased insulin mRNA, but this was not the case in GK rats, suggesting a possible failure of the PDK1/PKB cascade to increase insulin gene expression. On the other hand, expression of miR-124a2, previously described as inhibiting insulin gene expression (27), is upregulated in GK islets. Hence, we speculate that the antagonizing actions of miR-124a2 and miR-375 are counterbalanced, resulting in an unchanged insulin mRNA level in GK islets. An alternative explanation is that the lower expression in GK islets results from a prolonged intracellular deprivation of glucose signals, as in the case of the islets cultured in 5 mmol/l glucose for 72 h that progress to apoptosis.
According to recent studies (39,40), total β-cell mass is reduced in patients and animals with type 2 diabetes and contributes to the disease process. Notably, β-cell–specific PDK1 knockout in mice leads to diabetes as a result of a reduction in β-cell mass (14). Another in vitro study reported that decreased PDK1 in human glioblastoma cells obtained with antisense oligonucleotides or with RNA interference blocks cell proliferation (41). Here, we find that miR-375 downregulates PDK1 by directly targeting PDK1 mRNA and may therefore impact on cell proliferation given its key role in the PI 3-kinase/PKB cascade. In this context, several studies have revealed dysregulations in microRNA expression in human cancers (42), suggesting that microRNAs may act on cell proliferation. More directly related to our work, recent studies show that miR-375 is decreased in pancreatic cancer (43–45). Our results point to a similar antiproliferative action of miR-375, because we found that miR-375 attenuates cell viability and proliferation.
A first key finding of our work is that miR-375 is regulated by glucose, which is the central molecule in islet metabolism and physiology. A second important observation is that miR-375 inhibits glucose-induced INS-1E cell proliferation. This result is particularly interesting in the context of diabetes. Although a distinct disease process is responsible for type 1 and type 2 diabetes, in both cases, β-cell failure occurs. Developing new approaches to induce β-cells to replicate is a supereminent goal in diabetes research. Our data presented here reveal the pancreatic islet-specific microRNA, miR-375, as an important regulator of glucose-stimulated insulin gene expression and proliferation of pancreatic β-cells. Taking into account the fact that miR-375 also decreases insulin secretion (25), miR-375 emerges as a target that should be prioritized to enhance islet function and to combat β-cell failure.
Acknowledgments
A.E.O. has received a PhD fellowship from la Ligue Nationale Contre le Cancer (France). D.P. has received Inter-University Pole Attraction Program Grant IUAP P5/17 from the Belgian Science Policy. E.V.O. has received a grant from the European Foundation for the Study of Diabetes/Juvenile Diabetes Research Foundation/Novo-Nordisk Program in type 1 diabetes. This work has been supported by the Research Foundation Flanders (grant FWO-G.0183.05), the Institut National de la Santé et de la Recherche Médicale, the Université de Nice-Sophia-Antipolis, the Conseil Régional Provence-Alpes-Côte d’Azur, the Conseil Général des Alpes-Maritimes, and the European Community FP6 EUGENE2 (grant LSHM-CT-2004-512013).
We thank Professor Bernard Portha for providing us with the GK rats; Dr. Olivier Dumortier for help with rat islets isolation; and Nadine Gautier, Chantal Filloux, Rachel Paul, Joseph Murdaca, Cyndie Raemdock, and Elke De Vos for assistance. We are grateful to Drs. Mourad Naïmi and Jean Giudicelli for discussions and advice.
Published ahead of print at http://diabetes.diabetesjournals.org on 30 June 2008.
The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
See accompanying commentary, p. 2567.
REFERENCES
- 1.Stumvoll M, Goldstein BJ, van Haeften TW: Type 2 diabetes: principles of pathogenesis and therapy. Lancet 365: 1333–1346, 2005 [DOI] [PubMed] [Google Scholar]
- 2.Ling Z, Wang Q, Stange G, In’t Veld P, Pipeleers D: Glibenclamide treatment recruits β-cell subpopulation into elevated and sustained basal insulin synthetic activity. Diabetes 55: 78–85, 2006 [PubMed] [Google Scholar]
- 3.Pipeleers D, Kiekens R, Ling Z, Wilikens A, Schuit F: Physiologic relevance of heterogeneity in the pancreatic beta-cell population. Diabetologia 37 (Suppl. 2): S57–S64, 1994 [DOI] [PubMed] [Google Scholar]
- 4.Hugl SR, White MF, Rhodes CJ: Insulin-like growth factor I (IGF-I)-stimulated pancreatic beta-cell growth is glucose-dependent: synergistic activation of insulin receptor substrate-mediated signal transduction pathways by glucose and IGF-I in INS-1 cells. J Biol Chem 273: 17771–17779, 1998 [DOI] [PubMed] [Google Scholar]
- 5.Alonso LC, Yokoe T, Zhang P, Scott DK, Kim SK, O’Donnell CP, Garcia-Ocana A: Glucose infusion in mice: a new model to induce β-cell replication. Diabetes 56: 1792–1801, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rafiq I, da Silva Xavier G, Hooper S, Rutter GA: Glucose-stimulated preproinsulin gene expression and nuclear trans-location of pancreatic duodenum homeobox-1 require activation of phosphatidylinositol 3-kinase but not p38 MAPK/SAPK2. J Biol Chem 275: 15977–15984, 2000 [DOI] [PubMed] [Google Scholar]
- 7.MacFarlane WM, Read ML, Gilligan M, Bujalska I, Docherty K: Glucose modulates the binding activity of the beta-cell transcription factor IUF1 in a phosphorylation-dependent manner. Biochem J 303: 625–631, 1994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Srinivasan S, Bernal-Mizrachi E, Ohsugi M, Permutt MA: Glucose promotes pancreatic islet beta-cell survival through a PI 3-kinase/Akt-signaling pathway. Am J Physiol Endocrinol Metab 283: E784–E793, 2002 [DOI] [PubMed] [Google Scholar]
- 9.Taniguchi CM, Emanuelli B, Kahn CR: Critical nodes in signalling pathways: insights into insulin action. Nat Rev Mol Cell Biol 7: 85–96, 2006 [DOI] [PubMed] [Google Scholar]
- 10.Stephens L, Anderson K, Stokoe D, Erdjument-Bromage H, Painter GF, Holmes AB, Gaffney PR, Reese CB, McCormick F, Tempst P, Coadwell J, Hawkins PT: Protein kinase B kinases that mediate phosphatidylinositol 3,4,5-trisphosphate-dependent activation of protein kinase B. Science 279: 710–714, 1998 [DOI] [PubMed] [Google Scholar]
- 11.Alessi DR, James SR, Downes CP, Holmes AB, Gaffney PR, Reese CB, Cohen P: Characterization of a 3-phosphoinositide-dependent protein kinase which phosphorylates and activates protein kinase Balpha. Curr Biol 7: 261–269, 1997 [DOI] [PubMed] [Google Scholar]
- 12.Rintelen F, Stocker H, Thomas G, Hafen E: PDK1 regulates growth through Akt and S6K in Drosophila. Proc Natl Acad Sci U S A 98: 15020–15025, 2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lawlor MA, Mora A, Ashby PR, Williams MR, Murray-Tait V, Malone L, Prescott AR, Lucocq JM, Alessi DR: Essential role of PDK1 in regulating cell size and development in mice. EMBO J 21: 3728–3738, 2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hashimoto N, Kido Y, Uchida T, Asahara S, Shigeyama Y, Matsuda T, Takeda A, Tsuchihashi D, Nishizawa A, Ogawa W, Fujimoto Y, Okamura H, Arden KC, Herrera PL, Noda T, Kasuga M: Ablation of PDK1 in pancreatic beta cells induces diabetes as a result of loss of beta cell mass. Nat Genet 38: 589–593, 2006 [DOI] [PubMed] [Google Scholar]
- 15.Lee RC, Feinbaum RL, Ambros V: The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 75: 843–854, 1993 [DOI] [PubMed] [Google Scholar]
- 16.Kim VN: MicroRNA biogenesis: coordinated cropping and dicing. Nat Rev Mol Cell Biol 6: 376–385, 2005 [DOI] [PubMed] [Google Scholar]
- 17.Bartel DP: MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116: 281–297, 2004 [DOI] [PubMed] [Google Scholar]
- 18.Reinhart BJ, Slack FJ, Basson M, Pasquinelli AE, Bettinger JC, Rougvie AE, Horvitz HR, Ruvkun G: The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature 403: 901–906, 2000 [DOI] [PubMed] [Google Scholar]
- 19.Xu P, Vernooy SY, Guo M, Hay BA: The Drosophila microRNA Mir-14 suppresses cell death and is required for normal fat metabolism. Curr Biol 13: 790–795, 2003 [DOI] [PubMed] [Google Scholar]
- 20.Esau C, Kang X, Peralta E, Hanson E, Marcusson EG, Ravichandran LV, Sun Y, Koo S, Perera RJ, Jain R, Dean NM, Freier SM, Bennett CF, Lollo B, Griffey R: MicroRNA-143 regulates adipocyte differentiation. J Biol Chem 279: 52361–52365, 2004 [DOI] [PubMed] [Google Scholar]
- 21.Krutzfeldt J, Rajewsky N, Braich R, Rajeev KG, Tuschl T, Manoharan M, Stoffel M: Silencing of microRNAs in vivo with ‘antagomirs.’ Nature 438: 685–689, 2005 [DOI] [PubMed] [Google Scholar]
- 22.Esau C, Davis S, Murray SF, Yu XX, Pandey SK, Pear M, Watts L, Booten SL, Graham M, McKay R, Subramaniam A, Propp S, Lollo BA, Freier S, Bennett CF, Bhanot S, Monia BP: miR-122 regulation of lipid metabolism revealed by in vivo antisense targeting. Cell Metab 3: 87–98, 2006 [DOI] [PubMed] [Google Scholar]
- 23.Callis TE, Chen JF, Wang DZ: MicroRNAs in skeletal and cardiac muscle development. DNA Cell Biol 26: 219–225, 2007 [DOI] [PubMed] [Google Scholar]
- 24.Makeyev EV, Zhang J, Carrasco MA, Maniatis T: The MicroRNA miR-124 promotes neuronal differentiation by triggering brain-specific alternative pre-mRNA splicing. Mol Cell 27: 435–448, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Poy MN, Eliasson L, Krutzfeldt J, Kuwajima S, Ma X, Macdonald PE, Pfeffer S, Tuschl T, Rajewsky N, Rorsman P, Stoffel M: A pancreatic islet-specific microRNA regulates insulin secretion. Nature 432: 226–230, 2004 [DOI] [PubMed] [Google Scholar]
- 26.Plaisance V, Abderrahmani A, Perret-Menoud V, Jacquemin P, Lemaigre F, Regazzi R: MicroRNA-9 controls the expression of Granuphilin/Slp4 and the secretory response of insulin-producing cells. J Biol Chem 281: 26932–26942, 2006 [DOI] [PubMed] [Google Scholar]
- 27.Baroukh N, Ravier MA, Loder MK, Hill EV, Bounacer A, Scharfmann R, Rutter GA, Van Obberghen E: MicroRNA-124a regulates Foxa2 expression and intracellular signaling in pancreatic beta-cell lines. J Biol Chem 282: 19575–19588, 2007 [DOI] [PubMed] [Google Scholar]
- 28.Cheng AM, Byrom MW, Shelton J, Ford LP: Antisense inhibition of human miRNAs and indications for an involvement of miRNA in cell growth and apoptosis. Nucleic Acid Res 33: 1290–1297, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schmittgen TD, Jiang J, Liu Q, Yang L: A high-throughput method to monitor the expression of microRNA precursors. Nucleic Acid Res 32: e43, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jiang J, Lee EJ, Gusev Y, Schmittgen TD: Real-time expression profiling of microRNA precursors in human cancer cell lines. Nucleic Acid Res 33: 5394–5403, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chang TC, Mendell JT: MicroRNAs in vertebrate physiology and human disease. Annu Rev Genomics Hum Genet 8: 215–239, 2007 [DOI] [PubMed] [Google Scholar]
- 32.He A, Zhu L, Gupta N, Chang Y, Fang F: Overexpression of micro ribonucleic acid 29, highly up-regulated in diabetic rats, leads to insulin resistance in 3T3–L1 adipocytes. Mol Endocrinol 21: 2785–2794, 2007 [DOI] [PubMed] [Google Scholar]
- 33.Kloosterman WP, Lagendijk AK, Ketting RF, Moulton JD, Plasterk RH: Targeted inhibition of miRNA maturation with morpholinos reveals a role for miR-375 in pancreatic islet development. PLoS Biol 5: e203, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Keller DM, McWeeney S, Arsenlis A, Drouin J, Wright CV, Wang H, Wollheim CB, White P, Kaestner KH, Goodman RH: Characterization of pancreatic transcription factor Pdx-1 binding sites using promoter microarray and serial analysis of chromatin occupancy. J Biol Chem 282: 32084–32092, 2007 [DOI] [PubMed] [Google Scholar]
- 35.Lingohr MK, Briaud I, Dickson LM, McCuaig JF, Alarcon C, Wicksteed BL, Rhodes CJ: Specific regulation of IRS-2 expression by glucose in rat primary pancreatic islet beta-cells. J Biol Chem 281: 15884–15892, 2006 [DOI] [PubMed] [Google Scholar]
- 36.Hoorens A, Van de Casteele M, Kloppel G, Pipeleers D: Glucose promotes survival of rat pancreatic beta cells by activating synthesis of proteins which suppress a constitutive apoptotic program. J Clin Invest 98: 1568–1574, 1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ling Z, Kiekens R, Mahler T, Schuit FC, Pipeleers-Marichal M, Sener A, Kloppel G, Malaisse WJ, Pipeleers DG: Effects of chronically elevated glucose levels on the functional properties of rat pancreatic β-cells. Diabetes 45: 1774–1782, 1996 [DOI] [PubMed] [Google Scholar]
- 38.Martens GA, Cai Y, Hinke S, Stange G, Van de Casteele M, Pipeleers D: Glucose suppresses superoxide generation in metabolically responsive pancreatic beta cells. J Biol Chem 280: 20389–20396, 2005 [DOI] [PubMed] [Google Scholar]
- 39.Weir GC, Bonner-Weir S: Five stages of evolving β-cell dysfunction during progression to diabetes. Diabetes 53 (Suppl. 3): S16–S21, 2004 [DOI] [PubMed] [Google Scholar]
- 40.Butler AE, Janson J, Bonner-Weir S, Ritzel R, Rizza RA, Butler PC: β-Cell deficit and increased β-cell apoptosis in humans with type 2 diabetes. Diabetes 52: 102–110, 2003 [DOI] [PubMed] [Google Scholar]
- 41.Bilanges B, Stokoe D: Direct comparison of the specificity of gene silencing using antisense oligonucleotides and RNAi. Biochem J 388: 573–583, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA, Downing JR, Jacks T, Horvitz HR, Golub TR: MicroRNA expression profiles classify human cancers. Nature 435: 834–838, 2005 [DOI] [PubMed] [Google Scholar]
- 43.Szafranska AE, Davison TS, John J, Cannon T, Sipos B, Maghnouj A, Labourier E, Hahn SA: MicroRNA expression alterations are linked to tumorigenesis and non-neoplastic processes in pancreatic ductal adenocarcinoma. Oncogene 26: 4442–4452, 2007 [DOI] [PubMed] [Google Scholar]
- 44.Lee EJ, Gusev Y, Jiang J, Nuovo GJ, Lerner MR, Frankel WL, Morgan DL, Postier RG, Brackett DJ, Schmittgen TD: Expression profiling identifies microRNA signature in pancreatic cancer. Int J Cancer 120: 1046–1054, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bloomston M, Frankel WL, Petrocca F, Volinia S, Alder H, Hagan JP, Liu CG, Bhatt D, Taccioli C, Croce CM: MicroRNA expression patterns to differentiate pancreatic adenocarcinoma from normal pancreas and chronic pancreatitis. JAMA 297: 1901–1908, 2007 [DOI] [PubMed] [Google Scholar]