Abstract
OBJECTIVE—To document the transcriptome of the pancreatic islet during the early and late development of the mouse pancreas and highlight the qualitative and quantitative features of gene expression that contribute to the specification, growth, and differentiation of the major endocrine cell types. A further objective was to identify endocrine cell biomarkers, targets of diabetic autoimmunity, and regulatory pathways underlying islet responses to physiological and pathological stimuli.
RESEARCH DESIGN AND METHODS—mRNA expression profiling was performed by microarray analysis of e12.5–18.5 embryonic pancreas from neurogenin 3 (Ngn3)-null mice, a background that abrogates endocrine pancreatic differentiation. The intersection of this data with mRNA expression in isolated adult pancreatic islets and pancreatic endocrine tumor cell lines was determined to compile lists of genes that are specifically expressed in endocrine cells.
RESULTS—The data provided insight into the transcriptional and morphogenetic factors that may play major roles in patterning and differentiation of the endocrine lineage before and during the secondary transition of endocrine development, as well as genes that control the glucose responsiveness of the β-cells and candidate diabetes autoantigens, such as insulin, IA-2 and Slc30a8 (ZnT8). The results are presented as downloadable gene lists, available at https://www.cbil.upenn.edu/RADQuerier/php/displayStudy.php?study_id=1330, stratified by predictive scores of relative cell-type specificity.
CONCLUSIONS—The deposited data provide a rich resource that can be used to address diverse questions related to islet developmental and cell biology and the pathogenesis of type 1 and 2 diabetes.
The major endocrine and exocrine cell types of the pancreas share a common embryological origin from the foregut endoderm and possess common phenotypic characteristics related to the regulated secretion of polypeptides (1). Microarray-based gene expression analysis of the genes that are related by ontology and common pathways of metabolism and signaling (2–4) in the tissue is hampered by difficulty of separating endocrine from exocrine tissue at early embryological ages except by laborious procedures based on cell sorting that may distort the expression profiles. An alternative approach adopted here is to compare gene expression in fetal pancreas from E12.5–18.5 from wild-type and neurogenin 3 (Ngn3) knockout (−/−) mice: animals in which the ductal and exocrine programs proceed normally but islet endocrine cells fail to develop (5). Ngn3 is expressed transiently around E14.5 in a small population of endocrine progenitor cells (6), and Ngn3−/− mice fail to express key islet transcription factors Isl1, Pax4, Pax6, and NeuroD1 (5). Nevertheless, pancreatic exocrine cell lineage is correctly specified through the expression of Ptf1a and Prox1, and only minor changes in acinar cellular polarity and zymogen granule accumulation occur (5).
Data expression profiles from pancreata of Ngn3−/− versus wild-type animals were compared with normal adult mouse pancreatic islets and islet-derived tumor cell lines (7) to provide information on endocrine cell-type specificity of genes that are downregulated in Ngn3−/− animals. The example of Slc30a8 (ZnT8), a gene that has been associated with type 2 diabetes (8) and that encodes a type 1 diabetes autoantigen (9) is highlighted.
RESEARCH DESIGN AND METHODS
Embryonic pancreata were harvested from time-mated CD1 strain mouse intercrosses of Ngn3+/− females and males and stored individually in RNAlater (Ambion) at 4°C until genotyping by PCR (30 cycles; Ngn3 forward, 5′-cctcttctggctttcactac-3′ and reverse, 5′-ggagcgagagtttgatgtgg-3′; neomycin forward, 5′-gtcttgtcgatcaggatgatctc-3′ and reverse, 5′-caatatcacgggtagccaacgc-3′). Pancreatic islets were prepared from 6-month-old female CD1 mice by collagenase digestion and isolated by Ficoll/Histopaque gradients and handpicking (each preparation: six mice/1,000 islets). αTC1-6, βTC3′, MIN6-, and mPAC-tumor cell lines (10) were grown to 60% confluency in RPMI medium. Total RNA was extracted from tissues with TRIzol reagent (Invitrogen, Carlsbad, CA), purified by RNeasy columns (Qiagen, Valencia, CA). Biotin-labeled cRNA was synthesized by the Affymetrix protocol (Affymetrix, Santa Clara, CA) and hybridized to MGU74Av2 and/or MOE 430 2.0 microarrays. Raw microarray datasets are available through EPCON db (http://www.cbil.upenn.edu/RAD/php/displayStudy.php?study_id=1330).
Data were analyzed with GeneSpring 7.2 (Silicon Genetics, Redwood City, CA) and GC Robust Multi-array Average (GC RMA) used for normalization. ANOVA was applied to cell line data, and P values were corrected by Benjamini and Hochberg's procedure. P < 0.05 were considered significant. NetAffx (Affymetrix, Santa Clara, CA; http://www.affymetrix.com/analysis/index.affx) was used to extract a common probe set between MOE430 2.0 and MGU74Av2 platforms. Gene classification was performed with FatiGO (http://fatigo.bioinfo.cipf.es).
Quantitative RT-PCR (qRT-PCR) was performed on cDNA (5 ng) derived from total RNA by iScript cDNA synthesis kits (Bio-Rad Laboratories, Hercules, CA). Assays used FAM dye–labeled Taqman MGB probes and an ABI 7000 PCR instrument (Applied Biosystems, Foster City, CA) and were normalized to glyceraldehyde-3-phosphate dehydrogenase. Triplicate cycle threshold values were normalized to samples.
Mouse pancreata were fixed in 4% paraformaldehyde and embedded in optimal cutting temperature compound (Tissue Tek; Sakura Finetek, Torrance, CA) for immunofluorescence microscopy. Sections (6 μm) were blocked in tyramide signal amplification blocking solution (Zymed, Carlsbad, CA) incubated overnight with guinea pig anti-insulin, mouse anti-glucagon (Sigma, St Louis, MO), rat anti-somatostatin (Abcam, Cambridge, MA), or rabbit anti-ZnT8 aa268-369 (9). Secondary antibodies conjugated to Alexa 488, 1-amino-1-methyl-3(4)-aminomethylcyclohexane, cyanine 5, and cyanine 3 fluorophores (Jackson Immunoresearch Laboratories, West Grove, PA) were applied for 60 min and sections mounted in glycerol-based media. Images were acquired with an Olympus 1X70 microscope equipped with a Photometrics Quantix cooled monochromatic CCD camera (Kodak chip KAF1400).
RESULTS
Pancreatic microarray analyses were performed on wild-type and Ngn3−/− mice at E12.5, E15.5, and E18.5. The majority of the endocrine fate allocations occur between E13.5 and E15.5, when Ngn3 is maximally expressed (11,12); endocrine markers (insulin and glucagon) subsequently appear along with neuroendocrine markers, e.g., prohormone convertases and chromogranins. Thus, comparison of E12.5 with E15.5 identifies transcripts that characterize precursor cells, responses to the activation of Ngn3 expression, and the transition to the differentiated endocrine fate. At E12.5, 203 gene transcripts were downregulated >threefold in Ngn3−/− versus wild-type littermates and, at E15.5, 54 (Table 1 and supplementary Tables 2 and 3, available in an online appendix at http://dx.doi.org/10.2337/db07-1126). Fewer genes were upregulated: 53 at >threefold in E12.5 and 17 in E15.5 (data not shown).
TABLE 1.
Pancreas transcripts downregulated in Ngn3−/− mice
| Gene ontology term (biological process) | E12.5 threefold down | E15.5 threefold down | E18.5 threefold down |
|---|---|---|---|
| Regulation of cellular process | Atrx, Sfpq, Braf, Nrip1, Zfp458, Rest, Klf7, Neurod1, Zfp398, Cpsf6, Nfia, Zfp192, Ncoa7, Crim1, Meis1, Mbtps1, Hnrpab, Hdac2, Zfp207, Gas2l3, Fubp1, Onecut2, Ercc6, Rbm5, Rapgef5, Arid5b, D030022P06Rik, Gna13, Shprh, Scmh1, Foxp2, and Paip1 | Pax6, Neurod1, Fev, St18, Neurog3, Isl1, and Ins2 | Pitx2, Fgd4, Nr3c2, Rapgef1, Rcor2, Il2, Gtf2i, Zfpm2, Stk36, Jarid1d, Inpp5d, Elavl1, E130112L23Rik, Rasgrf1, Zfp580, Zw10, Dnmt3a, Igfbp1, Cideb, Pax6, Rora, Spock2, Gtf3c1, Mlxipl, Nr2f2, Ins2, Tcfec, Malt1, Hcfc1, Neurod1, Igsf1, Itsn1, Ikzf1, Grm2, Isl1, Apoa2, Cables1, Mapk8ip2, Smurf1, Hipk1, Rps6ka5, Sim1, A230083H22Rik, Psd, Gli3, Nf1, Khdrbs3, Klf9, Braf, Pknox2, Tgfbr1, Gata1, Etv1, Smarca2, Bcl2l2, Ahsg, Dlg5, Ptpn11, Eda, Iqgap1, Pde6h, Brdt, Adam10, C030010B13Rik, Gatad2b, Mmp9, Ccnc, Cacna1a, Mecp2, Pde6g, Zdhhc17, Mafb, Ighmbp2, Creb1, Cdc37l1, Edg2, Pdlim1, Lst1, Calcoco1, Nkx6-1, AI324046, BC031441, Rph3al, Pias1, Tcf7l2, Sox13, St18, Frzb, Sebox, Ciapin1, Eef1a2, Rbx1, Batf, Fgf9, Ccna1, Dhcr24, Nono, Foxn4, and Hoxd3 |
| Transport | Abcb10, Huwe1, Colec10, Mbtps1, Atp8a1, Gcg, Napg, Exoc5, and Kif23 | Gcg, Ghrl, Cadps, Syt13, Slc7a14, Ins1, Ins2, and Sytl4 | Kif3b, Fabp1, Col4a6, Atp6v0a1, Kcnn2, Atp2a3, Fndc5, Atp11c, Syt13, Ins1, Zw10, Atp6v1g2, Snap25, Ghrl, Timm22, Atp1b1, Slc4a1, Uqcrq, Scn4a, Cadps, Lrp11, Fars2, Slc30a8, Laptm5, Aplp1, Ins2, Snx26, Slc2a5, Itpr2, Malt1, Sv2b, Slc9a1, Sytl4, Itsn1, Grm2, Apoa2, Slco2b1, Slc16a9, Rab23, Rab21, Rab37, Camk2b, Anxa6, Slc16a10, Ahsg, Stxbp1, Uty, Ptpn11, Eda, Apoa1, Brdt, Slc7a14, Adam10, 1300017J02Rik, Col11a2, Ap3m2, Lin7b, Cacna1a, Gcg, Zdhhc17, Prkcb1, Kif13a, AI324046, Thoc4, Abcc8, Rph3al, Igf2r, Mon1b, Ap4b1, Afp, Slc35a5, Syt7, Trpm5, Scnn1b, Fgf9, Slc39a2, Foxn4, and Scg5 |
| System development | Atrx, Mtss1, Nrip1, Neurod1, Cxadr, Ppp3cb, Mbp, Rapgef5, and Gna13 | Pax6, Neurod1, Neurog3, Isl1, and Insm1 | Pitx2, Casq2, Pip5k1c, Rapgef1, Il2, Atp6v0a1, Inpp5d, Crybb1, Crybb3, Pax6, Spock2, Chrdl1, Nr2f2, Aplp1, Tnfaip2, Neurod1, Ikzf1, Isl1, Apoa2, Robo1, Cables1, Smurf1, Sim1, Rab23, Papss2, Gli3, Nf1, Hsd11b1, Scube1, Tgfbr1, Add2, Etv1, Arts1, Ahsg, Stxbp1, Ptpn11, Eda, Apoa1, Tmprss6, Mmp9, Mecp2, Tnfrsf11a, Ankrd17, Mafb, Creb1, Lst1, Ptprj, Nkx6-1, Epm2a, Mepe, Insm1, Afp, Ciapin1, Efnb2, Fgf9, Dhcr24, Lor, Slitrk6, Foxn4, and Hoxd3 |
| Cellular biosynthetic process | Gch1, St8sia3, Galnt7, Etnk1, Zdhhc2, and Paip1 | Gch1 | Gch1, Inpp5d, Fut10, Atp6v1g2, Fars2, Apoa2, Pank2, Zdhhc2, St8sia5, Mecr, Pigx, Zdhhc17, Dct, Nmt2, Dio2, Eif2s3, Eef1a2, Cps1, Umps, and Foxn4 |
| Cell cycle process | Bub3, Gas2l3, and Rbm5 | 0 | Zw10, Hcfc1, Cables1, A230083H22Rik, Camk2b, Ptpn11, Ccnc, Cdc37l1, Suv39h2, Rbx1, Fgf9, and Ccna1 |
| Cellular component organization and biogenesis | Mtss1, Bub3, Cbx5, Cxadr, Huwe1, Crim1, Mbtps1, Hdac2, Setd8, Pot1a, Napg, Gna13, Shprh, and Kif23 | Pax6, Cadps, 4930488E11Rik, and Sytl4 | Kif3b, Fgd4, Pip5k1c, Fndc5, Ush1c, Fmn2, Zw10, Dnmt3a, Igfbp1, Pax6, Rtel1, Timm22, Cadps, Lrp11, Aplp1, Malt1, Sytl4, Itsn1, Robo1, Rps6ka5, Fhod1, Etv1, Smarca2, Ahsg, Stxbp1, Ptpn11, Eda, Brdt, Adam10, 6330505N24Rik, Ap3m2, Tmsb10, Mecp2, Hist3h2a, Creb1, Lst1, Kif13a, AI324046, Thoc4, 4930488E11Rik, Rph3al, Ap4b1, Suv39h2, Hist3h2ba, Syt7, Fgf9, Dhcr24, Slitrk6, and Scg5 |
Data are stratified by developmental age and gene ontology (biological process) as defined by FatiGO (http://fatigo.bioinfo.cipf.es). Further information on each gene (http://www.ensembl.org/Mus_musculus/geneview) is accessible by clicking on the common gene name.
At E18.5, 645 transcripts were downregulated >3-fold in Ngn3−/− versus wild-type animals (supplementary Table 4), 290 >4-fold, and 35 >10-fold. Validation of select transcripts by qRTPCR demonstrated that glucagon (down 1,400-fold), insulin (down 400-fold), and ZnT8 (down 18-fold) were readily detectable in wild-type but not Ngn3−/− animals with 40 cycles of PCR (data not shown). Of the transcripts that were downregulated >fourfold, 46 mapped to transcripts highly expressed in adult islets (raw value >2,000 including ZnT8) and 28 to transcripts moderately expressed (500–2,000) (Table 2).
TABLE 2.
Gene transcripts downregulated in Ngn3−/− pancreas at E18.5 versus expression levels in adult CD1 mouse islets
| Genbank ID | Fold change E18.5 (wild type/Ngn3−/−) | Expression in adult islets | |
|---|---|---|---|
| Gcg | AF276754 | 1,364.0 | 54,221.21 |
| Iapp | BM569571 | 965.0 | 53,018.00 |
| Ins1 | NM_008386 | 405.0 | 48,271.76 |
| Ins2 | NM_008387 | 497.7 | 40,357.31 |
| Iapp | BB434117 | 805.0 | 38,126.00 |
| Scg5 | AK019337 | 5.5 | 34,706.20 |
| Ppy | NM_008918 | 7.8 | 32,143.54 |
| Chga | NM_007693 | 43.2 | 25,948.83 |
| Tmem27 | AI314694 | 66.5 | 21,253.26 |
| G6pc2 | Z47787 | 43.9 | 21,243.67 |
| Scg3 | NM_009130 | 31.7 | 20,934.01 |
| Sst | NM_009215 | 165.1 | 19,871.07 |
| Chgb | NM_007694 | 97.0 | 15,589.74 |
| Pyy | BC010821 | 64.3 | 14,889.29 |
| Pcsk2 | AI839700 | 23.6 | 13,963.12 |
| Pcsk2 | NM_008792 | 62.9 | 12,877.78 |
| Slc30a8 (ZnT8) | BB433667 | 18.6 | 9,643.99 |
| Iapp | BM569571 | 94.0 | 8,830.73 |
| Gja9 | BI737842 | 9.6 | 7,247.54 |
| BB336256 | 29.9 | 7,043.94 | |
| Insm1 | BB468410 | 12.6 | 6,996.37 |
| Neurod1 | BM116592 | 7.9 | 6,698.26 |
| AW493291 | 4.3 | 6,392.52 | |
| Abcc8 | BB515948 | 4.3 | 6,098.58 |
| Sytl4 | NM_013757 | 8.3 | 6,038.47 |
| 4930544G21Rik | AV327549 | 20.3 | 5,745.73 |
| BB349472 | 5.0 | 5,704.96 | |
| Papss2 | BF786072 | 4.1 | 3,892.62 |
| 4930488E11Rik | AW912417 | 4.8 | 3,417.22 |
| Lrp11 | BB435348 | 7.1 | 3,342.56 |
| BC018249 | 4.0 | 3,276.17 | |
| AI987662 | AV282151 | 8.3 | 3,202.88 |
| Syt13 | BB244585 | 6.7 | 3,183.86 |
| Sez6l2 | AW121511 | 5.6 | 3,154.09 |
| Isl1 | BQ176915 | 8.4 | 2,971.00 |
| Pcsk1n | AF181560 | 4.9 | 2,950.96 |
| Pax6 | BC011272 | 8.8 | 2,943.98 |
| BG065782 | 6.4 | 2,905.03 | |
| Papss2 | BF786072 | 5.6 | 2,790.13 |
| Ppp1r1a | NM_021391 | 7.8 | 2,740.16 |
| Gch1 | BB698398 | 4.4 | 2,584.94 |
| Neurod1 | BM116592 | 33.6 | 2,444.55 |
| Frzb | U91905 | 4.4 | 2,348.46 |
| Scgn | BC016093 | 7.5 | 2,159.75 |
| 1700086L19Rik | BI526033 | 12.0 | 2,089.92 |
| Rbp4 | U63146 | 4.3 | 2,026.31 |
| Isl1 | BQ176915 | 8.9 | 1,928.84 |
| Iapp | BB434117 | 11.0 | 1,829.83 |
| AI429745 | 6.5 | 1,820.01 | |
| Cadps | NM_012061 | 7.7 | 1,811.08 |
| AV307860 | 8.1 | 1,779.41 | |
| Syt7 | AV138438 | 4.7 | 1,772.53 |
| Atp2a3 | NM_016745 | 4.3 | 1,754.10 |
| Fkbp1b | NM_016863 | 5.7 | 1,730.78 |
| Mlxipl | AF245479 | 26.6 | 1,727.16 |
| Zdhhc2 | BB224658 | 4.1 | 1,564.37 |
| Nol4 | AV323033 | 4.9 | 1,427.26 |
| Slc2a5 | NM_019741 | 6.5 | 1,377.04 |
| Abcc8 | BF466569 | 5.3 | 1,337.43 |
| Disp2 | BB023743 | 19.3 | 1,320.44 |
| Nkx6–1 | AF357883 | 4.9 | 1,255.80 |
| Npy | NM_023456 | 6.2 | 1,220.19 |
| Insm1 | NM_016889 | 13.2 | 1,168.05 |
| Pcsk2 | BB357975 | 4.3 | 1,057.46 |
| Spock2 | BM117672 | 6.6 | 981.56 |
| Chic1 | Y11896 | 4.4 | 879.21 |
| Zcchc18 | BC017627 | 5.1 | 841.97 |
| Pcsk2 | AI839700 | 4.1 | 784.11 |
| BB262415 | 4.5 | 758.46 | |
| Camk2b | NM_007595 | 5.3 | 749.76 |
| Gtpbp4 | AI987834 | 5.3 | 681.42 |
| AK015642 | 7.0 | 625.43 | |
| BE948505 | 6.1 | 615.68 | |
| Fabp1 | NM_017399 | 4.2 | 562.39 |
| Upb1 | NM_133995 | 14.9 | 518.36 |
| Rimbp2 | BQ174470 | 4.6 | 491.47 |
| Pak3 | BQ175796 | 4.3 | 472.47 |
| BC026682 | NM_025590 | 6.0 | 415.94 |
| BC052055 | BB392984 | 5.1 | 336.12 |
| AV030118 | 7.0 | 303.42 | |
| BC038479 | AV367395 | 4.2 | 293.78 |
| Cplx2 | BE946238 | 4.0 | 292.95 |
| Ankrd40 | AK017451 | 4.5 | 289.60 |
| Baiap3 | BC022659 | 25.2 | 288.45 |
| BQ175636 | 4.5 | 263.94 | |
| Ush1c | NM_023649 | 4.0 | 251.60 |
| BB084132 | 4.9 | 245.63 | |
| 1100001E04Rik | BB367590 | 7.1 | 242.20 |
| 2410129H14Rik | BI153133 | 4.5 | 239.18 |
| BM213778 | 4.4 | 209.61 | |
| AV329790 | 4.4 | 201.61 | |
| Phyhipl | AI267048 | 5.3 | 171.28 |
| AI467606 | BB234337 | 4.3 | 161.34 |
| Nnat | AV218841 | 5.4 | 152.18 |
| Pak3 | BQ174935 | 4.5 | 148.61 |
| BC051142 | AV278321 | 4.7 | 137.10 |
| Akap10 | AK005325 | 7.6 | 122.61 |
| Stk32a | BB279083 | 4.1 | 117.41 |
| C8b | BC022129 | 4.1 | 113.72 |
| C77501 | 4.9 | 113.19 | |
| Smarca2 | BM230202 | 5.0 | 112.24 |
| Hist3h2a | AV297651 | 6.0 | 109.44 |
| H2-K1 | M58156 | 4.2 | 100.31 |
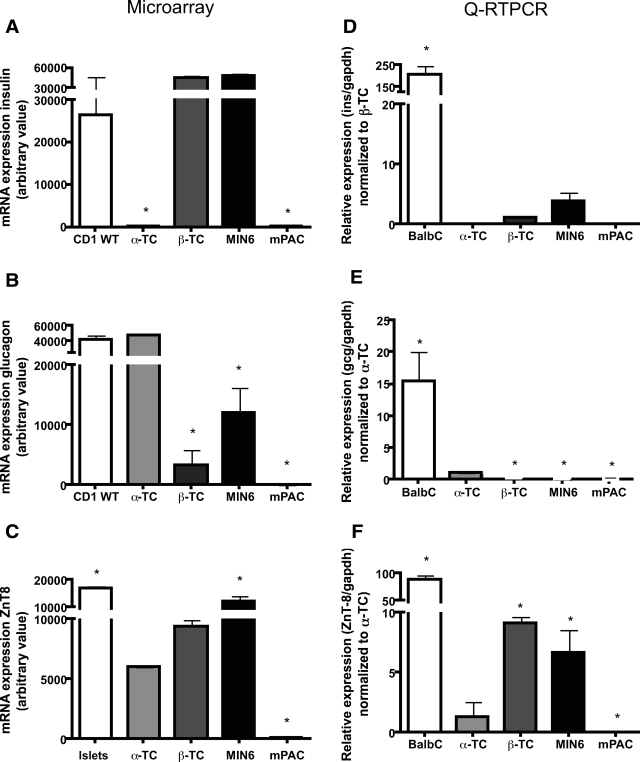
Comparison of αTC1.6- and βTC3-cell transcripts identified 1,554 transcripts that were >2-fold enriched in βTC3 versus αTC1.6 and 403 that were >10-fold enriched (supplementary Table 5). Conversely, 1,202 transcripts were >2-fold enriched in αTC1.6- versus βTC3-cell lines and 212 >10-fold (supplementary Table 6). Of the genes downregulated >fivefold in the Ngn3−/− mice, 74 were enriched in αTC-cells and 108 in βTC-cell lines (supplementary Tables 7 and 8). Another 1,396 transcripts were expressed in both αTC- and βTC-cell lines and 774 not detected. qRT-PCR analyses of islets and cell lines (Fig. 1A–F) showed that insulin transcripts were enriched in adult islets versus βTC- and not expressed in αTC- or mPAC-cell lines (Fig. 1D). By microarray analysis, insulin expression in islets and β-cell lines did not differ since signals were saturated (>25,000 arbitrary units). Glucagon transcript levels by qRTPCR were higher in islets than in αTC-cells and present at a low level in βTC3- and MIN6-cells but absent in mPAC-cells (Fig. 1E). By microarray analysis, the glucagon signals saturated in islets and αTC-cells but otherwise showed the same trend of expression. ZnT8 transcript levels by qRTPCR were 4- to 5-fold higher in β-cell lines than in αTC1.6-cells and 50- to 100-fold higher than in ductal cells (Fig. 1F). Microarray analysis showed ZnT8 expression in islets and α- and β-cell lines but not in ductal cells.
FIG. 1.
Expression of insulin, glucagon, and zinc transporter eight in islets and mouse tumor cell lines. A–C: Microarray analyses: the normalized intensity from data obtained on the MOE 430 2.0 Affymetrix chip is the mean ± SD from two arrays. D–F: qRT-PCR of insulin, glucagon, and ZnT8 mRNA. cDNA synthesized from islets and cells was analyzed with a 5′ nuclease assay and FAM dye–labeled TaQman MGB probes with two PCR primers. Endogenous glyceraldehyde-3-phosphate dehydrogenase (gapdh) was used for normalization. Data (means ± SD) are expressed relative to a control sample (listed on the y-axis). For statistical analysis, one-way ANOVA probability value (P < 0.05) was considered significant, followed by Student's t test (P < 0.05).
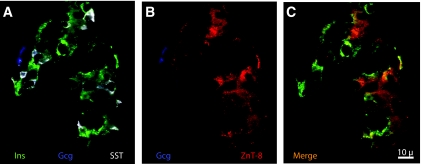
Immunohistochemical localization of ZnT8 in wild-type adult pancreas showed its expression in the majority of islet cells and absence in acinar and ductal tissue. It was principally colocalized with insulin and also found in a subpopulation of somatostatin-positive cells. Expression in α-cells was not detectable (Fig. 2). E18.5 Ngn3−/− pancreas tissue did not show any ZnT8, insulin, glucagon, or somatostatin expression (data not shown).
FIG. 2.
Immunofluorescent detection of ZnT8. E18.5 wild-type mouse pancreas incubated with guinea pig anti-insulin (Ins, green), rabbit anti-ZnT8 (red), mouse anti-glucagon (Gcg, blue), and rat anti-somatostatin (SST, white) antibodies were developed with secondary antibodies linked to Alexa 488, Cy3, 1-amino-1-methyl-3(4)-aminomethylcyclohexane, and Cy5, respectively. ZnT8 colocalized with insulin and somatostatin-expressing cells. Scale bar = 10 μm. (Please see http://dx.doi.org.10.2337/db07-1126 for a high-quality digital representation of this figure.)
DISCUSSION
The loss of expression of genes in the pancreas of Ngn3−/− mice provides a model to define the transcriptome of the endocrine pancreas and offers insight into the transcriptional and morphogenetic factors responsible for the patterning and differentiation of the endocrine lineage. Genes responsive to paracrine signaling from endocrine cells adjacent to exocrine or ductal cells might also have been revealed, although there was little evidence of this. As previously observed (5), expression of the pancreatic islet hormones (insulin, glucagon, somatostatin, pancreatic polypeptide, and ghrelin) were markedly diminished alongside genes known to be expressed in islets but not in acinar tissue (IAPP, ChgrA, ChgrB, Npy, Pyy, PCSK1, PCSK2, and IGRP/G6pc2). Several candidate transcriptional regulators of islet cell lineage development were likewise absent (NeuroD, MafB, Insm1, Myt1, Pax6, and Isl1).
At E12.5, there are a few endocrine cells that are immunoreactive to glucagon and occasional insulin and glucagon double-positive cells. During this phase, Robo1 (Dutt1 protein, orthologous to the Drosophila roundabout) and an expressed sequence tag 9430047L24Rik encoding Unc5c homolog are the two most endocrine-enriched transcripts. Robo1 encodes a molecule of the neural-cell adhesion molecule family that interacts with the extracellular ligand Slit (13–15) that has been implicated in migration of axons, myoblasts, and leukocytes in vertebrates and in lung development (16).
At E15.5, four transcripts notably absent in Ngn3−/− pancreas were Myt1, crystallin β2, secretogogin EF-hand, and 4930568N03Rik (Genbank accession no. AV323033). Microarray data defining the pancreatic development kinetics from E12.5 to E18.5 (K. Juhl, S. Sarkar, J. Jensen, J. Hutton, unpublished data) suggest that Myt1 is downstream of Ngn3, as previously documented (3,17). The AV323033 EST cluster is a component of the Unigene identification Mm.209896, which represents nucleolar protein four (Nol4), which is expressed in libraries from brain, eye, thymus, pancreas, endocrine, spinal cord, and male genitalia.
At E18.5, somatostatin and pancreatic polypeptide cells appear and islet formation is initiated. Genes notably absent in Ngn3−/− pancreas included MafB, Nkx6.1, Wbscr14, Nnat, Syt13, and Pcsk1n—genes that function in mature islet cells as transcription factors (MafB, Nkx6.1, and Wbscr14) (18) or in stimulus secretion coupling. Some transcripts downregulated at this point, however, are expressed at earlier times in wild-type and Ngn3−/− pancreas, notably Pitx2, Ramp2, Hmgn3, and Gtpbp4-pending, indicating that their cell specification is changing. Ramp2, for example, is a chaperone for the calcitonin receptor-like receptor that mediates adrenomedullin action on growth and differentiation where strong mesenchymal-epithelial interactions take place (19).
Comparison of αTC and βTC expression data with the list of 2,352 genes that were downregulated at any age in Ngn3−/− mice revealed 74 α-cell candidates including glucagon, Arx, Brn4, Spp1, Irx2, Rbp4, MafB, Car2, Tfpi, Vegfc, and Fev-pending and 108 β-cell candidates including Ins1&2, IAPP, Neuronatin, Pdx1, G6Pcrs, Npy, Prcad, Sepp1, Sytl4, Hpca, and Atp2a3. Genes that were expressed in both cell types included genes classifed as neuroendocrine (ChgrA and -B, Scgn2 and -3, Scgn, CPE, and Pcsk1 and -2) and transcription factors active in multiple pancreatic endocrine cells (Pax6, Isl1, NeuroD1, and Nkx2.2). In all, the Ngn3−/− model did a remarkably good job of predicting endocrine-specific and neuroendocrine genes, especially given that it reports on 5% of the pancreatic tissue.
At the time of the secondary transition, Ngn3−/−-downregulated transcripts are likely to reflect specific transcripts of the immediate precursors of endocrine cells and thus overlap with profiles derived from fluorescence-activated cell sorter–sorted Ngn3–enhanced green fluorescent protein+ cells (supplementary Tables 9 and 10) (3,20,21). Twenty-nine of the 190 transcripts of the MGU74Av2 dataset of Gu et al. (3) showed overlap including low-copy number transcription factors (MafB, Arx, Brn4, Ngn3, NeuroD1, Pax6, Myt1, and Zfp288). Of the remaining 161 transcripts, 112 were expressed at E12.5 and E15.5 irrespective of Ngn3 gene status, and four (Gnao, Gip, Sgne1, and Nkx2.2) were not found.
The above microarray datasets can be downloaded from an open access Web site: http://www.cbil.upenn.edu/RAD/php/displayStudy.php?study_id=1330. While the current study focuses on embryological development of the pancreas, these datasets can potentially be analyzed in other ways to provide an understanding of islet function in health and disease. Given the current interest in identifying biomarkers that could be used to isolate β-cells and their precursors or to image islet mass, it was of interest to review how effective the selection criteria used here to ascertain β-cell specificity would be in identifying a β-cell marker of lower abundance. The example is the insulin granule zinc transporter ZnT8 (Slc30a8), a type 1A diabetes autoantigen (9) and a gene associated with type 2 diabetes susceptibility (8). ZnT8 transcripts were decreased 18-fold in Ngn3−/− mice pancreas at E18.5 (Table 2), in βTC3- and MIN6-cells, and to a lesser extent in αTC-cells (Fig. 1A and F) but not in a mouse pancreatic ductal cell line (mPAC). Immunofluorescence microscopy showed colocalization with insulin and with a minor islet cell population that coincides with the δ-cell. A lower level of expression in the α-cell was indicated by qRTPCR and other studies (22) but could not be confirmed by immunohistochemistry. A similar strategy could apply the current datasets for the discovery of novel regulatory processes involved in islet metabolism, stimulus secretion coupling, gene transcription, and intracellular and intracellular signaling.
Supplementary Material
Acknowledgments
This study was supported by the Beta Cell Biology Consortium (U19 DK-61248-03), the University of Colorado at Denver Diabetes and Endocrinology Research Center (DERC) (National Institutes of Health Grant P30 DK57516), and a Juvenile Diabetes Research Foundation (JDRF) Center grant. J.J was the recipient of an American Diabetes Association career development award and K.J. a grantee of University of Copenhagen, DK Medical Sciences program, and Lundbeckfonden R7-A714-B576. S.A.S is supported by JDRF 1-2005-145, K01 DK080193, and a DERC Pilot and Feasibility grant. We thank the University of Denver at Colorado Affymetrix microarray support facility and Dr. D. Hanahan (University of California, San Francisco, San Francisco, CA) for provision of αTC1.6-, βTC3-, and mPAC-cell lines and Dr. G. Gradwohl and G. Mellitzer for sharing the Ngn−/− mouse.
Published ahead of print at http://diabetes.diabetesjournals.org on 3 July 2008.
K.J. is currently affiliated with Joslin Diabetes Center, Boston, Massachusetts, and J.J. is currently affiliated with Cleveland Clinic, Cleveland, Ohio.
The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
REFERENCES
- 1.Fishman MP, Melton DA: Pancreatic lineage analysis using a retroviral vector in embryonic mice demonstrates a common progenitor for endocrine and exocrine cells. Int J Dev Biol 46: 201–207, 2002 [PubMed] [Google Scholar]
- 2.Cardozo AK, Berthou L, Kruhoffer M, Orntoft T, Nicolls MR, Eizirik DL: Gene microarray study corroborates proteomic findings in rodent islet cells. J Proteome Res 2: 553–555, 2003 [DOI] [PubMed] [Google Scholar]
- 3.Gu G, Wells JM, Dombkowski D, Preffer F, Aronow B, Melton DA: Global expression analysis of gene regulatory pathways during endocrine pancreatic development. Development 131: 165–179, 2004 [DOI] [PubMed] [Google Scholar]
- 4.Kaestner KH, Lee CS, Scearce LM, Brestelli JE, Arsenlis A, Le PP, Lantz KA, Crabtree J, Pizarro A, Mazzarelli J, Pinney D, Fischer S, Manduchi E, Stoeckert CJ, Jr, Gradwohl G, Clifton SW, Brown JR, Inoue H, Cras-Meneur C, Permutt MA: Transcriptional program of the endocrine pancreas in mice and humans. Diabetes 52: 1604–1610, 2003 [DOI] [PubMed] [Google Scholar]
- 5.Gradwohl G, Dierich A, LeMeur M, Guillemot F: Neurogenin3 is required for the development of the four endocrine cell lineages of the pancreas. Proc Natl Acad Sci U S A 97: 1607–1611, 2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gu G, Dubauskaite J, Melton DA: Direct evidence for the pancreatic lineage: NGN3+ cells are islet progenitors and are distinct from duct progenitors. Development 129: 2447–2457, 2002 [DOI] [PubMed] [Google Scholar]
- 7.Powers AC, Efrat S, Mojsov S, Spector D, Habener JF, Hanahan D: Proglucagon processing similar to normal islets in pancreatic α-like cell line derived from transgenic mouse tumor. Diabetes 39: 406–414, 1990 [DOI] [PubMed] [Google Scholar]
- 8.Sladek R, Rocheleau G, Rung J, Dina C, Shen L, Serre D, Boutin P, Vincent D, Belisle A, Hadjadj S, Balkau B, Heude B, Charpentier G, Hudson TJ, Montpetit A, Pshezhetsky AV, Prentki M, Posner BI, Balding DJ, Meyre D, Polychronakos C, Froguel P: A genome-wide association study identifies novel risk loci for type 2 diabetes. Nature 445: 881–885, 2007 [DOI] [PubMed] [Google Scholar]
- 9.Wenzlau JM, Juhl K, Yu L, Moua O, Sarkar SA, Gottlieb P, Rewers M, Eisenbarth GS, Jensen J, Davidson HW, Hutton JC: The cation efflux transporter ZnT8 (Slc30A8) is a major autoantigen in human type 1 diabetes. Proc Natl Acad Sci U S A 104: 17040–17045, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Efrat S, Linde S, Kofod H, Spector D, Delannoy M, Grant S, Hanahan D, Baekkeskov S: Beta-cell lines derived from transgenic mice expressing a hybrid insulin gene-oncogene. Proc Natl Acad Sci U S A 85: 9037–9045, 1988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jensen J, Pedersen EE, Galante P, Hald J, Heller RS, Ishibashi M, Kageyama R, Guillemot F, Serup P, Madsen OD: Control of endodermal endocrine development by Hes-1. Nat Genet 24: 36–44, 2000 [DOI] [PubMed] [Google Scholar]
- 12.Pictet R, Rutter WJ, Steiner DF, Freinkel N: Development of the embryonic endocrine pancreas. In Handbook of Physiology, Section 7: Endocrinology. Vol. 1. Washington, DC, American Physiological Society, 1972, p. 25
- 13.Clark K, Hammond E, Rabbitts P: Temporal and spatial expression of two isoforms of the Dutt1/Robo1 gene in mouse development. FEBS Lett 523: 12, 2002 [DOI] [PubMed] [Google Scholar]
- 14.Jia L, Cheng L, Raper J: Slit/Robo signaling is necessary to confine early neural crest cells to the ventral migratory pathway in the trunk. Dev Biol 282: 411–421, 2005 [DOI] [PubMed] [Google Scholar]
- 15.Sundaresan V, Roberts I, Bateman A, Bankier A, Sheppard M, Hobbs C, Xiong J, Minna J, Latif F, Lerman M, Rabbitts P: The DUTT1 gene, a novel NCAM family member is expressed in developing murine neural tissues and has an unusually broad pattern of expression. Mol Cell Neurosci 11: 29–35, 1998 [DOI] [PubMed] [Google Scholar]
- 16.Greenberg JM, Thompson FY, Brooks SK, Shannon JM, Akeson AL: Slit and robo expression in the developing mouse lung. Dev Dyn 230: 350–360, 2004 [DOI] [PubMed] [Google Scholar]
- 17.Xu Y, Wang S, Zhang J, Zhao A, Stanger BZ, Gu G: The fringe molecules induce endocrine differentiation in embryonic endoderm by activating cMyt1/cMyt3. Dev Biol 297: 340–349, 2006 [DOI] [PubMed] [Google Scholar]
- 18.Towle HC: Glucose as a regulator of eukaryotic gene transcription. Trends Endocrinol Metab 16: 489–494, 2005 [DOI] [PubMed] [Google Scholar]
- 19.Carraro G, Albertin G, Bova S, Malendowicz LK, Belloni AS, Nussdorfer GG: Differential expression of adrenomedullin and its receptors in newborn and adult rat thymus. Int J Mol Med 10: 767–771, 2002 [PubMed] [Google Scholar]
- 20.White P, May CL, Lamounier RN, Brestelli JE, Kaestner KH: Defining pancreatic endocrine precursors and their descendants. Diabetes 57: 654–668, 2008 [DOI] [PubMed] [Google Scholar]
- 21.Ahnfelt-Ronne J, Hald J, Bodker A, Yassin H, Serup P, Hecksher-Sorensen J: Preservation of proliferating pancreatic progenitor cells by Delta-Notch signaling in the embryonic chicken pancreas. BMC Dev Biol 7: 63–70, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gyulkhandanyan AV, Lu H, Lee SC, Bhattacharjee A, Wijesekara N, Fox JE, MacDonald PE, Chimienti F, Dai FF, Wheeler MB: Investigation of transport mechanisms and regulation of intracellular Zn2+ in pancreatic alpha-cells. J Biol Chem 283: 10184–10197, 2008 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.