Abstract
OBJECTIVE—Identification of arterial genes and pathways altered in obesity and diabetes.
RESEARCH DESIGN AND METHODS—Aortic gene expression profiles of obese and diabetic db/db, high-fat diet–fed C57BL/6J, and control mice were obtained using mouse Affymetrix arrays. Neuronatin (Nnat) was selected for further analysis. To determine the function of Nnat, a recombinant adenovirus (Ad-Nnat) was used to overexpress the Nnat gene in primary endothelial cells and in the mouse aorta in vivo.
RESULTS—Nnat, a gene of unknown vascular function, was upregulated in the aortas of db/db and high-fat diet–fed mice. Nnat gene expression was increased in db/db mouse aorta endothelial cells. Nnat protein was localized to aortic endothelium and was selectively increased in the endothelium of db/db mice. Infection of primary human aortic endothelial cells (HAECs) with Ad-Nnat increased expression of a panel of nuclear factor-κB (NF-κB)-regulated genes, including inflammatory cytokines, chemokines, and cell adhesion molecules. Infection of mouse carotid arteries in vivo with the Ad-Nnat increased expression of vascular cell adhesion molecule 1 protein. Nnat activation of NF-κB and inflammatory gene expression in HAECs was mediated through pathways distinct from tumor necrosis factor-α. Nnat expression stimulated p38, Jun NH2-terminal kinase, extracellular signal–related kinase, and AKT kinase phosphorylation. Phosphatidylinositol 3-kinase and p38 inhibitors prevented Nnat-mediated activation of NF-κB–induced gene expression.
CONCLUSIONS—Nnat expression is increased in endothelial cells of obese and diabetic mouse blood vessels. The effects of Nnat on inflammatory pathways in vitro and in vivo suggest a pathophysiological role of this new gene in diabetic vascular diseases.
Epidemiological data has shown a strong association between diabetes and coronary heart disease (1,2). Although insulin-mediated improved glucose control reduced cardiovascular events in subjects with type 1 diabetes (3), less definitive information is available relating diabetes control and atherosclerosis prevention in type 2 diabetes. Recent findings suggest that hyperglycemia is associated with increased arterial wall inflammation (4) and increased expression of vascular inflammatory molecules, such as vascular cell adhesion molecule-1 (VCAM-1), intercellular adhesion molecule-1 (ICAM-1), and E-Selectin (SELE) (5,6); nuclear factor-κB (NF-κB) activation (7,8); and inflammatory cytokine production. Fundamental understanding of the effects of diabetes on arterial genes and pathways may contribute to the discovery of new strategies for the treatment of diabetic vascular diseases beyond blood glucose control.
Animal models of diabetes have been used to study the effects of hyperglycemia and insulin resistance at different stages of disease progression (9,10). We (11,12) and others (13,14) have demonstrated that mouse models of type 2 diabetes such as leptin receptor mutant db/db and diet-induced obesity mice have impaired vascular function. Kim et al. (15) demonstrated that high-fat diet feeding increases expression of markers of vascular inflammation in mouse thoracic aortas. Also, apolipoprotein E knockout db/db mice have increased VCAM-1 expression in aorta (16) and greater aortic sinus atherosclerosis (16,17). However, not all investigators have observed changes in expression of adhesion molecules in db/db mouse aortas (18).
A number of studies have focused on the effects of diabetes on vascular cells. Endothelial cells isolated from db/db aortas have increased inflammatory cytokine and chemokine expression and more monocyte adhesion (19–21). The reasons for the altered biology of these cells is thought by many to be due to hyperglycemia; increased glucose concentrations induce interleukin (IL)-6, IL-8, and monocyte chemotactic protein-1 (MCP-1) secretion and adhesion molecule expression in endothelial cells. IL-6/IL-6Rα complex can induce an inflammatory phenotype in endothelial cells, promoting SELE, ICAM-1, and VCAM-1 expression and monocyte adhesion (21,22).
In the current study, we performed gene expression profiling of aortas from two mouse models of type 2 diabetes to identify new genes and pathways that contribute to diabetic vascular diseases. We found that neuronatin (Nnat), a gene with unknown vascular function, was upregulated in the aortas of both db/db and high-fat diet–fed mice. Immunohistochemical studies localized Nnat to the vascular endothelium. To gain insight into the function of this molecule, the effects of adenovirus-induced Nnat expression in human aortic endothelial cells (HAECs) and mouse arteries were studied.
RESEARCH DESIGN AND METHODS
All procedures were approved by the institutional animal care and use committee. Male C57BL/6J (strain 000664), db/db (Leprdb/db strain 000642), and heterozygous littermate (db/+) mice were purchased from The Jackson Laboratories (Bar Harbor, ME). Mice were maintained in a temperature-controlled barrier facility with a 12-h light/dark cycle and were given free access to food and water. db/db mice were killed at 16–20 weeks of age. To induce obesity and diabetes, C57BL/6J mice were fed a high-fat TD03584 diet (Harlan Tekland, Indianapolis, IN) for 16–20 weeks starting at the age of 10 weeks. Control mice were fed regular chow. The high-fat diet contained 35% fat and 37% carbohydrate.
Mouse aorta endothelial cell isolation.
Endothelial cells were isolated by sorting with magnetic beads using anti–platelet-endothelial cell adhesion molecule-1 (CD31) biotin-conjugated antibody (Millipore). In brief, mice were anesthetized and perfused with 1,000 units/ml heparin in PBS. Aortas were isolated, stripped of perivascular fat, and incubated in RPMI containing 2 mg/ml collagenase 2 and 2 units/ml dispase for 30 min at 37°C. Cells were collected by centrifugation at 400g at 4°C and washed twice with MACS (magnetic cell sorting) buffer (Hanks’ balanced salt solution [Ca/Mg free], 2 mmol/l EDTA, and 0.5% BSA). The cells suspension was run through 70- and 40-μm cell strainers. Cells were incubated with CD31 antibody for 15 min, washed and incubated with streptavidin SA-Microbeads (Miltenyi Biotec) for another 15 min, washed again, and applied to an equilibrated column. After several washes, the column was removed from the magnetic holder, and cells were eluted with MACS buffer. Endothelial cells were collected by centrifugation at 400g and processed for RNA isolation.
Expression of adenovirus-encoded Nnat in endothelial cells.
HAECs were obtained from Cambrex Life Science. Cells were grown in endothelial cell growth medium EGM-2 to confluency and were infected with Ad-Nnat, Ad–green fluorescent protein (Ad-GFP), or Ad-Empty at multiplicity of infection (MOI) 10 unless indicated otherwise. Gene expression was assessed by real-time PCR, and proteins were analyzed by Western blot 48 h after infection. For treatment with tumor necrosis factor-α (TNF-α), TNF-α–blocking antibody, or kinase inhibitors, cells were treated 24 h after infection for another 24 h. Kinase inhibitors used in the study are 20 μmol/l U0126, 20 μmol/l SP600125, 5 μmol/l IKK-VII, 10 μmol/l SB202190, 50 μmol/l LY294002, and 0.2 μmol/l wortmannin.
Real-time PCR.
Primers for conventional real-time PCR were designed (supplementary Table S1, available in an online appendix at http://dx.doi.org/10.2337/db07-1746) or obtained from SuperArray Bioscience (Frederick, MD). Expression levels for individual genes were normalized to β-actin in mouse tissue and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) in HAECs. For RT2 Profiler PCR array, 1 μg RNA pool from HAECs infected with adenovirus was used for cDNA synthesis and analysis by human endothelial cell biology RT2 Profiler PCR array according to manufacturer's instructions (SuperArray Bioscience).
Western blot analysis.
Cell homogenates were made in buffer (10 mmol/l Tris-Cl, pH 7.4; 5 mmol/l EDTA; 150 mmol/l NaCl; 10% glycerol; and 1% Triton X-100) containing a protease inhibitor cocktail for mammalian tissues (Sigma-Aldrich, St. Louis, MO) and phosphatase inhibitors (Roche Applied Science, Indianapolis, IN). Antibodies used are as follows: inhibitor of κB (IκB)-α, ICAM-1, and VCAM-1 (Santa Cruz Biotechnology, Santa Cruz, CA); stress-activated protein kinase (SAPK)/Jun NH2-terminal kinase (JNK), p38, extracellular signal–related kinase 1/2 (ERK1/2), NF-κB activating kinase (NAK), p65, and Akt (Cell Signaling Technology); SELE (BioVision); β-actin (ABcam); and TNF-α (BioLegend).
Gene transfer into mouse carotid artery.
According to the method of Vassalli et al. (23), in vivo gene transfer was conducted using Ad-GFP or Ad-Nnat virus. Two days after the adenovirus infection, carotid arteries were isolated and fixed in 4% paraformaldehyde and processed for immunohistochemical analysis as described above. Intensity of staining was determined in sections from 4 to 7 Ad-Nnat–and Ad-GFP–infected mouse carotid arteries using Image Pro Plus software.
RESULTS
Gene expression profiling of aortas from diabetic mice.
Gene expression profiling was performed to identify differentially expressed genes in the aortas of diabetic mice. Metabolic data for db/db and high-fat diet–fed C67BL6/J mice are shown in Table 1. To create mice with comparable weights and insulin levels, we used 16- to 18-week-old db/db mice and ∼28-week-old high-fat diet–fed animals. Glucose levels were higher in db/db compared with high-fat diet–fed mice, but body weight (47 and 49 g, respectively) and insulin (912 and 775 pmol/l, respectively) were comparable between the two groups. As expected, cholesterol levels were greater in db/db than db/+ mice. Expression of 43 genes increased and expression of 3 genes decreased at least twofold in aortas of db/db mice when compared with nondiabetic db/+ controls (supplementary Table S2). In high-fat diet–fed mice, 85 aortic genes were upregulated, and 42 aortic genes were downregulated (supplementary Table S3). Analysis of gene expression across the two diabetic models revealed 10 genes that were differentially expressed in the aortas of diabetic mice (Table 2), but there were only 4 genes in which the direction of change was similar. These genes were Nnat, leptin, natriuretic peptide receptor 3, and lectin galactose binding, soluble 3. Among these common genes, the most strongly upregulated gene was Nnat. Nnat mRNA was upregulated 9-fold in the db/db and 2.5-fold in the high-fat diet–induced model. Therefore, we elected to focus our subsequent studies on identifying a role for Nnat in the aorta of diabetic mice.
TABLE 1.
Metabolic data for db/db and diet-induced obesity and diabetes models
| Mice | Weight (g) | Glucose (mmol/l) | Insulin (pmol/l) | TC (mg/dl) | TG (mg/dl) |
|---|---|---|---|---|---|
| Chow | 29.3 ± 0.57 | 7.4 ± 0.11 | 86 ± 5.2 | 132 ± 6.1 | 17 ± 1.9 |
| High fat | 49.3 ± 0.52* | 12.7 ± 0.83* | 775 ± 132.5* | 149 ± 8.9 | 16 ± 2.2 |
| db/+ | 28.2 ± 0.29 | 6.4 ± 0.22 | 121 ± 15.5 | 132 ± 8 | 32 ± 4 |
| db/db | 47.3 ± 0.54* | 21.2 ± 1.02* | 912 ± 141.1* | 194 ± 7* | 43 ± 7 |
Data are means ± SE. Blood from db/db mice was collected at 14–19 weeks of age (n = 18). C57BL6/J mice with diet-induced obesity and diabetes were studied after 6–20 weeks on mice on high-fat diet (n = 9). Triglyceride (TG) and total cholesterol (TC) measurements done were done at 9 weeks.
P < 0.0001 data comparison were between db/db vs. db/+ and high-fat vs. chow-fat mice.
TABLE 2.
Genes changed in two models of obesity and type 2 diabetes
| Gene name | Gene symbol | Fold change |
|
|---|---|---|---|
| db/db | DIO | ||
| Neuronatin | Nnat | 9.4 | 2.5 |
| Leptin | Lep | 4.6 | 3.7 |
| Natriuretic peptide receptor 3 | Npr3 | 2.3 | 4.9 |
| Lectin, galactose binding, soluble 3 | Lgals3 | 2.2 | 3.5 |
| Transthyretin | Ttr | 4.0 | 0.4 |
| Apolipoprotein A-II | Apoa2 | 3.8 | 0.4 |
| Fatty acid binding protein 1 | Fabp1 | 3.4 | 0.4 |
| Cytochrome P450, family 3, subfamily a, 11 | Cyp3a11 | 3.1 | 0.4 |
| Group-specific component | Gc | 3.1 | 0.4 |
| Serine (or cysteine) peptidase inhibitor, clade A, 1c | Serpina1c | 2.8 | 0.2 |
Fold change represent values obtained after data analysis with Wilcoxon test. DIO, diet-induced obesity and diabetes. See supplemental research design and methods in the online appendix for the details of the data analysis.
Nnat was upregulated in the aortic endothelium of diabetic mice.
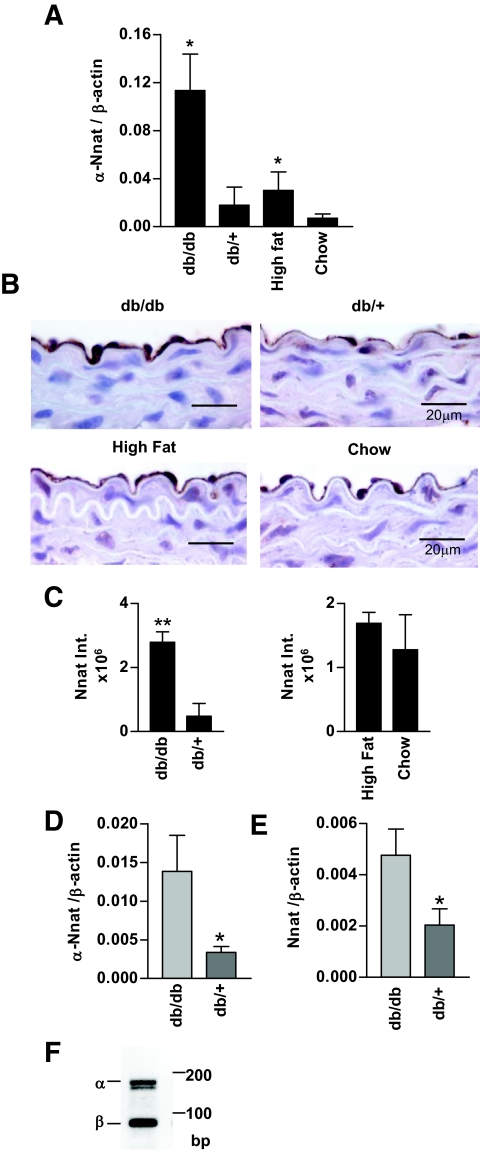
Real-time PCR was performed to quantify Nnat expression in the mouse aorta. Nnat mRNA expression was higher in the aorta of db/db and high-fat diet–fed diabetic mice when compared with littermate controls (Fig. 1A). Immunohistochemical staining of mouse aorta localized the Nnat protein in endothelial cells (Fig. 1B). Compared with control mice, the endothelium of db/db mice had more intense staining (Fig. 1B and C). The difference in Nnat staining was less prominent in the aortas from high-fat diet–fed compared with chow diet–fed control mice, and the difference was not statistically significant (Fig. 1B and C). Analysis of aorta sections stained with Nnat antibody revealed that not only endothelial cells but also perivascular fat stains positive for Nnat (supplementary Fig. S2).
FIG. 1.
Nnat expression in mouse aorta. A: Real-time PCR measurement of α-Nnat mRNA in the aortas of diabetic mice (C57BLKS/J db/db and high-fat diet–fed C57BL/6J) and their respective controls (C57BLKS/J db/+, and chow diet–fed C57BL/6J) (n = 4–6). Nnat expression levels were normalized to β-actin. B: Representative immunohistochemical staining of db/db versus db/+ (top) and high-fat diet–versus chow diet–fed (bottom) mouse aorta paraffin sections with Nnat antibody. Magnification ×100. C: Comparison of integrated intensities of Nnat staining determined using Image-Pro Plus software, n = 4. D and E: Real-time PCR data for α-Nnat (D) and total Nnat (E) expression in endothelial cells isolated form db/db or control db/+ mouse aortas, n = 5–6. F: Gel electrophoresis of PCR fragments amplified from endothelial cells RNA using primers for both α and β Nnat isoforms. **P < 0.01, *P < 0.05. (Please see http://dx.doi.org/10.2337/db07-1746 for a high-quality digital representation of this image.)
Nnat gene expression was increased in endothelial cells of db/db mouse aorta.
To reassure that the increase in Nnat is due to endothelial cell expression, we isolated mouse aorta endothelial cells using magnetic bead cell sorting with CD31 antibody. Endothelial cell isolation was confirmed by assessing endothelial-specific (endothelial nitric oxide [NO] synthase, vascular endothelial–cadherin), macrophage-specific (F4/80), and fibroblast- and smooth muscle cell–specific (Col3a) gene expression in CD31-positive and -negative cells (supplementary Fig. S2A). α-Nnat expression was increased approximately fourfold in the db/db endothelial cell preparations (Fig. 1D), whereas total nnat expression was increased only approximately twofold (Fig. 1E); the confirmation that this analysis detected both α- and β-isoforms of Nnat in mouse endothelial cells is shown in Fig. 1F. Because total Nnat expression changed less than α-Nnat, it is likely that β-Nnat expression did not change or decreased in these mice.
Nnat expression in cultured primary aortic endothelial cells.
The effects of Nnat expression were investigated in cultured HAECs. At baseline conditions, real-time PCR and Western blotting studies revealed low levels of transcript and no detectable protein in these cells (data not shown). To determine whether metabolic stimuli would induce Nnat expression, HAECs were cultured for 1–4 days in the presence of 25 mmol/l glucose, 100 ng/ml leptin, or 10 μg/ml insulin. These manipulations did not increase Nnat gene expression (data not shown). Because Nnat is an imprinted gene, it was possible that Nnat expression was repressed by DNA methylation. Treatment of HAECs with 5-Aza-2′-deoxycytidine (ADC), an inhibitor of DNA methyl transferase, increased Nnat mRNA levels (supplementary Figure S4A). Treatment of the cells with high-glucose insulin and leptin after induction of Nnat expression with ADC did not result in the further increase of Nnat (supplementary Figure S4B).
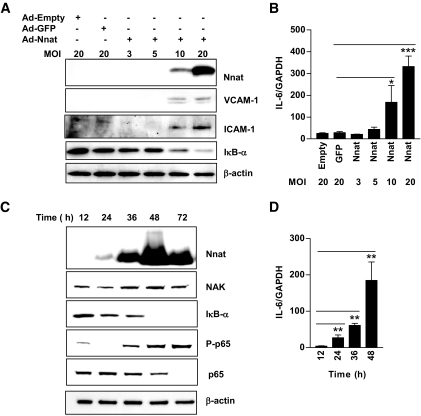
HAECs were infected with an Ad-Nnat to determine the downstream effects of Nnat expression. Western blotting revealed high levels of the Nnat protein in infected cells (Fig. 2A and C; see below). Because the function of Nnat in endothelium was unknown, we compared gene expression in endothelial cells infected with Ad-Nnat to that of an empty adenovirus control (Ad-Empty) using an endothelial cell biology RT2 Profiler PCR array. Of 84 endothelial-specific genes, 27 were differentially regulated in Ad-Nnat–infected cells. Twenty-one genes were increased, and six were decreased (supplementary Table S4). Among the increased genes, four major groups were apparent: 1) cytokines (IL-1β, IL-6, IL-7, IL-11, and TNF-α) and chemokines (CX3CL1 and CCL2); 2) adhesion molecules (ICAM-1, VCAM-1, and SELE); 3) growth factors (vascular endothelial growth factor, endothelial cell growth factor-1, and placental growth factor); and 4) extracellular matrix proteases/inhibitors (matrix metallopeptidase-1; serpin peptidase inhibitor, clade E, member-1; and plasminogen activator urokinase). The changes in TNF-α, IL-1β, IL-6, MCP-1 (CCL2), ICAM-1, VCAM-1, and SELE expression in Ad-Nnat–infected cells were verified by real-time PCR (Table 3). These results suggest that Nnat expression activates endothelial cells.
FIG. 2.
Nnat expression in HAECs activates NF-κB–dependent gene expression in time- and concentration-dependent manner. A: Western blots of Nnat, VCAM-1, ICAM-1, and IκB-α protein from endothelial cells infected with Ad-Empty, Ad-GFP, and increasing number of Ad-Nnat adenovirus per cell (MOI). B: Real-time PCR measurement of IL-6 mRNA expression in endothelial cells infected with Ad-Empty, Ad-GFP, or Ad-Nnat at different MOI. (n = 3). C: Endothelial cells were collected at various times after infection with Ad-Nnat and Western blots were probed with antibodies to Nnat, NAK, IκB-α, phospho-p65, p65, and β-actin. D: Time course of IL-6 mRNA expression after Ad-Nnat infection of endothelial cells (n = 3). IL-6 mRNA expression levels were normalized to GAPDH. ***P < 0.001, **P < 0.01, *P < 0.05. Data are a representative of three to four experiments.
TABLE 3.
Genes differentially expressed in HAECs after transduction of HAECs with Ad-Nnat
| Gene symbol | Gene name | Fold change | P value |
|---|---|---|---|
| Cytokine and chemokine | |||
| IL-6 | Interleukin 6 (interferon, β2) | 29.4 | <0.0001 |
| IL-1β | Interleukin 1β | 12 | <0.0001 |
| TNF | Tumor necrosis factor | 3.7 | <0.05 |
| Endothelial cell adhesion molecules and growth factors | |||
| MCP-1 | Chemokine (C-C motif) ligand 2 | 6.1 | <0.0001 |
| SELE | Selectin E | 221 | <0.001 |
| VCAM-1 | Vascular cell adhesion molecule 1 | 13 | <0.0001 |
| ICAM-1 | Intercellular adhesion molecule 1 (CD54) | 136 | <0.001 |
Data presented are representative of three to six experiments. Fold change in gene expression is determined as difference in mRNA levels for the gene of interest in Ad-Nnat–versus Ad-Empty–transduced HAECs. P values are determined using t test. n = 4.
Nnat expression activates NF-κB in endothelial cells.
Many genes induced by Nnat expression in HAECs are regulated by NF-κB. To study this, HAECs were infected with increasing numbers of Ad-Nnat particles per cell from MOI 3 to 20. At MOI 10–20, Nnat protein expression paralleled with IκB-α decrease and with ICAM-1 and VCAM-1 protein (Fig. 2A) and IL-6 mRNA increase (Fig. 2B). Time course analysis of Nnat expression also demonstrated that accumulation of Nnat protein was accompanied by an increase of immunoreactive NAK, a decrease in IκB-α, and an increase in phosphorylation of the p65 subunit of the NF-κB transcription complex (Fig. 2C). Increases in IL-6 accompanied the time-dependent increase in Nnat expression and activation of NF-κB (Fig. 2D). ICAM-1 and TNF-α mRNA expression also increased with time, and IL-1β and SELE were increased only after 48 h (data not shown).
Nnat- versus TNF-α–induced activation of NF-κB transcription factor.
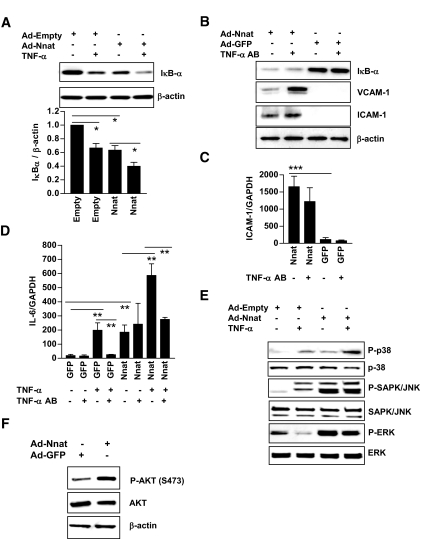
Additional experiments were performed to identify the signal transduction pathways by which Nnat activated NF-κB in endothelial cells. Because Nnat induced TNF-α mRNA expression (Table 3), we entertained the possibility that Nnat might induce inflammatory gene expression through stimulation of TNF-α release by the cell, leading to ligation of TNF-α cell surface receptors. Cells were first infected with Ad-Nnat to determine whether Nnat expression would result in activation of NF-κB in the presence of exogenous TNF-α. Treatment of Ad-Empty–infected HAECs with TNF-α decreased IκB-α protein level (Fig. 3A). Treatment of Nnat-expressing HAECs with TNF-α further decreased IκB-α protein.
FIG. 3.
TNF-α augments Nnat-induced activation of NF-κB. A: Western blot for IκB-α protein in HAECs infected with Ad-Nnat in the presence or absence of exogenous TNF-α (top). Quantitative densitometry of Western blot (bottom) for IκB-α protein expression was normalized to β-actin (n = 3). B: IκB-α and endothelial cell adhesion molecules VCAM-1 and ICAM-1 protein. C: ICAM-1 mRNA levels in endothelial cells infected with Ad-GFP or Ad-Nnat in the presence or absence of TNF-α neutralizing antibody. D: Effect of TNF-α blocking antibody on Nnat induced increase in cytokine IL-6 mRNA expression. Endothelial cells were infected with Ad-GFP or Ad-Nnat and treated with TNF-α or TNF-α neutralizing antibody. IL-6 and ICAM-1 expression levels were normalized to GAPDH. E: Western blot analysis of p38, SAPK/JNK1/2, and ERK1/2 MAPK phosphorylation in HAECs infected with Ad-Nnat or Ad-Empty in the presence or absence of exogenous TNF-α. F: Western blot analysis of Akt phosphorylation: P-AKT (S473), AKT, and β-actin. ***P < 0.001; **P < 0.01; *P < 0.05. Data are a representative of three to four experiments.
We then removed any cellular TNF-α using a neutralizing antibody. This did not block the Nnat-mediated decrease in IκB-α protein (Fig. 3B), increase in ICAM-1 mRNA, or increase in ICAM-1 and VCAM-1 protein expression (Fig. 3B and C). Expression of VCAM-1 was increased with antibody treatment; VCAM-1 shedding was decreased by TNF-α absence in the culture media (24).
IL-6 mRNA expression, another read-out for NF-κB activation, was increased by TNF-α, and this was further augmented by Ad-Nnat viral infection (Fig. 3D). TNF-α neutralizing antibody blocked the TNF-α induced increase in IL-6 gene expression but had no effect on Nnat induced IL-6 gene expression. These data indicate that Nnat activates the NF-κB transcription factor and induces inflammatory gene expression by a pathway that does not require cell surface TNF-α receptor activation.
Nnat expression in HAEC-activated kinases.
As we have shown above, Nnat expression increased NAK and phospho-p65 and decreased IκB-α protein. To learn what kinases were involved in the signal transduction from Nnat to NAK or p65 phosphorylation, we looked at mitogen-activated protein kinase (MAPK) activation and compared it with that of TNF-α. Nnat expression induced phosphorylation of p38, JNK, and ERK kinase (Fig. 3E). TNF-α treatment of HAECs induced phosphorylation of p38 and JNK kinase but decreased Nnat-induced ERK phosphorylation. Nnat expression in HAECs also induced Akt phosphorylation (Fig. 3F), suggesting a role in activation of phosphatidylinositol 3-kinase (PI 3-kinase).
Nnat induced signal transduction in endothelial cells.
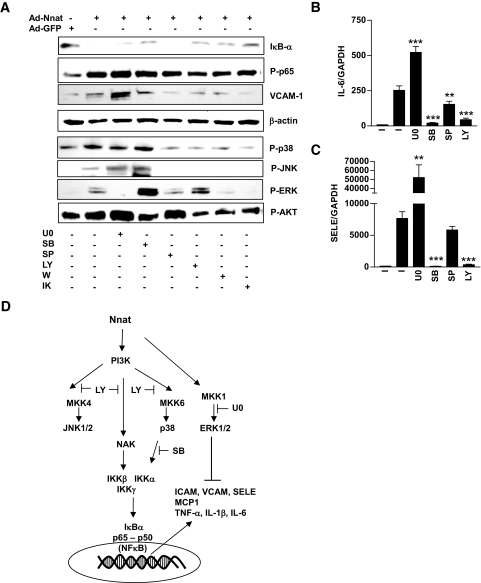
To test whether Nnat-induced kinase phosphorylation was involved in activation of NF-κB, we treated Ad-Nnat–infected endothelial cells with p38 (SB202190), JNK1,2,3 (SP600125), ERK1/2 kinase (U0126), PI 3-kinase (LY294002 and wortmannin), or IκB kinase (IKK) (IKK-VII) inhibitors (Fig. 4). SB202190, LY294002, wortmannin, and IKKVII blocked the Nnat-induced IκB-α degradation and increase in VCAM-1 protein. U0126 had the opposite effect; it further increased VCAM-1 expression. SP600125 deceased VCAM-1 protein expression without affecting IκB-α protein levels. LY294002 and wortmannin partially decreased phosphorylation of p65 subunit. SB202190, SP600125, and IKK-VII had no effect. Both LY294002 and SB202190 blocked (∼95%) the increase in IL-6 and SELE gene expression in Ad-Nnat–infected endothelial cells (Fig. 4B and C). U0126 had the opposite effect and increased IL-6 and SELE expression. SP600125 inhibitor only partially (∼40%) blocked the Nnat-mediated increase in IL-6 but not SELE expression. U0126 inhibited ERK phosphorylation and increased p-JNK and p-AKT. SB202190 decreased AKT and increased ERK phosphorylation. SP600125 decreased JNK and p38 phosphorylation without having an effect on others. This inhibitor is able to inhibit kinases upstream of p38 and JNK. We could speculate that partial inhibition of IL-6 but not SELE expression by this compound is a result of blocking kinase upstream from p38 but not JNK itself. LY294002 and wortmannin blocked Nnat-induced phosphorylation of NAK, p38, and JNK, suggesting that these kinases are downstream from PI 3-kinase. IKK-VII treatment blocked IκB-α degradation and did not effect p65 phosphorylation. On the other hand, IKK-VII inhibited phosphorylation of all three MAPKs. As summarized in Fig. 4D, Nnat effects on NF-κB appear to be mediated via PI 3-kinase and p38 and inhibited by ERK1/2.
FIG. 4.
Effect of kinase inhibitors on Nnat-induced NF-κB activation. A: Western blot analysis of Iκ B-α, Phospho-p65 (P-p65), VCAM-1, β-actin P-p38, P-JNK1/2, P-ERK1/2, and P-AKT protein levels in the presence of kinase inhibitors. IL-6 (B) and SELE (C) mRNA expression in HAECs infected with Ad-Nnat. Effect of kinase inhibitors on Nnat-induced increase in IL-6 and SELE expression levels, normalized to GAPDH. I, no inhibitor; U0, U0126; SB, SB202190; SP, SP600125; LY, LY294002; W, Wortmannin; and IK, IKK-VII. ***P < 0.001, **P < 0.01. Data are representative of three to four experiments. D: Schematic describing Nnat-induced signal transduction in endothelial cells. Inhibitors of PI 3-kinase and p38 completed blocked Nnat-induced NF-κB activation, JNK1/2 inhibition led to a partial block, and inhibition of ERK1/2 increased NF-κB activation.
Adenovirus-induced expression of Nnat in mouse carotid arteries.
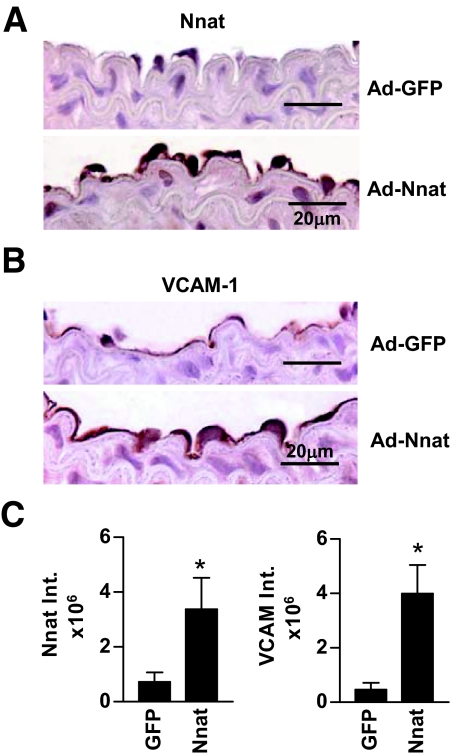
To test whether Nnat can induce adhesion molecule expression in endothelium in vivo, Ad-Nnat or Ad-GFP was injected into carotid arteries of C57BL/6J wild-type mice. Mouse arteries were harvested 2 days after infection, and arterial sections were processed for determination of immunoreactive Nnat and VCAM-1. Ad-Nnat increased the intensity of staining for the Nnat protein in the endothelium approximately fourfold. Staining for VCAM-1 increased by ninefold compared with staining of arteries that had been infected with Ad-GFP (Fig. 5). These data demonstrate that expression of Nnat in endothelium induces endothelial expression of adhesion molecules in vivo.
FIG. 5.
Adenovirus-mediated expression of Nnat in mouse carotid arteries induced expression of VCAM-1. Immunohistochemical staining of C57BL/6J mouse carotid arteries for Nnat (A) and VCAM-1 (B) protein after incubation of blood vessels with Ad-Nnat or Ad-GFP adenovirus. C: Integrated intensity of endothelial cell staining. n = 4–7; *P < 0.05. (Please see http://dx.doi.org/10.2337/db07-1746 for a high-quality digital representation of this image.)
DISCUSSION
In the present study, using Affymetrix arrays, we found that Nnat, a gene with unknown vascular functions, was upregulated in both db/db and high-fat diet–fed mouse aortas. Diabetes-associated changes in gene expression were quite different between the two mouse models. Although insulin resistance was present in both db/db and high-fat diet–fed mice, the db/db mouse has higher glucose levels. The degree of metabolic abnormalities, the duration of diabetes (longer in db/db), and the effect of the high-fat diet may be responsible for the differences in gene expression across the two diabetic models. Some gene expression changes found only in db/db aortas are known to be associated with obesity or diabetes. Angiopoietin-like protein 4 (ANGPTL4), which is increased in the db/db aorta, inhibits lipoprotein lipase activity in vitro (25). ANGPTL4 also markedly inhibits the proliferation, chemotaxis, and tubule formation of endothelial cells (26). Two genes, other than leptin and Nnat, were increased in both models of obesity and insulin resistance: lectin, galactose binding, soluble 3, galectin-3 (LGALS3) and natriuretic peptide receptor 3 (Npr3). LGALS3 produces a receptor that interacts with advanced glycation end products (27). Npr3 encodes for a receptor for natriuretic peptides, which are important in the maintenance of blood pressure and extracellular fluid volume (28). Another gene differentially expressed in this model is transthyretin. Blood levels of transthyretin are increased in serum of humans with type 2 diabetes (29).
Immunohistochemical staining revealed that Nnat was expressed on the vascular endothelium. The function of Nnat in vascular tissue is unknown. Previous studies identified Nnat mRNA in the developing rat brain (30). Nnat is an imprinted gene (31) and is expressed in neonatal mouse brain, eye, bladder, colon, and lung; mouse and human pituitary (32); and mouse pancreatic β-cell lines (33). Nnat expression in pancreas appears to be regulated by the β2 transcription factor, also known as NeuroD1. Knockdown of Nnat with small interfering RNA in the pancreatic β-cell line NIT1 deceased glucose-stimulated insulin secretion (34).
It is not clear why Nnat expression was increased in endothelial cells of obese and diabetic mice. Nnat is an imprinted gene because its expression should be increased by hypomethylation. Recent studies by several groups found changes in concentrations of S-adenosylmethionine (SAM) and S-adenosylhomocysteine (SAH) associated with diabetes in human subjects and in animal models (35,36). SAM is a universal donor of methyl groups. Increased intracellular SAH levels are associated with DNA hypomethylation in human umbilical vein endothelial cells (37). Patients with vascular disease had significantly higher plasma SAH concentrations and significantly lower plasma SAM-to-SAH ratios and genomic DNA methylation (38). Glycine N-methyltransferase (GNMT) is the enzyme that converts SAM to SAH. We found a dramatic increase in GNMT in aortas of db/db versus control mice (supplementary Table 2) and speculated that the elevated levels of this enzyme increased SAH levels and caused DNA hypomethylation and greater Nnat expression. We found that treatment of HAECs with DNA methyl transferase inhibitor ADC increased Nnat transcript expression. ADC also induced Nnat expression in HepG2 cells (39). It is interesting to note that the leptin gene, which was also dramatically increased in aortas, is regulated by promoter methylation (40) and could also have been induced by hypomethylation.
To investigate the function of Nnat in endothelial cells, Nnat was expressed in HAECs using adenovirus mediated gene transfer. Nnat expression induced a gene expression pattern characteristic of endothelial cell activation with increased inflammatory cytokines (TNF-α, IL-1β, and IL-6), endothelial cell adhesion molecules (ICAM-1, VCAM-1, and SELE), and chemokine (MCP-1) expression. Nnat expression also induced VCAM-1 expression in the mouse carotid artery. ICAM-1 expression was variable and did not show a statistically significant increase (data not shown). It is possible that the increased Nnat in the arteries of the diabetic mice increased inflammatory responses under certain conditions.
Cell adhesion molecule and inflammatory cytokine gene expression is regulated by NF-κB. Expression of Nnat in HAECs increased expression of NAK, decreased IκB-α protein, and increased phosphorylation of the p65 subunit, consistent with the activation of the NF-κB transcription complex. TNF-α treatment of endothelial cells activates JNK1/2 and p38 MAPK (41–43). In our study, TNF-α increased JNK1/2 and p38 and inhibited Nnat-induced ERK1/2 phosphorylation. To understand whether Nnat overexpression directly induced degradation of IκB-α or increased TNF-α triggered activation of other NF-κB–regulated genes, HAECs were transduced with Ad-Nnat and incubated in medium containing TNF-α antibody. TNF-α antibody did not block Nnat-induced activation of gene expression. Therefore, Nnat effects do not appear to involve extracellular TNF-α interaction with cell surface receptors.
We attempted to define the pathway whereby Nnat induced inflammatory gene expression. We showed that Nnat expression in HAECs induced phosphorylation of p38, JNK, ERK, and Akt. Increased Akt suggested involvement of PI 3-kinase in signal transduction from Nnat to NF-κB. PI 3-kinase inhibitors blocked Nnat-induced activation of IL-6 and SELE gene and VCAM-1 protein expression. SB202190, an inhibitor of p38 kinase, also blocked Nnat-induced expression of IL-6 and SELE genes. Shedding of VCAM-1 protein from cell membrane is regulated by p38 kinase (24); therefore, the seemingly paradoxical increase in VCAM-1 protein by SB202190 might be secondary to blocked shedding of this protein from the cell surface. As expected, VCAM-1 expression was blocked by an IKK inhibitor. Inhibition of JNK had no effect on SELE but decreased IL-6 mRNA and VCAM-1 protein expression.
NF-κB signaling can be inhibited at several levels, including phosphorylation of Iκ B-α and p65. Nnat-induced decrease in IκB-α protein level was reversed by IKK, p38, and PI 3-kinase inhibitors. Blocking IKK or p38 had no effect on phosphorylation of p65 subunit, whereas JNK1/2 and PI 3-kinase inhibitors decreased P-p65. Even though Nnat increased JNK phosphorylation, the role of this kinase in signal transduction from Nnat to NF-κB is not clear. Partial inhibition of gene expression by SP600125 could have resulted from decrease in p65 phosphorylation. It is also possible that JNK activates other transcription factors that work in concert with NF-κB.
In contrast, inhibition of ERK kinase augmented Nnat-induced activation of NF-κB–regulated gene (IL-6 and SELE) and protein (VCAM-1) expression. Several investigators found inhibitory effects of ERK on NF-κB transcription factor–regulated gene expression; ERK inhibits IKK activity (44), p65 subunit phosphorylation (45), and TATA binding protein phosphorylation (46).
In summary, using a gene discovery approach, we identified Nnat as a gene upregulated in the aortas of two mouse models of obesity and type 2 diabetes, db/db and high-fat–fed mice. The Nnat protein was localized to the aortic endothelium. Overexpression of Nnat in HAECs or in mouse carotid artery increased endothelial cell adhesion molecule expression. This effect of Nnat was mediated through PI 3-kinase/p38–dependent activation of NF-κB. Nnat is an imprinted gene, and its expression in diabetes might reflect alterations in the methylation status of endothelial cells. We conclude that Nnat is a candidate molecule involved in inflammatory pathways associated with obesity and insulin resistance. Experiments to alter Nnat gene expression in genetically modified mice are underway to assess whether Nnat expression will promote endothelial dysfunction and/or accelerate the development of atherosclerosis.
Supplementary Material
Acknowledgments
This work has received National Institutes of Health Grant HL073191 and funding from the Arlene and Arnold Goldstein Foundation.
Parts of this study were presented at the Arteriosclerosis, Thrombosis, and Vascular Biology Annual Conference, Chicago, Illinois, 19–21 April 2007.
Published ahead of print at http://diabetes.diabetesjournals.org on 30 June 2008.
H.M.D. is currently Director of Clinical Research, Cardiovascular Disease, Merck Research Laboratories, Rahway, New Jersey.
These studies were begun under the supervision of H.M.D. and completed with supervision from I.G.
The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
REFERENCES
- 1.Haffner SM, Lehto S, Ronnemaa T, Pyorala K, Laakso M: Mortality from coronary heart disease in subjects with type 2 diabetes and in nondiabetic subjects with and without prior myocardial infarction. N Engl J Med 339: 229–234, 1998 [DOI] [PubMed] [Google Scholar]
- 2.Grundy SM, Benjamin IJ, Burke GL, Chait A, Eckel RH, Howard BV, Mitch W, Smith SC Jr, Sowers JR: Diabetes and cardiovascular disease: a statement for healthcare professionals from the American Heart Association. Circulation 100: 1134–1146, 1999 [DOI] [PubMed] [Google Scholar]
- 3.Nathan DM, Lachin J, Cleary P, Orchard T, Brillon DJ, Backlund JY, O'Leary DH, Genuth S: Intensive diabetes therapy and carotid intima-media thickness in type 1 diabetes mellitus. N Engl J Med 348: 2294–2303, 2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Basta G, Schmidt AM, De Caterina R: Advanced glycation end products and vascular inflammation: implications for accelerated atherosclerosis in diabetes. Cardiovasc Res 63: 582–592, 2004 [DOI] [PubMed] [Google Scholar]
- 5.Lim SC, Caballero AE, Smakowski P, LoGerfo FW, Horton ES, Veves A: Soluble intercellular adhesion molecule, vascular cell adhesion molecule, and impaired microvascular reactivity are early markers of vasculopathy in type 2 diabetic individuals without microalbuminuria. Diabetes Care 22: 1865–1870, 1999 [DOI] [PubMed] [Google Scholar]
- 6.Bloomgarden ZT: Inflammation, atherosclerosis, and aspects of insulin action. Diabetes Care 28: 2312–2319, 2005 [DOI] [PubMed] [Google Scholar]
- 7.Rothlein R, Czajkowski M, O'Neill MM, Marlin SD, Mainolfi E, Merluzzi VJ: Induction of intercellular adhesion molecule 1 on primary and continuous cell lines by pro-inflammatory cytokines: regulation by pharmacologic agents and neutralizing antibodies. J Immunol 141: 1665–1669, 1988 [PubMed] [Google Scholar]
- 8.Kim I, Moon SO, Kim SH, Kim HJ, Koh YS, Koh GY: Vascular endothelial growth factor expression of intercellular adhesion molecule 1 (ICAM-1), vascular cell adhesion molecule 1 (VCAM-1), and E-selectin through nuclear factor-kappa B activation in endothelial cells. J Biol Chem 276: 7614–7620, 2001 [DOI] [PubMed] [Google Scholar]
- 9.Schreyer SA, Wilson DL, LeBoeuf RC: C57BL/6 mice fed high fat diets as models for diabetes-accelerated atherosclerosis. Atherosclerosis 136: 17–24, 1998 [DOI] [PubMed] [Google Scholar]
- 10.Goldberg IJ, Dansky HM: Diabetic vascular disease: an experimental objective. Arterioscler Thromb Vasc Biol 26: 1693–1701, 2006 [DOI] [PubMed] [Google Scholar]
- 11.Molnar J, Yu S, Mzhavia N, Pau C, Chereshnev I, Dansky HM: Diabetes induces endothelial dysfunction but does not increase neointimal formation in high-fat diet fed C57BL/6J mice. Circ Res 96: 1178–1184, 2005 [DOI] [PubMed] [Google Scholar]
- 12.Salzberg SP, Filsoufi F, Anyanwu A, von Harbou K, Karlof E, Carpentier A, Dansky HM, Adams DH: Increased neointimal formation after surgical vein grafting in a murine model of type 2 diabetes. Circulation 114: I302–I307, 2006 [DOI] [PubMed] [Google Scholar]
- 13.Howarth AG, Wiehler WB, Pannirselvam M, Jiang Y, Berger JP, Severson D, Anderson TJ, Triggle CR: A nonthiazolidinedione peroxisome proliferator-activated receptor gamma agonist reverses endothelial dysfunction in diabetic (db/db−/−) mice. J Pharmacol Exp Ther 316: 364–370, 2006 [DOI] [PubMed] [Google Scholar]
- 14.Lagaud GJ, Masih-Khan E, Kai S, van Breemen C, Dube GP: Influence of type II diabetes on arterial tone and endothelial function in murine mesenteric resistance arteries. J Vasc Res 38: 578–589, 2001 [DOI] [PubMed] [Google Scholar]
- 15.Kim F, Pham M, Luttrell I, Bannerman DD, Tupper J, Thaler J, Hawn TR, Raines EW, Schwartz MW: Toll-like receptor-4 mediates vascular inflammation and insulin resistance in diet-induced obesity. Circ Res 100: 1589–1596, 2007 [DOI] [PubMed] [Google Scholar]
- 16.Wendt T, Harja E, Bucciarelli L, Qu W, Lu Y, Rong LL, Jenkins DG, Stein G, Schmidt AM, Yan SF: RAGE modulates vascular inflammation and atherosclerosis in a murine model of type 2 diabetes. Atherosclerosis 185: 70–77, 2006 [DOI] [PubMed] [Google Scholar]
- 17.Wu KK, Wu TJ, Chin J, Mitnaul LJ, Hernandez M, Cai TQ, Ren N, Waters MG, Wright SD, Cheng K: Increased hypercholesterolemia and atherosclerosis in mice lacking both ApoE and leptin receptor. Atherosclerosis 181: 251–259, 2005 [DOI] [PubMed] [Google Scholar]
- 18.San Martin A, Du P, Dikalova A, Lassegue B, Aleman M, Gongora MC, Brown K, Joseph G, Harrison DG, Taylor WR, Jo H, Griendling KK: Reactive oxygen species-selective regulation of aortic inflammatory gene expression in type 2 diabetes. Am J Physiol Heart Circ Physiol 292: H2073–H2082, 2007 [DOI] [PubMed] [Google Scholar]
- 19.Hatley ME, Srinivasan S, Reilly KB, Bolick DT, Hedrick CC: Increased production of 12/15 lipoxygenase eicosanoids accelerates monocyte/endothelial interactions in diabetic db/db mice. J Biol Chem 278: 25369–25375, 2003 [DOI] [PubMed] [Google Scholar]
- 20.Srinivasan S, Hatley ME, Reilly KB, Danziger EC, Hedrick CC: Modulation of PPARalpha expression and inflammatory interleukin-6 production by chronic glucose increases monocyte/endothelial adhesion. Arterioscler Thromb Vasc Biol 24: 851–857, 2004 [DOI] [PubMed] [Google Scholar]
- 21.Kaplanski G, Marin V, Montero-Julian F, Mantovani A, Farnarier C: IL-6: a regulator of the transition from neutrophil to monocyte recruitment during inflammation. Trends Immunol 24: 25–29, 2003 [DOI] [PubMed] [Google Scholar]
- 22.Modur V, Li Y, Zimmerman GA, Prescott SM, McIntyre TM: Retrograde inflammatory signaling from neutrophils to endothelial cells by soluble interleukin-6 receptor alpha. J Clin Invest 100: 2752–2756, 1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vassalli G, Agah R, Qiao R, Aguilar C, Dichek DA: A mouse model of arterial gene transfer: antigen-specific immunity is a minor determinant of the early loss of adenovirus-mediated transgene expression. Circ Res 85: e25–e32, 1999 [PubMed] [Google Scholar]
- 24.Singh RJ, Mason JC, Lidington EA, Edwards DR, Nuttall RK, Khokha R, Knauper V, Murphy G, Gavrilovic J: Cytokine stimulated vascular cell adhesion molecule-1 (VCAM-1) ectodomain release is regulated by TIMP-3. Cardiovasc Res 67: 39–49, 2005 [DOI] [PubMed] [Google Scholar]
- 25.Yoshida K, Shimizugawa T, Ono M, Furukawa H: Angiopoietin-like protein 4 is a potent hyperlipidemia-inducing factor in mice and inhibitor of lipoprotein lipase. J Lipid Res 43: 1770–1772, 2002 [DOI] [PubMed] [Google Scholar]
- 26.Ito Y, Oike Y, Yasunaga K, Hamada K, Miyata K, Matsumoto S, Sugano S, Tanihara H, Masuho Y, Suda T: Inhibition of angiogenesis and vascular leakiness by angiopoietin-related protein 4. Cancer Res 63: 6651–6657, 2003 [PubMed] [Google Scholar]
- 27.Mercer N, Ahmed H, McCarthy AD, Etcheverry SB, Vasta GR, Cortizo AM: AGE-R3/galectin-3 expression in osteoblast-like cells: regulation by AGEs. Mol Cell Biochem 266: 17–24, 2004 [DOI] [PubMed] [Google Scholar]
- 28.Pitzalis MV, Sarzani R, Dessi-Fulgheri P, Iacoviello M, Forleo C, Lucarelli K, Pietrucci F, Salvi F, Sorrentino S, Romito R, Guida P, Rappelli A, Rizzon P: Allelic variants of natriuretic peptide receptor genes are associated with family history of hypertension and cardiovascular phenotype. J Hypertens 21: 1491–1496, 2003 [DOI] [PubMed] [Google Scholar]
- 29.Sundsten T, Eberhardson M, Goransson M, Bergsten P: The use of proteomics in identifying differentially expressed serum proteins in humans with type 2 diabetes. Proteome Sci 4: 22, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Joseph R, Dou D, Tsang W: Molecular cloning of a novel mRNA (neuronatin) that is highly expressed in neonatal mammalian brain. Biochem Biophys Res Commun 201: 1227–1234, 1994 [DOI] [PubMed] [Google Scholar]
- 31.Williamson CM, Beechey CV, Ball ST, Dutton ER, Cattanach BM, Tease C, Ishino F, Peters J: Localisation of the imprinted gene neuronatin, Nnat, confirms and refines the location of a second imprinting region on mouse chromosome 2. Cytogenet Cell Genet 81: 73–78, 1998 [DOI] [PubMed] [Google Scholar]
- 32.Usui H, Morii K, Tanaka R, Tamura T, Washiyama K, Ichikawa T, Kumanishi T: cDNA cloning and mRNA expression analysis of the human neuronatin: high level expression in human pituitary gland and pituitary adenomas. J Mol Neurosci 9: 55–60, 1997 [DOI] [PubMed] [Google Scholar]
- 33.Arava Y, Adamsky K, Ezerzer C, Ablamunits V, Walker MD: Specific gene expression in pancreatic β-cells: cloning and characterization of differentially expressed genes. Diabetes 48: 552–556, 1999 [DOI] [PubMed] [Google Scholar]
- 34.Chu K, Tsai MJ: Neuronatin, a downstream target of β2/NeuroD1 in the pancreas, is involved in glucose-mediated insulin secretion. Diabetes 54: 1064–1073, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Poirier LA, Brown AT, Fink LM, Wise CK, Randolph CJ, Delongchamp RR, Fonseca VA: Blood S-adenosylmethionine concentrations and lymphocyte methylenetetrahydrofolate reductase activity in diabetes mellitus and diabetic nephropathy. Metabolism 50: 1014–1018, 2001 [DOI] [PubMed] [Google Scholar]
- 36.Ratnam S, Wijekoon EP, Hall B, Garrow TA, Brosnan ME, Brosnan JT: Effects of diabetes and insulin on betaine-homocysteine S-methyltransferase expression in rat liver. Am J Physiol Endocrinol Metab 290: E933–E939, 2006 [DOI] [PubMed] [Google Scholar]
- 37.Castro R, Rivera I, Martins C, Struys EA, Jansen EE, Clode N, Graca LM, Blom HJ, Jakobs C, de Almeida IT: Intracellular S-adenosylhomocysteine increased levels are associated with DNA hypomethylation in HUVEC. J Mol Med 83: 831–836, 2005 [DOI] [PubMed] [Google Scholar]
- 38.Castro R, Rivera I, Struys EA, Jansen EE, Ravasco P, Camilo ME, Blom HJ, Jakobs C, Tavares de Almeida I: Increased homocysteine and S-adenosylhomocysteine concentrations and DNA hypomethylation in vascular disease. Clin Chem 49: 1292–1296, 2003 [DOI] [PubMed] [Google Scholar]
- 39.Dannenberg LO, Edenberg HJ: Epigenetics of gene expression in human hepatoma cells: expression profiling the response to inhibition of DNA methylation and histone deacetylation. BMC Genomics 7: 181, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Melzner I, Scott V, Dorsch K, Fischer P, Wabitsch M, Bruderlein S, Hasel C, Moller P: Leptin gene expression in human preadipocytes is switched on by maturation-induced demethylation of distinct CpGs in its proximal promoter. J Biol Chem 277: 45420–45427, 2002 [DOI] [PubMed] [Google Scholar]
- 41.Read MA, Whitley MZ, Gupta S, Pierce JW, Best J, Davis RJ, Collins T: Tumor necrosis factor alpha-induced E-selectin expression is activated by the nuclear factor-kappaB and c-JUN N-terminal kinase/p38 mitogen-activated protein kinase pathways. J Biol Chem 272: 2753–2761, 1997 [DOI] [PubMed] [Google Scholar]
- 42.Goebeler M, Kilian K, Gillitzer R, Kunz M, Yoshimura T, Brocker EB, Rapp UR, Ludwig S: The MKK6/p38 stress kinase cascade is critical for tumor necrosis factor-alpha-induced expression of monocyte-chemoattractant protein-1 in endothelial cells. Blood 93: 857–865, 1999 [PubMed] [Google Scholar]
- 43.Ritchie E, Saka M, Mackenzie C, Drummond R, Wheeler-Jones C, Kanke T, Plevin R: Cytokine upregulation of proteinase-activated-receptors 2 and 4 expression mediated by p38 MAP kinase and inhibitory kappa B kinase beta in human endothelial cells. Br J Pharmacol 150: 1044–1054, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Maeng YS, Min JK, Kim JH, Yamagishi A, Mochizuki N, Kwon JY, Park YW, Kim YM, Kwon YG: ERK is an anti-inflammatory signal that suppresses expression of NF-kappaB-dependent inflammatory genes by inhibiting IKK activity in endothelial cells. Cell Signal 18: 994–1005, 2006 [DOI] [PubMed] [Google Scholar]
- 45.Yeh PY, Yeh KH, Chuang SE, Song YC, Cheng AL: Suppression of MEK/ERK signaling pathway enhances cisplatin-induced NF-kappaB activation by protein phosphatase 4-mediated NF-kappaB p65 Thr dephosphorylation. J Biol Chem 279: 26143–26148, 2004 [DOI] [PubMed] [Google Scholar]
- 46.Carter AB, Hunninghake GW: A constitutive active MEK → ERK pathway negatively regulates NF-kappa B-dependent gene expression by modulating TATA-binding protein phosphorylation. J Biol Chem 275: 27858–27864, 2000 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.