Abstract
OBJECTIVE— Genome-wide association studies have identified common variants in CDKAL1, CDKN2A/B, IGF2BP2, SLC30A8, HHEX/IDE, EXT2, and LOC387761 loci that significantly increase the risk of type 2 diabetes. We aimed to replicate these observations in a population-based cohort of Chinese Hans and examine the associations of these variants with type 2 diabetes and diabetes-related phenotypes.
RESEARCH DESIGN AND METHODS— We genotyped 17 single nucleotide polymorhisms (SNPs) in 3,210 unrelated Chinese Hans, including 424 participants with type 2 diabetes, 878 with impaired fasting glucose (IFG), and 1,908 with normal fasting glucose.
RESULTS— We confirmed the associations between type 2 diabetes and variants near CDKAL1 (odds ratio 1.49 [95% CI 1.27–1.75]; P = 8.91 × 10−7) and CDKN2A/B (1.31 [1.12–1.54]; P = 1.0 × 10−3). We observed significant association of SNPs in IGF2BP2 (1.17 [1.03–1.32]; P = 0.014) and SLC30A8 (1.12 [1.01–1.25]; P = 0.033) with combined IFG/type 2 diabetes. The SNPs in CDKAL1, IGF2BP2, and SLC30A8 were also associated with impaired β-cell function estimated by homeostasis model assessment of β-cell function. When combined, each additional risk allele from CDKAL1-rs9465871, CDKN2A/B-rs10811661, IGF2BP2-rs4402960, and SLC30A8-rs13266634 increased the risk for type 2 diabetes by 1.24-fold (P = 2.85 × 10−7) or for combined IFG/type 2 diabetes by 1.21-fold (P = 6.31 × 10−11). None of the SNPs in EXT2 or LOC387761 exhibited significant association with type 2 diabetes or IFG. Significant association was observed between the HHEX/IDE SNPs and type 2 diabetes in individuals from Shanghai only (P < 0.013) but not in those from Beijing (P > 0.33).
CONCLUSIONS— Our results indicate that in Chinese Hans, common variants in CDKAL1, CDKN2A/B, IGF2BP2, and SLC30A8 loci independently or additively contribute to type 2 diabetes risk, likely mediated through β-cell dysfunction.
The rapid increase in prevalence of type 2 diabetes has been a major public health challenge worldwide, including China. The total number of people with diabetes in China is estimated to increase from 20.8 million in 2000 to 42.3 million in 2030 (1). Besides the important contribution of environmental factors, including changes in dietary patterns and lifestyle, genetic determinants also play a major role in type 2 diabetes susceptibility. Over the past decade, serious efforts have been put into the search for type 2 diabetes susceptibility genes, but progress has been slower than anticipated (2,3). Although common variants in a few genes including PPARG, KCNJ11, and TCF7L2 have been convincingly replicated in individuals with European ancestry, relatively few studies have been conducted in Chinese, and, so far, no variants have been unambiguously confirmed as diabetes susceptibility loci in Chinese. However, recent advances in genome-wide association studies (GWASs) have revived the initial optimism and accelerated the discovery of diabetes susceptibility genes (4–6).
The first GWAS, conducted in a French case-control cohort, confirmed TCF7L2 as a major type 2 diabetes susceptibility gene and identified four novel loci consistently associated with type 2 diabetes (7). These loci are located in chromosomal regions that harbor several genes involved in β-cell function or development, including a variant in the SLC30A8 (zinc transporter solute carrier family 30 member 8) gene, variants located in a linkage disequilibrium (LD) block that contains the IDE (insulin-degrading enzyme), KIF11 (kinesin family member 11), and the HHEX (hematopoietically expressed homeobox) genes, as well as variants in another LD block that contains genes encoding EXT2 (exostosin 2). A fourth locus mapped to a hypothetical gene LOC387761 on chromosome 11. Four subsequent GWASs (8–12), performed in European case-control studies, confirmed the SLC30A8 and HHEX/IDE genes as type 2 diabetes susceptibility loci. Furthermore, additional variants in several new gene regions were also identified, including single nucleotide polymorhisms (SNPs) in the CDKAL1 gene, which encodes the CDK5 regulatory subunit associated protein 1-like 1; in the CDKN2A/B genes, which encode the cyclin-dependent kinase inhibitor p15INK4a and p16INK4b; in the IGF2BP2 gene, which encodes the IGF-2 mRNA binding protein 2; and a variant in a region of chromosome 11, not known to contain any genes. Most of these newly identified loci are suggested to play a role in the regulation of insulin production and β-cell function (5,7,9,12–15). It is unclear whether these variants have the same effect in Chinese populations, which have a different genetic background and lower diabetes prevalence compared with European populations (16–18).
Although case-control studies provide a useful design for the discovery of susceptibility loci, they are limited in providing insight into the mechanisms through which genetic variants exert their effect on the risk of type 2 diabetes. Population-based cohort studies with detailed measures of diabetes-related traits, however, might unravel the physiopathology that underlies the association between the newly discovered genetic variants and diabetes. The purpose of this study is to examine whether these novel variants are individually or collectively associated with type 2 diabetes and related traits in a population-based Chinese Han cohort including 3,210 unrelated individuals from Beijing and Shanghai.
RESEARCH DESIGN AND METHODS
The study sample consisted of 3,210 individuals (1,423 men and 1,787 women) from the Study on Nutrition and Health of Aging Population in China. The study population, design, and protocols of this population-based cohort study have been previously described (19). Briefly, all participants were unrelated Chinese Hans, aged 50–70 years, with at least 20 years residence in Beijing or Shanghai. Among them, 424 participants had type 2 diabetes (267 had previously diagnosed type 2 diabetes and 157 had screen-detected and treatment-naive type 2 diabetes), 878 participants had impaired fasting glucose (IFG) (all 878 were screen detected and treatment naive), and 1,908 participants had normal fasting glucose (NFG). Type 2 diabetes was defined by either 1999 World Health Organization criteria (20) or previously diagnosed type 2 diabetes. NFG and IFG were defined as fasting glucose <5.6 mmol/l (100 mg/dl) and 5.6 mmol/l (100 mg/dl) less than or equal to fasting glucose <7.0mmol/l (126 mg/dl), respectively. The study was conducted simultaneously in both Beijing and Shanghai from March to June 2005. The participants were recruited from two urban districts (400 participants for each district) and one rural district (800 participants), representing people with high to low socioeconomic status, using a multistage sampling method in each city. All participants were selected randomly from the eligible candidates listed in the residential registration record. One person from each household was allowed to participate, and at least 40% of the total participants were men in each district. Individuals with the following conditions were excluded from the study: 1) severe psychological disorders, physical disabilities, cancer, cardiovascular disease, Alzheimer's disease, or dementia, within 6 months; or 2) currently diagnosed with tuberculosis, AIDS, and other communicable diseases. All participants attended a physical examination, during which standard anthropometric measurements and overnight fasting blood samples were collected. Glucose was measured enzymatically on an automatic analyzer (Hitachi 7080; Hitachi, Tokyo, Japan) with reagents purchased from Wako Pure Chemical Industries (Osaka, Japan). Fasting insulin was determined by radioimmunoassay (Linco Research, St. Charles, MO). A1C concentrations were measured by turbidometric immunoassay in red blood cells on the Hitachi 7080 Analyzer using reagents from Roche Diagnostics (Indianapolis, IN). Homeostasis model assessment (HOMA) of insulin sensitivity (HOMA-S) and β-cell function (HOMA-B) were estimated using Levy's computer model (21). Written informed consent was obtained from all participants, and study protocols were approved by the institutional review board of the Institute for Nutritional Sciences. The phenotypic characteristics of the population are shown in Table 1.
TABLE 1.
Characteristics of the study population
| All samples | Beijing | Shanghai | P | |
|---|---|---|---|---|
| n (% male) | 3,210 (44.3) | 1,574 (45.2) | 1,636 (43.5) | |
| Age (years) | 58.6 ± 6.0 | 58.3 ± 5.9 | 58.9 ± 6.0 | 0.0095 |
| BMI (kg/m2) | 24.2 (22.0–26.6) | 25.1 (22.8–27.4) | 23.5 (21.3–25.9) | <0.0001 |
| Fasting glucose (mmol/l) | 5.84 ± 1.74 | 6.16 ± 1.96 | 5.53 ± 1.42 | <0.0001 |
| A1C (%) | 5.99 ± 1.10 | 6.08 ± 1.22 | 5.90 ± 0.96 | <0.0001 |
| Fasting insulin (pmol/l) | 82.2 (59.4–112.2) | 81.0 (57.6–110.7) | 84.0 (61.8–114.0) | 0.0777 |
| HOMA-B (%) | 110.3 ± 47.0 | 100.1 ± 44.9 | 120.0 ± 46.9 | <0.0001 |
| HOMA-S (%) | 63.7 (47.1–86.9) | 64.0 (47.3–89.5) | 63.5 (46.9–85.1) | 0.0454 |
| IFG (%) | 878 (27.4) | 579 (36.8) | 299 (18.3) | <0.0001 |
| Type 2 diabetes (%) | 424 (13.2) | 272 (17.3) | 152 (9.3) | <0.0001 |
Data are means ± SD, median (interquartile range), or n (%), unless otherwise indicated. P represents significance of the differences between individuals from Beijing and from Shanghai.
Genotyping.
Genomic DNA was extracted from peripheral blood leukocytes by the salting-out procedure (available at http://humgen.wustl.edu/hdk_lab_manual/dna/dna2.html). SNP genotyping was performed with the GenomeLab SNPstream Genotyping System (Beckman Coulter), according to the manufacturer's protocol. Seventeen SNPs previously reported to be associated with type 2 diabetes by at least one of the GWASs (7–12) were successfully genotyped in our population. These include SNPs near CDKAL1 (rs10946398, rs7754840, rs7756992, and rs9465871), HHEX/IDE (rs1111875, rs5015480, and rs7923837), EXT2 (rs1113132, rs11037909, and rs3740878), CDKN2A/B (rs10811661 and rs564398), IGF2BP2 (rs4402960 and rs1470579), SLC30A8 (rs13266634) (R325W), LOC387761 (rs7480010), and an intergenic SNP (rs9300039) in chromosome 11. The genotyping success rate was >97.1%, and the concordance rate was >99% based on 12% duplicate samples (n = 384). Samples with ambiguous base calling were genotyped again. Genotype frequencies of all 17 SNPs were consistent with Hardy-Weinberg equilibrium (P > 0.01), and most of the minor allele frequencies observed in this study were comparable with those in the HapMap CHB (Chinese Han in Beijing) sample (online appendix Table 1 [available at http://dx.doi.org/10.2337/db08-0047]). Genotypic distributions were similar in Beijing and Shanghai populations (P > 0.05), except for the three HHEX SNPs (P = 0.041, 0.003, and 0.005 for rs1111875, rs5015480, and rs7923837, respectively).
Statistical analyses.
Hardy-Weinberg equilibrium was tested using a likelihood ratio test. LD between SNPs was estimated using Haploview version 3.2 (available at http://www.broad.mit.edu/mpg/haploview). The association between each SNP and the risk of type 2 diabetes and IFG was examined using logistic regression. Generalized linear regression was applied to study the associations between each SNP and type 2 diabetes–related quantitative traits. Participants with known diabetes or receiving glucose-lowering treatment (n = 267) were excluded from the type 2 diabetes–related quantitative trait analyses. All association analyses assumed an additive effect of the risk allele and were adjusted for sex, age, BMI (where appropriate), and geographical region (Shanghai versus Beijing). BMI, insulin, and HOMA-S were log transformed before analyses, and the data were presented as geometric means. Likelihood ratio tests were used to examine genotype distribution in Beijing and Shanghai. Because of a significant difference in genotype distribution of the three HHEX/IDE SNPs (P < 0.05) and in diabetes prevalence between the Shanghai and Beijing participants (P < 0.0001), analyses for these SNPs were performed for Shanghai and Beijing separately.
Gene-gene interactions were assessed by including the respective interaction terms of pairwise SNPs in logistic regressions using the maximum likelihood estimation. The combined effect of multiple SNPs on the risk of type 2 diabetes and/or IFG was determined by logistic regression after categorizing the participants into groups according to the number of the risk alleles they carried. Participants with one or no risk alleles served as the reference group. Bonferroni correction was used to adjust for multiple testing in the quantitative trait analyses. Association analyses were performed with SAS version 9.1 (SAS Institute, Cary, NC). Meta-analyses were conducted with Stata (version 9.2; Stata, College Station, TX). Cochran's Q test was performed to assess heterogeneity among different groups. Power calculations were performed using Quanto software (available at http://hydra.usc.edu/gxe/), and the power shown in online appendix Table 1 was calculated for association between each SNP and type 2 diabetes using the odds ratios (7–12) reported in the original studies and sample size and minor allele frequencies in our own study.
RESULTS
We first examined the association with the risk of type 2 diabetes and IFG (Table 2). The four CDKAL1 SNPs spanned two LD blocks (r2 = 1.0 for rs7754840 and rs10946398 and 0.96 for rs7756992 and rs9465871) and were each significantly associated with type 2 diabetes (odds ratios ranged between 1.38 and 1.49; P < 1.9 × 10−5) and with combined IFG/type 2 diabetes (between 1.20 and 1.22; P < 0.0013). The CDKN2A/B rs10811661 variant was also associated with type 2 diabetes (odds ratio 1.31 [95% CI 1.12–1.54]; P = 0.001) and combined IFG/type 2 diabetes (1.26 [1.13–1.41]; P = 2.76 × 10−5). The second CDKN2A/B SNP (rs564398), which was not in LD with rs10811661 (r2 = 0), was not associated with type 2 diabetes or combined type 2 diabetes/IFG. The two SNPs in IGF2BP2 (r2 = 0.83) and the SLC30A8 SNP (rs13266634) showed modest association with combined IFG/type 2 diabetes (odds ratios between 1.12 and 1.17; P = 0.013–0.033) but not with type 2 diabetes alone.
TABLE 2.
Associations with type 2 diabetes or IFG and type 2 diabetes combined
| SNP identification | Gene | Major/minor allele* | Type 2 diabetes vs. normal |
Type 2 diabetes and IFG vs. normal |
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Minor allele frequency |
Odds ratio (95% CI) | P(add) | Minor allele frequency |
Odds ratio (95% CI) | P(add) | |||||
| Case | Control | Case | Control | |||||||
| All samples | ||||||||||
| rs10946398 | CDKAL1 | A/C | 0.500 | 0.409 | 1.47 (1.25–1.73) | 2.32 × 10−6 | 0.457 | 0.409 | 1.20 (1.07–1.33) | 0.0012 |
| rs7754840 | CDKAL1 | G/C | 0.501 | 0.407 | 1.49 (1.27–1.75) | 8.91 × 10−7 | 0.459 | 0.407 | 1.22 (1.10–1.36) | 0.0003 |
| rs7756992 | CDKAL1 | G/A | 0.426 | 0.497 | 1.38 (1.17–1.62) | 9.35 × 10−5 | 0.454 | 0.497 | 1.21 (1.09–1.35) | 0.0004 |
| rs9465871 | CDKAL1 | C/T | 0.415 | 0.493 | 1.41 (1.21–1.66) | 1.80 × 10−5 | 0.449 | 0.493 | 1.21 (1.09–1.35) | 0.0003 |
| rs10811661 | CDKN2A/B | T/C | 0.418 | 0.483 | 1.31 (1.12–1.54) | 0.0010 | 0.432 | 0.483 | 1.26 (1.13–1.41) | 2.76 × 10−5 |
| rs564398 | CDKN2A/B | T/C | 0.131 | 0.128 | 1.07 (0.84–1.26) | 0.59 | 0.135 | 0.128 | 0.99 (0.85–1.16) | 0.92 |
| rs4402960 | IGF2BP2 | G/T | 0.264 | 0.241 | 1.14 (0.95–1.35) | 0.16 | 0.263 | 0.241 | 1.17 (1.03–1.32) | 0.014 |
| rs1470579 | IGF2BP2 | A/C | 0.272 | 0.246 | 1.15 (0.97–1.38) | 0.11 | 0.268 | 0.246 | 1.17 (1.03–1.32) | 0.013 |
| rs13266634 | SLC30A8 | C/T | 0.417 | 0.432 | 1.09 (0.93–1.27) | 0.28 | 0.411 | 0.432 | 1.12 (1.01–1.25) | 0.033 |
| rs1113132 | EXT2 | C/G | 0.390 | 0.418 | 1.12 (0.96–1.32) | 0.15 | 0.410 | 0.418 | 1.04 (0.93–1.15) | 0.53 |
| rs11037909 | EXT2 | T/C | 0.381 | 0.418 | 1.16 (0.99–1.36) | 0.07 | 0.406 | 0.418 | 1.04 (0.94–1.16) | 0.43 |
| rs3740878 | EXT2 | A/G | 0.405 | 0.431 | 1.11 (0.95–1.31) | 0.19 | 0.418 | 0.431 | 1.06 (0.95–1.18) | 0.29 |
| rs7480010 | LOC387761 | A/G | 0.223 | 0.225 | 0.98 (0.80–1.18) | 0.79 | 0.230 | 0.225 | 1.00 (0.88–1.13) | 0.97 |
| rs9300039 | Unknown | C/A | 0.279 | 0.274 | 0.96 (0.80–1.14) | 0.62 | 0.265 | 0.274 | 1.06 (0.94–1.19) | 0.34 |
| Beijing | ||||||||||
| rs1111875 | HHEX | A/G | 0.306 | 0.309 | 1.00 (0.81–1.25) | 0.94 | 0.279 | 0.309 | 0.89 (0.76–1.04) | 0.13 |
| rs5015480 | HHEX | T/C | 0.202 | 0.185 | 1.13 (0.88–1.46) | 0.33 | 0.173 | 0.185 | 0.94 (0.78–1.13) | 0.52 |
| rs7923837 | HHEX | A/G | 0.244 | 0.231 | 1.09 (0.86–1.39) | 0.48 | 0.229 | 0.231 | 1.01 (0.84–1.20) | 0.95 |
| Shanghai | ||||||||||
| rs1111875 | HHEX | A/G | 0.376 | 0.276 | 1.64 (1.25–2.15) | 0.0004 | 0.294 | 0.276 | 1.10 (0.92–1.32) | 0.30 |
| rs5015480 | HHEX | T/C | 0.218 | 0.138 | 1.79 (1.30–2.47) | 0.0003 | 0.183 | 0.138 | 1.43 (1.15–1.78) | 0.0013 |
| rs7923837 | HHEX | A/G | 0.252 | 0.186 | 1.45 (1.08–1.94) | 0.0131 | 0.231 | 0.186 | 1.30 (1.07–1.58) | 0.0089 |
Odds ratios represent the effects of risk alleles. The P values were adjusted for age, sex, BMI, and region (where appropriate).
Alleles in bold are the risk alleles for type 2 diabetes identified by previous studies, while alleles underlined are the risk alleles for type 2 diabetes or IFG observed in this study. All analyses were based on an additive model, in which individuals homozygous for the nonrisk alleles were coded as 0, heterozygous individuals were coded as 1, and individuals homozygous for the risk alleles were coded as 2.
The three EXT2 variants were in complete LD (r2 = 1.0) and occurred less frequently in our population (58%) than in European populations (70%). These variants, as well as those in chromosome 11 (rs7480010 and rs9300039, r2 = 0.037), were not associated with type 2 diabetes or IFG. Analyses for the three SNPs in the HHEX/IDE LD block were performed separately in Shanghai and Beijing populations, as the difference in genotype distribution and prevalence of type 2 diabetes and IFG could lead to spurious associations due to population stratification (Table 2). All three HHEX/IDE SNPs were significantly associated with type 2 diabetes in Shanghai participants, with rs5015480 and rs7923837 also associated with combined IFG/type 2 diabetes. Meta-analyses suggested that the associations exhibited significant heterogeneity for SNPs rs1111875 (P = 0.006) and rs5015480 (P = 0.028) between Beijing and Shanghai populations.
We next examined the association between genetic variants and type 2 diabetes–related quantitative traits (glucose, A1C, insulin, HOMA-B, HOMA-S, and BMI) to investigate whether these variants conferred risk of type 2 diabetes through their effects on any of these intermediate traits (Table 3). Consistent with the case-control analyses, the SNPs that showed significant evidence for association with diabetes-related phenotypes were those that were also associated with type 2 diabetes or IFG, except for CDKN2A/B rs10811661 and LOC387761 rs7480010. All four CDKAL1 SNPs were significantly associated with A1C (P values 0.036–0.0096) and HOMA-B (P values 0.024–0.0009). The SNPs (rs7756992 and rs9465871) in the second LD block of this locus also showed significant association with fasting glucose levels (P < 0.04). Interestingly, the allele of SLC30A8 SNP rs13266634 that increases the risk of combined IFG/type 2 diabetes was significantly associated with lower BMI (P = 0.0087) and marginally associated with decreased HOMA-B (P = 0.05). Only the associations of CDKAL1-rs10946398, rs7754840, and IGF2BP2-rs4402960 with HOMA-B remained significant after Bonferroni correction for multiple testing (P = 0.0014, 0.05/36 tests).
TABLE 3.
Associations with type 2 diabetes–related quantitative traits
| SNP identification (major/minor allele) | n | Glucose (mmol/l)* |
A1C (%)* |
Insulin (pmol/l)† |
HOMA-B (%)* |
HOMA-S (%)† |
BMI (kg/m2)† |
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Means ± SE | P‡ | Means ± SE | P‡ | Means ± SE | P‡ | Means ± SE | P‡ | Means ± SE | P‡ | Means ± SE | P§ | ||
| CDKAL1 | |||||||||||||
| rs10946398 (A/C) Genotype | |||||||||||||
| AA | 972 | 5.53 ± 0.04 | 5.78 ± 0.03 | 80.4 ± 1.3 | 115.9 ± 1.4 | 65.4 ± 1.0 | 24.1 ± 0.1 | ||||||
| AC | 1,429 | 5.62 ± 0.03 | 0.10 | 5.85 ± 0.02 | 0.036 | 78.7 ± 1.0 | 0.11 | 112.8 ± 1.1 | 0.0009¶ | 66.6 ± 0.8 | 0.10 | 24.2 ± 0.1 | 0.16 |
| CC | 516 | 5.62 ± 0.05 | 5.85 ± 0.04 | 77.1 ± 1.7 | 99.4 ± 1.8 | 68.2 ± 1.4 | 23.8 ± 0.1 | ||||||
| rs7754840 (G/C) Genotype | |||||||||||||
| GG | 975 | 5.52 ± 0.04 | 5.77 ± 0.03 | 80.4 ± 1.3 | 115.8 ± 1.4 | 65.5 ± 1.0 | 24.2 ± 0.1 | ||||||
| GC | 1,443 | 5.61 ± 0.03 | 0.09 | 5.85 ± 0.02 | 0.034 | 79.1 ± 1.0 | 0.08 | 113.3 ± 1.1 | 0.0011¶ | 66.3 ± 0.8 | 0.07 | 24.2 ± 0.1 | 0.10 |
| CC | 514 | 5.62 ± 0.05 | 5.85 ± 0.04 | 76.6 ± 1.6 | 107.9 ± 1.9 | 68.7 ± 1.4 | 23.8 ± 0.2 | ||||||
| rs7756992 (G/A) Genotype | |||||||||||||
| GG | 774 | 5.68 ± 0.04 | 5.88 ± 0.03 | 79.2 ± 1.4 | 109.6 ± 1.5 | 66.7 ± 1.1 | 24.0 ± 0.1 | ||||||
| GA | 1,455 | 5.56 ± 0.03 | 0.035 | 5.82 ± 0.02 | 0.011 | 79.0 ± 1.0 | 0.62 | 114.1 ± 1.1 | 0.024 | 66.6 ± 0.8 | 0.84 | 24.2 ± 0.1 | 0.69 |
| AA | 681 | 5.55 ± 0.05 | 5.77 ± 0.03 | 78.2 ± 1.5 | 114.6 ± 1.6 | 66.8 ± 1.2 | 24.1 ± 0.1 | ||||||
| rs9465871 (C/T) Genotype | |||||||||||||
| CC | 805 | 5.67 ± 0.04 | 5.88 ± 0.03 | 78.8 ± 1.4 | 109.9 ± 1.5 | 66.5 ± 1.1 | 24.0 ± 0.1 | ||||||
| CT | 1,422 | 5.57 ± 0.03 | 0.035 | 5.82 ± 0.02 | 0.0096 | 78.9 ± 1.0 | 0.72 | 113.5 ± 1.1 | 0.0065 | 66.8 ± 0.8 | 0.65 | 24.2 ± 0.1 | 0.59 |
| TT | 688 | 5.54 ± 0.04 | 5.77 ± 0.03 | 79.5 ± 1.5 | 115.9 ± 1.6 | 65.7 ± 1.2 | 24.1 ± 0.1 | ||||||
| CDKN2A/B | |||||||||||||
| rs10811661 (T/C) Genotype | |||||||||||||
| TT | 813 | 5.60 ± 0.04 | 5.81 ± 0.03 | 78.3 ± 1.3 | 110.7 ± 1.5 | 67.2 ± 1.1 | 24.0 ± 0.1 | ||||||
| TC | 1,489 | 5.59 ± 0.03 | 0.48 | 5.84 ± 0.02 | 0.73 | 80.3 ± 1.0 | 0.70 | 113.9 ± 1.1 | 0.09 | 65.5 ± 0.8 | 0.85 | 24.0 ± 0.1 | 0.39 |
| CC | 620 | 5.55 ± 0.05 | 5.82 ± 0.03 | 77.2 ± 1.5 | 114.4 ± 1.7 | 67.8 ± 1.3 | 24.2 ± 0.1 | ||||||
| rs564398 (T/C) Genotype | |||||||||||||
| TT | 2,173 | 5.58 ± 0.03 | 5.82 ± 0.02 | 78.6 ± 0.8 | 112.5 ± 0.9 | 66.9 ± 0.7 | 24.0 ± 0.1 | ||||||
| TC | 694 | 5.62 ± 0.05 | 0.55 | 5.83 ± 0.03 | 0.73 | 80.9 ± 1.5 | 0.22 | 114.5 ± 1.6 | 0.31 | 65.8 ± 1.1 | 0.19 | 24.2 ± 0.1 | 0.51 |
| CC | 40 | 5.58 ± 0.19 | 5.92 ± 0.13 | 78.4 ± 6.1 | 113.4 ± 6.7 | 67.3 ± 5.0 | 23.5 ± 0.5 | ||||||
| IGF2BP2 | |||||||||||||
| rs4402960 (G/T) Genotype | |||||||||||||
| GG | 1,635 | 5.54 ± 0.03 | 5.81 ± 0.02 | 80.3 ± 1.0 | 115.4 ± 1.1 | 65.5 ± 0.8 | 24.1 ± 0.1 | ||||||
| GT | 1,108 | 5.64 ± 0.04 | 0.037 | 5.85 ± 0.02 | 0.33 | 70.0 ± 1.0 | 0.06 | 110.2 ± 1.3 | 0.0005¶ | 67.8 ± 1.0 | 0.07 | 24.1 ± 0.1 | 0.31 |
| TT | 173 | 5.67 ± 0.09 | 5.83 ± 0.06 | 77.5 ± 2.9 | 108.0 ± 3.2 | 67.7 ± 2.4 | 23.8 ± 0.3 | ||||||
| rs1470579 (A/C) Genotype | |||||||||||||
| AA | 1,597 | 5.54 ± 0.03 | 5.80 ± 0.02 | 80.0 ± 1.0 | 115.3 ± 1.1 | 65.8 ± 0.8 | 24.2 ± 0.1 | ||||||
| AC | 1,143 | 5.64 ± 0.04 | 0.029 | 5.85 ± 0.02 | 0.11 | 78.2 ± 1.1 | 0.17 | 111.0 ± 1.3 | 0.0026 | 67.1 ± 0.9 | 0.20 | 24.0 ± 0.1 | 0.23 |
| CC | 167 | 5.66 ± 0.10 | 5.86 ± 0.06 | 77.1 ± 2.9 | 108.2 ± 3.3 | 68.1 ± 2.5 | 23.9 ± 0.3 | ||||||
| SLC30A8 | |||||||||||||
| rs13266634 (C/T) Genotype | |||||||||||||
| CC | 960 | 5.64 ± 0.04 | 5.84 ± 0.03 | 77.0 ± 1.2 | 110.1 ± 1.4 | 67.8 ± 1.0 | 23.9 ± 0.1 | ||||||
| CT | 1,411 | 5.57 ± 0.03 | 0.18 | 5.82 ± 0.02 | 0.61 | 80.3 ± 1.0 | 0.25 | 114.6 ± 1.1 | 0.05 | 65.6 ± 0.8 | 0.28 | 24.2 ± 0.1 | 0.0087 |
| TT | 531 | 5.56 ± 0.05 | 5.82 ± 0.04 | 78.6 ± 1.7 | 113.7 ± 1.8 | 66.5 ± 1.4 | 24.3 ± 0.2 | ||||||
| EXT2 | |||||||||||||
| rs1113132 (C/G) Genotype | |||||||||||||
| CC | 989 | 5.58 ± 0.04 | 5.82 ± 0.03 | 79.4 ± 1.2 | 112.3 ± 1.4 | 66.4 ± 1.0 | 24.1 ± 0.1 | ||||||
| CG | 1,422 | 5.60 ± 0.03 | 0.97 | 5.83 ± 0.02 | 0.85 | 79.3 ± 1.0 | 0.26 | 114.0 ± 1.1 | 0.89 | 66.0 ± 0.8 | 0.37 | 24.1 ± 0.1 | 0.73 |
| GG | 506 | 5.57 ± 0.05 | 5.82 ± 0.04 | 76.7 ± 1.7 | 111.3 ± 1.9 | 68.4 ± 1.4 | 24.0 ± 0.2 | ||||||
| rs11037909 (T/C) Genotype | |||||||||||||
| TT | 996 | 5.60 ± 0.04 | 5.82 ± 0.03 | 79.8 ± 1.2 | 112.4 ± 1.4 | 65.9 ± 1.0 | 24.2 ± 0.1 | ||||||
| TC | 1,390 | 5.59 ± 0.03 | 0.73 | 5.83 ± 0.02 | 0.99 | 79.0 ± 1.0 | 0.20 | 114.0 ± 1.1 | 0.92 | 66.3 ± 0.8 | 0.23 | 24.1 ± 0.1 | 0.29 |
| CC | 509 | 5.58 ± 0.05 | 5.82 ± 0.04 | 77.1 ± 1.7 | 111.5 ± 1.9 | 68.1 ± 1.4 | 24.0 ± 0.2 | ||||||
| rs3740878 (A/G) Genotype | |||||||||||||
| TT | 931 | 5.59 ± 0.04 | 5.81 ± 0.03 | 79.9 ± 1.3 | 113.0 ± 1.4 | 65.8 ± 1.0 | 24.2 ± 0.1 | ||||||
| TC | 1,409 | 5.60 ± 0.03 | 0.83 | 5.83 ± 0.02 | 0.84 | 79.2 ± 1.0 | 0.18 | 113.4 ± 1.1 | 0.78 | 66.4 ± 0.8 | 0.24 | 24.0 ± 0.1 | 0.39 |
| CC | 518 | 5.57 ± 0.05 | 5.82 ± 0.04 | 76.9 ± 1.6 | 112.1 ± 1.9 | 67.9 ± 1.4 | 24.0 ± 0.2 | ||||||
| LOC387761 | |||||||||||||
| rs7480010 (A/G) Genotype | |||||||||||||
| AA | 1,718 | 5.59 ± 0.03 | 5.82 ± 0.02 | 79.9 ± 0.9 | 113.9 ± 1.0 | 65.7 ± 0.7 | 24.1 ± 0.1 | ||||||
| AG | 1,034 | 5.56 ± 0.04 | 0.57 | 5.83 ± 0.03 | 0.97 | 78.3 ± 1.2 | 0.025 | 112.8 ± 1.3 | 0.08 | 67.1 ± 1.0 | 0.028 | 24.2 ± 0.1 | 0.87 |
| GG | 145 | 5.60 ± 0.10 | 5.82 ± 0.07 | 72.1 ± 2.9 | 106.1 ± 3.5 | 72.0 ± 2.8 | 23.8 ± 0.3 | ||||||
| Intergenic | |||||||||||||
| rs9300039 (C/A) Genotype | |||||||||||||
| CC | 1,550 | 5.62 ± 0.03 | 5.84 ± 0.02 | 79.0 ± 1.0 | 112.5 ± 1.1 | 66.6 ± 0.8 | 24.1 ± 0.1 | ||||||
| CA | 1,130 | 5.57 ± 0.04 | 0.07 | 5.81 ± 0.02 | 0.14 | 78.8 ± 1.1 | 0.99 | 112.9 ± 1.3 | 0.23 | 66.4 ± 0.9 | 0.87 | 24.1 ± 0.1 | 0.61 |
| AA | 212 | 5.46 ± 0.08 | 5.76 ± 0.06 | 79.2 ± 2.6 | 117.5 ± 3.0 | 66.2 ± 2.1 | 24.1 ± 0.2 | ||||||
Data are means ± SE or
geometric means ± SE, unless otherwise indicated. Alleles in bold are the risk alleles for type 2 diabetes identified by previous studies while alleles underlined are the risk alleles for type 2 diabetes or IFG observed in this study.
Adjusted for age, sex, region, and BMI.
Adjusted for age, sex, and region.
The associations remained significant after Bonferroni correction for multiple tests, and the Bonferroni corrected cutoff P value is 0.0014 (0.05/36 tests).
To examine whether the associations for the CDKAL1 variants were independent, we performed additional multiple regression analyses that included all four CDKAL1 SNPs in one model. Results showed that none of the four SNPs remained significant (P ≥ 0.17). Next, we tested whether the two CDKAL1 “pairs” (rs7754840 and rs7756992 were chosen to represent each of the pairs) were independent from each other for the associations with type 2 diabetes or related quantitative traits in multiple regression models with both rs7754840 and rs7756992 genotypes in the model, with age, sex, region, and BMI (where appropriate) as covariates. The results revealed that the association seems to be driven by rs7754840, for the associations with type 2 diabetes, BMI, and HOMA-B, or by rs7756992, for the association with A1C, but interestingly, rs7754840 and rs7756992 seem to have independent effects on the associations with HOMA-S or insulin (online appendix Table 2).
We also performed a meta-analysis with the data from the previously published studies (10–12), including those from Japanese, Korean, and Hong Kong Chinese populations (22–25), to assess the heterogeneity between Caucasians and Asians for the CDKAL1 and CDKN2A/B loci (rs7754840 and rs10811661 were chosen to represent each of them, respectively). The results showed that for the CDKN2A/B loci (rs10811661), the heterogeneity between Caucasians and Asians did not reach significance (P = 0.059), while a significant heterogeneity was observed between Caucasians and Asians (P = 8.872 × 10−6) for the CDKAL1 loci (rs7754840) (online appendix Fig. 1), and this is consistent with the recent finding reported by Ng et al. (25).
Although we did not observe the association among the LOC387761 SNP rs7480010 and type 2 diabetes or IFG, we found that the allele that increased the diabetes risk in European populations was modestly associated (P < 0.03) with increased insulin sensitivity (HOMA-S) and lower fasting insulin levels. Furthermore, despite a strong association between the CDKN2A/B SNP rs10811661 and type 2 diabetes, no association was observed with any of the diabetes-related quantitative traits. The intergenic SNP rs9300039 and the three EXT2 SNPs (rs3740878, rs11037909, and rs1113132) were not associated with any of the diabetes-related quantitative traits.
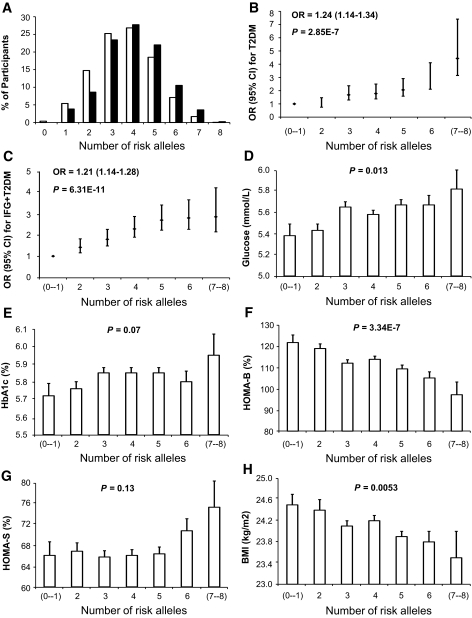
We found no evidence of multiplicative gene-gene interactions among the main SNPs (rs9465871, rs10811661, rs4402960, and rs13266634) in each of the CDKAL1, CDKN2A/B, IGF2BP2, and SLC30A8 genes. A significantly higher proportion of participants with type 2 diabetes carry increasing numbers of risk alleles, compared with participants with NFG (Fig. 1A). In combined analysis, each additional risk allele increased the risk of type 2 diabetes by 1.24-fold (P = 2.85 × 10−7) (Fig. 1B) and combined IFG/type 2 diabetes by 1.21-fold (P = 6.31 × 10−11) (Fig. 1C). Participants harboring seven or all eight risk alleles had a 4.44-fold increased risk for type 2 diabetes (P = 5 × 10−4) compared with those with one or no risk alleles (Fig. 1B). Consistently, participants with increasing numbers of risk alleles tended to have increased fasting levels of plasma glucose (P = 0.013) (Fig. 1D) and A1C (P = 0.07) (Fig. 1E), as well as decreased HOMA-B values (P = 3.34 × 10−7) (Fig. 1F). Of note, participants with increasing numbers of risk alleles tended to have significantly lower BMI (P = 5.3 × 10−3) (Fig. 1F), which is consistent with previous results found for the CDKAL1 and SLC30A8 polymorphisms (Table 3).
FIG. 1.
Combined effects of increasing numbers of the risk alleles from CDKAL1-rs9465871, CDKN2A/B-rs10811661, IGF2BP2-rs4402960, and SLC30A8-rs13266634. A: The risk allele distribution in the participants with NFG and participants with type 2 diabetes. □, control; ▪, type 2 diabetes. Each additional risk allele increased the risk of type 2 diabetes by 1.24-fold (P = 2.85 × 10−7) (B) and of IFG and diabetes combined by 1.21-fold (P = 6.31 × 10−11) (C). B: Participants harboring seven or all eight risk alleles had a 4.44-fold increased risk for type 2 diabetes (P = 5 × 10−4) compared with the reference group. Consistently, participants with increasing numbers of risk alleles tended to have increased fasting levels of plasma glucose (P = 0.013) (D) and A1C (P = 0.07) (E), as well as decreased HOMA-B values (P = 3.34 × 10−7) (F) and lower BMI (P = 5.3 × 10−3) (H), but showed no association with plasma insulin (P = 0.13) (G).
DISCUSSION
In this study of Chinese Hans, we replicated associations with several diabetes susceptibility variants recently identified through GWASs in white Europeans (7–12). Variants in CDKAL1, CDKN2A/B, IGF2BP2, SLC30A8, and HHEX loci were significantly associated with the risk of type 2 diabetes or combined IFG/type 2 diabetes. Furthermore, variants in CDKAL1 and IGF2BP2 were strongly associated with β-cell function estimated by HOMA-B.
The risk alleles of the CDKAL1 variants increased diabetes risk by ∼1.4-fold. These associations were stronger than those observed in individuals of European Ancestry (8–10,12) (online appendix Table 1), and CDKAL1 risk allele frequencies are also substantially higher in Chinese (43–55%) than Europeans (15–31%). Moreover, significant heterogeneity between Caucasians and Asians was found for the CDKAL1 loci (rs7754840) in the meta-analysis that combined the data from the previous studies in white Europeans, Japanese, Korean, Hong Kong Chinese, and our study (P = 8.872 × 10−6) (online appendix Fig. 1), while no significant heterogeneity was observed among the Asians (P = 0.369). These observations suggest that these CDKAL1 variants might play an even more important role in diabetes susceptibility in Chinese. The risk allele of the first pair of CDKAL1 variants was strongly associated with reduced β-cell function (HOMA-B) and increased A1C levels, while the second pair of CDKAL1 variants showed an association with impaired β-cell function (HOMA-B) and higher glucose levels, as well as with increased A1C. The results from additional multiple regression analyses suggest that the four SNPs most likely represent the effects of a single CDKAL1 locus. However, none of these four SNPs stands out as being the variant driving the association. Therefore, we assume that none of them is likely to be the causal variant, but presumably they are in moderate to high LD with the causal SNP and are therefore less consistently associated with the traits of interest. This region would benefit from a detailed fine mapping to identify possible causal variants in future studies. These results support previous findings (9,13,26) that the four CDKAL1 SNPs confer the risk of type 2 diabetes through reduced insulin secretion, although the causal SNP is yet to be identified.
We also observed significant association between CDKN2A/B rs10811661 and type 2 diabetes and IFG with a slightly higher odds ratio (∼1.3) than that observed in Europeans (∼1.20) (10–12). The risk allele is twice as prevalent in Chinese Hans (46%) as in Europeans (21%). However, we did not observed significant heterogeneity between Caucasians and Asians in the meta-analysis with data from the previously published studies (P = 0.059). Interestingly, none of the diabetes-related traits showed an association with CDKN2A/B rs10811661. The second CDKN2A/B variant, rs564398, which is less frequent in Chinese Hans (13%) than in Europeans (38%), was not associated with type 2 diabetes or any related traits.
The association between variants in IGF2BP2 and type 2 diabetes was not significant, although the odds ratios were similar to those observed in European populations (∼1.15), suggesting that our study may not have been sufficiently powered. Indeed, assuming an additive model and a minor allele frequency of 25%, we had <50% power to detect previously reported odds ratios at P < 0.05. We did, however, find a significant association with combined IFG/type 2 diabetes. The associations with HOMA-B suggest that IGF2BP2 confer type 2 diabetes risk through a reduced β-cell function. Similarly, we found no association between the SLC30A8 rs13266634 variant and type 2 diabetes, while an association with combined IFG/type 2 diabetes reached borderline significance. Interestingly, the risk allele that increased diabetes risk in Europeans was also associated with a lower BMI in this population.
We also failed to find any evidence for association between type 2 diabetes and the SNPs in EXT2 (rs3740878, rs11037909, and rs1113132) and the intergenic SNP rs9300039, despite ∼80% power to detect previously reported effect estimates (7). Although these SNPs exhibited marginal associations with type 2 diabetes in the original study (7), they were largely negative in the subsequent four GWASs and other replication studies in samples from U.K. (8,12), Finnish (10,11), Swedish (10), Icelandic (9), German (27), and Japanese (23) populations. Therefore, the original associations for these SNPs were either population specific or overestimated due to the “winner's curse” (28,29), but the consistent lack of replication suggests that these findings were more likely false-positives. Meta-analyses or studies with larger sample sizes will be required to draw definitive conclusions. Although there was no association between rs7480010 (LOC387761) and type 2 diabetes or IFG, the allele conferring risk of diabetes in Europeans was associated with increased insulin sensitivity and showed a tendency toward a reduced β-cell function as well. For the three SNPs in HHEX/IDE gene region, the associations with type 2 diabetes or IFG were observed only in Shanghai individuals in whom each risk allele resulted in 1.45- to 1.79-fold increased diabetes risk, suggesting that geographical stratification may exist in our population for these SNPs and their roles in type 2 diabetes susceptibility. However, given the relatively small sample size, we cannot rule out sampling bias. This observation needs to be confirmed in larger studies.
We found no evidence of pairwise synergistic gene-gene interactions on type 2 diabetes and the related phenotypes among CDKAL1-rs9465871, CDKN2A/B-rs10811661, IGF2BP2-rs4402960, and SLC30A8-rs13266634. In joint analyses, the risk of type 2 diabetes was increased by 1.24-fold for each additional risk allele, and participants with seven or all eight risk alleles (3.8%) had a 4.44-fold increased risk of type 2 diabetes (P = 5 × 10−4) compared with those with one or no risk allele. These results are consistent with those reported by Scott et al. (11), who examined combined effects of 10 risk variants in a GWAS of European populations. Compared with Scott's study, the advantage of our study is that our data are based on the general population. However, a replication in larger population is required to examine whether combinations of risk alleles from these variants have good predictive and diagnostic potential in Chinese Hans.
In conclusion, we replicated the association of type 2 diabetes with the SNPs in CDKAL1 and CDKN2A/B genes and confirmed that the SNPs in SLC30A8 and IGF2BP2 were associated with the risk of combined IFG/type 2 diabetes. Most of these SNPs were also associated with the impaired β-cell function. Importantly, the risk variants in CDKAL1, CDKN2A/B, IGF2BP2, and SLC30A8 appear to act in an additive manner to increase the risk of type 2 diabetes and related phenotypes. These results provide solid evidence for the notion that these variants individually or collectively contribute to the risk of type 2 diabetes in the Chinese Han population, possibly by impairing β-cell function or reducing insulin secretion.
Supplementary Material
Acknowledgments
This study was funded by research grants from the National Natural Science Foundation of China (30571562), a grant from the Knowledge Innovation Program Pilot Project of the Chinese Academy of Sciences (KSCX2-YW-R-73), the National Basic Research Program of China (973 program no. 2006CB503902), the Major Projects of Knowledge Innovation Program (KSCX2-YW-R-116 and KSCX1-YW-02), and the Shanghai-Unilever Research Development Fund (CH-2006-0941).
We are grateful to all participants of the Nutrition and Health of Aging Population in China. We also thank our colleagues at the laboratory and the local Centers for Disease Control and Prevention staff of Beijing and Shanghai for their assistance with data collection.
Published ahead of print at http://diabetes.diabetesjournals.org on 15 July 2008.
Y.W. and H.L. contributed equally to this article.
The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
REFERENCES
- 1.Boutayeb A, Boutayeb S: The burden of non communicable diseases in developing countries. Int J Equity Health 4:2, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Das SK, Elbein SC: The genetic basis of type 2 diabetes. Cell Sci 2:100–131, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Owen KR, McCarthy MI: Genetics of type 2 diabetes. Curr Opin Genet Dev 17:239–244, 2007 [DOI] [PubMed] [Google Scholar]
- 4.Frayling TM: A new era in finding type 2 diabetes genes: the unusual suspects. Diabet Med 24:696–701, 2007 [DOI] [PubMed] [Google Scholar]
- 5.Frayling TM: Genome-wide association studies provide new insights into type 2 diabetes aetiology. Nat Rev Genet 8:657–662, 2007 [DOI] [PubMed] [Google Scholar]
- 6.Zeggini E: A new era for type 2 diabetes genetics. Diabet Med 24:1181–1186, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sladek R, Rocheleau G, Rung J, Dina C, Shen L, Serre D, Boutin P, Vincent D, Belisle A, Hadjadj S, Balkau B, Heude B, Charpentier G, Hudson TJ, Montpetit A, Pshezhetsky AV, Prentki M, Posner BI, Balding DJ, Meyre D, Polychronakos C, Froguel P: A genome-wide association study identifies novel risk loci for type 2 diabetes. Nature 445:881–885, 2007 [DOI] [PubMed] [Google Scholar]
- 8.The Wellcome Trust Case Control Consortium: Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature 447:661–678, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Steinthorsdottir V, Thorleifsson G, Reynisdottir I, Benediktsson R, Jonsdottir T, Walters GB, Styrkarsdottir U, Gretarsdottir S, Emilsson V, Ghosh S, Baker A, Snorradottir S, Bjarnason H, Ng MC, Hansen T, Bagger Y, Wilensky RL, Reilly MP, Adeyemo A, Chen Y, Zhou J, Gudnason V, Chen G, Huang H, Lashley K, Doumatey A, So WY, Ma RC, Andersen G, Borch-Johnsen K, Jorgensen T, van Vliet-Ostaptchouk JV, Hofker MH, Wijmenga C, Christiansen C, Rader DJ, Rotimi C, Gurney M, Chan JC, Pedersen O, Sigurdsson G, Gulcher JR, Thorsteinsdottir U, Kong A, Stefansson K: A variant in CDKAL1 influences insulin response and risk of type 2 diabetes. Nat Genet 39:770–775, 2007 [DOI] [PubMed] [Google Scholar]
- 10.Diabetes Genetics Initiative of Broad Institute of Harvard and MIT, Lund University, and Novartis Institutes of BioMedical Research, Saxena R, Voight BF, Lyssenko V, Burtt NP, de Bakker PI, Chen H, Roix JJ, Kathiresan S, Hirschhorn JN, Daly MJ, Hughes TE, Groop L, Altshuler D, Almgren P, Florez JC, Meyer J, Ardlie K, Bengtsson Boström K, Isomaa B, Lettre G, Lindblad U, Lyon HN, Melander O, Newton-Cheh C, Nilsson P, Orho-Melander M, Råstam L, Speliotes EK, Taskinen MR, Tuomi T, Guiducci C, Berglund A, Carlson J, Gianniny L, Hackett R, Hall L, Holmkvist J, Laurila E, Sjögren M, Sterner M, Surti A, Svensson M, Svensson M, Tewhey R, Blumenstiel B, Parkin M, Defelice M, Barry R, Brodeur W, Camarata J, Chia N, Fava M, Gibbons J, Handsaker B, Healy C, Nguyen K, Gates C, Sougnez C, Gage D, Nizzari M, Gabriel SB, Chirn GW, Ma Q, Parikh H, Richardson D, Ricke D, Purcell S: Genome-wide association analysis identifies loci for type 2 diabetes and triglyceride levels. Science 316:1331–1336, 2007 [DOI] [PubMed] [Google Scholar]
- 11.Scott LJ, Mohlke KL, Bonnycastle LL, Willer CJ, Li Y, Duren WL, Erdos MR, Stringham HM, Chines PS, Jackson AU, Prokunina-Olsson L, Ding CJ, Swift AJ, Narisu N, Hu T, Pruim R, Xiao R, Li XY, Conneely KN, Riebow NL, Sprau AG, Tong M, White PP, Hetrick KN, Barnhart MW, Bark CW, Goldstein JL, Watkins L, Xiang F, Saramies J, Buchanan TA, Watanabe RM, Valle TT, Kinnunen L, Abecasis GR, Pugh EW, Doheny KF, Bergman RN, Tuomilehto J, Collins FS, Boehnke M: A genome-wide association study of type 2 diabetes in Finns detects multiple susceptibility variants. Science 316:1341–1345, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zeggini E, Weedon MN, Lindgren CM, Frayling TM, Elliott KS, Lango H, Timpson NJ, Perry JR, Rayner NW, Freathy RM, Barrett JC, Shields B, Morris AP, Ellard S, Groves CJ, Harries LW, Marchini JL, Owen KR, Knight B, Cardon LR, Walker M, Hitman GA, Morris AD, Doney AS; Wellcome Trust Case Control Consortium (WTCCC), McCarthy MI, Hattersley AT: Replication of genome-wide association signals in UK samples reveals risk loci for type 2 diabetes. Science 316:1336–1341, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Palmer ND, Goodarzi MO, Langefeld CD, Ziegler J, Norris JM, Haffner SM, Bryer-Ash M, Bergman RN, Wagenknecht LE, Taylor KD, Rotter JI, Bowden DW: Quantitative trait analysis of type 2 diabetes susceptibility loci identified from whole genome association studies in the Insulin Resistance Atherosclerosis Family Study. Diabetes 57:1093–1100, 2008 [DOI] [PubMed] [Google Scholar]
- 14.Staiger H, Machicao F, Stefan N, Tschritter O, Thamer C, Kantartzis K, Schafer SA, Kirchhoff K, Fritsche A, Haring HU: Polymorphisms within novel risk loci for type 2 diabetes determine beta-cell function. PLoS ONE 2:e832, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grarup N, Rose CS, Andersson EA, Andersen G, Nielsen AL, Albrechtsen A, Clausen JO, Rasmussen SS, Jorgensen T, Sandbaek A, Lauritzen T, Schmitz O, Hansen T, Pedersen O: Studies of association of variants near the HHEX, CDKN2A/B and IGF2BP2 genes with type 2 diabetes and impaired insulin release in 10,705 Danish subjects validation and extension of genome-wide association studies. Diabetes 56:3105–3111, 2007 [DOI] [PubMed] [Google Scholar]
- 16.The International HapMap Consortium: A haplotype map of the human genome. Nature 437:1299–1320, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.The International HapMap Consortium: A second generation human haplotype map of over 3.1 million SNPs. Nature 449:851–861, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wild S, Roglic G, Green A, Sicree R, King H: Global prevalence of diabetes: estimates for the year 2000 and projections for 2030. Diabetes Care 27:1047–1053, 2004 [DOI] [PubMed] [Google Scholar]
- 19.Ye X, Yu Z, Li H, Franco OH, Liu Y, Lin X: Distributions of C-reactive protein and its association with metabolic syndrome in middle-aged and older Chinese people. J Am Coll Cardiol 49:1798–1805, 2007 [DOI] [PubMed] [Google Scholar]
- 20.World Health Organization: Definition, Diagnosis and Classification of Diabetes Mellitus and Its Complications: Reports of a WHO Consultation. Geneva, World Health Org., 1999
- 21.Levy JC, Matthews DR, Hermans MP: Correct homeostasis model assessment (HOMA) evaluation uses the computer program (Letter). Diabetes Care 21:2191–2192, 1998 [DOI] [PubMed] [Google Scholar]
- 22.Omori S, Tanaka Y, Takahashi A, Hirose H, Kashiwagi A, Kaku K, Kawamori R, Nakamura Y, Maeda S: Association of CDKAL1, IGF2BP2, CDKN2A/B, HHEX, SLC30A8, and KCNJ11 With Susceptibility to type 2 diabetes in a Japanese population. Diabetes 57:791–795, 2008 [DOI] [PubMed] [Google Scholar]
- 23.Horikoshi M, Hara K, Ito C, Shojima N, Nagai R, Ueki K, Froguel P, Kadowaki T: Variations in the HHEX gene are associated with increased risk of type 2 diabetes in the Japanese population. Diabetologia 50:2461–2466, 2007 [DOI] [PubMed] [Google Scholar]
- 24.Horikawa Y, Miyake K, Yasuda K, Enya M, Hirota Y, Yamagata K, Hinokio Y, Oka Y, Iwasaki N, Iwamoto Y, Yamada Y, Seino Y, Maegawa H, Kashiwagi A, Yamamoto K, Tokunaga K, Takeda J, Kasuga M: Replication of genome-wide association studies of type 2 diabetes susceptibility in Japan. J Clin Endocrinol Metab, 2008. [Epub ahead of print] [DOI] [PubMed]
- 25.Ng MCY, Park KS, Oh B, Tam CHT, Cho YM, Shin HD, Lam VKL, Ma RCW, So WY, Cho YS, Kim HL, Lee HK, Chan JCN, Cho NH: Implication of genetic variants near TCF7L2, SLC30A8, HHEX, CDKAL1, CDKN2A/B, IGF2BP2, and FTO in type 2 diabetes and obesity in 6,719 Asians. Diabetes, 2008 [DOI] [PMC free article] [PubMed]
- 26.Pascoe L, Tura A, Patel SK, Ibrahim IM, Ferrannini E, Zeggini E, Weedon MN, Mari A, Hattersley AT, McCarthy MI, Frayling TM, Walker M: Common variants of the novel type 2 diabetes genes, CDKAL1 and HHEX/IDE, are associated with decreased pancreatic β-cell function. Diabetes 56:3101–3104, 2007 [DOI] [PubMed] [Google Scholar]
- 27.Schulze MB, Al-Hasani H, Boeing H, Fisher E, Doring F, Joost HG: Variation in the HHEX-IDE gene region predisposes to type 2 diabetes in the prospective, population-based EPIC-Potsdam cohort. Diabetologia 50:2405–2407, 2007 [DOI] [PubMed] [Google Scholar]
- 28.Lohmueller KE, Pearce CL, Pike M, Lander ES, Hirschhorn JN: Meta-analysis of genetic association studies supports a contribution of common variants to susceptibility to common disease. Nat Genet 33:177–182, 2003 [DOI] [PubMed] [Google Scholar]
- 29.Ioannidis JP, Trikalinos TA, Ntzani EE, Contopoulos-Ioannidis DG: Genetic associations in large versus small studies: an empirical assessment. Lancet 361:567–571, 2003 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.