Abstract
OBJECTIVE— Association between genetic variants at the FTO locus and obesity has been consistently observed in populations of European ancestry and inconsistently in non-Europeans. The aim of this study was to examine the effects of FTO variants on obesity and type 2 diabetes in Southeast Asian populations.
RESEARCH DESIGN AND METHODS— We examined associations between nine previously reported FTO single nucleotide polymorphisms (SNPs) with obesity, type 2 diabetes, and related traits in 4,298 participants (2,919 Chinese, 785 Malays, and 594 Asian Indians) from the 1998 Singapore National Health Survey (NHS98) and 2,996 Malays from the Singapore Malay Eye Study (SiMES).
RESULTS— All nine SNPs exhibited strong linkage disequilibrium (r2 = 0.6–0.99), and minor alleles were associated with obesity in the same direction as previous studies with effect sizes ranging from 0.42 to 0.68 kg/m2 (P < 0.0001) in NHS98 Chinese, 0.65 to 0.91 kg/m2 (P < 0.02) in NHS98 Malays, and 0.52 to 0.64 kg/m2 (P < 0.0001) in SiMES Malays after adjustment for age, sex, smoking, alcohol consumption, and exercise. The variants were also associated with type 2 diabetes, though not after adjustment for BMI (with the exception of the SiMES Malays: odds ratio 1.17–1.22; P ≤ 0.026).
CONCLUSIONS— FTO variants common among European populations are associated with obesity in ethnic Chinese and Malays in Singapore. Our data do not support the hypothesis that differences in allele frequency or genetic architecture underlie the lack of association observed in some populations of Asian ancestry. Examination of gene-environment interactions involving variants at this locus may provide further insights into the role of FTO in the pathogenesis of human obesity and diabetes.
A recent genome-wide association study for type 2 diabetes using a U.K.-based population revealed a novel locus associated with BMI: the fat mass–and obesity-related gene (FTO) on chromosome 16 (1). The representative single-nucleotide polymorphism (SNP), rs9939609, was confirmed to be associated with elevated BMI after replication in more than 38,000 study participants of European ancestry. Further replication of this association has been observed in several populations of distinctly European ancestry (2–6). However, this association is inconsistent in populations of non-European ancestry. A study in Japanese showed an association between variants at this locus and obesity (7) that was not observed in African Americans (6) or Han Chinese (8). The aims of this study were 1) to determine the associations between previously identified obesity-associated SNPs at the FTO locus with obesity and type 2 diabetes in Chinese, Malays, and Asian-Indians and 2) to examine whether any associations were modulated by exercise.
RESEARCH DESIGN AND METHODS
This study utilized data from two cross-sectional studies: the 1998 Singapore National Health Survey (NHS98) (4,723 subjects) and the Singapore Malay Eye Study (SiMES) (3,280 subjects). NHS98 is a population-based, cross-sectional study of Chinese, Malays, and Asian Indians, aged between 18 and 69 years, that has previously been described (9,10). An interviewer-administered questionnaire was used to capture data on sociodemographic factors, smoking, and alcohol consumption. The level of physical activity was categorized into three groups: those who regularly exercised, defined as participation in any form of sports for at least 20 min for 3 or more days per week; those who occasionally exercised (<3 days per week); and those who did not exercise. BMI and blood pressure were measured for all subjects. Waist circumferences were measured at the narrowest part of the body below the costal margin, and hip circumference was measured at the widest part of the body below the waist. Fasting blood samples were drawn for measurement of serum lipids, glucose, and insulin after a 10-h overnight fast. Type 2 diabetes was defined as fasting glucose ≥7.0 mmol/l, 2 h postchallenge glucose (2HPG) ≥11.1 mmol/l, or self-reported type 2 diabetes. Impaired fasting glucose/impaired glucose tolerance was diagnosed if 6.0 mmol/l < fasting glucose <7.0 mmol/l or 7.8 mmol/l < 2HPG <11.1 mmol/l.
SiMES is a population-based, cross-sectional epidemiological study of Malay adults, aged between 40 and 79 years, that has previously been described (11–14). Serum lipids and glucose were measured in nonfasting venous samples. Type 2 diabetes was defined as random glucose >11.1 mmol/l or self-reported type 2 diabetes (additional information regarding the methods of NHS98 and SiMES can be found in an online appendix, available at http://dx.doi.org/10.2337/db08-0214).
Genotyping.
Genotype data were available for 4,298 NHS98 subjects, comprising 2,919 Chinese (1,331 male and 1,588 female), 785 Malays (377 male and 408 female), and 594 Asian Indians (284 male and 310 female). In SiMES, genotype data were available for 2,996 subjects (1,442 male and 1,554 female).
10 FTO SNPs that have previously been described (1–6,8) were selected for this study (rs9939609, rs8050136, rs1421085, rs17817449, rs7193144, rs1121980, rs9940128, rs9939973, rs9926289, and rs9930506). However, rs9930506 failed assay design and was not genotyped.
Genotyping of the remaining nine SNPs was carried out using the Sequenom MassARRAY platform (Sequenom, San Diego, CA). All nine SNPs passed the genotyping call-rate threshold (>95%) with an average call rate of 97.7% in NHS98 samples and 99.5% in SiMES samples. Genotyping success rates for individual SNPs are listed in Table 1 of the online appendix.
TABLE 1.
Clinical characteristics of the NHS98 and SiMES study populations
| NHS 98 |
SiMES |
|||
|---|---|---|---|---|
| Chinese | Malay | Indian | Malay | |
| n | 2,919 | 785 | 594 | 2,996 |
| Male (%) | 45.6 | 47.9 | 47.8 | 48.1 |
| Age (years) | 37.9 ± 12.2 | 38.9 ± 12.5 | 40.6 ± 11.9 | 58.6 ± 11.0 |
| BMI (kg/m2) | 22.7 ± 3.71 | 25.5 ± 4.96 | 25.1 ± 4.60 | 26.3 ± 5.11 |
| Waist-to-hip ratio | 0.82 ± 0.07 | 0.83 ± 0.07 | 0.85 ± 0.07 | NA |
| Waist circumference (cm) | 78.1 ± 10.6 | 82.6 ± 11.9 | 85.1 ± 11.5 | NA |
| HDL cholesterol (mmol/l) | 1.42 ± 0.37 | 1.30 ± 0.33 | 1.14 ± 0.30 | 1.35 ± 0.33 |
| LDL cholesterol (mmol/l) | 3.38 ± 0.95 | 3.86 ± 1.08 | 3.69 ± 1.03 | 3.54 ± 1.00 |
| Triglycerides (mmol/l) | 1.40 ± 1.19 | 1.67 ± 1.28 | 1.68 ± 1.36 | 1.60 ± 1.32 |
| Total cholesterol (mmol/l) | 5.41 ± 1.04 | 5.81 ± 1.15 | 5.51 ± 1.10 | 5.62 ± 1.16 |
| Fasting plasma glucose (mmol/l) | 5.62 ± 1.30 | 6.09 ± 2.23 | 6.23 ± 2.17 | NA |
| 2HPG (mmol/l) | 6.65 ± 2.76 | 7.37 ± 3.55 | 7.63 ± 4.07 | NA |
| Systolic blood pressure (mmHg) | 120. ± 16.3 | 124. ± 19.3 | 121. ± 17.1 | 147. ± 23.7 |
| Diastolic blood pressure (mmHg) | 73.7 ± 11.2 | 76.0 ± 12.0 | 73.6 ± 12.0 | 79.7 ± 11.2 |
| Hypertension | 18.6 | 25.9 | 20.2 | 68.5 |
| Glucose tolerance (%) | ||||
| Normal | 2,179 (74.7) | 473 (60.2) | 362 (60.9) | 2,288 (76.4)* |
| IFG/IGT | 515 (17.6) | 201 (25.6) | 118 (19.8) | NA |
| Type 2 diabetes | 224 (7.7) | 111 (14.1) | 114 (19.2) | 708 (23.6) |
| Currently smoking (%) | 12.4 | 22.9 | 14.6 | 20.2 |
| Regularly exercise (%)† | 14.7 | 17.6 | 22.8 | NA |
| Consume alcohol (%)‡ | 45.1 | 7.3 | 33.3 | 1.6 |
Data are means ± SD or n (%) unless otherwise indicated.
SiMES participants with no diabetes;
regular exercise defined as participation in any form of sports for at least 20 min for 3 or more days per week;
individuals who consume at least 1 alcoholic beverage per month. IFG/IGT, impaired fasting glucose/impaired glucose tolerance; NA, not applicable.
Statistical analysis.
Minor allele frequency (MAF), deviation from Hardy-Weinberg equilibrium (HWE), and linkage disequilibrium (reported as r2) were estimated for the nine SNPs using Haploview (15).
All measurement variables that were skewed were normalized by natural logarithmic transformation. Estimated means were subsequently backtransformed for presentation in the tables. Power calculations were carried out using QUANTO software (http://hydra.usc.edu/gxe). Based on an MAF of 0.12, as found in Chinese from Beijing and Shanghai (8), our study had 89% power (α level 0.05) to detect an effect size between 0.4 and 0.5 kg/m2 in BMI from NHS98 Chinese and SiMES Malays, similar to effect sizes observed in populations of European ancestry (0.4–0.66 kg/m2) (16).
Multiple linear regression analyses were performed to study the associations between individual SNPs with obesity, blood pressure, and lipid levels. A general inheritance model was fitted and an additive model used based on observed effects. Individuals were assigned as 0, 1, or 2 according to their number of minor/risk alleles, which correspond to the risk alleles associated with obesity in European populations. Initial analysis did not reveal statistically significant heterogeneity between the sexes (P > 0.1); hence, subsequent analyses were performed with the data from sexes combined with summary indexes adjusted for sex. All analyses were stratified by ethnic group. Logistic regression was used to examine the association between SNPs and categorical outcomes. To test the hypothesis that exercise may modify the effect of FTO variants, the interaction variable (SNP/exercise) was included into the regression models. The likelihood ratio test was used to estimate the P values for interaction by comparing the regression models with and without the interaction term. Where stated, adjustment for age, sex, BMI, current smoking, alcohol intake, and regular exercise was carried out by adding these variables to the model. Meta-analysis was performed to determine the pooled effects across all four populations studied using the inverse variance–weighted method. A test of heterogeneity of effects between populations was carried out using Cochran's test of heterogeneity. These statistical analyses were performed using STATA (version 9.1 for Windows). We also carried out haplotype-based analyses, the methods for which are described in the online appendix.
RESULTS
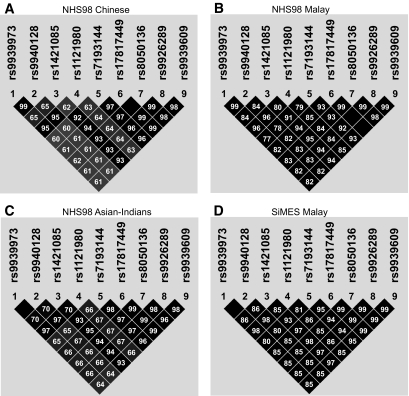
Table 1 shows the clinical characteristics of the two study populations. Allele frequencies for the nine genotyped SNPs and test for HWE deviation are listed in online appendix Table 1. The MAFs for all nine SNPs were higher in Asian Indians (0.33–0.43) than in Malays (0.28–0.33) or Chinese (0.12–0.18). None of the SNPs showed significant deviation from HWE. Figure 1 illustrates the linkage disequilibrium between the nine FTO SNPs in the different ethnic populations. A high degree of linkage disequilibrium was observed between the SNPs, with similar patterns in all three ethnic groups in our population. In addition, the linkage disequilibrium structure of our population showed similarity to that of the European population (CEU population of HapMap [online appendix Fig. 1]).
FIG. 1.
Linkage disequilibrium (r2) between the nine FTO SNPs in the NHS98 and SiMES populations.
Table 2 shows associations between the nine SNPs with obesity, impaired fasting glucose/impaired glucose tolerance, and type 2 diabetes. All nine FTO SNPs were associated with increased BMI with an effect size, per risk allele, of 0.42–0.68 kg/m2 (P < 0.0001) in NHS98 Chinese, 0.65–0.91 kg/m2 (P < 0.02) in NHS98 Malays, and 0.52–0.64 kg/m2 (P < 0.0001) in SiMES Malays. FTO variants were also associated with an increased risk of type 2 diabetes in the NHS98 Chinese (odds ratio 1.32–1.42; P ≤ 0.049), NHS98 Malays (1.52–1.63; P ≤ 0.028) and SiMES Malays (1.20–1.24; P ≤ 0.007). However, these associations were abolished after adjustment for BMI, except in the SiMES Malays (1.17–1.22; P ≤ 0.026). No statistically significant associations were observed in Asian Indians.
TABLE 2.
Association between nine FTO SNPs with obesity, risk of type 2 diabetes, and impaired fasting glucose/impaired glucose tolerance (IFG/IGT) in the NHS98 and SiMES populations
| SNP | rs9939973 | rs9940128 | rs1421085 | rs1121980 | rs7193144 | rs17817449 | rs8050136 | rs9926289 | rs9939609 |
|---|---|---|---|---|---|---|---|---|---|
| NHS98 Chinese (n = 2,919) | |||||||||
| ΔBMI per risk allele present (kg/m2) | 0.43 | 0.42 | 0.68 | 0.43 | 0.63 | 0.67 | 0.68 | 0.68 | 0.66 |
| P*/P† | <0.0001/<0.0001 | 0.001/<0.0001 | <0.0001/<0.0001 | <0.0001/<0.0001 | <0.0001/<0.0001 | <0.0001/<0.0001 | <0.0001/<0.0001 | <0.0001/<0.0001 | <0.0001/<0.0001 |
| Glucose tolerance | |||||||||
| OR (95% CI)* for type 2 diabetes vs. NGT | 1.33 (1.01–1.74) | 1.32 (1.00–1.72) | 1.42 (1.04–1.93) | 1.19 (0.91–1.53) | 1.42 (1.04–1.92) | 1.39 (1.01–1.90) | 1.40 (1.02–1.90) | 1.40 (1.02–1.90) | 1.37 (1.00–1.86) |
| P*/P† | 0.036/0.132 | 0.043/0.15 | 0.024/0.128 | 0.187/0.434 | 0.025/0.102 | 0.039/0.178 | 0.036/0.168 | 0.035/0.163 | 0.049/0.212 |
| OR (95% CI) for IFG/IGT vs. NGT* | 1.10 (0.91–1.32) | 1.09 (0.90–1.31) | 1.15 (0.92–1.42) | 1.08 (0.90–1.29) | 1.10 (0.89–1.37) | 1.14 (0.91–1.41) | 1.14 (0.91–1.40) | 1.14 (0.92–1.41) | 1.12 (0.89–1.38) |
| P*/P† | 0.323/0.527 | 0.338/0.546 | 0.197/0.503 | 0.368/0.702 | 0.366/0.729 | 0.241/0.578 | 0.25/0.611 | 0.223/0.568 | 0.313/0.684 |
| NHS98 Malay (n = 785) | |||||||||
| ΔBMI per risk allele present (kg/m2) | 0.83 | 0.82 | 0.90 | 0.65 | 0.82 | 0.91 | 0.86 | 0.88 | 0.89 |
| P*/P† | 0.001/0.002 | 0.002/0.002 | 0.001/0.001 | 0.011/0.015 | 0.002/0.002 | 0.001/0.001 | 0.001/0.002 | 0.001/0.001 | 0.001/0.001 |
| Glucose tolerance | |||||||||
| OR (95% CI)* for type 2 diabetes vs. NGT | 1.61 (1.10–2.33) | 1.60 (1.09–2.32) | 1.59 (1.09–2.32) | 1.57 (1.08–2.25) | 1.52 (1.04–2.21) | 1.63 (1.11–2.38) | 1.53 (1.04–2.24) | 1.60 (1.09–2.32) | 1.57 (1.08–2.29) |
| P*/P† | 0.013/0.082 | 0.014/0.084 | 0.016/0.095 | 0.016/0.071 | 0.027/0.12 | 0.011/0.074 | 0.028/0.129 | 0.015/0.092 | 0.018/0.104 |
| OR (95% CI) for IFG/IGT vs. NGT* | 1.34 (1.03–1.74) | 1.35 (1.03–1.74) | 1.22 (0.93–1.58) | 1.37 (1.06–1.77) | 1.21 (0.93–1.57) | 1.25 (0.95–1.63) | 1.20 (0.91–1.56) | 1.24 (0.95–1.61) | 1.22 (0.93–1.59) |
| P*/P† | 0.028/0.075 | 0.025/0.067 | 0.144/0.331 | 0.015/0.033 | 0.154/0.331 | 0.103/0.243 | 0.186/0.392 | 0.113/0.267 | 0.136/0.318 |
| NHS98 Asian-Indians (n = 594) | |||||||||
| ΔBMI per risk allele present (kg/m2) | 0.24 | 0.23 | 0.03 | 0.28 | 0.12 | 0.10 | 0.04 | 0.11 | 0.10 |
| P*/P† | 0.352/0.333 | 0.36/0.347 | 0.913/0.781 | 0.267/0.247 | 0.658/0.458 | 0.715/0.527 | 0.877/0.719 | 0.674/0.487 | 0.7/0.532 |
| Glucose tolerance | |||||||||
| OR (95% CI)* for type 2 diabetes vs. NGT | 0.84 (0.59–1.19) | 0.85 (0.59–1.20) | 0.87 (0.60–1.25) | 0.94 (0.66–1.31) | 0.93 (0.64–1.32) | 0.85 (0.58–1.23) | 0.87 (0.60–1.24) | 0.88 (0.61–1.27) | 0.96 (0.67–1.37) |
| P*/P† | 0.341/0.356 | 0.349/0.365 | 0.467/0.564 | 0.711/0.74 | 0.677/0.766 | 0.393/0.438 | 0.445/0.53 | 0.51/0.543 | 0.84/0.914 |
| OR (95% CI) for IFG/IGT vs. NGT* | 0.95 (0.69–1.29) | 0.96 (0.70–1.30) | 0.90 (0.65–1.24) | 1.05 (0.77–1.41) | 0.88 (0.63–1.21) | 0.85 (0.60–1.17) | 0.91 (0.66–1.25) | 0.87 (0.62–1.20) | 0.87 (0.63–1.20) |
| P*/P† | 0.754/0.707 | 0.775/0.732 | 0.515/0.509 | 0.774/0.895 | 0.432/0.404 | 0.327/0.294 | 0.576/0.565 | 0.411/0.378 | 0.419/0.369 |
| SiMES Malay | |||||||||
| ΔBMI per risk allele present (kg/m2) | 0.55 | 0.55 | 0.64 | 0.52 | 0.62 | 0.64 | 0.63 | 0.61 | 0.64 |
| P*/P† | <0.0001/<0.0001 | 0.001/<0.0001 | <0.0001/<0.0001 | <0.0001/<0.0001 | <0.0001/<0.0001 | <0.0001/<0.0001 | <0.0001/<0.0001 | <0.0001/<0.0001 | <0.0001/<0.0001 |
| Glucose tolerance | |||||||||
| OR (95% CI)* for type 2 diabetes vs. NGT | 1.23 (1.08–1.40) | 1.24 (1.08–1.40) | 1.20 (1.05–1.37) | 1.24 (1.08–1.40) | 1.21 (1.05–1.37) | 1.20 (1.05–1.37) | 1.20 (1.05–1.37) | 1.20 (1.05–1.37) | 1.21 (1.05–1.38) |
| P*/P† | 0.001/0.004 | 0.001/0.004 | 0.007/0.025 | 0.001/0.003 | 0.005/0.019 | 0.007/0.026 | 0.007/0.023 | 0.006/0.021 | 0.005/0.019 |
Adjusted for age and sex;
adjusted for age, sex, current smoking, exercise (except in the SiMES population), BMI (for risk of type 2 diabetes), education, and alcohol consumption. NGT, normal glucose tolerance.
We next examined the association between FTO SNPs with obesity-related traits. Table 3 summarizes the association between rs9939609 and these traits. We chose rs9939609 as the representative SNP in our study because it was the index SNP in the original study (1) and had one of the strongest associations with BMI. In our study, rs9939609 also showed highly significant association with waist circumference in both NHS98 Chinese (P < 0.0001) and NHS98 Malay (P = 0.001); waist circumference data were not available for SiMES Malays. Borderline association was also observed with LDL cholesterol and triglyceride, the latter only after adjustment for BMI, in NHS98 Chinese samples. Details of the associations for the other SNPs are listed in online appendix Table 2. Meta-analysis of the association between this SNP and BMI for all four populations in our study was also performed. No significant heterogeneity was observed between the four populations, and the pooled effect size was BMI 0.62 kg/m2 (95% CI 0.45–0.80; P = 2 × 10−12). Haplotype-based analyses recapitulated findings from individual SNP analyses; these results are shown in online appendix Table 3.
TABLE 3.
Association of FTO SNP rs9939609 with obesity-related traits in the NHS98 and SiMES populations
| 1998 Singapore National Health Study (NHS98) |
Singapore Malay Eye Study (SiMES) |
|||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Chinese (n = 2,919) |
Malay (n = 785) |
Asian Indians (n = 594) |
SiMES Malay (n = 2,996) |
|||||||||||||||||
| 0 | 1 | 2 | P* | P† | 0 | 1 | 2 | P* | P† | 0 | 1 | 2 | P* | P† | 0 | 1 | 2 | P* | P† | |
| HDL cholesterol (mmol/l) | 1.42 | 1.42 | 1.42 | 0.735 | 0.127 | 1.31 | 1.29 | 1.27 | 0.244 | 0.792 | 1.15 | 1.14 | 1.14 | 0.832 | 0.741 | 1.36 | 1.34 | 1.33 | 0.164 | 0.785 |
| LDL cholesterol (mmol/l) | 3.36 | 3.44 | 3.52 | 0.024 | 0.298 | 3.87 | 3.86 | 3.84 | 0.787 | 0.248 | 3.70 | 3.70 | 3.70 | 0.787 | 0.939 | 3.52 | 3.55 | 3.58 | 0.251 | 0.508 |
| Triglycerides (mmol/l) | 1.18 | 1.18 | 1.17 | 0.813 | 0.037 | 1.37 | 1.44 | 1.52 | 0.062 | 0.522 | 1.42 | 1.43 | 1.44 | 0.876 | 0.833 | 1.25 | 1.26 | 1.27 | 0.752 | 0.955 |
| Waist (cm) | 77.84 | 79.09 | 80.35 | <0.0001 | <0.0001 | 81.70 | 83.56 | 85.42 | 0.001 | 0.002 | 84.97 | 85.17 | 85.38 | 0.74 | 0.775 | NA | — | — | ||
| Waist-to-hip ratio | 0.82 | 0.83 | 0.83 | 0.184 | 0.174 | 0.83 | 0.83 | 0.84 | 0.198 | 0.202 | 0.86 | 0.85 | 0.85 | 0.584 | 0.597 | NA | — | — | ||
| Systolic BP (mmHG) | 120.6 | 121.0 | 121.4 | 0.49 | 0.641 | 124.3 | 125.4 | 126.5 | 0.236 | 0.835 | 120.6 | 121.7 | 122.9 | 0.194 | 0.175 | 147.0 | 147.4 | 147.7 | 0.541 | 0.949 |
| Diastolic BP (mmHG) | 73.6 | 74.2 | 74.7 | 0.179 | 0.79 | 75.7 | 76.7 | 77.7 | 0.108 | 0.995 | 72.7 | 74.1 | 75.6 | 0.039 | 0.027 | 79.9 | 79.8 | 79.6 | 0.56 | 0.308 |
Genotypes 0, 1, and 2 represent numbers of risk/minor alleles, which correspond with risk alleles in previous studies in Europeans.
Adjusted for age and sex;
adjusted for age, sex, current smoking, exercise (except in SiMES), BMI (where appropriate), education, and alcohol consumption. BP, blood pressure; IFG/IGT, impaired fasting glucose/impaired glucose tolerance; NA, not applicable; NGT, normal glucose tolerance.
We also examined the interaction between rs9939609 and physical activity in relation to BMI (online appendix Fig. 2). Although it appeared that rs9939609 had a smaller effect on BMI in those who exercised regularly, the interaction was not statistically significant (P = 0.248).
DISCUSSION
In our study, FTO variants showed associations with obesity and type 2 diabetes in Chinese and Malays living in Singapore. Similar effects were not observed in Asian Indian samples from the NHS98 cohort. However, it should be noted that given the relatively small sample size (n = 594) for this ethnic group, we had only 40% power to detect changes in BMI of 0.5 kg/m2 for this population. Furthermore, meta-analysis showed no heterogeneity of effect between populations. We also examined the association between these SNPs and other obesity-related traits. Other than a strong association with waist circumference, only borderline associations were observed with LDL cholesterol and triglycerides. A recent study by Freathy et al. (17) showed associations with LDL cholesterol in the same direction as that observed in our study. Their study also suggested that much larger sample sizes than are currently available are required to detect the effect of these genetic variants on secondary traits related to obesity. Given the multiple associations tested, we also cannot exclude the possibility that these could represent false-positive findings.
Results from our NHS98 Chinese subjects are in line with findings in a Japanese study (7) but in contrast to those of a recent study by Li et al. (8) describing the lack of association between FTO variants and obesity in Han Chinese resident in China. Our study had a similarly large sample size (n = 2,919 for NHS98 Chinese vs. n = 3,210 for Han Chinese) and, as described in research design and methods, was more than adequately powered to detect an effect size on BMI similar to that observed in populations of European ancestry. The authors suggested that the lower MAFs and differences in genetic architecture at this locus between Europeans and Chinese may have contributed to the lack of association in other ethnic groups. It has been suggested that population differences in the patterns of association such as these may occur as a result of an evolutionary divergence that might reflect a history of negative selection against the FTO risk alleles in African and Chinese populations, as has been suggested in relation to variants at the TCF7L2 locus (18). However, it seems unlikely that any of these hypotheses provide an explanation for these different findings. The MAFs of FTO SNPs examined were very similar between the Chinese in our population and the Han Chinese from Beijing and Shanghai, yet we observed a strong association with obesity. Importantly, we studied two of the three SNPs tested by Li et al. in Chinese Hans (rs8050136 and rs9939609), both of which were strongly associated with obesity traits. Furthermore, comparisons of linkage disequilibrium patterns between our study populations and European (CEU) populations from HapMap (excluding rs9926289, where no data were available based on NCBI Build36) revealed no major differences in this region. The nine SNPs tested were correspondingly in strong linkage disequilibrium with each other in our Chinese (r2 = 0.60–0.99), Malay (r2 = 0.77–0.99 in NHS98 and r2 = 0.80–0.99 in SiMES), and Asian Indian (r2 = 0.64–0.99) samples, as they were in the CEU population from HapMap (r2 = 0.83–0.96 [online appendix Fig. 1]).
The common form of obesity is a multifactorial condition thought to develop from an intricate interplay of genes and environmental factors such as dietary habits and levels of physical activity. The occurrence of gene-gene and gene-environment factors would, therefore, make it difficult to clearly elucidate the role of specific genetic variants in obesity risk (19). Recent studies have already recognized the significance of environmental modulation in variants of LIPC, APOA5, and PPARG with metabolic traits (20). Likewise, possible explanations for the differences in associations seen in our population compared with the findings of Li et al. (8) could lie in different exposures to environmental or lifestyle factors between the populations. For example, Andreason et al. (4) reported that physical activity attenuated the effects of the FTO variants on obesity. This may be relevant given that studies in Shanghai Chinese by Lee et al. (21) and Jurj et al. (22) have reported that an average of 35% of subjects participated in regular exercise compared with 14.7% of the NHS98 Singapore Chinese population. While our study found no significant interaction with physical activity, our study may have been underpowered to detect these interactions (assuming an MAF of 12%, we only had 40% power at an α level of 0.05). Furthermore, recent findings related to the association between ROBO1 variants and obesity have emphasized the importance of age-gene interactions that may result in nonreplication (23). The differences in average age between the two Chinese populations (37.9 ± 12.2 years in NHS98 Chinese vs. 58.6 ± 6.0 years in Han Chinese) may have contributed to the discrepancies. Although this is an interesting hypothesis, it should be noted that the FTO associations with BMI showed no heterogeneity across populations of European ancestry with greatly varying mean ages (1).
Interestingly, adjustment for BMI diminished but did not abolish the association with type 2 diabetes among SiMES Malays. Perhaps this indicates a direct effect of FTO with type 2 diabetes, which, to the best of our knowledge, has not been observed in other studies. Another possibility could relate to residual confounding by obesity. It is well known that compared with Caucasians of similar BMI, Asians have different levels of adiposity and, thus, risks of type 2 diabetes (24). Consequently, adjusting for BMI may not fully account for the confounding effects of adiposity on the risk of type 2 diabetes in the SiMES Malays.
In conclusion, we have found that variants at the FTO locus are associated with obesity in ethnic Chinese and Malays living in Singapore. In addition, statistically significant associations with type 2 diabetes were observed in Chinese and Malays. These two ethnic groups represent a large proportion of the population living in Southeast Asia, a region wherein a dramatic increase in the burden of diabetes is anticipated over the next several decades (25). Our findings make it unlikely that differences in allele frequency or genetic architecture underlie the lack of association reported between these variants and obesity-related traits in Chinese Hans (8). However, it is still possible that varied linkage disequilibrium structures and lower MAFs could reduce power to detect associations in other populations. Given these findings, it seems important to explore interactions between these genetic variants with lifestyle factors (e.g., physical activity) to better elucidate possible gene-environmental interactions that may underlie population differences.
Supplementary Material
Published ahead of print at http://diabetes.diabetesjournals.org on 3 July 2008.
J.T.T. and R.D. contributed equally to this study.
The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
REFERENCES
- 1.Frayling TM, Timpson NJ, Weedon MN, Zeggini E, Freathy RM, Lindgren CM, Perry JR, Elliott KS, Lango H, Rayner NW, Shields B, Harries LW, Barrett JC, Ellard S, Groves CJ, Knight B, Patch AM, Ness AR, Ebrahim S, Lawlor DA, Ring SM, Ben-Shlomo Y, Jarvelin MR, Sovio U, Bennett AJ, Melzer D, Ferrucci L, Loos RJ, Barroso I, Wareham NJ, Karpe F, Owen KR, Cardon LR, Walker M, Hitman GA, Palmer CN, Doney AS, Morris AD, Smith GD, Hattersley AT, McCarthy MI: A common variant in the FTO gene is associated with body mass index and predisposes to childhood and adult obesity. Science 316:889–894, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dina C, Meyre D, Gallina S, Durand E, Korner A, Jacobson P, Carlsson LM, Kiess W, Vatin V, Lecoeur C, Delplanque J, Vaillant E, Pattou F, Ruiz J, Weill J, Levy-Marchal C, Horber F, Potoczna N, Hercberg S, Le Stunff C, Bougneres P, Kovacs P, Marre M, Balkau B, Cauchi S, Chevre JC, Froguel P: Variation in FTO contributes to childhood obesity and severe adult obesity. Nat Genet 39:724–726, 2007 [DOI] [PubMed] [Google Scholar]
- 3.Hinney A, Nguyen TT, Scherag A, Friedel S, Bronner G, Muller TD, Grallert H, Illig T, Wichmann HE, Rief W, Schafer H, Hebebrand J: Genome wide association (GWA) study for early onset extreme obesity supports the role of fat mass and obesity associated gene (FTO) variants. PLoS ONE 2:e1361, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Andreasen CH, Stender-Petersen KL, Mogensen MS, Torekov SS, Wegner L, Andersen G, Nielsen AL, Albrechtsen A, Borch-Johnsen K, Rasmussen SS, Clausen JO, Sandbaek A, Lauritzen T, Hansen L, Jorgensen T, Pedersen O, Hansen T: Low physical activity accentuates the effect of the FTO rs9939609 polymorphism on body fat accumulation. Diabetes 57:95–101, 2008 [DOI] [PubMed] [Google Scholar]
- 5.Peeters A, Beckers S, Verrijken A, Roevens P, Peeters P, Van Gaal L, Van Hul W: Variants in the FTO gene are associated with common obesity in the Belgian population. Mol Genet Metab 93:481–484, 2008 [DOI] [PubMed] [Google Scholar]
- 6.Scuteri A, Sanna S, Chen WM, Uda M, Albai G, Strait J, Najjar S, Nagaraja R, Orru M, Usala G, Dei M, Lai S, Maschio A, Busonero F, Mulas A, Ehret GB, Fink AA, Weder AB, Cooper RS, Galan P, Chakravarti A, Schlessinger D, Cao A, Lakatta E, Abecasis GR: Genome-wide association scan shows genetic variants in the FTO gene are associated with obesity-related traits. PLoS Genet 3:e115, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Omori S, Tanaka Y, Takahashi A, Hirose H, Kashiwagi A, Kaku K, Kawamori R, Nakamura Y, Maeda S: Association of CDKAL1, IGF2BP2, CDKN2A/B, HHEX, SLC30A8, and KCNJ11 with susceptibility to type 2 diabetes in a Japanese population. Diabetes 57:791–795, 2008 [DOI] [PubMed] [Google Scholar]
- 8.Li H, Wu Y, Loos RJ, Hu FB, Liu Y, Wang J, Yu Z, Lin X: Variants in the fat mass–and obesity-associated (FTO) gene are not associated with obesity in a Chinese Han population. Diabetes 57:264–268, 2008 [DOI] [PubMed] [Google Scholar]
- 9.Tai ES, Ordovas JM, Corella D, Deurenberg-Yap M, Chan E, Adiconis X, Chew SK, Loh LM, Tan CE: The TaqIB and -629C>A polymorphisms at the cholesteryl ester transfer protein locus: associations with lipid levels in a multiethnic population: the 1998 Singapore National Health Survey. Clin Genet 63:19–30, 2003 [DOI] [PubMed] [Google Scholar]
- 10.Cutter J, Tan BY, Chew SK: Levels of cardiovascular disease risk factors in Singapore following a national intervention programme. Bull World Health Organ 79:908–915, 2001 [PMC free article] [PubMed] [Google Scholar]
- 11.Shankar A, Leng C, Chia KS, Koh D, Tai ES, Saw SM, Lim SC, Wong TY: Association between body mass index and chronic kidney disease in men and women: population-based study of Malay adults in Singapore. Nephrol Dial Transplant 23:1910–1918, 2008 [DOI] [PubMed] [Google Scholar]
- 12.Su DH, Wong TY, Wong WL, Saw SM, Tan DT, Shen SY, Loon SC, Foster PJ, Aung T: Diabetes, hyperglycemia, and central corneal thickness: the Singapore Malay Eye Study. Ophthalmology 115:964–968, 2008 [DOI] [PubMed] [Google Scholar]
- 13.Foong AW, Saw SM, Loo JL, Shen S, Loon SC, Rosman M, Aung T, Tan DT, Tai ES, Wong TY: Rationale and methodology for a population-based study of eye diseases in Malay people: the Singapore Malay eye study (SiMES). Ophthalmic Epidemiol 14:25–35, 2007 [DOI] [PubMed] [Google Scholar]
- 14.Wong TY, Chong EW, Wong WL, Loo JL, Shen S, Loon SC, Rosman M, Aung T, Tan DTH, Tai ES, Saw SM: Prevalence and causes of visual impairment and blindness in an urban Malay Population: the Singapore Malay Eye Study (SiMES). Arch Ophthalmology 126:1091–1099, 2008 [DOI] [PubMed] [Google Scholar]
- 15.Barrett JC, Fry B, Maller J, Daly MJ: Haploview: analysis and visualization of linkage disequilibrium and haplotype maps. Bioinformatics 21:263–265, 2005 [DOI] [PubMed] [Google Scholar]
- 16.Loos RJ, Bouchard C: FTO: the first gene contributing to common forms of human obesity. Obes Rev 9:246–250, 2008 [DOI] [PubMed] [Google Scholar]
- 17.Freathy RM, Timpson NJ, Lawlor DA, Pouta A, Ben-Shlomo Y, Ruokonen A, Ebrahim S, Shields B, Zeggini E, Weedon MN, Lindgren CM, Lango H, Melzer D, Ferrucci L, Paolisso G, Neville MJ, Karpe F, Palmer CN, Morris AD, Elliott P, Jarvelin MR, Smith GD, McCarthy MI, Hattersley AT, Frayling TM: Common variation in the FTO gene alters diabetes-related metabolic traits to the extent expected given its effect on BMI. Diabetes 57:1419–1426, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Helgason A, Palsson S, Thorleifsson G, Grant SF, Emilsson V, Gunnarsdottir S, Adeyemo A, Chen Y, Chen G, Reynisdottir I, Benediktsson R, Hinney A, Hansen T, Andersen G, Borch-Johnsen K, Jorgensen T, Schafer H, Faruque M, Doumatey A, Zhou J, Wilensky RL, Reilly MP, Rader DJ, Bagger Y, Christiansen C, Sigurdsson G, Hebebrand J, Pedersen O, Thorsteinsdottir U, Gulcher JR, Kong A, Rotimi C, Stefansson K: Refining the impact of TCF7L2 gene variants on type 2 diabetes and adaptive evolution. Nat Genet 39:218–225, 2007 [DOI] [PubMed] [Google Scholar]
- 19.Marti A, Moreno-Aliaga MJ, Hebebrand J, Martinez JA: Genes, lifestyles and obesity. Int J Obes Relat Metab Disord 28 (Suppl 3):S29–S36, 2004 [DOI] [PubMed] [Google Scholar]
- 20.Grarup N, Andersen G: Gene-environment interactions in the pathogenesis of type 2 diabetes and metabolism. Curr Opin Clin Nutr Metab Care 10:420–426, 2007 [DOI] [PubMed] [Google Scholar]
- 21.Lee SA, Xu WH, Zheng W, Li H, Yang G, Xiang YB, Shu XO: Physical activity patterns and their correlates among Chinese men in Shanghai. Med Sci Sports Exerc 39:1700–1707, 2007 [DOI] [PubMed] [Google Scholar]
- 22.Jurj AL, Wen W, Gao YT, Matthews CE, Yang G, Li HL, Zheng W, Shu XO: Patterns and correlates of physical activity: a cross-sectional study in urban Chinese women. BMC Public Health 7:213, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lasky-Su J, Lyon HN, Emilsson V, Heid IM, Molony C, Raby BA, Lazarus R, Klanderman B, Soto-Quiros ME, Avila L, Silverman EK, Thorleifsson G, Thorsteinsdottir U, Kronenberg F, Vollmert C, Illig T, Fox CS, Levy D, Laird N, Ding X, McQueen MB, Butler J, Ardlie K, Papoutsakis C, Dedoussis G, O'Donnell CJ, Wichmann HE, Celedon JC, Schadt E, Hirschhorn J, Weiss ST, Stefansson K, Lange C: On the replication of genetic associations: timing can be everything! Am J Hum Genet 82:849–858, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yoon KH, Lee JH, Kim JW, Cho JH, Choi YH, Ko SH, Zimmet P, Son HY: Epidemic obesity and type 2 diabetes in Asia. Lancet 368:1681–1688, 2006 [DOI] [PubMed] [Google Scholar]
- 25.Zimmet P, Alberti KG, Shaw J: Global and societal implications of the diabetes epidemic. Nature 414:782–787, 2001 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.