Abstract
Lysine acetylation has emerged as a major posttranslational modification for histones. Cross-regulation between this and other modifications is crucial in modulating chromatin-based transcriptional control and shaping inheritable epigenetic programs. In addition to histones, many other nuclear proteins and various cytoplasmic regulators are subject to lysine acetylation. This review focuses on recent findings pertinent to acetylation of non-histone proteins and emphasizes how this modification might crosstalk with phosphorylation, methylation, ubiquitination, sumoylation, proline isomerization, and others to form code-like multisite modification programs for dynamic control of cellular signaling under diverse conditions.
Introduction
Post-translational modification (PTM) is crucial for regulating the functions of many eukaryotic proteins. Among the prominent PTMs are Ser, Thr and Tyr phosphorylation; Lys ubiquitination, sumoylation and neddylation; Lys acetylation; Lys and Arg methylation; and Pro isomerization. The Lys side chain is thus a target of different PTMs, but these PTMs are mutually exclusive on the same Lys and generate a great potential for cross-regulation (Fig. 1A). First identified in histones 40 years ago, Lys acetylation is now known to occur in over 80 transcription factors, many other nuclear regulators, and various cytoplasmic proteins (Glozak et al., 2005). As a result, this PTM is not only crucial in the nucleus, but also appears to be important for regulating different cytoplasmic processes, including cytoskeleton dynamics, energy metabolism, endocytosis, autophagy, and even signaling from the plasma membrane (Matthias et al., 2008; Yang and Seto, 2008).
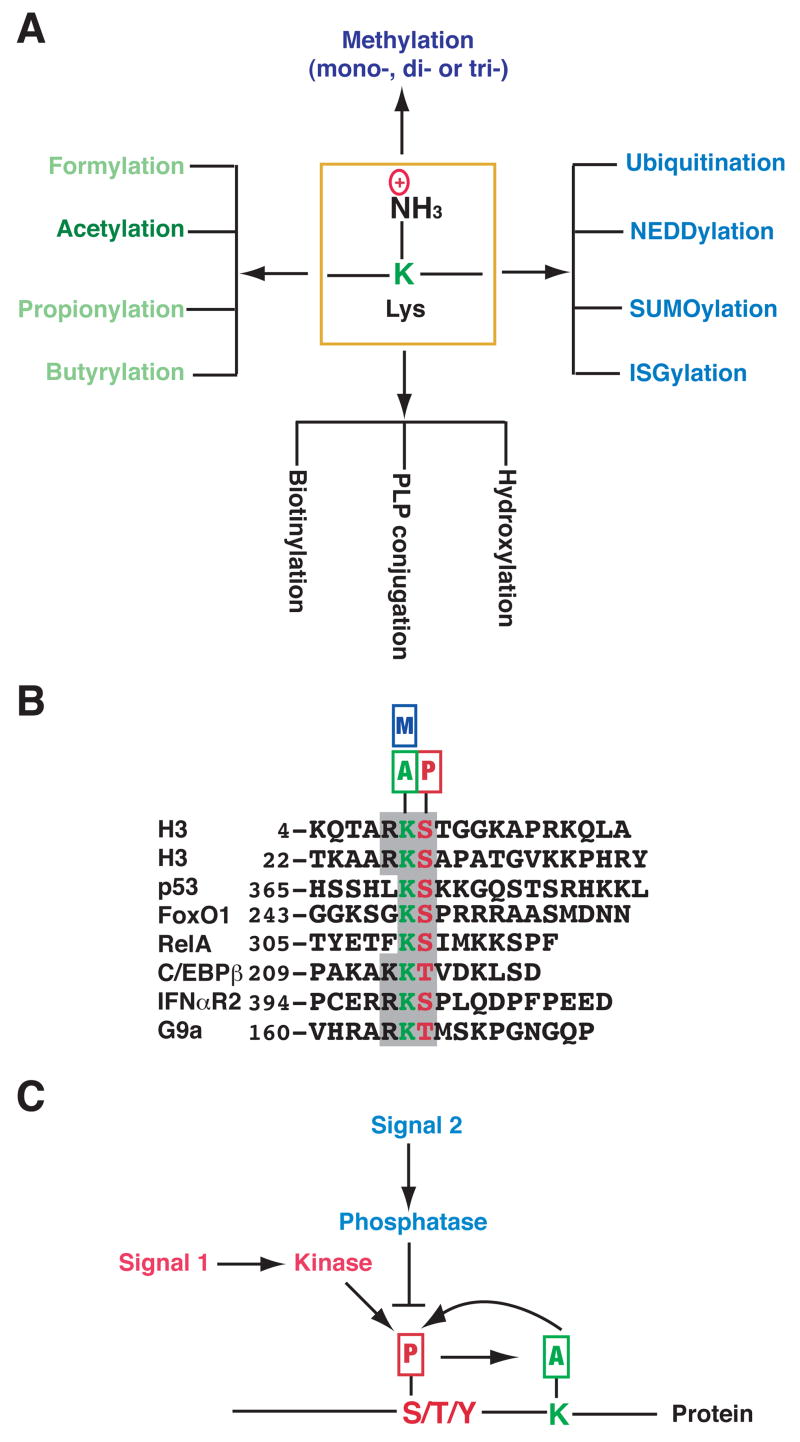
Figure 1. Multiplicity of Lys modifications and signal conversion from phosphorylation to acetylation.
(A) The side chain of a Lys (K) residue can be modified by acetylation and other covalent modifications. Except for hydroxylation, all listed modifications target the ε-amino group. Formylation, propionylation and butyrylation are three recently discovered modifications that are structurally similar to acetylation; however, it is not known if formylation occurs enzymatically. Aside from formylation, hydroxylation and PLP (pyridoxal 5′-phosphate, a vitamin B6 derivative) conjugation, all listed modifications are reversible. Acetylation of a Lys residue precludes further modifications by others, and vice versa.
(B) Alignment of sequences surrounding KS dipeptides from histone H3, p53 and several other proteins. The KS dipeptides are highlighted in color. Small rectangles with the letters A, P and M denote acetylation, phosphorylation and methylation, respectively. IFNαR2 (interferon α receptor 2) acetylation at Lys 399 and phosphorylation at Ser 400 might crosstalk with one another (Tang et al., 2007). It remains to be established whether similar interplay occurs in p53, FoxO1, RelA (NF-κB subunit) (Perkins, 2006), and C/EBPβ (CCAAT/enhancer binding protein β) (Hasselgren, 2007). Methylation of the highlighted Lys residues has been documented only for histone H3, p53 (Huang et al., 2006b) and G9a (Sampath et al., 2007). It is unclear whether the Ser in p53 is phosphorylated or the Lys in G9a is acetylated.
(C) Cartoon illustrating crosstalk between phosphorylation and acetylation. Different signals act on Ser (S), Thr (T) or Tyr (Y) phosphorylation, which in turn affects acetylation of a neighboring Lys. Acetylation might also regulate phosphorylation. The Lys can be adjacent to or far away from the phosphorylation site, which can be either N-terminal or C-terminal from the acetylation site.
In addition to acetylation, other PTMs such as methylation, ubiquitination and phosphorylation have been identified in histones (Kouzarides, 2007; Ruthenburg et al., 2007). Different PTMs form a complex regulatory program with characteristics of a sophisticated language (Strahl and Allis, 2000; Berger, 2007), and such a program is fundamental to normal development and disease pathogenesis (Margueron et al., 2005; Bhaumik et al., 2007). Like histones, many non-histone proteins are subject to acetylation and other PTMs, raising an important question: can we extrapolate and apply what we have learned about histone modifications to non-histone proteins? The simple answer is yes. For example, acetylation of histone H3 at Lys 9 and 27 crosstalks with phosphorylation of Ser 10 and 28, respectively (Fig. 1B) (Latham and Dent, 2007). Similar crosstalk might also occur in other proteins (Fig. 1B). As phosphorylation is often the first wave of PTMs in response to cellular stimuli, the crosstalk provides an effective means to convert phosphorylation-based signals to acetylation-based actions (Fig. 1C).
Recent studies reiterate that in some non-histone proteins, acetylation mingles with phosphorylation, methylation, ubiquitination, sumoylation and other PTMs to form complex regulatory programs. Here we select a few representative examples to illustrate the complexity of such programs. We begin with an overview of Lys acetyltransferases (KATs) and deacetylases (KDACs) along with a short description of KAT autoacetylation. We then examine how acetylation modulates signaling pathways important for nuclear transcriptional control and describe how this PTM is involved in regulation of cytoplasmic proteins. Concurrently, we speculate on how relative sequence conservation could shed light on the occurrence and impact of PTMs. To understand how acetylation might crosstalk with other PTMs, we consider the protein modification code, a concept similar to the hotly debated ‘histone code’(Ruthenburg et al., 2007). We end with a discussion of potential directions for future research.
Lys acetyltransferases and deacetylases
Lys acetylation was initially identified in histones, so many KATs and KDACs are often referred to as histone acetyl transferases (HATs) and deacetylases (HDACs), respectively. There are three major groups of HATs: GNATs (Gcn5-related N-acetyltransferases), p300 (E1A-associated protein of 300 kDa)/CBP (CREB-binding protein), and MYST proteins (Allis et al., 2007; Lee and Workman, 2007). Unlike the GNAT and MYST families, the p300/CBP group is unique to metazoans (Fig. 1) (Goodman and Smolik, 2000). Known HDACs are divided into the Rpd3/Hda1 and sirtuin families. In humans, the former contains HDAC1, 2, 3 and 8 (class I, similar to yeast Rpd3); HDAC4, 5, 6, 7, 9 and 10 (class II, homologs of yeast Hda1); and HDAC11 (class IV) (Yang and Seto, 2008). Within the sirtuin family, there are seven members in humans (SIRT1-7, related to yeast Sir2; also known as class III HDACs) (Haigis and Guarente, 2006). As HDAC1, 2 and 3 are catalytic subunits of multiprotein complexes, it is important to consider whether a deacetylase itself or one of its complexes is the enzyme targeting a given substrate. Similar consideration should also be made for GCN5, PCAF and the MYST family of acetyltransferases, all of which form multiprotein complexes (Lee and Workman, 2007).
Autoacetylation of p300 and CBP
Reminiscent of phosphorylation of kinases and phosphatases, acetyltransferases and deacetylases themselves are acetylated. Prominent examples are p300 and CBP, which function as coactivators for a variety of transcription factors (Fig. 2A) and play key roles in diverse physiological and pathological processes (Goodman and Smolik, 2000). They possess modular domains conserved from C. elegans to humans. p300 and CBP are heavily autoacetylated (Fig. 2) in an intermolecular fashion (Karanam et al., 2006), suggesting that dimerization triggers autoacetylation and activation. A cluster of 12 acetylated sites is located within a trypsin-sensitive loop, forms a charged patch, and serves as an activation switch (Fig. 2B) (Thompson et al., 2004). An attractive model is that DNA-binding proteins recruit p300 and bring two or more p300 molecules into physical proximity, leading to intermolecular acetylation and enzymatic activation. Consistent with this idea, many transcription factors stimulate the acetyltransferase activity of p300 and CBP. Moreover, recruitment to DNA is important for p300 activation (Dornan et al., 2003). This model is reminiscent of the way in which ligands activate membrane receptors.
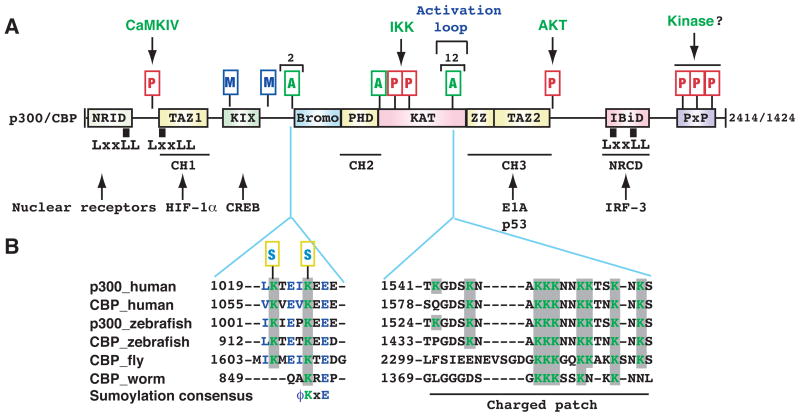
Figure 2. Autoacetylation and other modifications of p300 and CBP.
(A) Domain organization of human p300 and CBP. As mammalian p300 and CBP are functionally interchangeable in many in vitro assays, the term p300/CBP is used to refer to one or the other. Acetylation, phosphorylation, methylation and sumoylation sites are denoted with small rectangles containing the letters A, P, M and S, respectively. Within the activation loop of p300 and perhaps also CBP, there are 12 acetylation sites. The responsible kinases are indicated for the phosphorylation sites. Representative transcription factors that use p300 and CBP as coactivators are listed along with arrows pointing to the respective binding domains. Domain abbreviation: Bromo, bromodomain; CH1, Cys- & His-rich domain 1; IBiD, IRF-3 binding domain; KIX, kinase-inducing domain (KID) binding region; NRCD, nuclear receptor coactivator binding domain (synonymous to IBiD); NRID, nuclear receptor interacting domain; PxP, Pro-rich domain (no known function); TAZ1, transcriptional adaptor zinc finger; ZZ, zinc finger near the dystrophin WW domain.
(B) Regional sequence comparison of p300 and CBP from human and other species. The alignment was generated and shaded using ClustalW of MacVector 7.2 (Accelrys), using GenBank accession numbers Q09472, 119943104, 125830995, 125850731, 24640865, and 17552710. According to the new nomenclature system for histone-modifying enzymes (Allis et al., 2007), mammalian CBP and p300 are referred to as KAT3A and KAT3B, respectively. In the sumoylation consensus, φ is preferably a bulky residue (e.g., Leu, Ile or Val). Note that Lys 1057 of mouse CBP (1056 in humans) has been shown to be sumoylated (Kuo et al., 2005).
In addition, acetylation occurs at the tandem sumoylation sites upstream from the p300 bromodomain (Fig. 2B) to block sumoylation and relieve transcriptional repression (Girdwood et al., 2003). Both sites are conserved in homologs from zebrafish to humans (Fig. 2B). p300 and CBP are phosphorylated and methylated at multiple sites (Fig. 2A) (Legube and Trouche, 2003). It will be interesting to decipher how acetylation coordinates with phosphorylation, sumoylation, methylation and ubiquitination to regulate p300 and CBP activities in a concerted fashion.
p53 acetylation in the DNA damage response and beyond
The genomic guardian and tumor suppressor p53 is one of the most studied transcription factors (Vogelstein et al., 2000; Vousden and Lane, 2007). Its gene is commonly mutated in different cancers (Soussi et al., 2006). In response to various stimuli, p53 induces cell cycle arrest, promotes apoptosis, and affects other cellular programs including senescence and differentiation. A very unstable protein, p53 is modified by ubiquitination, acetylation, and other PTMs at multiple sites (Fig. 3A) and thus constitutes a paradigm for understanding how acetylation crosstalks with other modifications under diverse cellular conditions (Appella and Anderson, 2000; Brooks and Gu, 2003; Toledo and Wahl, 2006). Among acetylated non-histone proteins, p53 is the most extensively characterized, and its regulation by acetylation is thus discussed in detail here.
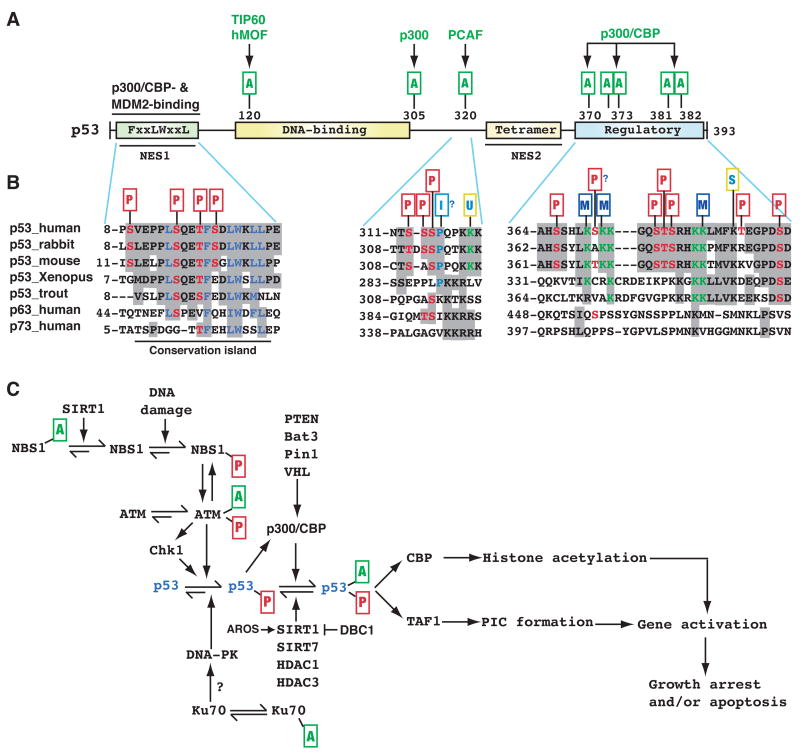
Figure 3. Regulation of p53 modifications in response to DNA damage.
(A) Domain organization of human p53. Residues 8–28 harbor overlapping binding sites for p300/CBP and the ubiquitin ligase MDM2, whereas within residues 369–393 there is a cluster of five Lys residues acetylated by p300/CBP. These residues are also targets of ubiquitination and neddylation (Brooks and Gu, 2003). Other acetylation sites are shown along with the responsible KATs. Unlike Lys372, methylation of Lys 370 or 382 inhibits p53 activity. NES, nuclear export signal; tetramer, tetramerization domain; regulatory, regulatory region.
(B) Sequences encompassing residues 8–28, 311–321 and 369–393 of human p53 are compared with the corresponding regions of orthologs and paralogs. p53 contains numerous phosphorylation sites (Toledo and Wahl, 2006), but only those located within these three regions are shown. PTMs are marked as in Fig. 2, and the small rectangle with letter I denotes proline isomerization. Sequence analysis was performed as in Fig. 2B, using GenBank accession numbers ABM86630, 126722898, 148747262, NP_0010811568, P25035, 31543818, and 4885645. Residues 10–27 are highly conserved from zebrafish to humans and are also more conserved than the flanking sequences, so this region forms a small ‘conservation island’ (Yang and Grégoire, 2006). In comparison, residues 311–321 and 369–393 are less conserved.
(C) In DNA damage responses, Lys acetylation targets not only p53 itself but also various components of the signaling pathways that control p53 activation. Exactly how acetylation of upstream components affect p53 regulation remains to be established. AROS, active regulator of SIRT1; Bat3, HLA B-associated transcript 3 (also known as Scythe); DBC1 (deleted in breast cancer 1); PIC, transcriptional pre-initiation complex; Pin1, protein interacting with NIMA (never in mitosis A)-1; TAF1, TBP-associated factor 1; VHL, von Hippel-Lindau gene product. See text for more details.
The enzymes for p53 acetylation
A decade ago, p300 was first shown to acetylate a cluster of 5 Lys at the C-terminal regulatory region (Fig. 3A) (Gu and Roeder, 1997). UV- and ionizing radiation (IR)-induced DNA damage triggers acetylation by p300 and PCAF (Sakaguchi et al., 1998; Liu et al., 1999). In addition, oncogenic Ras and a variety of stress-inducing agents promote p53 acetylation (Pearson et al., 2000; Ito et al., 2001). Interestingly, a phosphorylation-acetylation cascade activates p53 in response to DNA damage (Sakaguchi et al., 1998). p53 phosphorylation (e.g., at Ser residues N-terminal to a conserved LxxLL motif) promotes its association with p300, which in turn acetylates the Lys residues in the C-terminal region (Fig. 3A–B) (Appella and Anderson, 2000; Brooks and Gu, 2003). PCAF acetylates Lys 320 (Fig. 3A–B). In comparison, TIP60 (HIV Tat-interactive protein of 60 kDa) and MOF (males absent of the first)—two members of the MYST family—acetylate Lys 120 located at the DNA-binding domain (Fig. 3A) (Sykes et al., 2006; Tang et al., 2006). Lys 120 acetylation activates pro-apoptotic target genes but has minimal effects on cell cycle progression, suggesting that this PTM is a key switch for apoptotic and cell cycle control.
Four deacetylases (HDAC1, HDAC3, SIRT1 and SIRT7) have been shown to deacetylate p53 (Brooks and Gu, 2003; Zeng et al., 2006; Vakhrusheva et al., 2008). Consistent with these findings, inhibitors that target classical HDACs and sirtuins elevate p53 acetylation. HDAC1 deacetylates multiple Lys residues. In vitro assays indicate that SIRT1 targets Lys 382, but gene inactivation analysis revealed that SIRT1 also acts on Lys 320 and 373 (Cheng et al., 2003). Thus, site specificity for the deacetylases has not been as clearly defined as for the acetyltransferases (Fig. 3A).
Upstream control of p53 acetylation
Many upstream regulators of p53 are also regulated by acetylation (Fig. 3C). TIP60 acetylates and activates ATM kinase in response to DNA damage (Sun et al., 2005). This modification occurs on a single residue, K3016. Although this Lys is not conserved in DNA-PK (an ATM family member), some evidence suggests that TIP60 also targets this kinase (Jiang et al., 2006). Upstream from ATM (Fig. 3C), NBS1 (Nijmegen breakage syndrome 1) is acetylated (Yuan et al., 2007). SIRT1 associates with the MRN nuclease complex and maintains NBS1 in a hypoacetylated state, poised for IR-induced phosphorylation. Moreover, Ku70, an important regulator of DNA-PK, is acetylated (Cohen et al., 2004). Although it remains unclear whether Ku70 acetylation plays a role in DNA-PK regulation, HDAC inhibitors up-regulate the acetylation and affect DNA double-strand break formation (Chen et al., 2007).
Mechanistic impact of p53 acetylation
Acetylation not only neutralizes the positive charge of the Lys side chain, but also impairs its ability to form hydrogen bonds. p53 acetylation directs both ‘loss-of-function’ and ‘gain-of-function’, via multiple mechanisms. This PTM might enhance specific DNA binding (Prives and Manley, 2001; Brooks and Gu, 2003). It might also inhibit non-specific DNA (and perhaps RNA) binding by the p53 C-terminal domain (Friedler et al., 2005). In addition, acetylation forms docking sites for recruitment of transcriptional coactivators. For example, Lys 382 acetylation increases p53 affinity for the CBP bromodomain (Mujtaba et al., 2004), whereas di-acetylation of Lys 373 and 382 facilitates interaction with the tandem bromodomains of TAF1, a TFIID subunit (Fig. 3C) (Li et al., 2007a). Moreover, the p53 acetylation sites are ubiquitination targets, so acetylation blocks ubiquitination. Likewise, PCAF-mediated Lys 320 acetylation precludes ubiquitination by E4F1 (Fig. 3B) (Le Cam et al., 2006). Interestingly, both acetylation and ubiquitination activate p53 although the affected target genes are different. Ubiquitination of Lys 370, 372, 373, 381 and 382 directs p53 for nuclear export and proteasomal degradation (Brooks and Gu, 2003). Therefore, acetylation of these residues is expected to stabilize p53 and promote its nuclear localization. Recent studies revealed that Lys 370, 372 and 382 can also be methylated (Chuikov et al., 2004; Huang et al., 2006b; Shi et al., 2007), indicating that acetylation would inhibit methylation. p53 Lys acetylation may also crosstalk with neighboring PTMs in a manner similar to the interaction between histone H3 acetylation and phosphorylation (Fig. 1B) (Latham and Dent, 2007). For example, p53 acetylation at Lys 370 and 372 might communicate with Ser 371 phosphorylation (Fig. 3A). Strikingly, a recent study revealed that Lys 372 methylation is required for p53 acetylation at multiple sites (Kurash et al., 2008). The methylation likely recruits TIP60 via its chromodomain and promotes Lys 120 acetylation, which might then form docking sites to enlist PCAF and p300 for acetylation at the C-terminal region (Fig. 3A–B). Therefore, the mechanistic impact of p53 acetylation is truly multifaceted.
Physiological relevance of p53 acetylation
As most studies regarding p53 acetylation were performed in vitro using cell-based and biochemical assays, the relevance of the obtained results to human pathophysiology is an important issue. As the acetylation sites are conserved in between human and mouse p53 (Fig. 3A), two groups have investigated this issue using knock-in mice (Toledo and Wahl, 2006). One group engineered a strain expressing the mutant that contain the C-terminal 7 Lys residues replaced with Arg (7KR) (Krummel et al., 2005). The homozygous mice were viable and phenotypically normal. The 7KR mutant protein was activated more easily than wild-type p53 in thymocytes and embryonic fibroblasts, suggesting that acetylation of the Lys residues fine-tunes p53 function. Another group generated mice expressing the p53 mutant with six of the seven Lys changed to Arg (6KR) (Feng et al., 2005). In support of what was observed with the 7KR mice, p53 stability remained unaffected in the 6KR mice. As the Lys residues are also targets of negative regulation by ubiquitination, neddylation and methylation (Brooks and Gu, 2003; Huang et al., 2006b; Shi et al., 2007), Arg substitution removes both negative and positive regulation. Arg is the most common substitute for Lys in mutation studies, but their side chain structures are quite different. Both issues complicate interpretation of these knock-in mouse phenotypes.
p53 Lys 320 (317 in mice) has also been investigated using a similar strategy (Chao et al., 2006). Homozygous K317R mutant mice had normal life span, leaving p53-dependent gene expression largely unaffected except for increased p53-dependent apoptosis in thymocytes. Lys 320 (317 in mice) can also be ubiquitinated (Le Cam et al., 2006). It also remains possible that additional modifications occur at this position. Thus, as with the 6KR and 7KR mice, an important issue is how to interpret the resulting phenotypes. Interestingly, the p300 and PCAF acetylation sites are not mutated in human cancers (Soussi et al., 2006; Toledo and Wahl, 2006), raising the question of what roles acetylation at these sites plays in vivo. Unlike these sites, Lys 120 (117 in mice) is mutated in human cancers and conserved from C. elegans to humans (Tang et al., 2006). Moreover, its acetylation lies at the nexus of apoptotic and cell cycle control (Sykes et al., 2006; Tang et al., 2006), so it will be interesting to determine if K117R mutant mice display more dramatic phenotypes.
Overall, since the discovery of p53 acetylation 11 years ago, numerous studies have been performed and have revealed quite unexpected complexity. These studies set a good example and provide valuable lessons for investigating acetylation of other proteins. One realization is that, as with histone acetylation, p53 acetylation does not act alone but forms an integral part of a complex multisite modification program.
Acetylation in signal transduction from the plasma membrane
As with p53 acetylation in the DNA damage response (Fig. 3C), acetylation of other transcription factors is important for transmitting various cues from cellular signaling networks. In this section, we describe roles of acetylation in two signaling pathways (Fig. 4).
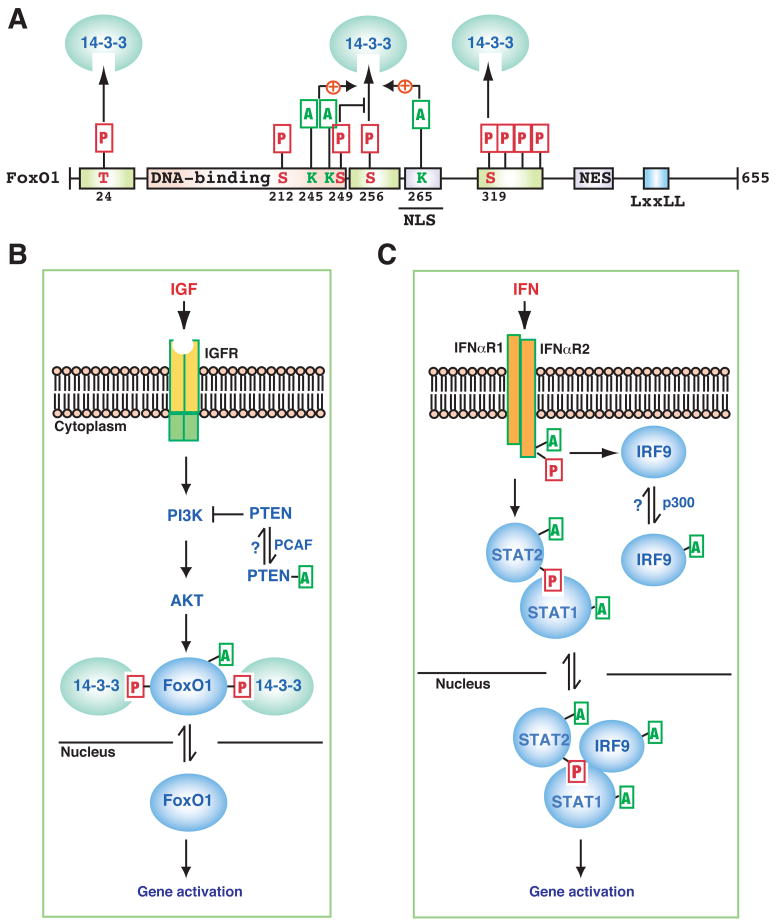
Figure 4. Lys acetylation in IGF and IFN signaling.
(A) Domain organization of human FoxO1. In addition to a highly conserved DNA-binding domain in the middle, FoxO1 possesses six conserved motifs, three of which are phosphorylated for 14-3-3 binding and nuclear export. The critical Thr (T) and Ser (S) residues are indicated along with their positions. In addition, a nuclear localization signal (NLS), a nuclear export signal (NES), and an LxxLL motif are conserved. (B–C) Schematics showing how Lys acetylation modulates pathways that transduce signals from insulin-like growth factor (IGF, B) and interferon (IFN, C) to the nucleus. IGFR, IGF receptor; IFNαR1, IFNα receptor 1. See text for details.
FoxO acetylation in insulin-like growth factor (IGF) signaling
In mammals, the O group of the forkhead-box transcription factors are comprised of four members (FoxO1, 3, 4 and 6) that share sequence similarity in DNA-binding domains and other functional motifs (Fig 4A) (van der Horst and Burgering, 2007). In mammals, IGF activates AKT, which then phosphorylates FoxO transcription factors at three sites (e.g., Thr 24, Ser 256 and 319 of FoxO1, Fig. 4A–B) and stimulates 14-3-3 binding, leading to nuclear export (Fig. 4B). CBP acetylates FoxO1 at Lys 245, 248 and 265 (Fig. 4A) (Matsuzaki et al., 2005). As with p53, the impact of FoxO1 acetylation is also multifaceted: this PTM promotes FoxO1 Ser 256 phosphorylation and stimulates cytoplasmic retention, which is similar to the positive effects of histone H3 acetylation at Lys 9 or 14 on 14-3-3 binding to Ser 10 (Walter et al., 2008; Winter et al., 2008). As Ser 256 phosphorylation stimulates the interaction of FoxO1 with the ubiquitin ligase Skp2 (van der Horst and Burgering, 2007), acetylation might indirectly regulate ubiquitination. As Lys 245 and 248 are located within the DNA-binding domain and Lys 265 is embedded within the nuclear localization signal (Fig. 4A), acetylation might attenuate DNA binding and impede nuclear import. Consistent with this idea, SIRT1- or SIRT2-mediated deacetylation retains FoxO1 in the nucleus (Jing et al., 2007; van der Horst and Burgering, 2007). Furthermore, Lys 248 is adjacent to and may crosstalk with Ser 249 (Fig. 1B), and Cdk1-mediated phosphorylation of this Ser promotes FoxO1 nuclear localization perhaps by interfering with 14-3-3 binding to Ser 256 (Fig. 4A) (Huang et al., 2006a; Yuan et al., 2008). Upstream from AKT (Fig. 4B), PCAF acetylates PTEN at two residues in the catalytic cleft, impairs phosphatase activity, and stimulates PI3K signaling (Okumura et al., 2006). Therefore, acetylation modulates IGF signaling to FoxO-dependent transcription from different angles.
Lys acetylation in interferon (IFN) signaling
As an important part of the initial antiviral defense, IFN is rapidly induced to bind and activate its receptors, which in turn trigger phosphorylation, dimerization and nuclear translocation of STAT proteins (Schindler et al., 2007). A recent study revealed an unexpected signaling cascade involving acetylation of multiple components of type I IFN signaling pathways (Fig. 4C) (Tang et al., 2007). Upon binding to IFNα, type I IFN receptor 2 (IFNαR2) phosphorylates Ser 364 and 384 within its proline-rich domain, which then recruits CBP to acetylate the receptor at Lys 399; HDAC6 deacetylase might reverse the modification. As CBP is nuclear (Goodman and Smolik, 2000), this cytoplasmic activity suggests that its subcellular localization might be regulated. Mass spectrometry revealed that Ser 400 of IFNαR2 is phosphorylated (Fig. 1B). Together with Lys 399 acetylation, the phosphorylation forms a docking site to recruit IRF9 (IFN regulatory factor 9) for acetylation, thereby stimulating its dimerization and DNA binding affinity (Fig. 4C). STAT2 is then acetylated at Lys 390, thereby promoting its heterodimerization with STAT1. In addition, STAT3 acetylation is known to promote its homodimerization and nuclear localization (Yuan et al., 2005). Acetylation of STAT1 and STAT3 also affects NF-κB signaling (Kramer et al., 2006; Nadiminty et al., 2006). Overall, the acetylation cascade triggered by IFN (Fig. 4C) is reminiscent of many phosphorylation-based signaling pathways, raising the possibility that similar acetylation cascades might be employed in other signaling schemes.
Acetylation in nuclear hormone signaling
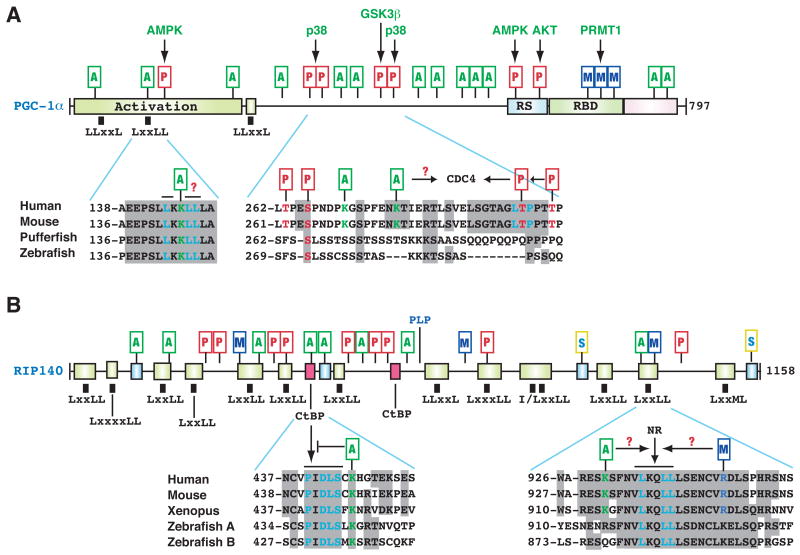
Unlike plasma membrane receptors, nuclear hormone receptors mainly act within the nucleus to regulate gene expression. Lys acetylation has been reported for androgen, progesterone, estrogen and liver X receptors (Li et al., 2007b; Whittle et al., 2007). The latter is acetylated at KLxxLL, a consensus motif similar to that in steroid receptor coactivator 3 (Chen et al., 1999; Li et al., 2007b). Other nuclear receptor coregulators are also acetylated, including p300/CBP (Fig. 2), PGC-1α (peroxisome proliferator-activated receptor gamma (PPARγ)-coactivator 1α), and RIP140 (receptor interacting protein 140) (Lonard and O’Malley B, 2007; Rodgers et al., 2008; White et al., 2008). PGC-1α and RIP140 harbor various PTMs with the potential for active crosstalk (Fig. 5) and are thus highlighted here.
Figure 5. Acetylation and other PTMs in PGC-1α and RIP140.
(A) Domain organization of human PGC-1α, with conserved domains shown in colored boxes. One LxxLL and two LLxxL motifs are depicted by small black boxes. These motifs are evolutionarily conserved. Within the LxxLL motif, the acetylatable Lys is highlighted in green. PTMs are maked as in Fig. 2. Activation, transcriptional activation domain; RS, Arg- and Ser-rich domain; RBD, RNA-binding domain.
(B) Domain organization of human RIP140. Conserved domains are shown in small colored boxes. Small black boxes indicate the rough positions of Leu-rich motifs. RIP140 contains two conserved CtBP-binding motifs and double sumoylation sites. NR, nuclear receptor. Sequence analyses were performed as in Fig. 2B, using GenBank accession numbers 7019499, AAH66868, CAG02304, and AAY15212 (for PGC-1α); CAA59108, NP_032761, NP_001083708, 125838185, and 125828075 (for RIP140). See text for details.
Acetylation of the coactivator PGC-1α
Initially identified as a cofactor for PPARγ, PGC-1α is now known to regulate distinct biological programs via its role as a coactivator for different nuclear receptors and other transcription factors including FoxO1 (Fig. 4B) (Lin et al., 2005; Rodgers et al., 2008). Like many coactivators, PGC-1α has a modular domain organization (Fig. 5A). Its N-terminal and C-terminal parts are conserved from pufferfish to humans: both domains show some sequence similarity to two other family members, PGC-1β and PRC (Lin et al., 2005). Within PGC-1α, one LxxLL and two LLxxL motifs (Fig. 5A) mediate nuclear receptor interaction. GCN5 acetylates PGC-1α, thereby inhibiting its coactivator activity, whereas SIRT1-mediated deacetylation reverses this inhibition (Rodgers et al., 2005; Lerin et al., 2006). PGC-1α acetylation sites span the entire protein (Fig. 5A), raising the question of how the modification exerts its effect. One site, Lys 146, lies within the LxxLL motif, so acetylation of this Lys might selectively affect nuclear receptors’ binding to this motif. Several kinases, including AMPK, p38, GSK3β, and AKT, phosphorylate different sites on PGC-1α to regulate its stability and coactivator activity (Fig. 5A) (Olson et al., 2008; Rodgers et al., 2008). Thr 299 phosphorylation by p38 primes GSK3β-mediated phosphorylation of Thr 295, thereby generating a docking site for the ubiquitin ligase CDC4 (Olson et al., 2008). Whether acetylation of Lys 271, 278 or other Lys residues (Fig. 5A) nearby affects this process is an issue awaiting investigation. In addition, PRMT1 (protein arginine methyltransferase 1) methylates PGC-1α and upregulates its coactivator activity (Rodgers et al., 2008). It will be interesting to determine how acetylation interacts with other PTMs for coordinated PGC-1α regulation.
Acetylation of the corepressor RIP140
Like PGC-1α, RIP140 is highly modular and contains small domains conserved from zebrafish to humans (Fig. 5B) (White et al., 2008). Within these domains lie many LxxLL and LLxxL motifs, as well as two binding sites for the NADH-responsive corepressor CtBP (E1A C-terminal binding protein). One of the two sites conforms to the consensus sequence PxDLSxK, which is similar to the CtBP-binding sites in E1A and several other proteins; acetylation of the Lys impairs CtBP binding (Vo et al., 2001). A recent mass spectrometric analysis identified seven new acetylation sites (Fig. 5B) (Huq and Wei, 2005). One site lies near an LxxLL motif and may therefore affect nuclear receptor interaction. RIP140 also undergoes phosphorylation, methylation, PLP conjugation, and sumoylation (Fig. 5B) (Gupta et al., 2005; Rytinki and Palvimo, 2008), so it will be important to determine how acetylation and other PTMs coordinate to direct RP140 regulation.
Acetylation of non-nuclear proteins
Lys acetylation has also been found in many cytoplasmic enzymes, including acetyl-CoA synthase (Starai et al., 2002; Hallows et al., 2006; Schwer et al., 2006), glutamate dehydrogenase (Lombard et al., 2007), nitric oxide synthase (Mattagajasingh et al., 2007), and other metabolic enzymes (Kim et al., 2006). As shown for acetyl-CoA and nitric oxide synthases, acetylation of these cytoplasmic enzymes might function as a simple on/off switch and is thus not covered in detail here. Instead, we discuss how acetylation might interact with other PTMs in the regulation of α-tubulin, cortactin, and Hsp90 (heat shock protein 90).
α-tubulin acetylation
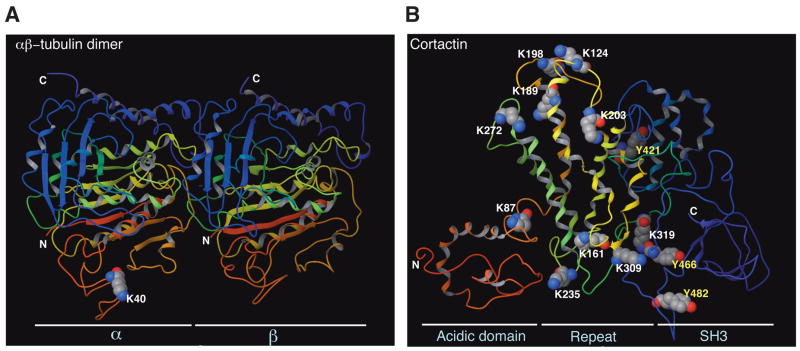
α- and β-tubulins form heterodimers and serve as building blocks for microtubules. α-tubulin was among the first acetylated non-histone proteins to be discovered (Westermann and Weber, 2003). The acetylation site was mapped to Lys 40, which is absent in β-tubulin and is located on the lumenal side of microtubules (Fig. 6A). α-tubulin acetylation is widespread and present in organisms from protists to vertebrates, but not in yeast. Other PTMs, including phosphorylation, polyglutamylation, polyglycylation, tyrosination, nitrotyrosination, and aspartate isomerization, target the C-terminal 10 residues located outside microtubules (Fig. 6A) (Westermann and Weber, 2003). This physical separation makes the interplay of acetylation with C-terminal tubulin modifications unlikely. The potential functional consequence of tubulin acetylation has been a subject of debate, but two groups independently reported that acetylation directs the formation of a docking site for motor proteins to facilitate cargo transport on microtubules (Reed et al., 2006; Dompierre et al., 2007). This PTM could therefore serve as a simple landmark. HDAC6 is the most active tubulin deacetylase (Hubbert et al., 2002; Matsuyama et al., 2002), as supported by the recent observation that mouse Hdac6 inactivation led to α-tubulin hyperacetylation (Zhang et al., 2008). The identity of the responsible acetyltransferase(s) remains elusive, and it remains unclear how cell signalling might regulate this PTM.
Figure 6. Acetylation sites on 3D structures of tubulin and cortactin.
(A) Ribbon representation of an α/β-tubulin heterodimer. Electron diffraction data was obtained from Protein Data Bank (PDB ID 1tub) for preparation of the 3D structure using the Maestro program (version 8). N and C mark the N- and C-terminal ends, respectively. Whereas Lys 40 of α-tubulin faces the lumenal side, the C-terminal ends of α- and β-tubulin are outside from microtubules.
(B) Ribbon representation of the cortactin structure. Electron diffraction data was obtained from Protein Data Bank (PDB ID 2F9X). The 3D structure was generated along with acetylation sites and three phospho-Tyr residues colored as in (A). Lys 161, 235, 309 and 319 form one charged patch, whereas Lys 124, 189, 198, 203, and 272 form another. Albeit over 200 residues apart in the primary sequence, Lys 203 and Tyr 421 are close to one another at the 3D level. Similarly, Lys 309 and 319 are adjacent to Tyr 466 in the 3D structure. Physical proximity at the 3D level suggests potential crosstalk between Lys acetylation and Tyr phosphorylation, possibly linking cellular signaling to regulation of reversible acetylation.
Cortactin acetylation
Cortactin acetylation was recently reported (Zhang et al., 2007). Originally identified as a Src kinase substrate, cortactin regulates cell motility. It contains an N-terminal acidic domain, six and a half 37-residue repeats, and a C-terminal SH3 domain (Fig. 6B). Cortactin undergoes acetylation at up to ten residues, five of which are conserved from Drosophila to humans (Zhang et al., 2007). Mutation of the acetylation sites located in the repeat region affects the ability to bind F-actin and consequently alters cell motility. The sites are separated in the primary sequence but form two ‘charged patches’ at the 3D level (Fig. 6B). Importantly, the acetylation of a particular Lys is not important; rather the total number of acetylated residues is critical for inhibiting the affinity for F-actin. HDAC6-mediated deacetylation relieves this inhibition. Cortactin is also extensively phosphorylated, with nine phospho-Tyr situated between the proline-rich region and the SH3 domain (Fig. 6B). Src targets primarily Tyr 421, 466, and 482 to inhibit F-actin cross-linking activity (Huang et al., 1998). This inhibition is dependent on the number of phosphorylatable Tyr residues. At the 3D level, Tyr 421 is near Lys 203, whereas Tyr 466 is next to Lys 309 and 319 (Fig. 6B), suggesting that cortactin acetylation and phosphorylation might crosstalk with one another.
Acetylation of the molecular chaperone Hsp90
Hsp90 activity is essential for the regulation of many signaling proteins and has become a promising chemotherapeutic target (Pearl and Prodromou, 2006). It comprises a family of well-conserved chaperones constituting 1–2% of cellular proteins even under non-stress conditions. Inducible Hsp90α and constitutive Hsp90β are the best characterized family members. They share 86% sequence identity and comprise three structural domains: a conserved N-terminal domain for nucleotide binding (also the site for drug binding), a middle segment for client protein recognition, and a C-terminal domain essential for Hsp90 dimerization (Pearl and Prodromou, 2006). A ‘charged linker’ connects the N-terminal domain to the middle segment. The first indication of Hsp90 acetylation came from a study in which treatment of mammalian cells with an HDAC inhibitor increased Hsp90 acetylation and reduced its interaction with client proteins (Yu et al., 2002). Later studies revealed that HDAC6 binds Hsp90 and reverses its acetylation (Bali et al., 2005; Kovacs et al., 2005). Accordingly, HDAC6 depletion leads to Hsp90 hyperacetylation and compromises its chaperone roles in glucocorticoid receptor maturation (Kovacs et al., 2005) and in signaling by the leukemic tyrosine kinase Bcr-Abl (Bali et al., 2005). One acetylation site of Hsp90α was mapped to Lys 294, located in the middle region and C-terminal from the charged linker (Scroggins et al., 2007), and studies indicate that the presence of at least one acetylation site (Chen et al., 2005; Scroggins et al., 2007). Lys 294 substitution affects the association with co-chaperones and client proteins, including v-Src, ErbB2, p53 and HIF-1α (Scroggins et al., 2007). Many kinases and transcription factors require Hsp90 for maturation and proper function (Pearl and Prodromou, 2006), and its acetylation could therefore affect many signaling pathways (e.g, those featured in Figs 3B & 4). Hsp90 is also phosphorylated, ubiquitinated and S-nitrosylated. Some phosphorylation sites lie near Lys 294, so acetylation could potentially regulate Hsp90 phosphorylation (or vice versa).
Lys acetylation and the protein modification code
It is clear now that Lys acetylation occurs in proteins from across the nucleus to the plasma membrane. Acetylation patterns and the mechanistic impact vary from one protein to another (Glozak et al., 2005). As discussed above for p53 and the other selected examples (Figs 2–6), effects are multifaceted even for one protein. In general, different acetylation events can be roughly categorized into three groups. First, acetylation occurs at one or a few residues and thus functions as a simple on/off switch. For example, acetylation of acetyl-CoA synthase (Starai et al., 2002) and nitric oxide synthase (Mattagajasingh et al., 2007) inactivates the enzymes, whereas α-tubulin acetylation (Fig. 6A) generates a simple mark for cargo transport. Acetylation of PTEN (Okumura et al., 2006), liver X receptors (Li et al., 2007b), Hsp90 (Scroggins et al., 2007), and perhaps many mitochondrial proteins (Kim et al., 2006; Lombard et al., 2007) also fall within this category. Second, acetylation occurs at clusters of Lys residues that form ‘charged patches.’ The number of acetylated residues rather than acetylation of an individual Lys is important for the functional impact. In this case, multisite acetylation exerts gauge-like effects or provides a fail-safe mechanism. For example, cortactin is acetylated at ~10 residues in its repeat domain (Fig. 6B) (Zhang et al., 2007). Each repeat is sufficient for F-actin binding and contains at least one Lys for acetylation. Another example is p300 autoacetylation at a cluster of residues within its activation loop (Fig. 2A–B) (Thompson et al., 2004).
In a third category, acetylation interacts with other PTMs and imparts effects in a site-specific manner. For example, diverse PTMs occur in many transcription factors, including p53 (Fig. 3A), FoxO1 (Fig. 4A), PGC-1α (Fig. 5A) and RIP140 (Fig. 5B). The PTMs form a dynamic layer of molecular information beyond the amino acid sequences. It is thus intriguing to ask if there are any general rules for interpreting this rather complicated layer of information. Related to this, a ‘posttranslational code’ model was proposed for p53 (Appella and Anderson, 2000; Chuikov et al., 2004), and the ‘histone code’ hypothesis has been widely discussed (Strahl and Allis, 2000; Ruthenburg et al., 2007; Winter et al., 2008). The latter has been extended to cover p300/CBP (Legube and Trouche, 2003), α-tubulin (Westermann and Weber, 2003), the C-terminal domain of RNA polymerase II (Buratowski, 2003; Corden, 2007), and nuclear receptor coregulators (Lonard and O’Malley B, 2007). Conceptually, a general term like ‘PTM code’, ‘protein modification code’ or ‘protein code’ can be used to describe proteins with multisite PTMs. These terms will likely face the scrutiny and debate that the ‘histone code’ hypothesis has received (Schreiber and Bernstein, 2002; Kouzarides, 2007; Ruthenburg et al., 2007; Turner, 2007). The meaning of the word ‘code’ here is clearly more complicated than that in the ‘genetic code’, but recent studies clearly indicate that multisite PTMs are somewhat codified and act in sequential and combinatory manners (Corden, 2007; Sampath et al., 2007; Kurash et al., 2008; Walter et al., 2008; Winter et al., 2008). As in histones (Berger, 2007), multisite modifications of p53 (Fig. 3) and other tightly regulated proteins (Figs 4–5) form sophisticated programs for ‘intramolecular and intermolecular signaling’ (Yang, 2005). Within these programs, different PTMs interact with one another and various combinations yield distinct outcomes. Dependent on the context, Lys acetylation could interplay with other PTMs in an agonistic or antagonistic manner. Diverse signaling cues act through the programs for coordinated regulation of protein functions. The complexity of such a program in a given protein is proportional to its biological importance and the complexity of the corresponding organism (akin to a complicated traffic control system for a busy intersection in a big city). Therefore, Lys acetylation along with other PTMs exerts diverse cellular effects in a context-dependent manner.
Conclusion and future directions
In the past decade, Lys acetylation has been found in a variety of proteins, ranging from many nuclear regulators to cytosolic proteins, mitochondrial enzymes, and plasma membrane-associated receptors. As emphasized herein (Figs 2–6), acetylation of some non-histone proteins occurs at multiple sites and crosstalks with phosphorylation, methylation, sumoylation, ubiquitination and other PTMs to form dynamic regulatory programs. Many acetyl-proteins are key components of different signaling pathways (Figs 3B & 4B–C). Instead of relying solely on phosphorylation, signaling pathways likely are controlled by the coordinated actions of phosphorylation, acetylation, and other PTMs. Thus, acetylation diversifies cellular signaling networks. Owing to biased candidate approaches and/or the fact that there are more acetyl-proteins present in higher organisms, most known acetylated non-histone proteins are mammalian. Although Lys acetylation occurs in bacteria, many non-histone acetylation events may have been acquired during evolution. In support of this idea, α-tubulin Lys 40 is not conersved in yeast, and most p53 acetylation sites are absent in fly and worm homologs (Fig. 3).
At least six research trends will enhance our understanding of Lys acetylation. First, proteomic survey by mass spectrometry will continue to identify new acetyl-proteins. Second, systematic mapping of acetylation sites in known acetylated proteins by mass spectrometry will reveal additional sites, as recently shown for RIP140 (Fig. 5B) (Huq and Wei, 2005). This approach has also yielded surprises concerning histone modifications (Hyland et al., 2005) and surely will lead to unexpected discoveries about non-histone acetylation. Third, the mouse knock-in strategy will be more frequently used to examine the biological consequences of individual modification sites, as exemplified by the studies of p53 acetylation (Toledo and Wahl, 2006). Fourth, gene inactivation has been used to analyze nearly all known HDACs and some HATs in mouse development. Together with mass spectrometry, these mutant mice will be invaluable for identifying new substrates for HATs and HDACs, as shown for Sirt3−/− mice (Lombard et al., 2007). A similar approach should also be applicable to other model organisms. Fifth, it will be important to map out signaling pathways that regulate reversible acetylation. One relevant question is how this modification interacts with other PTMs (e.g., sequence of occurrence and kinetics of duration) and forms dynamic programs for regulating cellular function under diverse conditions. Finally, studies of non-histone protein acetylation will shed light on the pathogenesis—and thus diagnosis, therapy and prevention—of different diseases. It will be interesting to examine how single nucleotide polymorphisms (SNPs) and different genetic variations affect acetylation and other PTM patterns among individuals. Small-molecule HDAC inhibitors and activators have emerged as promising therapeutic agents for cancer, heart diseases, diabetes, and neurodegenerative disorders, so studies of Lys acetylation would identify non-histone targets for KAT- and KDAC-modulating compounds and illuminate new avenues to improve the efficacy of related therapeutic agents.
In summary, Lys acetylation has emerged as a major PTM for over one hundred proteins. Within the acetyl-proteome, functional impact of this PTM is context-dependent and varies from protein to protein. As in histones (Strahl and Allis, 2000; Margueron et al., 2005; Berger, 2007; Latham and Dent, 2007) and in the representative examples illustrated here (Figs 2–6), Lys acetylation interplays actively with other PTMs—agonistically or antagonistically—to form codified ‘intramolecular signaling’ programs that are crucial for governing functions of various nuclear and cytoplasmic proteins.
Acknowledgments
We thank Shen-Shu Sung for help with molecular modeling and three anonymous reviewers for constructive suggestions. We wish to indicate that some research papers cannot be referred here due to strict space constraints. Our laboratories are supported by grants from the NIH, AHA, and the Kaul Foundation (to E.S.) and from the Canadian agencies CIHR and NCIC (to X.J.Y.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Allis CD, Berger SL, Cote J, Dent S, Jenuwien T, Kouzarides T, Pillus L, Reinberg D, Shi Y, Shiekhattar R, et al. New nomenclature for chromatin-modifying enzymes. Cell. 2007;131:633–636. doi: 10.1016/j.cell.2007.10.039. [DOI] [PubMed] [Google Scholar]
- Appella E, Anderson CW. Signaling to p53: breaking the posttranslational modification code. Pathol Biol (Paris) 2000;48:227–245. [PubMed] [Google Scholar]
- Bali P, Pranpat M, Bradner J, Balasis M, Fiskus W, Guo F, Rocha K, Kumaraswamy S, Boyapalle S, Atadja P, et al. Inhibition of histone deacetylase 6 acetylates and disrupts the chaperone function of heat shock protein 90: a novel basis for antileukemia activity of histone deacetylase inhibitors. J Biol Chem. 2005;280:26729–26734. doi: 10.1074/jbc.C500186200. [DOI] [PubMed] [Google Scholar]
- Berger SL. The complex language of chromatin regulation during transcription. Nature. 2007;447:407–412. doi: 10.1038/nature05915. [DOI] [PubMed] [Google Scholar]
- Bhaumik SR, Smith E, Shilatifard A. Covalent modifications of histones during development and disease pathogenesis. Nat Struct Mol Biol. 2007;14:1008–1016. doi: 10.1038/nsmb1337. [DOI] [PubMed] [Google Scholar]
- Brooks CL, Gu W. Ubiquitination, phosphorylation and acetylation: the molecular basis for p53 regulation. Curr Opin Cell Biol. 2003;15:164–171. doi: 10.1016/s0955-0674(03)00003-6. [DOI] [PubMed] [Google Scholar]
- Buratowski S. The CTD code. Nat Struct Biol. 2003;10:679–680. doi: 10.1038/nsb0903-679. [DOI] [PubMed] [Google Scholar]
- Chao C, Wu Z, Mazur SJ, Borges H, Rossi M, Lin T, Wang JY, Anderson CW, Appella E, Xu Y. Acetylation of mouse p53 at lysine 317 negatively regulates p53 apoptotic activities after DNA damage. Mol Cell Biol. 2006;26:6859–6869. doi: 10.1128/MCB.00062-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B, Piel WH, Gui L, Bruford E, Monteiro A. The HSP90 family of genes in the human genome: insights into their divergence and evolution. Genomics. 2005;86:627–637. doi: 10.1016/j.ygeno.2005.08.012. [DOI] [PubMed] [Google Scholar]
- Chen CS, Wang YC, Yang HC, Huang PH, Kulp SK, Yang CC, Lu YS, Matsuyama S, Chen CY. Histone deacetylase inhibitors sensitize prostate cancer cells to agents that produce DNA double-strand breaks by targeting Ku70 acetylation. Cancer Res. 2007;67:5318–5327. doi: 10.1158/0008-5472.CAN-06-3996. [DOI] [PubMed] [Google Scholar]
- Chen H, Lin RJ, Xie W, Wilpitz D, Evans RM. Regulation of hormone-induced histone hyperacetylation and gene activation via acetylation of an acetylase. Cell. 1999;98:675–686. doi: 10.1016/s0092-8674(00)80054-9. [DOI] [PubMed] [Google Scholar]
- Cheng HL, Mostoslavsky R, Saito S, Manis JP, Gu Y, Patel P, Bronson R, Appella E, Alt FW, Chua KF. Developmental defects and p53 hyperacetylation in Sir2 homolog (SIRT1)-deficient mice. Proc Natl Acad Sci USA. 2003;100:10794–10799. doi: 10.1073/pnas.1934713100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuikov S, Kurash JK, Wilson JR, Xiao B, Justin N, Ivanov GS, McKinney K, Tempst P, Prives C, Gamblin SJ, et al. Regulation of p53 activity through lysine methylation. Nature. 2004;432:353–360. doi: 10.1038/nature03117. [DOI] [PubMed] [Google Scholar]
- Cohen HY, Lavu S, Bitterman KJ, Hekking B, Imahiyerobo TA, Miller C, Frye R, Ploegh H, Kessler BM, Sinclair DA. Acetylation of the C Terminus of Ku70 by CBP and PCAF Controls Bax-Mediated Apoptosis. Mol Cell. 2004;13:627–638. doi: 10.1016/s1097-2765(04)00094-2. [DOI] [PubMed] [Google Scholar]
- Corden JL. Seven ups the code. Science. 2007;318:1735–1736. doi: 10.1126/science.1152624. [DOI] [PubMed] [Google Scholar]
- Dompierre JP, Godin JD, Charrin BC, Cordelieres FP, King SJ, Humbert S, Saudou F. Histone deacetylase 6 inhibition compensates for the transport deficit in Huntington’s disease by increasing tubulin acetylation. J Neurosci. 2007;27:3571–3583. doi: 10.1523/JNEUROSCI.0037-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dornan D, Shimizu H, Perkins ND, Hupp TR. DNA-dependent acetylation of p53 by the transcription coactivator p300. J Biol Chem. 2003;278:13431–13441. doi: 10.1074/jbc.M211460200. [DOI] [PubMed] [Google Scholar]
- Feng L, Lin T, Uranishi H, Gu W, Xu Y. Functional analysis of the roles of posttranslational modifications at the p53 C terminus in regulating p53 stability and activity. Mol Cell Biol. 2005;25:5389–5395. doi: 10.1128/MCB.25.13.5389-5395.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedler A, Veprintsev DB, Freund SM, von Glos KI, Fersht AR. Modulation of binding of DNA to the C-terminal domain of p53 by acetylation. Structure. 2005;13:629–636. doi: 10.1016/j.str.2005.01.020. [DOI] [PubMed] [Google Scholar]
- Girdwood D, Bumpass D, Vaughan OA, Thain A, Anderson LA, Snowden AW, Garcia-Wilson E, Perkins ND, Hay RT. p300 transcriptional repression is mediated by SUMO modification. Mol Cell. 2003;11:1043–1054. doi: 10.1016/s1097-2765(03)00141-2. [DOI] [PubMed] [Google Scholar]
- Glozak MA, Sengupta N, Zhang X, Seto E. Acetylation and deacetylation of non-histone proteins. Gene. 2005;363:15–23. doi: 10.1016/j.gene.2005.09.010. [DOI] [PubMed] [Google Scholar]
- Goodman RH, Smolik S. CBP/p300 in cell growth, transformation, and development. Genes Dev. 2000;14:1553–1577. [PubMed] [Google Scholar]
- Gu W, Roeder RG. Activation of p53 sequence-specific DNA binding by acetylation of the p53 C-terminal domain. Cell. 1997;90:595–606. doi: 10.1016/s0092-8674(00)80521-8. [DOI] [PubMed] [Google Scholar]
- Gupta P, Huq MD, Khan SA, Tsai NP, Wei LN. Regulation of co-repressive activity of and HDAC recruitment to RIP140 by site-specific phosphorylation. Mol Cell Proteomics. 2005;4:1776–1784. doi: 10.1074/mcp.M500236-MCP200. [DOI] [PubMed] [Google Scholar]
- Haigis MC, Guarente LP. Mammalian sirtuins--emerging roles in physiology, aging, and calorie restriction. Genes Dev. 2006;20:2913–2921. doi: 10.1101/gad.1467506. [DOI] [PubMed] [Google Scholar]
- Hallows WC, Lee S, Denu JM. Sirtuins deacetylate and activate mammalian acetyl-CoA synthetases. Proc Natl Acad Sci USA. 2006;103:10230–10235. doi: 10.1073/pnas.0604392103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasselgren PO. Ubiquitination, phosphorylation, and acetylation--triple threat in muscle wasting. J Cell Physiol. 2007;213:679–689. doi: 10.1002/jcp.21190. [DOI] [PubMed] [Google Scholar]
- Huang C, Liu J, Haudenschild CC, Zhan X. The role of tyrosine phosphorylation of cortactin in the locomotion of endothelial cells. J Biol Chem. 1998;273:25770–25776. doi: 10.1074/jbc.273.40.25770. [DOI] [PubMed] [Google Scholar]
- Huang H, Regan KM, Lou Z, Chen J, Tindall DJ. CDK2-dependent phosphorylation of FOXO1 as an apoptotic response to DNA damage. Science. 2006a;314:294–297. doi: 10.1126/science.1130512. [DOI] [PubMed] [Google Scholar]
- Huang J, Perez-Burgos L, Placek BJ, Sengupta R, Richter M, Dorsey JA, Kubicek S, Opravil S, Jenuwein T, Berger SL. Repression of p53 activity by Smyd2-mediated methylation. Nature. 2006b;444:629–632. doi: 10.1038/nature05287. [DOI] [PubMed] [Google Scholar]
- Hubbert C, Guardiola A, Shao R, Kawaguchi Y, Ito A, Nixon A, Yoshida M, Wang XF, Yao TP. HDAC6 is a microtubule-associated deacetylase. Nature. 2002;417:455–458. doi: 10.1038/417455a. [DOI] [PubMed] [Google Scholar]
- Huq MD, Wei LN. Post-translational modification of nuclear co-repressor receptor-interacting protein 140 by acetylation. Mol Cell Proteomics. 2005;4:975–983. doi: 10.1074/mcp.M500015-MCP200. [DOI] [PubMed] [Google Scholar]
- Hyland EM, Cosgrove MS, Molina H, Wang D, Pandey A, Cottee RJ, Boeke JD. Insights into the role of histone H3 and histone H4 core modifiable residues in Saccharomyces cerevisiae. Mol Cell Biol. 2005;25:10060–10070. doi: 10.1128/MCB.25.22.10060-10070.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito A, Lai CH, Zhao X, Saito S, Hamilton MH, Appella E, Yao TP. p300/CBP-mediated p53 acetylation is commonly induced by p53-activating agents and inhibited by MDM2. EMBO J. 2001;20:1331–1340. doi: 10.1093/emboj/20.6.1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang X, Sun Y, Chen S, Roy K, Price BD. The FATC domains of PIKK proteins are functionally equivalent and participate in the Tip60-dependent activation of DNA-PKcs and ATM. J Biol Chem. 2006;281:15741–15746. doi: 10.1074/jbc.M513172200. [DOI] [PubMed] [Google Scholar]
- Jing E, Gesta S, Kahn CR. SIRT2 regulates adipocyte differentiation through FoxO1 acetylation/deacetylation. Cell Metab. 2007;6:105–114. doi: 10.1016/j.cmet.2007.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karanam B, Jiang L, Wang L, Kelleher NL, Cole PA. Kinetic and mass spectrometric analysis of p300 histone acetyltransferase domain autoacetylation. J Biol Chem. 2006;281:40292–40301. doi: 10.1074/jbc.M608813200. [DOI] [PubMed] [Google Scholar]
- Kim SC, Sprung R, Chen Y, Xu Y, Ball H, Pei J, Cheng T, Kho Y, Xiao H, Xiao L, et al. Substrate and functional diversity of lysine acetylation revealed by a proteomics survey. Mol Cell. 2006;23:607–618. doi: 10.1016/j.molcel.2006.06.026. [DOI] [PubMed] [Google Scholar]
- Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- Kovacs JJ, Murphy PJ, Gaillard S, Zhao X, Wu JT, Nicchitta CV, Yoshida M, Toft DO, Pratt WB, Yao TP. HDAC6 regulates Hsp90 acetylation and chaperone-dependent activation of glucocorticoid receptor. Mol Cell. 2005;18:601–607. doi: 10.1016/j.molcel.2005.04.021. [DOI] [PubMed] [Google Scholar]
- Kramer OH, Baus D, Knauer SK, Stein S, Jager E, Stauber RH, Grez M, Pfitzner E, Heinzel T. Acetylation of Stat1 modulates NF-kappaB activity. Genes Dev. 2006;20:473–485. doi: 10.1101/gad.364306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krummel KA, Lee CJ, Toledo F, Wahl GM. The C-terminal lysines fine-tune p53 stress responses in a mouse model but are not required for stability control or transactivation. Proc Natl Acad Sci USA. 2005;102:10188–10193. doi: 10.1073/pnas.0503068102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo HY, Chang CC, Jeng JC, Hu HM, Lin DY, Maul GG, Kwok RP, Shih HM. SUMO modification negatively modulates the transcriptional activity of CREB-binding protein via the recruitment of Daxx. Proc Natl Acad Sci USA. 2005;102:16973–16978. doi: 10.1073/pnas.0504460102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurash JK, Lei H, Shen Q, Marston WL, Granda BW, Fan H, Wall D, Li E, Gaudet F. Methylation of p53 by Set7/9 mediates p53 acetylation and activity in vivo. Mol Cell. 2008;29:392–400. doi: 10.1016/j.molcel.2007.12.025. [DOI] [PubMed] [Google Scholar]
- Latham JA, Dent SY. Cross-regulation of histone modifications. Nat Struct Mol Biol. 2007;14:1017–1024. doi: 10.1038/nsmb1307. [DOI] [PubMed] [Google Scholar]
- Le Cam L, Linares LK, Paul C, Julien E, Lacroix M, Hatchi E, Triboulet R, Bossis G, Shmueli A, Rodriguez MS, et al. E4F1 is an atypical ubiquitin ligase that modulates p53 effector functions independently of degradation. Cell. 2006;127:775–788. doi: 10.1016/j.cell.2006.09.031. [DOI] [PubMed] [Google Scholar]
- Lee KK, Workman JL. Histone acetyltransferase complexes: one size doesn’t fit all. Nat Rev Mol Cell Biol. 2007;8:284–295. doi: 10.1038/nrm2145. [DOI] [PubMed] [Google Scholar]
- Legube G, Trouche D. Regulating histone acetyltransferases and deacetylases. EMBO Rep. 2003;4:944–947. doi: 10.1038/sj.embor.embor941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerin C, Rodgers JT, Kalume DE, Kim SH, Pandey A, Puigserver P. GCN5 acetyltransferase complex controls glucose metabolism through transcriptional repression of PGC-1alpha. Cell Metab. 2006;3:429–438. doi: 10.1016/j.cmet.2006.04.013. [DOI] [PubMed] [Google Scholar]
- Li AG, Piluso LG, Cai X, Gadd BJ, Ladurner AG, Liu X. An acetylation switch in p53 mediates holo-TFIID recruitment. Mol Cell. 2007a;28:408–421. doi: 10.1016/j.molcel.2007.09.006. [DOI] [PubMed] [Google Scholar]
- Li X, Zhang S, Blander G, Tse JG, Krieger M, Guarente L. SIRT1 deacetylates and positively regulates the nuclear receptor LXR. Mol Cell. 2007b;28:91–106. doi: 10.1016/j.molcel.2007.07.032. [DOI] [PubMed] [Google Scholar]
- Lin J, Handschin C, Spiegelman BM. Metabolic control through the PGC-1 family of transcription coactivators. Cell Metab. 2005;1:361–370. doi: 10.1016/j.cmet.2005.05.004. [DOI] [PubMed] [Google Scholar]
- Liu L, Scolnick DM, Trievel RC, Zhang HB, Marmorstein R, Halazonetis TD, Berger SL. p53 sites acetylated in vitro by PCAF and p300 are acetylated in vivo in response to DNA damage. Mol Cell Biol. 1999;19:1202–1909. doi: 10.1128/mcb.19.2.1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombard DB, Alt FW, Cheng HL, Bunkenborg J, Streeper RS, Mostoslavsky R, Kim J, Yancopoulos G, Valenzuela D, Murphy A, et al. Mammalian Sir2 homolog SIRT3 regulates global mitochondrial lysine acetylation. Mol Cell Biol. 2007;27:8807–8814. doi: 10.1128/MCB.01636-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lonard DM, O’Malley BW. Nuclear receptor coregulators: judges, juries, and executioners of cellular regulation. Mol Cell. 2007;27:691–700. doi: 10.1016/j.molcel.2007.08.012. [DOI] [PubMed] [Google Scholar]
- Margueron R, Trojer P, Reinberg D. The key to development: interpreting the histone code? Curr Opin Genet Dev. 2005;15:163–176. doi: 10.1016/j.gde.2005.01.005. [DOI] [PubMed] [Google Scholar]
- Matsuyama A, Shimazu T, Sumida Y, Saito A, Yoshimatsu Y, Seigneurin-Berny D, Osada H, Komatsu Y, Nishino N, Khochbin S, et al. In vivo destabilization of dynamic microtubules by HDAC6-mediated deacetylation. EMBO J. 2002;21:6820–6831. doi: 10.1093/emboj/cdf682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuzaki H, Daitoku H, Hatta M, Aoyama H, Yoshimochi K, Fukamizu A. Acetylation of Foxo1 alters its DNA-binding ability and sensitivity to phosphorylation. Proc Natl Acad Sci USA. 2005;102:11278–11283. doi: 10.1073/pnas.0502738102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattagajasingh I, Kim CS, Naqvi A, Yamamori T, Hoffman TA, Jung SB, DeRicco J, Kasuno K, Irani K. SIRT1 promotes endothelium-dependent vascular relaxation by activating endothelial nitric oxide synthase. Proc Natl Acad Sci USA. 2007;104:14855–14860. doi: 10.1073/pnas.0704329104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthias P, Yoshida M, Khochbin S. HDAC6 a new cellular stress surveillance factor. Cell Cycle. 2008;7:7–10. doi: 10.4161/cc.7.1.5186. [DOI] [PubMed] [Google Scholar]
- Mujtaba S, He Y, Zeng L, Yan S, Plotnikova O, Sachchidanand Sanchez R, Zeleznik-Le NJ, Ronai Z, Zhou MM. Structural mechanism of the bromodomain of the coactivator CBP in p53 transcriptional activation. Mol Cell. 2004;13:251–263. doi: 10.1016/s1097-2765(03)00528-8. [DOI] [PubMed] [Google Scholar]
- Nadiminty N, Lou W, Lee SO, Lin X, Trump DL, Gao AC. Stat3 activation of NF-{kappa}B p100 processing involves CBP/p300-mediated acetylation. Proc Natl Acad Sci USA. 2006;103:7264–7269. doi: 10.1073/pnas.0509808103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okumura K, Mendoza M, Bachoo RM, DePinho RA, Cavenee WK, Furnari FB. PCAF modulates PTEN activity. J Biol Chem. 2006;281:26562–26568. doi: 10.1074/jbc.M605391200. [DOI] [PubMed] [Google Scholar]
- Olson BL, Hock MB, Ekholm-Reed S, Wohlschlegel JA, Dev KK, Kralli A, Reed SI. SCFCdc4 acts antagonistically to the PGC-1alpha transcriptional coactivator by targeting it for ubiquitin-mediated proteolysis. Genes Dev. 2008;22:252–264. doi: 10.1101/gad.1624208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearl LH, Prodromou C. Structure and mechanism of the Hsp90 molecular chaperone machinery. Annu Rev Biochem. 2006;75:271–294. doi: 10.1146/annurev.biochem.75.103004.142738. [DOI] [PubMed] [Google Scholar]
- Pearson M, Carbone R, Sebastiani C, Cioce M, Fagioli M, Saito S, Higashimoto Y, Appella E, Minucci S, Pandolfi PP, Pelicci PG. PML regulates p53 acetylation and premature senescence induced by oncogenic Ras. Nature. 2000;406:207–210. doi: 10.1038/35018127. [DOI] [PubMed] [Google Scholar]
- Perkins ND. Post-translational modifications regulating the activity and function of the nuclear factor kappa B pathway. Oncogene. 2006;25:6717–6730. doi: 10.1038/sj.onc.1209937. [DOI] [PubMed] [Google Scholar]
- Prives C, Manley JL. Why is p53 acetylated? Cell. 2001;107:815–818. doi: 10.1016/s0092-8674(01)00619-5. [DOI] [PubMed] [Google Scholar]
- Reed NA, Cai D, Blasius TL, Jih GT, Meyhofer E, Gaertig J, Verhey KJ. Microtubule acetylation promotes kinesin-1 binding and transport. Curr Biol. 2006;16:2166–2172. doi: 10.1016/j.cub.2006.09.014. [DOI] [PubMed] [Google Scholar]
- Rodgers JT, Lerin C, Gerhart-Hines Z, Puigserver P. Metabolic adaptations through the PGC-1alpha and SIRT1 pathways. FEBS Lett. 2008;582:46–53. doi: 10.1016/j.febslet.2007.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodgers JT, Lerin C, Haas W, Gygi SP, Spiegelman BM, Puigserver P. Nutrient control of glucose homeostasis through a complex of PGC-1alpha and SIRT1. Nature. 2005;434:113–118. doi: 10.1038/nature03354. [DOI] [PubMed] [Google Scholar]
- Ruthenburg AJ, Li H, Patel DJ, Allis CD. Multivalent engagement of chromatin modifications by linked binding modules. Nat Rev Mol Cell Biol. 2007;8:983–994. doi: 10.1038/nrm2298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rytinki MM, Palvimo JJ. Sumoylation modulates the transcription repressor function of receptor interacting protein 140. J Biol Chem. 2008 doi: 10.1074/jbc.M709359200. Epub. [DOI] [PubMed] [Google Scholar]
- Sakaguchi K, Herrera JE, Saito S, Miki T, Bustin M, Vassilev A, Anderson CW, Appella E. DNA damage activates p53 through a phosphorylation-acetylation cascade. Genes Dev. 1998;12:2831–2841. doi: 10.1101/gad.12.18.2831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampath SC, Marazzi I, Yap KL, Krutchinsky AN, Mecklenbrauker I, Viale A, Rudensky E, Zhou MM, Chait BT, Tarakhovsky A. Methylation of a histone mimic within the histone methyltransferase G9a regulates protein complex assembly. Mol Cell. 2007;27:596–608. doi: 10.1016/j.molcel.2007.06.026. [DOI] [PubMed] [Google Scholar]
- Schindler C, Levy DE, Decker T. JAK-STAT signaling: from interferons to cytokines. J Biol Chem. 2007;282:20059–20063. doi: 10.1074/jbc.R700016200. [DOI] [PubMed] [Google Scholar]
- Schreiber SL, Bernstein BE. Signaling network model of chromatin. Cell. 2002;111:771–778. doi: 10.1016/s0092-8674(02)01196-0. [DOI] [PubMed] [Google Scholar]
- Schwer B, Bunkenborg J, Verdin RO, Andersen JS, Verdin E. Reversible lysine acetylation controls the activity of the mitochondrial enzyme acetyl-CoA synthetase 2. Proc Natl Acad Sci USA. 2006;103:10224–10229. doi: 10.1073/pnas.0603968103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scroggins BT, Robzyk K, Wang D, Marcu MG, Tsutsumi S, Beebe K, Cotter RJ, Felts S, Toft D, Karnitz L, et al. An acetylation site in the middle domain of Hsp90 regulates chaperone function. Mol Cell. 2007;25:151–159. doi: 10.1016/j.molcel.2006.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi X, Kachirskaia I, Yamaguchi H, West LE, Wen H, Wang EW, Dutta S, Appella E, Gozani O. Modulation of p53 function by SET8-mediated methylation at lysine 382. Mol Cell. 2007;27:636–646. doi: 10.1016/j.molcel.2007.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soussi T, Ishioka C, Claustres M, Beroud C. Locus-specific mutation databases: pitfalls and good practice based on the p53 experience. Nat Rev Cancer. 2006;6:83–90. doi: 10.1038/nrc1783. [DOI] [PubMed] [Google Scholar]
- Starai VJ, Celic I, Cole RN, Boeke JD, Escalante-Semerena JC. Sir2-dependent activation of acetyl-CoA synthetase by deacetylation of active lysine. Science. 2002;298:2390–2392. doi: 10.1126/science.1077650. [DOI] [PubMed] [Google Scholar]
- Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- Sun Y, Jiang X, Chen S, Fernandes N, Price BD. A role for the Tip60 histone acetyltransferase in the acetylation and activation of ATM. Proc Natl Acad Sci USA. 2005;102:13182–13187. doi: 10.1073/pnas.0504211102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sykes SM, Mellert HS, Holbert MA, Li K, Marmorstein R, Lane WS, McMahon SB. Acetylation of the p53 DNA-binding domain regulates apoptosis induction. Mol Cell. 2006;24:841–851. doi: 10.1016/j.molcel.2006.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang X, Gao JS, Guan YJ, McLane KE, Yuan ZL, Ramratnam B, Chin YE. Acetylation-dependent signal transduction for type I interferon receptor. Cell. 2007;131:93–105. doi: 10.1016/j.cell.2007.07.034. [DOI] [PubMed] [Google Scholar]
- Tang Y, Luo J, Zhang W, Gu W. Tip60-dependent acetylation of p53 modulates the decision between cell-cycle arrest and apoptosis. Mol Cell. 2006;24:827–839. doi: 10.1016/j.molcel.2006.11.021. [DOI] [PubMed] [Google Scholar]
- Thompson PR, Wang D, Wang L, Fulco M, Pediconi N, Zhang D, An W, Ge Q, Roeder RG, Wong J, et al. Regulation of the p300 HAT domain via a novel activation loop. Nat Struct Mol Biol. 2004;11:308–315. doi: 10.1038/nsmb740. [DOI] [PubMed] [Google Scholar]
- Toledo F, Wahl GM. Regulating the p53 pathway: in vitro hypotheses, in vivo veritas. Nat Rev Cancer. 2006;6:909–923. doi: 10.1038/nrc2012. [DOI] [PubMed] [Google Scholar]
- Turner BM. Defining an epigenetic code. Nat Cell Biol. 2007;9:2–6. doi: 10.1038/ncb0107-2. [DOI] [PubMed] [Google Scholar]
- Vakhrusheva O, Smolka C, Gajawada P, Kostin S, Boettger T, Kubin T, Braun T, Bober E. Sirt7 Increases Stress Resistance of Cardiomyocytes and Prevents Apoptosis and Inflammatory Cardiomyopathy in Mice. Circ Res. 2008 doi: 10.1161/CIRCRESAHA.107.164558. epub. [DOI] [PubMed] [Google Scholar]
- van der Horst A, Burgering BM. Stressing the role of FoxO proteins in lifespan and disease. Nat Rev Mol Cell Biol. 2007;8:440–450. doi: 10.1038/nrm2190. [DOI] [PubMed] [Google Scholar]
- Vo N, Fjeld C, Goodman RH. Acetylation of nuclear hormone receptor-interacting protein RIP140 regulates binding of the transcriptional corepressor CtBP. Mol Cell Biol. 2001;21:6181–6188. doi: 10.1128/MCB.21.18.6181-6188.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogelstein B, Lane D, Levine AJ. Surfing the p53 network. Nature. 2000;408:307–310. doi: 10.1038/35042675. [DOI] [PubMed] [Google Scholar]
- Vousden KH, Lane DP. p53 in health and disease. Nat Rev Mol Cell Biol. 2007;8:275–283. doi: 10.1038/nrm2147. [DOI] [PubMed] [Google Scholar]
- Walter W, Clynes D, Tang Y, Marmostein R, Mellor J, Berger SL. 14-3-3 interaction with histone H3 involves dual modification pattern of phosphoacetylation. Mol Cell Biol. 2008 doi: 10.1128/MCB.01457-07. Epub. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westermann S, Weber K. Post-translational modifications regulate microtubule function. Nat Rev Mol Cell Biol. 2003;4:938–947. doi: 10.1038/nrm1260. [DOI] [PubMed] [Google Scholar]
- White R, Morganstein D, Christian M, Seth A, Herzog B, Parker MG. Role of RIP140 in metabolic tissues: Connections to disease. FEBS Lett. 2008;582:39–45. doi: 10.1016/j.febslet.2007.11.017. [DOI] [PubMed] [Google Scholar]
- Whittle JR, Powell MJ, Popov VM, Shirley LA, Wang C, Pestell RG. Sirtuins, nuclear hormone receptor acetylation and transcriptional regulation. Trends Endocrinol Metab. 2007;18:356–364. doi: 10.1016/j.tem.2007.07.007. [DOI] [PubMed] [Google Scholar]
- Winter S, Simboeck E, Fischle W, Zupkovitz G, Dohnal I, Mechtler K, Ammerer G, Seiser C. 14-3-3 Proteins recognize a histone code at histone H3 and are required for transcriptional activation. EMBO J. 2008;27:88–99. doi: 10.1038/sj.emboj.7601954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang XJ. Multisite protein modification and intramolecular signaling. Oncogene. 2005;24:1653–1662. doi: 10.1038/sj.onc.1208173. [DOI] [PubMed] [Google Scholar]
- Yang XJ, Grégoire S. A recurrent phospho-sumoyl switch in transcriptional repression and beyond. Mol Cell. 2006;23:779–786. doi: 10.1016/j.molcel.2006.08.009. [DOI] [PubMed] [Google Scholar]
- Yang XJ, Seto E. The Rpd3/Hda1 family of lysine deacetylases: from bacteria and yeast to mice and men. Nat Rev Mol Cell Biol. 2008;9:206–218. doi: 10.1038/nrm2346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X, Guo ZS, Marcu MG, Neckers L, Nguyen DM, Chen GA, Schrump DS. Modulation of p53, ErbB1, ErbB2, and Raf-1 expression in lung cancer cells by depsipeptide FR901228. J Natl Cancer Inst. 2002;94:504–513. doi: 10.1093/jnci/94.7.504. [DOI] [PubMed] [Google Scholar]
- Yuan Z, Becker EB, Merlo P, Yamada T, DiBacco S, Konishi Y, Schaefer EM, Bonni A. Activation of FOXO1 by Cdk1 in cycling cells and postmitotic neurons. Science. 2008;319:1665–1668. doi: 10.1126/science.1152337. [DOI] [PubMed] [Google Scholar]
- Yuan Z, Zhang X, Sengupta N, Lane WS, Seto E. SIRT1 regulates the function of the Nijmegen breakage syndrome protein. Mol Cell. 2007;27:149–162. doi: 10.1016/j.molcel.2007.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan ZL, Guan YJ, Chatterjee D, Chin YE. Stat3 dimerization regulated by reversible acetylation of a single lysine residue. Science. 2005;307:269–273. doi: 10.1126/science.1105166. [DOI] [PubMed] [Google Scholar]
- Zeng L, Xiao Q, Margariti A, Zhang Z, Zampetaki A, Patel S, Capogrossi MC, Hu Y, Xu Q. HDAC3 is crucial in shear- and VEGF-induced stem cell differentiation toward endothelial cells. J Cell Biol. 2006;174:1059–1069. doi: 10.1083/jcb.200605113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Yuan Z, Zhang Y, Yong S, Salas-Burgos A, Koomen J, Olashaw N, Parsons JT, Yang XJ, Dent SR, et al. HDAC6 modulates cell motility by altering the acetylation level of cortactin. Mol Cell. 2007;27:197–213. doi: 10.1016/j.molcel.2007.05.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Kwon S, Yamaguchi T, Cubizolles F, Rousseaux S, Kneissel M, Cao C, Li N, Cheng HL, Chua K, et al. Mice lacking histone deacetylase 6 have hyperacetylated tubulin but are viable and develop normally. Mol Cell Biol. 2008;28:1688–1701. doi: 10.1128/MCB.01154-06. [DOI] [PMC free article] [PubMed] [Google Scholar]