Abstract
Traditional production of therapeutic glycoproteins relies on mammalian cell culture technology. Glycoproteins produced by mammalian cells invariably display N-glycan heterogeneity resulting in a mixture of glycoforms the composition of which varies from production batch to production batch. However, extent and type of N-glycosylation has a profound impact on the therapeutic properties of many commercially relevant therapeutic proteins making control of N-glycosylation an emerging field of high importance. We have employed a combinatorial library approach to generate glycoengineered Pichia pastoris strains capable of displaying defined human-like N-linked glycans at high uniformity. The availability of these strains allows us to elucidate the relationship between specific N-linked glycans and the function of glycoproteins. The aim of this study was to utilize this novel technology platform and produce two human-like N-linked glycoforms of recombinant human lactoferrin (rhLF), sialylated and non-sialylated, and to evaluate the effects of terminal N-glycan structures on in vitro secondary humoral immune responses. Lactoferrin is considered an important first line defense protein involved in protection against various microbial infections. Here, it is established that glycoengineered P. pastoris strains are bioprocess compatible. Analytical protein and glycan data are presented to demonstrate the capability of glycoengineered P. pastoris to produce fully humanized, active and immunologically compatible rhLF. In addition, the biological activity of the rhLF glycoforms produced was tested in vitro revealing the importance of N-acetylneuraminic (sialic) acid as a terminal sugar in propagation of proper immune responses.
Keywords: Recombinant human lactoferrin, Pichia pastoris expression system, Humanized N-linked glycoforms, Humoral immune responses
Introduction
Compared to the mammalian cell culture technology currently in use for the production of most therapeutic glycoproteins, the methylotrophic yeast Pichia pastoris has the capability of yielding up to 20 times more product. However, glycoproteins derived from P. pastoris as well as other fungal and yeast expression systems display fungal-type high mannose N-linked glycans. These glycans are believed to contribute to reduce the half-life of the glycoprotein in vivo and may be immunogenic, thus limiting the potential therapeutic value of fungal-derived glycoproteins [15].
Human N-glycosylation is a multi-step process localized to the secretory pathway of cells. The complex metabolic engineering endeavor of replicating the mammalian glycosylation machinery in yeast requires the cloning and functional expression of a large number of foreign glycosylation pathway enzymes in the host strains. Each enzyme catalyzes a reaction yielding the substrate for the subsequent enzyme. Thus, each enzyme must be properly targeted and must function at high efficiency in its respective location in the secretory pathway. The application of a combinatorial library approach has been essential to generate P. pastoris strains harboring combinations of mannosidases, glycosyltransferases, GlcNAc/Gal transporters, and Gal epimerase [24]. Over the past years we have created a library of glycoengineered P. pastoris strains each capable of displaying defined N-linked glycans at high uniformity [5, 8, 12, 17, 24]. This growing library of strains has the potential to allow elucidation of the relationship between specific N-linked glycans and the function of glycoproteins.
Lactoferrin (LF), an iron-binding glycoprotein is found in most mammalian exocrine secretions, including milk, tears, saliva, bronchial and intestinal secretions and also in the secondary granules of neutrophils. It is considered a first line defense protein involved in protection against microbial infections [26, 31] and prevention of systemic inflammation [2, 3, 21]. More recently, lactoferrin has been implicated in immunoregulatory functions [19, 42, 44, 46], as a modulator of vaccine function [18], and containing chemoprotective activity [1]. The primary structure of human LF is characterized by a single polypeptide chain containing 692 amino acids organized in two highly homologous lobes, designated the N- and C-lobe, each capable of binding one ferric ion (Fe+++). The low-density lipoprotein receptor-related protein-1 and -2 (LRP1 and LRP2) are considered primary LF receptors. Although members of the LRP family are generally considered as endocytic receptors, LRP1 can also function as a signaling receptor [29]. The key to understanding the molecular basis of LF various activities is thought to reside in part according to patterns of glycosylation [37]. There are three possible N-linked glycosylation sites in hLF, one at Asn138, a second site at Asn479, and a third site at Asn624; differential utilization of these sites results in distinct glycosylation variants [30, 35]. Human LF glycans are the N-acetyllactosaminic type, α1-3-fucosylated on the N-acetylglucosamine residue linked to the peptide chain. Unlike the milk-derived LF, the neutrophilic form is not fucosylated, and the difference in molecular structure and function of the two forms is not fully understood [23, 32].
The aim of this study was to produce two glycoforms of recombinant human lactoferrin (rhLF): sialylated and non-sialylated, and evaluate the effects of terminal N-glycan structures on in vitro secondary humoral immune responses suppressed by methotrexate (MXT). To this end glycoengineered P. pastoris capable of producing a highly uniform N-glycan structure with terminal galactose (Gal2GlcNAc2Man3GlcNAc2) was selected and employed for in vitro sialylation to create terminally sialylated N-glycan (Sia2Gal2GlcNAc2Man3GlcNAc2) of rhLF. Here, it is demonstrated that LF with specific human N-glycan structures can be produced in glycoengineered lines of the yeast P. pastoris and the two major LF glycoforms produced in this expression system exhibit a significantly different ability to overcome the suppressive action of methotrexate in the secondary, humoral immune response to sheep erythrocytes in mice.
Experimental procedures
Strains, culture conditions and reagents
Escherichia coli strain TOP10 was used for recombinant DNA work. P. pastoris yAS309 [24] was used for generation of rhLF producing strains. Protein expression studies were done at room temperature in a 96-well plate format with buffered glycerol-complex medium (BMGY) consisting of 1% yeast extract, 2% peptone, 100 mM potassium phosphate buffer, pH 6.0, 1.34% yeast nitrogen base, 4×10−5% biotin, and 1% glycerol as a growth medium. The induction media were buffered methanol-complex medium (BMMY) consisting of 0.5% methanol and buffered dextrose-complex medium (BMDY) consisting of 2% dextrose, respectively. hLF standard purified from human milk was purchased from Sigma (St. Louis, MO). Restriction and modification enzymes were from New England BioLabs (Beverly, MA). All chemicals were ACS-grade, and CMP-sialic acid and trypsin were purchased from Sigma (St. Louis, MO), recombinant rat α 2,6-(N)-sialyltransferase from Calbiochem (San Diego, CA), 2-aminobenzamide (2-AB) dye from Aldrich (St. Louis, MO). A polyclonal antibody against P. pastoris-derived host cell proteins (anti-HCP) was generated as follows; the culture supernatant from P. pastoris lacking hLF construct was purified in a similar fashion as described in the capturing step of rhLF. The purified proteins in PBS were used for polyclonal antibody generation in rabbits (Rockland Immunochemical Inc., Boyertown, PA).
Expression constructs and generation of production strains
For the signal sequence study, pPICZA (Invitrogen, Carlsbad, CA) was digested with EcoRI and KpnI, and the resulting pPICZA was ligated with 11 different signal sequences (EcoRI and blunt ended) and the codon-optimized hLF cDNA (blunt and KpnI ended) (provided by PharmaReview Corporation, Houston, TX). For the promoter study, PpAOX1 promoter was replaced with the PpGAPDH promoter in pBK422 at BglII and EcoRI sites. All expression constructs were sequence verified. For the generation of hLF production strains, PmeI-digested DNAs were transformed into yAS309 by electroporation, according to the Pichia expression kit handbook from Invitrogen. See Table 1 for designated plasmids and strains.
Table 1.
Strains and plasmids used in this study
| Strain and plasmid | Description | Source |
|---|---|---|
| TOP10 | E. coli (F-mcrA Δ(mrr-hsdRMS-mcrBC) ϕ80lacZΔM15 ΔlacX74 recA1 araD139 Δ(ara-leu)7697 galU galK rpsL (StrR) endA1 nupG) | Invitrogen (Carlsbad, CA) |
| yAS309 | Glycoengineered Pichia pastoris | Li et al. [24] |
| BK422 | pBK842 in yAS309 | This study |
| BK427 | pBK850 in yAS309 | This study |
| pPICZAa | PpAOX1 promoter, ZeoR | Invitrogen |
| pGAPZAa | PpGAPDH promoter, ZeoR | Invitrogen |
| pBK833 | S. cerevisiae α-mating factor pre+hLF in pPICZA | This study |
| pBK834 | α-amylase signal sequence (ss)+hLF in pPICZA | This study |
| pBK835 | Glucoamylase ss+hLF in pPICZA | This study |
| pBK836 | Human serum albumin ss+hLF in pPICZA | This study |
| pBK837 | Inulinase ss+hLF in pPICZA | This study |
| pBK838 | Invertase ss+hLF in pPICZA | This study |
| pBK839 | P. pastoris KAR2 ss+hLF in pPICZA | This study |
| pBK840 | S. cerevisiae killer toxin 1 ss+hLF in pPICZA | This study |
| pBK841 | P. pastoris phosphotase 1 ss+hLF in pPICZA | This study |
| pBK842 | S. cerevisiae α-mating factor prepro+hLF in pPICZA | This study |
| pBK843 | Chicken lysozyme ss+hLF in pPICZA | This study |
| pBK850 | S. cerevisiae α-mating factor preproKR+hLF in pGAPZA | This study |
Identical signal sequence used with different promoter sequences.
Detection of rhLF
Proteins were separated by 4–20% gradient SDS-PAGE and then electroblotted onto nitrocellulose membrane (Schleicher & Schuell Inc., Keene, NH). The membrane was probed with rabbit anti-human LF antibody (anti-hLF) (1:1000) (Sigma, St. Louis, MO) and followed by goat anti-rabbit IgG antibody conjugated with horseradish peroxidase (1:4,000) (Pierce, Rockford, IL). The results were visualized using an ImmunoPure Metal Enhanced DAB Substrate Kit (Pierce, Rockford, IL).
Fermentation
A seed culture was prepared by adding 1 ml of thawed cells to a 2 L baffled flask containing 400 ml of 4% BMGY medium. When an OD600 of 20±5 was reached, the seed culture was transferred to the production fermenter. The systems was controlled by Applikon 1030 Bio-controller with closed loop control of pH, temperature, dissolved oxygen concentration and foam control as described earlier [24]. The pH was maintained at 6.0 throughout the fermentation. Fermentation runs were carried out in 15 L (12 L working) autoclavable glass bioreactors from Applikon (Foster City, CA).
Protein purification
Primary clarification was performed at 4°C for 15 min at 13,000×g by centrifugation in a Sorvall Evolution RC (Kendo, Asheville, NC) followed by the microfiltration and diafiltration steps using a 0.1 μm cut-off 3600 cm2 PES hollow fibre cartridge (CFP-1-E-8A) (GE Amersham, Pittsburgh, PA) and 5×0.1 m2 Pellicon 2 Mini 10 kDa NMWCO regenerated cellulose ultrafiltration cassettes (A screen) (Millipore, Billerica, MA), respectively. Protease inhibitors pepstatin A and chymostatin (Sigma) were added to the supernatant after filtration steps in a concentration of 5 and 3 μg/ml respectively. After the ultrafiltration/diafiltration/filtration steps, P. pastoris-derived rhLF was purified by two chromatographic steps; cation exchange chromatography using SP Sepharose Fast Flow followed by Heparin Sepharose 6 Fast Flow chromatography (GE Healthcare, Piscataway, NJ). Briefly, SP Sepharose resin was equilibrated with 50 mM Tris–HCl, pH 8.0 while the supernatant media was adjusted at the same pH and conductivity around 5 mS/cm by the diafiltration procedure. The elution was done with 10 column volume (CV) of a gradient of 0–1 M NaCl in the same buffer. rhLF containing fractions were pooled and dialyzed against 50 mM Tris–HCl, pH 7.5 overnight. As the final purification step the affinity resin, Heparin Sepharose 6 Fast Flow, was used and the column was equilibrated with 50 mM Tris–HCl (pH 7.5). The pooled and dialyzed protein from SP Sepharose was loaded on Heparin Sepharose and washed in three steps. The first wash was done with 2 CV of the same buffer, followed by the second wash with 10 CV of a detergent buffer (10 mM CHAPS, 10 mM EDTA in 50 mM Tris–HCl, pH 7.5) to decrease endotoxin levels. The last wash was carried out with a 10 CV of 50 mM Tris–HCl (pH 7.5). The protein was eluted with a 10 CV of a gradient of 0–1 M NaCl. A fraction of the pooled protein was dialyzed against PBS (pH 7.2) and stored at 4°C as a final product of nonsialylated hLF. The other fraction was dialyzed against 50 mM MES, pH 6.5 for the preparation of the in vitro sialylation. The in vitro sialylated rhLF was purified using Heparin Sepharose 6 Fast Flow and dialyzed against PBS (pH 7.2), and stored at 4°C.
Size exclusion chromatography
Proteins were separated with a BioRad BioSil SEC250 column (Hercules, CA) using a mobile phase of 100 mM sodium phosphate, pH 6.8, 150 mM NaCl and 0.05% sodium azide at 0.5 ml/min and detected at 280 nm.
Reverse-phase HPLC
Proteins were separated with a Phenomenex Jupiter 5 μ C4 300 Å column (Torrance, CA) using a 1.0 ml/min 39-min linear gradient from 95% to 30% buffer A (0.1% TFA in water). Buffer B was 0.08% TFA in acetonitrile. Temperature was maintained at 80°C using a column oven. A Hitachi diode array detector monitoring at 220 nm was used for detection.
MALDI-TOF analysis of glycans
N-glycans were released and separated from hLF as described earlier [8, 12]. Molecular weight was determined by using a Voyager linear matrix-assisted laser desorption/ionization time of flight (MALDI-TOF) mass spectrometer (Applied Biosystems) with delayed extraction.
N-glycan structure analysis
2-Aminobenzamide (2-AB) labeling was used to quantify N-glycan structures. A solution of 5% 2-AB dye and 6.3% sodium cyanoborohydride was prepared in 1:4 glacial acetic acid/DMSO. Five microliters of this solution was added to dried glycan samples, mixed, and incubated for 2–3 h at 65°C. Each sample was applied to wells of a 96-well lysate plate (Promega Cat# A2241, Madison, WI) and then washed and pre-wetted with acetonitrile and adsorbed for 10–15 min; wells were then washed with 1 ml acetonitrile followed by three 1 ml 96% acetonitrile/4% water washes. Glycans were eluted three times with 0.4 ml water and dried in a centrifugal vacuum for 24 h. Labeled glycans were then separated by HPLC using a flow rate of 1.0 ml/min with a Prevail CHO ES 5-micron bead, amino-bound column using a 50-min linear gradient of 80% to 40% buffer A (100% acetonitrile). Buffer B consisted of 50 mM ammonium formate pH 4.4. Sialylated glycans were separated using a 30-min 80–40% Buffer A linear gradient with an additional 30-min gradient bringing buffer A from 40% to 0%. Labeled glycans were detected and quantified against standards using a fluorescence detector with an excitation of 330 nm and an emission at 420 nm.
Trypsin peptide mapping
Two milligrams of 0.25 mg/ml rhLF was diluted 1:2 with 10 M guanidine HCl pH 7.8. To this mixture, dithiothreitol (DTT) was added to a final concentration of 10 mM and incubated at 37°C for 1 h. Iodoacetic acid in Tris base neutralized with NH4OH was added to a final concentration of 40 mM and incubated in the dark for 1 h. The sample was then buffer-exchanged and concentrated with a Vivaspin 50,000 MWCO concentrator to roughly 1 mg/ml in 100 mM Tris–HCl, pH 7.4. The sample was subsequently split into four aliquots with each receiving 2.6 μg of trypsin in 10 mM HCl. Samples were incubated overnight at 37°C then boiled for 5 min. Two aliquots were deglycosylated with 5 μl of PNGase F for 30 min at 37°C. Peptides were resolved with a Phenomenex Jupiter 4 μ Proteo 90 Å column using an 87-min linear gradient of 95% to 55% Buffer A (0.1% TFA in water) at 1.0 ml/min. Buffer B was 0.08% TFA in acetonitrile. Temperature was maintained at 30°C using a column oven. A Hitachi diode array detector monitoring at 220 nm was used for detection. One minute fractions were collected from the column into a 96-well plate and dried.
Nanospray mass spectrometry
Digested peptides were dissolved in 50 μl 30% acetonitrile, 0.1% formic acid. Fractions with peptides that appeared on the HPLC trace after PNGase F digest (RT=55, 56, 63, 64, 65, 66 min) were transferred to a 96-well Abgene sample plate and sealed with adhesive foil. A Thermo Finnigan LTQ mass spectrometer was set to data-dependent triple play mode to analyze the top five most intense peaks for 2.9 min through Advion Triversa Nanomate. Data were analyzed using SEQUEST (Thermo Finnigan, Waltham, MA).
In vitro sialylation
Two hundred microunits sialyltransferase and 3.875 mg CMP-sialic acid (final concentration; 52.5 μM) was added to 31 mg of hLF in 50 mM MES pH 6.5, 10 mM MnCl2, 0.73 μM chymostatin, and 52 ng/l pepstatin and incubated overnight at 25°C. Sialylated rhLF was dialyzed against 20 mM Tris (pH 8.0) overnight followed by purification using heparin Sepharose Fast Flow 6.
Endotoxin assay
Endotoxin levels were assayed using a limulus amebocyte lysate endochrome assay kit from Charles River Laboratories according to the manufacturer's instructions (Charleston, SC).
Enzyme-linked immunosorbent assay
A high-protein binding 96-well plate (Costar) was coated with 100 μl/well of rabbit anti-human LF antibody (Sigma) diluted 1:5,000 in PBS (EMD Biosciences, San Diego, CA) overnight at 4°C. Antibody was aspirated and the plate was blocked for 1 h at room temperature with 200 μl of 3% BSA in PBS. Blocking solution was aspirated and replaced with 100 μl of serial dilutions of commercially available human colostrum hLF (Sigma) and samples to be assayed. Standard hLF was diluted two-fold serially in PBS from 100 to 0.1 ng/ml; fermenter samples were typically diluted 1:100 and then two-fold serially to 1:100,000. Standards and samples were incubated for 1 h at room temperature then aspirated and washed three times with 300 μl/well of 0.05% Tween 20 in PBS using a manifold plate washer. Wash buffer was aspirated and then 100 μl/well of HRP-conjugated anti-hLF (Jackson Immunoresearch, West Grove, PA) diluted 1:5,000 in PBS was added and incubated for 1 h at room temperature. Wells were washed, 100 μl/well of 3,3′,5,5′-tetramethylbenzidine (Sigma) was added and the final reaction terminated with 1 MH2SO4 after which absorbance at 450 nm was measured.
Protein assays
Protein concentration was estimated using the method of Bradford as described [6]. Protein assay reagents were from Pierce Biotechnology (Rockford, IL). Bovine serum albumin (Pierce Biotechnology) and human milk lactoferrin (Sigma) were used as standards.
Mice
Twelve-week-old CBA male and female mice were used for the studies. All in vivo experiments were conducted under animal ethics committee approved guidelines.
The secondary humoral immune response in vitro
Mice were primed with intraperitoneal administration of 0.2 ml 1% sheep red blood cells (SRBC) suspension. Splenocytes were isolated after 4 days and single cell suspensions were prepared in culture medium consisting of RPMI 1640, supplemented with 10% fetal calf serum, glutamine, sodium pyruvate, 2-mercaptoethanol and antibiotics. The cells were incubated in 24-well culture plates (5×106/ml/well) with 50 μl 0.005% SRBC. Lactoferrins were added to the cells cultures in concentration of 1 μg/ml at the initiation of culture, MTX at concentration of 0.25–0.5 mM, after 24 h, and sialic acid (0.5 mM) at 30 min before addition of LF. The antibody against mouse sialoadhesin (Serotec, rat anti-mouse CD169, clone 3D6.112, final dilution 1:250) was added to the cell cultures 1 h before LF. After 4 days the number of AFC was determined by the method of local hemolysis in agar [27]. The results are shown as mean values of AFC number from five wells±SE, calculated per 106 viable cells.
Statistics
The differences across groups were determined by analysis of variance after testing homogeneity of variance by Levene's test. Individual grades were then compared using the Tukey's test for multiple comparisons. The data are expressed as: mean, mean±SE (standard error) and mean±SD (standard deviation). Differences were considered significant when p<0.05. The statistical analysis was performed using STATISTICA 6.0 for Windows.
Results
Optimization of recombinant human lactoferrin expression in glycoengineered P. pastoris
The expression of rhLF was optimized by such factors as codon usage, signal sequence, promoter, pH, FeCl3, and induction time. Previously, DNA codon optimization has successfully been applied to improve expression of heterologous proteins in yeast. In order to improve the translational efficiency of rhLF in P. pastoris, a codon-optimized nucleotide sequence encoding hLF was synthesized based on the original sequence (Fig. 1). Amino acid codons were selected based on a P. pastoris codon usage table (GlycoFi™ proprietary).
Fig. 1.
DNA sequence encoding the mature human lactoferrin
A codon-optimized hLF cDNA was fused to 11 different signal sequences (pBK833–pBK850 in Table 1) to identify the optimal secretion sequence facilitating translocation of rhLF into the secretory pathway and ultimately the culture medium. rhLF-producing strains representing 11 signal sequences were generated by transforming eleven expression constructs into yAS309. The efficiency of each signal sequence was evaluated by Western blot of the culture supernatants. Out of the signal sequences tested, S. cerevesiae alpha mating factor prepro (ScαMFppKR: pBK842) was selected in which ScαMFppKR was engineered to contain a Kex2p cleavage site (KR) at C-terminal of the signal sequence to facilitate processing of the protein prior to secretion of mature rhLF. The resulting strain was designated BK422. BK422 was further optimized to maximize rhLF production.
The pH of medium can have an impact on the overall production of the protein of interest by increasing the activity of specific proteases secreted from the host strain as well as influencing protein stability. In order to minimize proteolysis and to enhance protein stability, protein production was tested at different pH values in the induction medium (BMMY) ranging from 6.0 to 7.5. The pH of growth medium (BMGY) was maintained at 6.0. At pH over 6.5, proteolytic degradation of rhLF was observed. The optimal pH of the induction medium was found to be 6.0.
hLF is an iron-binding glycoprotein and is structurally organized into two lobes. Each lobe binds one Fe3+ ion. In some cases the expression levels of hLF have been influenced by supplementation of FeCl3 [36]. In order to identify the optimal concentration of FeCl3, the induction medium was supplemented with FeCl3 in the range of 0.05–2.0 mM final concentration. In our study, FeCl3 supplementation had little effect on the product yield in the range from 0.05 to 0.5 mM whereas concentrations above 0.5 mM displayed a negative impact. Thus, FeCl3 supplementation was not required in our induction medium at pH 6.0.
Two different P. pastoris (Pp) promoters were evaluated for their ability to drive rhLF expression. An inducible promoter from P. pastoris: alcohol oxidase 1 (PpAOX1) and one constitutive promoter derived from the glyceraldehyde-3-phosphate dehydrogenase gene of P. pastoris (PpGAPDH) were evaluated for their ability to drive hLF expression. The AOX1 promoter is tightly regulated at the transcription level, that is, it is repressed in the presence of glucose, whereas it is strongly induced in the methanol medium. rhLF was induced at 0.5% methanol under AOX1 promoter and at 2% dextrose under GAPDH promoter. The proteolytic degradation of rhLF was more pronounced under GAPDH promoter (BK427) than AOX1 promoter-driven protein expression (BK422). The AOX1 promoter-driven expression displayed the least amount of proteolysis and it was used to produce rhLF in a bioreactor.
A time course study was performed to determine the optimal induction time. Methanol was added to the culture every 20 h to maintain rhLF induction during 4-day fermentation. Samples were taken at different time-points during fermentation. Over the course of 4 days of induction, product yield continued to increase. However, an increased amount of proteolysis was also observed. An induction time of about 2 days was determined as an optimum.
Production of recombinant lactoferrin in a bioreactor
rhLF production run was carried out with BK422 in a 15 L bioreactor described in the Experimental procedures section. rhLF was induced for 38 h and the product yield was 99.8 mg/l as measured by ELISA using hLF standard (Sigma). Proteolytic degradation was further reduced by supplementing the fermentation media with the protease inhibitors pepstatin A and chymostatin during induction. A total 841 mg rhLF was subjected to microfiltration followed by ultrafiltration/diafiltration steps (see the “Experimental procedures” section). After a series of filtration steps, the product recovery was 183 mg of rhLF (21% recovery).
Purification of recombinant lactoferrin
rhLF was purified using SP Sepharose and Heparine Sepharose 6. SP Sepharose captured rhLF effectively and was easily scalable from 300 μl to 160 ml of resin. Heparin Sepharose 6 was able to separate isoforms of hLF and chosen for the final purification step to further improve the protein purity.
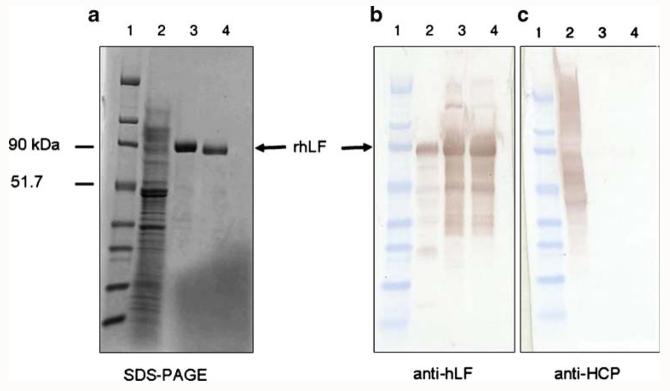
Figure 2 shows the SDS-PAGE and Western blot analysis of rhLF purified from the fermentation supernatant. rhLF was successfully purified by SP Sepharose (Fig. 2a,b and c). Western blot analysis using an anti-HCP antibody demonstrated that SP Sepharose is a good capturing step to obtain a highly pure protein free from host cell contaminants (Fig. 2c). As an advantage, passage over Heparin Sepharose significantly reduced endotoxin levels.
Fig. 2.
SDS-PAGE with Coomassie blue stain (a) and Western blot with antibodies αhLF (b) and αHCP (c) of samples at different purification steps. Lane 1 broad range molecular weight standards, lane 2 rhLF fermentation supernatant, lane 3 SP Sepharose Fast Flow eluant, lane 4 Heparin Sepharose 6 Fast Flow eluant. αHCP antibody against Pichia host cell proteins
Generation of sialylated bi-antennary rhLF by in vitro sialylation
In vitro sialylation of rhLF was carried out as described in the Experimental procedures section, with purity reconfirmed by SDS-PAGE of the non-sialylated and sialylated rhLF. The protein N-terminal sequence was directly compared, indicating that the in vitro sialylation process did not affect the integrity of the N-terminus of the protein. The endotoxin levels were measured and determined to be 4 EU/mg (sialylated) and 3 EU/mg (non-sialylated), respectively.
The purity of sialylated and non-sialylated rhLF samples was confirmed by reverse phase HPLC (data not shown). Size exclusion chromatography was also performed, demonstrating similarity in molecular weights of both molecules. Prominent peaks at m/z of 79,373.5 and 81,250.86 were observed by MALDI-TOF (Fig. 3a). A second pair of peaks at 39,731.4 and 40,646.1 suggests doubly-charged species of 79.3 and 81.2 kDa respectively, correlating with the masses of doubly- and triply-glycosylated rhLF (79.6 and 81.2 kDa, respectively). MALDI-TOF analysis of sialylated rhLF (Fig. 3b) showed a broader peak at about 81,000 m/z with the doubly charged spectra having a m/z of ∼40,000. The greater m/z ratio is most likely due to the presence of sialic acid in this sample.
Fig. 3.
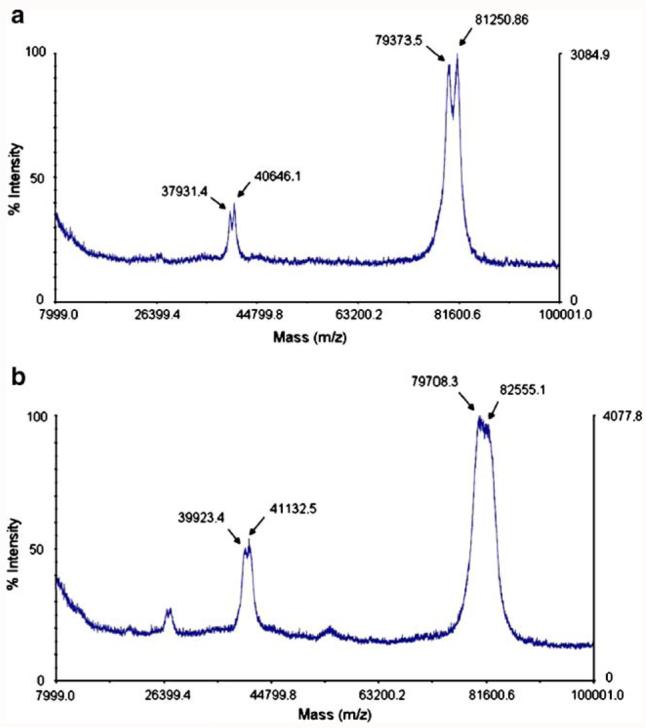
MALDI-TOF spectra of non-sialylated (a) and sialylated (b) rhLFs. Protein samples were diluted 1:1 with sinapinic acid in 0.1% trifluoroacetic acid and 50% acetonitrile, then spotted on a MALDI plate and examined using positive mode on an Applied Biosystems Bioanalyzer MALDI-TOF instrument
MALDI-TOF analysis was used to assess the N-linked glycan profile after enzymatic release from rhLF. Analysis of N-linked glycans released from non-sialylated rhLF in the positive ion mode showed a predominant mass of 1,666.33 indicative of an afucosylated biantennary complex glycan with terminal galactose or Gal2GlcNAc2Man3GlcNAc2 (Fig. 4a). Additionally, the human glycoforms GalGlcNAc2Man3GlcNAc2 (1504.08) and Man5GlcNAc2 (1,258.82) were also observed, although less prominent. N-glycan of sialylated rhLF was analyzed by MALDI-TOF in the negative ion mode and showed a dominant cluster of ions in the range of 2,229.03–2,444.43 consistent with a mass describing Sia2 Gal2GlcNAc2Man3GlcNAc2 and associated cation adducts (Fig. 4b) Since MALDI-TOF provides a qualitative/semi-quantitative assessment of N-glycosylation, we employed a normal phase-HPLC analysis to quantify the relative abundance of specific N-linked glycan structures. Non-sialylated rhLF was determined to contain 50.99% Gal2GlcNAc2 Man3GlcNAc2 (GS5.0) with other human-type glycans comprising GlcNAc2Man3GlcNAc2 (GS4.0), GalGlcNAc2-Man3GlcNAc2 (GS4.5), GalGlcNAcMan5 and GlcNAcMan5 hybrids, and Man5GlcNAc2 (GS2.0). N-glycosylation analysis of the sialylated rhLF sample contained 46.2% terminally bisialylated N-linked glycans and 15.9% monosialylated N-linked glycans, where most of GalGlcNAc2Man3GlcNAc2 (GS4.5) and Gal2GlcNAc2Man3GlcNAc2 (GS5.0) glycans were converted into SiaGal2GlcNAc2Man3GlcNAc2 (GS5.5) and Sia2Gal2GlcNAc2Man3GlcNAc2 (GS6.0). However, GlcNAc2Man3GlcNAc2 (GS4.0) and high mannose containing structures content from sialylated rhLF remained similar to non-sialylated rhLF because they are not substrates for rat α 2,6-(N)-sialyltransferase.
Fig. 4.
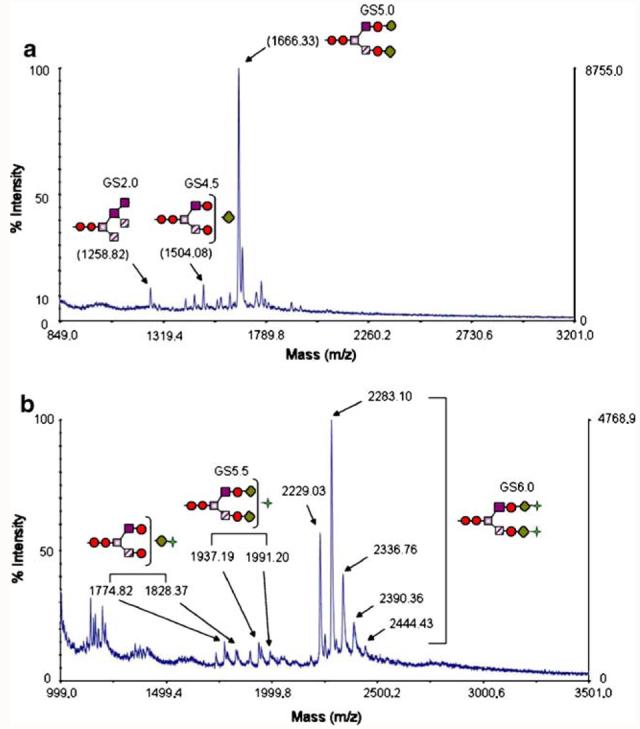
MALDI spectra of released glycan structures. Glycans were removed from rhLF with PNGase F and subjected to MALDI-TOF analysis in the positive ion mode for non-sialylated rhLF (a) and the negative ion mode for sialylated rhLF (b). The major peak observed in the non-sialylated rhLF N-glycan spectrum corresponds to Gal2GlcNAc2Man3GlcNAc2 (GS5.0), whereas the major peak in the sialylated rhLF N-glycan spectrum corresponds to Sia2Gal2GlcNAc2Man3GlcNAc2 (GS6.0). Red circles GlcNac, grey squares Man, violet squares Man, diagonal-striped squares Man, green diamond Gal, green four-pointed star Sia
Recently, it has been described that glycosylation of hLF derived from mammalian cells occurs predominantly at two sites (N138 and N479) in approximately 85% of all hLF molecules. Glycosylation at a single site (N479) or at all three sites (N138, N479, and N624) occurs in only approximately 5% and 9% of hLF, respectively [35]. In order to determine N-glycosylation occupancy of each site, rhLF was digested with trypsin. One half of the sample was deglycosylated with PNGase F with the other one-half remaining untreated. Both samples were subjected to HPLC analysis. Whole chromatograms for each sample were overlaid in order to identify glycopeptides and the same peptides lacking glycan. Figure 5 shows that the N-glycan occupancy was identified on chromatogram between 52 and 67 min of retention time. To confirm the peptide sequences, HPLC fractions were collected and subjected to MS/MS analysis. For fractions from the untreated sample, PNGase F treatment was performed prior to MS/MS analysis in order to elucidate the glycopeptide sequences. The mixture of glycopeptides (N138 and N479) was identified, but the glycopeptide containing N629 was not resolved well. It is likely that the N629 glycopeptide's signal was buried because of co-elution with other peptides. However, based on the peptide mapping, it was confirmed that all three N-linked sites of rhLF were predominantly occupied with N-glycans, whereas it has been shown that only 9% of hLF was N-glycosylated at all three sites when expressed in mammalian cell-derived systems.
Fig. 5.
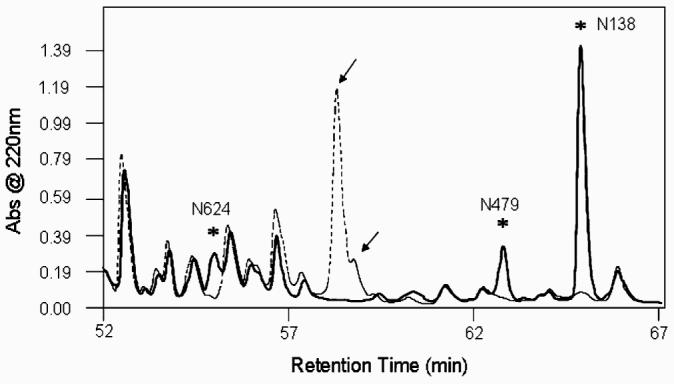
Peptide mapping of non-sialylated rhLF. The dotted chromatogram displays the chromatographic UV trace for tryptic peptides from glycosylated rhLF; the solid line corresponds to the trace for deglycosylated tryptic peptides. Fractions from the glycosylated sample were then treated with PNGase F and run on liquid chromatography-coupled mass spectrometer via nanospray to identify peptide sequences. Fragments corresponding to deglycosylated and non-glycosylated N-glycosylation sites (N138, N479, N624) were identified in both samples and peaks containing these fractions were indicated with asterisks. Glycopeptides (N138 and N479) were identified in peaks marked with arrows
Reconstituting action of recombinant lactoferrin in the secondary humoral immune response in mice suppressed by methotrexate
It was previously demonstrated that bovine lactoferrin (bLF) can reduce methotrexate (MTX)-induced suppression of secondary humoral immune responses in vitro to sheep erythrocytes (SRBC) [1]. Here both sialylated and non-sialylated rhLF were tested in vitro to demonstrate effects on the secondary humoral immune response. Human, milk-derived LF was used as a reference protein. The results (Fig. 6) demonstrated that sialylated rhLF was able to restore the number of antibody-forming cells in cultures treated with MTX. The non-sialylated rhLF was ineffective in the reduction of MTX-induced suppression of the secondary humoral immune response. Milk-derived LF was either moderately active or not active at all (data not shown). Since the results indirectly indicate the importance of sialic acid in mediation of the biological activity, a moderate concentration of free sialic acid was added to the cell cultures to block rhLF interaction with receptor (Fig. 7a). The results demonstrate that the addition of sialic acid significantly reduced the ability of sialylated rhLF to restore the MTX-mediated suppression. In addition, when monoclonal antibody to CD169, directed against the murine sialoadhesin, was added to the culture, the sialylated rhLF was unable to reverse the MTX-mediated suppression (Fig. 7b). This demonstrates a requirement of rhLF to bind to sialoadhesin for functional activity.
Fig. 6.
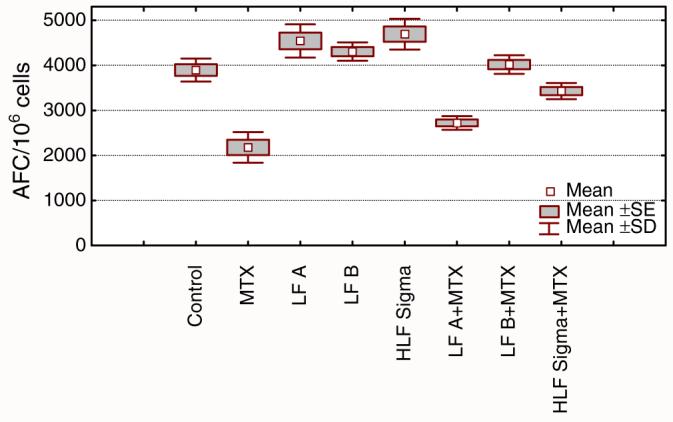
Reconstituting effect of lactoferrin on the secondary humoral immune response in mice suppressed by methotrexate. Splenocytes isolated from sheep red blood cell (SRBC) primed mice were incubated with SRBC, alone in the presence of methotrexate (MTX). Antibody forming cells (AFC) were evaluated after 4 days. Cells were cultured in the presence of non-sialylated LF (LF A), sialylated LF (LF B), or milk-derived LF (HLF Sigma). All LF concentrations were 1 μg/ml. The results are shown as mean values of AFC number from five wells±SE, calculated per 106 viable cells. Statistical analysis of groups: control vs. LF A NS; control vs. LF B NS; control vs. HLF Sigma p=0.0068; control vs. MTX p=0.0001; control vs. LF A+MTX p=0.0001; control vs. LF B+MTX NS; control vs. HLF Sigma+MTX NS; MTX vs. LF A+MTX NS; MTX vs. LF B+MTX p=0.0001; MTX vs. HLF Sigma+MTX p=0.0001 (ANOVA)
Fig. 7.
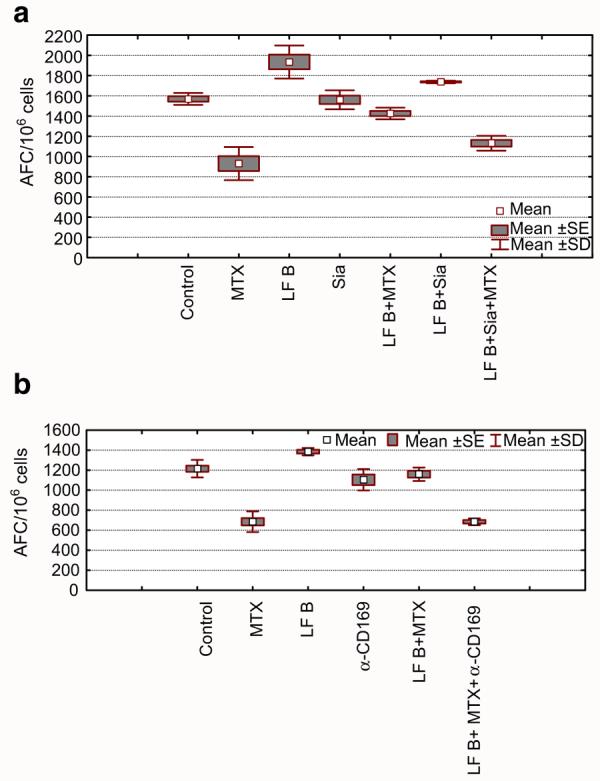
The activity of human recombinant lactoferrin in the secondary immune response in vitro is abolished by addition of sialic acid. Splenocytes isolated from sheep red blood cell (SRBC) primed mice were incubated with SRBC, alone in the presence of methotrexate (MTX). Antibody forming cells (AFC) were evaluated after 4 days. Cells were cultured in the presence of sialylated LF (LF B). Free sialic acid was added alone, or in combination with LF (a). Alternatively, anti-sialoadhesin monoclonal antibody CD169 (sialoadhesin) was added before LF (1:250) (b). a Control vs. LF B p=0.0001; control vs MTX p=0.0001; control vs. Sia NS; control vs. LF B+Sia NS; control vs. LF B+MTX NS; control vs LF B+Sia+MTX p=0.0001; MTX vs. LF B+MTX p=0.0001; MTX vs LF B+Sia+MTX NS; LF B+MTX vs. LF B+Sia+MTX p=0.0023 (ANOVA). b Control vs.LF B p=0.0291; control vs. MTX p=0.0001; control vs. Ab NS; control vs. LF B+Ab+MTX p=0.0001; LF B vs. LF B+Ab p=0.0001; MTX vs. LF B+MTX p=0.0001; MTX vs. LF B+Ab+MTX NS; LF B+MTX vs. LF B+Ab+MTX p=0.0001 (ANOVA)
Discussion
GlycoFi™ has developed glycoengineered P. pastoris strains capable of producing glycoproteins with humanized N-glycans [5, 16, 17]. In contrast to mammalian cells, glycoengineered yeasts allow the production of glycoproteins with highly homogenous N-linked glycans. The use of glycoengineered yeast permits the exploration of the structure-function relationships and can improve efficacy of therapeutic glycoproteins. In this study, rhLF was produced at a high level in glycoengineered P. pastoris that predominantly consists of a humanized glycan, Gal2GlcNAc2Man3GlcNAc2. A subsequent in vitro sialylation of rhLF (glycoform: Sia2Gal2GlcNAc2Man3GlcNAc2) was further explored for reconstitution activity in a splenocyte model of immune-mediated recovery of suppressed responses.
Optimization of culture conditions and genetic improvements allowed for high-level expression of rhLF. Signal sequences play an important role in translocating proteins into the secretory pathway and they have shown their different preferences for proteins. Based on the screening of signal sequence library, the signal sequences from A. oryzae (glucoamylase), K. maxianus (inulinase), and P. pastoris (KAR2), and S. cerevisiae (αMFppKR) demonstrated higher rhLF levels secreted in the culture supernatants as well as increased intracellular rhLF accumulation. The optimal signal sequence (ScαMFppKR) was found not only to facilitate its secretion, but also to minimize intracellular accumulation. In addition, N-terminal sequencing of rhLF confirmed the correct processing of the signal sequence (ScαMFppKR).
Several promoters have been developed for heterologous protein expression using the Pichia system. The AOX1 and GAPDH promoters are widely used for protein production. Under optimized culture conditions, the PpAOX1 promoter performed better than the PpGAPDH promoter since more degradation of rhLF was observed under the PpGAPDH promoter. This indicates that the inducible promoter (PpAOX1) provides better control, most likely due to the shorter production phase, than the PpGAPDH process.
The purification of rhLF was performed using two chromatographic steps after filtration. The SP Sepharose step proved to be an excellent capture and purification step as shown in Fig. 2. In addition, the removal of host cell proteins was very efficient as determined by Western blot analysis. The second step proved to be crucial for the reduction of endotoxin levels to a value <5 EU/mg, equally successful with sialylated and non-sialylated protein. The combination of SP Sepharose and Heparin Sepharose enabled greater than 95% purification of rhLF with no isoforms observed as seen with the commercial standard. Analytical data support that the purified rhLF should be sufficient and acceptable as a homogenous molecule to proceed with defined in vivo testing.
The sequence of hLF contains three putative N-linked glycosylation sites at positions Asn138, Asn479 and Asn624. It has been demonstrated that Asn138 and Asn479 are preferentially N-glycosylated in LF produced in human kidney-derived 293(S) cell lines, indicating site-specific heterogeneity [35]. However, all three N-linked sites of rhLF from our glycoengineered P. pastoris are predominantly occupied as characterized by peptide mapping and LC-MS (Fig. 5). This indicates that a significant portion of Asn624 located at the C-lobe is glycosylated in P. pastoris compared with that of mammalian cells. It has been reported that Asn624 is not glycosylated in human milk and leukocyte LFs [13, 32]. In addition, glycoengineered P. pastoris do not display core-fucosylation of N-linked glycans. Non-fucosylated hLF is a characteristic of human neutrophilic leukocytes whereas human milk-derived LF contains fucose residues at the N-acetylglucosamine residue [13]. Sialylated rhLF derived from our glycoengineered P. pastoris demonstrates that the glycoform (afucosylated biantennary: Sia2Gal2GlcNAc2Man3GlcNAc2) is structurally the same as one of the glycoforms typically found in leucocytes.
Lactoferrin in humans is considered a first-line defense protein involved in a multitude of innate and adaptive immune responses. There are two primary forms of human LF, one contained in exocrine secretions including milk, tears, saliva, bronchial and intestinal secretions. The other form is present in the secondary granules of neutrophils. While the secreted form is thought to be involved in the host defense against microbial infection at mucosal sites, granulocytic lactoferrin has notable immunomodulatory function [22]. Also, the two forms of human LF are identical in their amino acid sequence, but different in sugar moiety. The granulocytic LF is not fucosylated and this seemingly predisposes this form to transduce certain signals that do not require fucose-specific receptors such as mannose receptor exhibiting equal affinity to fucose and mannose [38]. Here, it was demonstrated that the activity of rhLF depends on the terminal N-acetylneuraminic acid. This would suggest that reconstruction of MTX-inhibited secondary humoral immune response requires interaction of LF with cells expressing neuraminic dependent receptors. The prime candidate for such a receptor is sialoadhesin, a sialic acid-binding receptor, found on murine tissue macrophages [10, 14, 28] and dendritic cells [4], and perhaps the CD22 marker on B cells that bears sequence similarity to sialoadhesin [20]. Nevertheless, the use of specific anti-sialoadhesin antibody, with no apparent cross-reactivity with CD22, makes such a possibility very unlikely. The reconstructive effect of LF on the secondary humoral response in MTX-suppressed splenocytes [1] was further explored. MTX, an antagonist of folic acid synthesis, causes apoptosis of activated cells primarily in the G1-and S-cycle phases [25]. Interestingly, LF could partially reconstitute the cellular immune response in MTX-treated mice, but not the primary humoral immune response in vivo [1]. It can further be hypothesized that memory T helper cells, responsible for development of the secondary immune response are less prone to apoptosis [40], coinciding well with lactoferrin's anti-apoptotic properties [9, 34, 45]. LF could prevent MTX-induced suppression in several ways, including the induction of specific cytokines in sialoadhesin-positive splenocytes. Mediators such as IL-6 [33], IL-12 [39], and TGF-beta [7] induce anti-apoptotic responses and LF indeed increases the production of such mediators [11, 19, 41]. Because LF has the ability to accelerate the function of the immune system cells (reviewed in 43), it opens new perspectives for the systemic application of recombinant human LF in many immune disorders. Furthermore, both the protein and sugar moiety are compatible with their natural counterparts, thus potentially limiting immunogenicity inherent in most biological therapeutics.
In conclusion, glycoengineered P. pastoris was used to produce fully humanized and immunologically compatible rhLF. The primary immune characteristic of rhLF showed differential effects of sialylated and non-sialylated rhLF on restoration of the secondary humoral immune response to SRBC in mouse splenocytes. This suggests that the terminal N-acetylneuraminic acid plays an important role in propagation of proper immune responses. Further studies are underway to demonstrate additional actions of recombinant human LF, in vitro and in vivo, and elucidate the function of terminal sugars in defined innate and adaptive immune functions.
Acknowledgments
This research was supported by the National Institutes of Health: R42AI051050-02 and R41GM079810-01. We thank Teresa Mitchell for the technical assistant and Bing Gong for the critical reading of the manuscript.
Abbreviations
- Man
mannose
- Gal
galactose
- GlcNAc
N-acetylglucosamine
- Sia
sialic acid
- hLF
human lactoferrin
- rhLF
recombinant human lactoferrin
- anti-hLF
anti-human LF antibody
- anti-HCP
anti-host cell protein antibody
- CV
column volume
- AFC
antibody forming colonies
- MALDI-
matrix-assisted laser desorption/ionization
- TOF
time of flight
- MS/MS
tandem mass spectrometry
References
- 1.Artym J, Zimecki M, Kruzel ML. Effect of lactoferrin on the methotrexate-induced suppression of the cellular and humoral immune response in mice. Anticancer Res. 2004;24:3831–3836. [PubMed] [Google Scholar]
- 2.Baveye S, Elass E, Mazurier J, Spik G, Legrand D. Lactoferrin: a multifunctional glycoprotein involved in the modulation of the inflammatory process. Clin. Chem. Lab. Med. 1999;37:281–286. doi: 10.1515/CCLM.1999.049. [DOI] [PubMed] [Google Scholar]
- 3.Bayens RD, Bezwoda WR. Lactoferrin and the inflammatory response. Adv. Exp. Med. Biol. 1994;357:133–141. doi: 10.1007/978-1-4615-2548-6_13. [DOI] [PubMed] [Google Scholar]
- 4.Berney C, Herren S, Power CA, Gordon S, Martinez-Pomares L, Kosco-Vilbois MH. A member of the dendritic cell family that enters B cell follicles and stimulates primary antibody responses identified by a mannose receptor fusion protein. J. Exp. Med. 1999;190:851–860. doi: 10.1084/jem.190.6.851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bobrowicz P, Davidson RC, Li H, Potgieter TI, Nett JH, Hamilton SR, Stadheim TA, Miele RG, Bobrowicz B, Mitchell T, Rausch S, Renfer E, Wildt S. Engineering of an artificial glycosylation pathway blocked in core oligosaccharide assembly in the yeast Pichia pastoris: production of complex humanized glycoproteins with terminal galactose. Glycobiology (Oxf.) 2004;14:757–766. doi: 10.1093/glycob/cwh104. [DOI] [PubMed] [Google Scholar]
- 6.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 7.Cerwenka A, Swain SL. TGF-beta1: immunosuppressant and viability factor for T lymphocytes. Microbes Infect. 1999;1:1291–1296. doi: 10.1016/s1286-4579(99)00255-5. [DOI] [PubMed] [Google Scholar]
- 8.Choi BK, Bobrowicz P, Davidson RC, Hamilton SR, Kung DH, Li H, Miele RG, Nett JH, Wildt S, Gerngross TU. Use of combinatorial genetic libraries to humanize N-linked glycosylation in the yeast Pichia pastoris. Proc. Natl. Acad. Sci. U. S. A. 2003;100:5022–5027. doi: 10.1073/pnas.0931263100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cornish J, Callon KE, Naot D, Palmano KP, Banovic T, Bava U, Watson M, Lin JM, Tong PC, Chen Q, Chan VA, Reid HE, Fazzalari N, Baker HM, Baker EN, Haggarty NW, Grey AB, Reid IR. Lactoferrin is a potent regulator of bone cell activity and increases bone formation in vivo. Endocrinology. 2004;145:4366–4374. doi: 10.1210/en.2003-1307. [DOI] [PubMed] [Google Scholar]
- 10.Crocker PR, Kelm S, Dubois C, Martin B, McWilliam AS, Shotton DM, Paulson JC, Gordon S. Purification and properties of sialoadhesin, a sialic acid-binding receptor of murine tissue macrophages. EMBO J. 1991;10:1661–1669. doi: 10.1002/j.1460-2075.1991.tb07689.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Curran CS, Demick KP, Mansfield JM. Lactoferrin activates macrophages via TLR4-dependent and -independent signaling pathways. Cell. Immunol. 2006;242:23–30. doi: 10.1016/j.cellimm.2006.08.006. [DOI] [PubMed] [Google Scholar]
- 12.Davidson RC, Nett JH, Renfer E, Li H, Stadheim TA, Miller BJ, Miele RG, Hamilton SR, Choi BK, Mitchell TI, Wildt S. Functional analysis of the ALG3 gene encoding the Dol-P-Man: Man5GlcNAc2-PP-Dol mannosyltransferase enzyme of P. pastoris. Glycobiology (Oxf.) 2004;14:399–407. doi: 10.1093/glycob/cwh023. [DOI] [PubMed] [Google Scholar]
- 13.Derisbourg P, Wieruszeski JM, Montreuil J, Spik G. Primary structure of glycans isolated from human leucocyte lactotransferrin. Absence of fucose residues questions the proposed mechanism of hyposideraemia. Biochem. J. 1990;269:821–825. doi: 10.1042/bj2690821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Frei K, Steger C, Samorapoompichit P, Lucas T, Forster O. Expression and function of sialoadhesin in rat alveolar macrophages. Immunol. Lett. 2000;71:167–170. doi: 10.1016/s0165-2478(99)00180-7. [DOI] [PubMed] [Google Scholar]
- 15.Gerngross TU. Advances in the production of human therapeutic proteins in yeasts and filamentous fungi. Nat. Biotechnol. 2004;22:1409–1414. doi: 10.1038/nbt1028. [DOI] [PubMed] [Google Scholar]
- 16.Hamilton SR, Bobrowicz P, Bobrowicz B, Davidson RC, Li H, Mitchell T, Nett JH, Rausch S, Stadheim TA, Wischnewski H, Wildt S, Gerngross TU. Production of complex human glycoproteins in yeast. Science. 2003;301:1244–1246. doi: 10.1126/science.1088166. [DOI] [PubMed] [Google Scholar]
- 17.Hamilton SR, Davidson RC, Sethuraman N, Nett JH, Jiang Y, Rios S, Bobrowicz P, Stadheim TA, Li H, Choi BK, Hopkins D, Wischnewski H, Roser J, Mitchell T, Strawbridge RR, Hoopes J, Wildt S, Gerngross TU. Humanization of yeast to produce complex terminally sialylated glycoproteins. Science. 2006;313:1441–1443. doi: 10.1126/science.1130256. [DOI] [PubMed] [Google Scholar]
- 18.Hwang SA, Kruzel ML, Actor JK. Lactoferrin augments BCG vaccine efficacy to generate T helper response and subsequent protection against challenge with virulent Mycobacterium tuberculosis. Int. Immunopharmacol. 2005;5:591–599. doi: 10.1016/j.intimp.2004.11.006. [DOI] [PubMed] [Google Scholar]
- 19.Hwang SA, Wilk KM, Bangale YA, Kruzel ML, Actor JK. Lactoferrin modulation of IL-12 and IL-10 response from activated murine leukocytes. Med. Microbiol. Immunol. 2007;196:171–180. doi: 10.1007/s00430-007-0041-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kelm S, Pelz A, Schauer R, Filbin MT, Tang S, de Bellard ME, Schnaar RL, Mahoney JA, Hartnell A, Bradfield P, et al. Sialoadhesin, myelin-associated glycoprotein and CD22 define a new family of sialic acid-dependent adhesion molecules of the immunoglobulin superfamily. Curr. Biol. 1994;4:965–972. doi: 10.1016/s0960-9822(00)00220-7. [DOI] [PubMed] [Google Scholar]
- 21.Kruzel ML, Harari Y, Mailman D, Actor JK, Zimecki M. Differential effects of prophylactic, concurrent and therapeutic lactoferrin treatment on LPS-induced inflammatory responses in mice. Clin. Exp. Immunol. 2002;130:25–31. doi: 10.1046/j.1365-2249.2002.01956.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kruzel ML, Zimecki M. Lactoferrin and immunologic dissonance: clinical implications. Arch. Immunol. Ther. Exp. 2002;50:399–410. [PubMed] [Google Scholar]
- 23.Legrand D, Salmon V, Coddeville B, Benaissa M, Plancke Y, Spik G. Structural determination of two N-linked glycans isolated from recombinant human lactoferrin expressed in BHK cells. FEBS Lett. 1995;365:57–60. doi: 10.1016/0014-5793(95)00441-b. [DOI] [PubMed] [Google Scholar]
- 24.Li H, Sethuraman N, Stadheim TA, Zha D, Prinz B, Ballew N, Bobrowicz P, Choi BK, Cook WJ, Cukan M, Houston-Cummings NR, Davidson R, Gong B, Hamilton SR, Hoopes JP, Jiang Y, Kim N, Mansfield R, Nett JH, Rios S, Strawbridge R, Wildt S, Gerngross TU. Optimization of humanized IgGs in glycoengineered Pichia pastoris. Nat. Biotechnol. 2006;24:210–215. doi: 10.1038/nbt1178. [DOI] [PubMed] [Google Scholar]
- 25.Loginov AV, Uteshev BS, Livshits MA. [Mathematical modelling of the action of methotrexate on the kinetics of B-lymphocyte proliferation during the primary response] Farmakol. Toksikol. 1987;50:58–70. [PubMed] [Google Scholar]
- 26.Lonnerdal B, Iyer S. Lactoferrin: molecular structure and biological function. Annu. Rev. Nutr. 1995;15:93–110. doi: 10.1146/annurev.nu.15.070195.000521. [DOI] [PubMed] [Google Scholar]
- 27.Mishell RI, Dutton RW. Immunization of dissociated spleen cell cultures from normal mice. J. Exp. Med. 1967;126:423–442. doi: 10.1084/jem.126.3.423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nakamura K, Yamaji T, Crocker PR, Suzuki A, Hashimoto Y. Lymph node macrophages, but not spleen macrophages, express high levels of unmasked sialoadhesin: implication for the adhesive properties of macrophages in vivo. Glycobiology (Oxf.) 2002;12:209–216. doi: 10.1093/glycob/12.3.209. [DOI] [PubMed] [Google Scholar]
- 29.Naot D, Grey A, Reid IR, Cornish J. Lactoferrin—a novel bone growth factor. Clin. Med. Res. 2005;3:93–101. doi: 10.3121/cmr.3.2.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Samyn-Petit B, Wajda Dubos JP, Chirat F, Coddeville B, Demaizieres G, Farrer S, Slomianny MC, Theisen M, Delannoy P. Comparative analysis of the site-specific N-glycosylation of human lactoferrin produced in maize and tobacco plants. Eur. J. Biochem. 2003;270:3235–3242. doi: 10.1046/j.1432-1033.2003.03706.x. [DOI] [PubMed] [Google Scholar]
- 31.Sanchez L, Calvo M, Brock JH. Biological role of lactoferrin. Arch. Dis. Child. 1992;67:657–661. doi: 10.1136/adc.67.5.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Spik G, Coddeville B, Montreuil J. Comparative study of the primary structures of sero-, lacto- and ovotransferrin glycans from different species. Biochimie. 1988;70:1459–1469. doi: 10.1016/0300-9084(88)90283-0. [DOI] [PubMed] [Google Scholar]
- 33.Takeda K, Kaisho T, Yoshida N, Takeda J, Kishimoto T, Akira S. Stat3 activation is responsible for IL-6-dependent T cell proliferation through preventing apoptosis: generation and characterization of T cell-specific Stat3-deficient mice. J. Immunol. 1998;161:4652–4660. [PubMed] [Google Scholar]
- 34.van't Land B, van Beek NM, van den Berg JJ, M'Rabet L. Lactoferrin reduces methotrexate-induced small intestinal damage, possibly through inhibition of GLP-2-mediated epithelial cell proliferation. Dig. Dis. Sci. 2004;49:425–433. doi: 10.1023/b:ddas.0000020497.35250.93. [DOI] [PubMed] [Google Scholar]
- 35.van Berkel PH, van Veen HA, Geerts ME, de Boer HA, Nuijens JH. Heterogeneity in utilization of N-glycosylation sites Asn624 and Asn138 in human lactoferrin: a study with glycosylation-site mutants. Biochem. J. 1996;319(Pt 1):117–122. doi: 10.1042/bj3190117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang SH, Yang TS, Lin SM, Tsai MS, Wu SC, Mao SJ. Expression, characterization, and purification of recombinant porcine lactoferrin in Pichia pastoris. Protein Expr. Purif. 2002;25:41–49. doi: 10.1006/prep.2001.1607. [DOI] [PubMed] [Google Scholar]
- 37.Wei Z, Nishimura T, Yoshida S. Characterization of glycans in a lactoferrin isoform, lactoferrin-a. J. Dairy Sci. 2001;84:2584–2590. doi: 10.3168/jds.S0022-0302(01)74712-1. [DOI] [PubMed] [Google Scholar]
- 38.Weis WI, Taylor ME, Drickamer K. The C-type lectin superfamily in the immune system. Immunol. Rev. 1998;163:19–34. doi: 10.1111/j.1600-065x.1998.tb01185.x. [DOI] [PubMed] [Google Scholar]
- 39.Yoo JK, Cho JH, Lee SW, Sung YC. IL-12 provides proliferation and survival signals to murine CD4+ T cells through phosphatidylinositol 3-kinase/Akt signaling pathway. J. Immunol. 2002;169:3637–3643. doi: 10.4049/jimmunol.169.7.3637. [DOI] [PubMed] [Google Scholar]
- 40.Zielinski CC, Stuller I, Dorner F, Potzi P, Muller C, Eibl MM. Impaired primary, but not secondary, immune response in breast cancer patients under adjuvant chemotherapy. Cancer. 1986;58:1648–1652. doi: 10.1002/1097-0142(19861015)58:8<1648::aid-cncr2820580812>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- 41.Zimecki M, Artym J, Chodaczek G, Kocieba M, Kruzel M. Effects of lactoferrin on the immune response modified by the immobilization stress. Pharmacol. Rep. 2005;57:811–817. [PubMed] [Google Scholar]
- 42.Zimecki M, Kocieba M, Kruzel M. Immunoregulatory activities of lactoferrin in the delayed type hypersensitivity in mice are mediated by a receptor with affinity to mannose. Immunobiology. 2002;205:120–131. doi: 10.1078/0171-2985-00115. [DOI] [PubMed] [Google Scholar]
- 43.Zimecki M, Kruzel ML. Milk-derived proteins and peptides of potential therapeutic and nutritive value. J. Exp. Ther. Oncol. 2007;6:89–106. [PubMed] [Google Scholar]
- 44.Zimecki M, Mazurier J, Spik G, Kapp JA. Human lactoferrin induces phenotypic and functional changes in murine splenic B cells. Immunology. 1995;86:122–127. [PMC free article] [PubMed] [Google Scholar]
- 45.Zimecki M, Mazurier J, Spik G, Kapp JA. Lactoferrin (LF) lowers IgM and interleukin 2 receptor expression on WEHI 231 cells and decreases anti-IgM antibody-induced cell death. Pol. J. Immunol; VIII Meeting of the Polish Immunological Society; Wroclaw, Poland. 1995. p. 324. [Google Scholar]
- 46.Zimecki M, Miedzybrodzki R, Mazurier J, Spik G. Regulatory effects of lactoferrin and lipopolysaccharide on LFA-1 expression on human peripheral blood mononuclear cells. Arch. Immunol. Ther. Exp. 1999;47:257–264. [PubMed] [Google Scholar]