On April 21, thousands of visitors invaded Louisville, KY, including >100 scientists from around the globe. Most of the visitors were there to enjoy the festivities that were in progress as a prelude to the Kentucky Derby. In addition to the Derby celebration, the scientists were there for the First International Meeting on Quadruplex DNA, the first-ever full conference on the roles of quadruplex DNA in biology and chemistry. The field is relatively new. It is quite promising for helping scientists understand cancer and perhaps other diseases, and for the development of new types of anticancer drugs. The meeting opened on Saturday evening with a reception on the 16th floor of the historic Brown Hotel in downtown Louisville. The conference attendees had the pleasant choices of catching up with friends, enjoying the food and drink, or gazing through the large windows of the reception area as the world’s largest fireworks display exploded over the nearby Ohio River (Figure 1). The excitement of being part of the first meeting in this area, and the excellent talks in several different quadruplex research areas, held the interest of the conference group until its end at midday on April 24. No heated discussions took place during the conference sessions; however, areas clearly exist in this relatively new field in which significantly different views must be reconciled.
Figure 1.


The historic Brown Hotel meeting site and sidewalk clock (left). The “Thunder Over Louisville” Kentucky Derby fireworks (right), which could be seen from the Quadruplex Meeting opening reception.
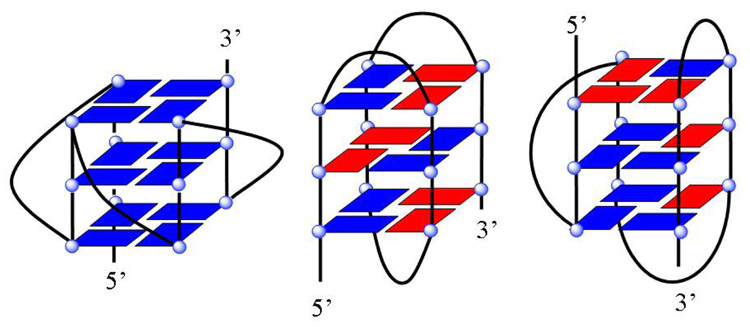
The conference was organized by Jonathan B. (Brad) Chaires (University of Louisville) and Laurence Hurley (University of Arizona), with extensive support from the James Graham Brown Cancer Center. The program included 23 speakers, five sessions, and a two-day poster session with >60 posters. The five sessions were Quadruplex Structure and Stability; Molecular Modeling, Assembly, Dynamics; Human Telomeres; G-Quadruplexes and Biology; and Quadruplex Drug Targets and Therapeutics. Current key research in quadruplex structure, biology, and drug targeting were also covered. DNA quadruplexes are formed from one or more strands of guanine-rich DNA that can fold or associate into a four-strand structure that is stabilized by H-bonds and ion interactions among four G bases in a structural unit (Figure 2, panel a). The four Gs, which can be syn or anti, create a planar structural unit with a total of eight Hoogsteen H-bonds. The planar tetrad of Gs can stack with other similar units to form a quadruplex conformation. The H-bonded four-G unit creates a central space that serves as a metal ion coordination site to form a complete stable tetramer-metal ion unit of G4 DNA. The optimum metal ion under physiological conditions is K+, and quadruplex stability depends on ion type and concentration. The quadruplex strands can be in parallel or antiparallel orientations and, with different sequences of loop bases, a large number of different quadruplex structures is possible (Figure 2), many of which were discussed in detail at the meeting. The loop sequences that connect the G4 planar units in a parallel or antiparallel orientation can have a pronounced effect on the overall quadruplex conformation (Figure 2, panel b).
Figure 2.
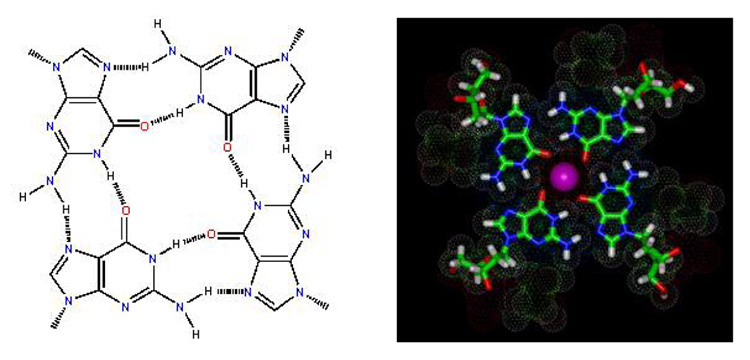
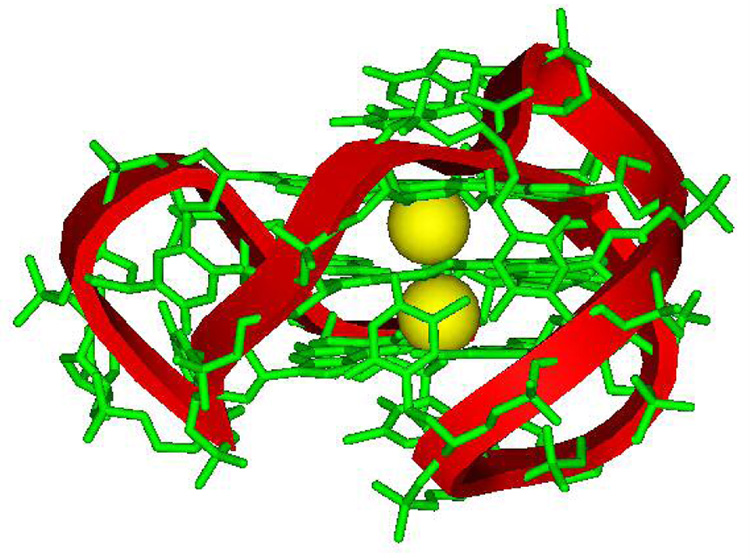
a) H-bonded, four-G stacking unit of quadruplexes: in the model on the right, a K+ ion can be seen (pink) coordinated to the G carbonyl oxygens (red) that point to the center of the four-G tetramer unit that forms the core structure of G4 DNA quadruplexes. Other atoms in the model are green, C; blue, N; and white, H. b) Possible folding models for the human telomere G4 DNA structure. The G bases in the G4 units can be either anti (blue) or syn (red) as shown for a parallel (left) or propeller, a basket (center), or hybrid (right) conformation. c) Molecular model of a human telomere hybrid structure (5) with bases in green, backbone sugars and phosphate shown as a ribbon, and coordinated K+ as yellow spheres. The model is for the major human telomere conformation found in the mix of species in K+ solution (2, 3, 5–9).
Quadruplex DNA was discovered in 1962 at the National Institutes of Health (NIH) by David Davies and coworkers. Davies presented a historical background in the opening address of the conference. In the original research, his group proposed an H-bonding scheme, basically as shown in Figure 2, panel a, as well as the four-G stacking arrangement that forms each quadruplex helical unit. The discovery resulted from several of those unexpected observations that have led to many scientific breakthroughs; these involved G-base association into a quadruplex, and gel formation from a concentrated solution of GMP.
G4 DNA engendered little interest in the years that followed, until the realization that cancer cells could perhaps be selectively targeted through their telomeres. Telomeres protect chromosomes from end-to-end fusion and against the loss of coding DNA at the ends of chromosomes during replication. Dividing cells undergo a progressive loss of ~100 DNA base pairs at each cell division because of the inability of DNA polymerase to copy the final base pairs of the lagging strand. Somatic cells start with a protective sequence of repeated -TTAGGG-single-strand telomere DNA that is progressively lost during cell replication. Four of these sequence repeats can form an intramolecular quadruplex with TTA connecting loops in the human telomere, as illustrated in Figure 2, panels b and c. Most cancer, but not somatic, cells have the telomerase gene activated so that they make the enzyme and can regenerate their telomeres during multiple cell replications. The meeting coincided with the 10-year anniversary of a publication by the groups of Hurley and Stephen Neidle (University of London) (1). It was the first to clearly show that the telomerase enzyme could be effectively inhibited by compounds that induce its G-rich recognition sequence to fold into a stable unimolecular quadruplex structure. The quadruplex–drug complex could not serve as an appropriate recognition structure for telomerase activity. This publication, and others that followed, showed the importance of G-rich DNA sequences in cancer biology and have generated increasing excitement about quadruplex DNA. It should be emphasized that it is not necessary for a compound to target an existing cellular quadruplex to have biological activity. Induction of a quadruplex or change of a quadruplex conformation on binding may be the most powerful method to exert the desired biological effect. In addition to the telomere quadruplex and telomerase inhibition, targeting other cellular quadruplexes for development of new anticancer drugs was a theme of several talks and posters at the meeting.
Some idea of the geographical diversity of quadruplex research is illustrated by the fact that the four speakers after Davies in the opening session have laboratories in the U.K., France, the U.S., and Singapore. Various models have been proposed for how a sequence of G-rich DNA can fold to create a four-strand quadruplex (several examples are in Figure 2). A key goal of the first session and the conference as a whole was to organize and expand our understanding of how quadruplexes fold and how their stability is affected by loop sequence, solvent conditions, drugs, and protein binding. Folding to a G4 structure is a complex problem for the single-strand G-sequences of telomeres, but it becomes even more difficult for duplex G-rich regions in chromosomes that must separate from the complementary C-strand before they can form a quadruplex structure (the structure and interactions of the resulting C-strand are an important developing research area). Presentations by Anh Tuan Phan (Nanyang Technological University, Singapore), Jean-Louis Mergny (Muséum National d’Histoire Naturelle, France), and Richard Shafer (University of California at San Francisco) highlighted experimental findings on quadruplex conformations. Mateus Webba da Silva (University of Ulster, U.K.) pointed out that some general rules for classifying and predicting quadruplex structures are now being generated.
Phan presented some basic principles of quadruplex structure and an overview of NMR structural studies on several important G4 sequences obtained in the laboratory of Dinshaw Patel (Memorial Sloan-Kettering Cancer Center) (2–4). A major problem that structural biologists in the quadruplex field have faced is the structural diversity of many G-rich sequences, including that of the telomere. The human telomere at biologically relevant K+ concentrations, for example, is a mixture of species that can change with solution conditions, base modifications, flanking, and loop base sequence. Efforts to isolate the major conformational species have focused on modification of sequences that flank the basic G-quadruplex sequence and stack on the ends of the quadruplex structure as well as on modified bases that can lock a particular base into the syn or anti conformation. The bases in Figure 2, panel b, for example, have syn and anti orientations at different positions and can be favored by modified bases in either the syn or anti conformation at those positions. Phan and Patel (2–4), Hiroshi Sugiyama (Kyoto University, Japan) (5, 6), and Danzhou Yang (University of Arizona) (7–9) found similarly folded hybrid structures for different modifications of the human telomere sequence in K+ (Figure 2, panel b, right, and Figure 2, panel c). Phan described NMR structural work over a number of years on modified sequences of the telomere and other important quadruplexes that have resulted in detailed models of quadruplex conformation. To test the various quadruplex conformational models, Shafer created variants of the human telomere sequence where riboguanosine (rG) was substituted in place of the natural deoxy sugar (10). This substitution is a powerful method for locking the conformation of the nucleoside into the anti conformation and can be used to evaluate syn/anti patterns such as those shown in Figure 2, panel b. His results emphasized the diversity of possible conformations of the telomere quadruplex and showed that depending on the rG substitution position, conformations from fully parallel to antiparallel/hybrid species could be obtained for the telomere.
Speakers and audience members pointed out that as the rules and additional types of quadruplex structures become established, it is necessary to develop a standard system of reference and systematic nomenclature to describe quadruplexes and their complexes. Such a system is essential to make all experimental and theoretical results for quadruplexes readily available for the entire field. Mergny described the problem of switching from a duplex to a quadruplex structure and provided some kinetics insight into the process (11, 12). This is a critical point for the biology of quadruplexes and for the development of quadruplex targeting drugs that must induce or trap a transient quadruplex conformation. A different approach to quadruplex dynamics was described by Jiri Sponer (Academy of Sciences of the Czech Republic). He discussed advances and limitations of computer simulations of DNA quadruplexes and reported on an improved force field for the AMBER software package. Sponer cautioned about the limits of quantitative molecular modeling for complex systems such as DNA quadruplexes (13, 14). Cautionary comments were also made by Mergny and others about the use of the standard telomeric repeat amplification protocol (TRAP) assay for the evaluation of telomerase activity changes after the addition of drugs. The development of appropriate assays for quadruplex-targeting drugs is an area where much work must be done and standardization is needed.
The second session of the meeting described some new directions in G-quadruplex research. In addition to the important role of quadruplex DNAs in chromosomal telomeres, much evidence now shows that they can act as switches that “turn on” or “turn off” particular genes by using a duplex–quadruplex transition. The scientists at the conference were excited about this area because the quadruplex conformational switches have been shown in vitro to regulate oncogenes that are critical for the development of cancer (15, 16). This is clearly a promising new possibility for the design of anticancer drugs. Shankar Balasubramanian (University of Cambridge, U.K.) described bioinformatics methods to search for sequences that have the potential to form quadruplexes in chromosomal DNA (17). The results indicate that 43% of all human genes have a G-quadruplex-forming sequence in their promoters, a 6-fold enrichment over the genome average. Some of the sequences correspond to nuclease hypersensitive sites. This suggests that they have the ability to form nonduplex structures, including quadruplexes, which could play an important role in the regulation of gene expression (18). In addition to biological roles, G-quadruplexes can provide scaffolds for supramolecular assembly. Jeffery Davis (University of Maryland), for example, reported on the construction of an artificial ion channel using lipophilic guanine derivatives (19) and, following cross-linking using olefin metathesis, demonstrated selective ion transport by the structure. Naoki Sugimoto (Konan University, Japan) presented results on modified quadruplexes that can lead to metal-ion-dependent nanomachines and devices (20). He described new results on the use of G-quadruplex motifs to construct logic gates (21). The use of G4 DNA with modified bases to create specific molecular structures for nanoresearch is very promising. It seems certain that new ideas, devices, nanomotors, and applications in this area will appear in the next few years.
The human telomere quadruplex continues to be a major research topic; it was the subject of the third session of the conference, which continued the discussion of the telomere structure from the first session. Yang presented exciting new NMR structural results from her laboratory on the telomere in K+. As described in the first session, the telomere quadruplex is known to be a mixture of species in K+ solution, and the Yang group has used flanking sequence modifications to favor specific species for structural analysis. She reported structural comparison of two different stable hybrid folds that together can explain the observed mix of species that has complicated the understanding of the human telomere quadruplex (7–9). These results, along with structures from Phan and Patel (2, 3), are elucidating the long-evasive structure of the human telomere in a K+ environment. Sugiyama’s laboratory has used 8-BromoG to lock individual units of the human telomere into the syn conformation in an approach that is a complement to the rG method to lock the anti conformation described by Shafer (5, 6). This method does not require flanking sequence modifications, and the original telomere sequence can be studied at high resolution. Results with 8-BromoG substitutions from the Sugiyama laboratory offered early evidence for the major hybrid fold shown in Figure 2, panels b and c (5) and provided a high-resolution NMR structure for the telomere (6). Sugiyama noted that hybrid quadruplex units can stack into superhelical structures with a chiral cleft between the units. He and his group designed and studied chiral helicene-type “wedge” molecules that provide enantioselective binding to the superhelical quadruplexes. This extension of design methods to larger quadruplex assemblies offers the potential to design new compounds with increased affinity and specificity. The theme of induction or stabilization of a quadruplex structure in telomere DNA to inhibit telomerase for new anticancer drug development was also continued in this session. Neidle and coworkers in both talks and posters presented results for the design of new types of quadruplex-targeting agents. The large relatively planar surface of the quadruplex units forms a unique stacking surface at the ends of each G4 DNA. Several groups are using this surface to design G4 selective binding compounds. The Neidle group uses a combination of crystallography and molecular modeling to design the agents and help them to understand their G4 interactions (22–23). Neidle reported on a new modeling approach that uses two linked, separately folded telomere units to evaluate drug interactions in a larger context that may provide a better representation for in vivo interactions. The critical question of the mechanism of biological action of compounds that stabilize quadruplexes was the subject of the presentation by Jean-Francois Riou (University of Reims, France). He noted that the majority of cancers have telomerase activated for telomere length maintenance, and as a result, compounds that lock the telomere DNA into a G4 structure, which is not a template for telomerase, have excellent potential for development as telomerase inhibitors and anticancer agents. The Riou laboratory has collected clear evidence for in vivo telomerase inhibition in cancer cells by G4 binding compounds with anticancer activity. He suggested that this is a probable mechanism of action for some compounds (23). In addition, he described a possible compound-induced telomere uncapping–protein release mechanism for the activity of G4 targeting compounds. These results are very important in the developing understanding of the biological mechanisms of quadruplex binding molecules and how they selectively affect cancer cell function (24).
The fourth session of the conference focused on the biological functions of quadruplexes. Even a short time ago, no strong scientific evidence existed that DNA quadruplexes actually had a cellular function. The results in this session are illustrative of recent, dramatic advances in quadruplex biology. Brad Johnson (University of Pennsylvania) noted that even though sequences with quadruplex-forming potential are rarer in yeast than in humans, the simplicity of yeast makes it a good system in which to probe the in vivo function of quadruplexes. He presented several lines of evidence that quadruplexes play a role in transcriptional regulation in yeast. Nancy Maizels (University of Washington) pointed out that the double-helical structure of DNA can be significantly modified when DNA is “working”, in transcription or replication, for example, compared with when it’s simply storing information. She described several important G-rich DNA sequences that have the potential to form quadruplexes as part of their essential cellular activities. The Maizels laboratory has collected results from electron microscopy and other methods that strongly support formation of G-loops and G4 DNA quadruplex structures during transcription of G-rich DNA sequences, such as immunoglobulin switch regions and oncogenes that are targets of genomic instability. This direct evidence for quadruplex formation as part of normal cell biology is again an important step in the proof of biological relevance of quadruplex structures and of their potential as therapeutic targets (25–28). The results from the Maizels laboratory provide important ideas and models for the development of compounds to induce or trap quadruplex DNA conformations in cells. The presentations of Hans Joachim Lipps (UniversityWittern/Herdecke, Germany) and Daniela Rhodes (MRC) continued the discussion of possible biological roles of quadruplexes. They described collaborative work showing that telomere end-binding proteins (TEBPs) control the formation of G-quadruplexes in vivo. They proposed a mechanistic model for cell-cycle-dependent telomere conformation by TEBPs (29, 30).
The final meeting session continued the focus on quadruplexes as drug targets and included their use as therapeutic agents. A guanine-rich DNA 26mer, AS1411, originally designed as a triple-helix-forming oligomer (AGRO100), has been found to bind the protein nucleolin, which is overexpressed on the surface of many tumor cells. AS1411 forms a stable G-quadruplex and functions essentially as a nucleolin-specific aptamer. Many aptamers form quadruplexes, and AS1411 may be the first in a series of quadruplex drugs that have highly specific interactions with cancer cell receptors (31, 32). AS1411 is the first aptamer tested in humans, and its physical and biological properties, as well as its very promising and exciting phase I clinical-trial results, were described by Donald Miller and Paula Bates (University of Louisville). The aptamer has potent antitumor activity and very low toxicity, and it will go to phase II clinical trials soon. Other G-quadruplex structures may be related to human disorders, such as fragile X syndrome, which is known to be caused by an expansion of CGG repeat units. Michael Fry (Technion–Israel Institute of Technology) reported that the intermediately expanded (CGG) repeat in the messenger RNA of fragile X syndrome carriers forms a G-quadruplex that retards translation. Destabilization of the RNA secondary structure by quadruplex-disrupting hnRNPs alleviated the block to translation (33).
Throughout the talks and posters, scientists presented strategies for targeting four-strand G4 DNA structures to stabilize or disrupt them, depending on the type of gene and the desired biological outcome. Many drugs designed to target telomeres could also function by inducing or trapping promoter quadruplexes. The long-term goal, however, is to develop compounds that can selectively interact with specific quadruplex structures. Edwin Lewis (Northern Arizona University) described detailed biophysical studies on multi-stoichiometry drug–G4 DNA complexes that require participation of different regions of the c-MYC and bcl-2 quadruplexes (34). These detailed studies, a collaboration between the Lewis and Hurley laboratories, are providing information for our understanding of the energetic basis of drug–quadruplex interactions. This information is an essential complement to quadruplex structural results in the efforts to understand quadruplex interactions and design new drugs to selectively target quadruplexes. Jeffrey Whitten (Cylene Pharmaceuticals) presented the latest clinical and mechanistic results for CX-3543, the first drug that appears to act by targeting DNA quadruplexes. CX-3543 is derived from the fluoroquinolone class of drugs and appears to selectively target ribosomal DNA quadruplex-forming sequences and to inhibit ribosomal RNA synthesis in cancer cells. CX-3543 may function through disruption of the nucleolin–ribosomal DNA interaction that is especially critical in cancer cells. Initial human trials have produced promising anticancer activity with low nonspecific toxicity. The fact that a promising drug is in phase II clinical trials, despite the relatively short time that quadruplex DNA targeting has been investigated, is very encouraging. Several groups have compounds that are approaching the clinical-trial stage.
W. David Wilson (Georgia State University) presented an overview of biosensor–surface plasmon resonance studies from his laboratory on a broad range of quadruplex interactive compounds (35). His laboratory has characterized the interaction of DB832 (prepared by the laboratory of David Boykin), a new type of quadruplex-targeting compound. DB832 has high stoichiometry and an exciton-induced CD spectrum on binding with the human telomere G-quadruplex, and these observations suggest that it does not bind by end stacking. The majority of quadruplex-targeting compounds discovered to date have extended, intercalator-like, aromatic surfaces and bind by stacking on one or both ends of quadruplex G4 structures. In contrast, DB832 may be a paradigm for the design of compounds that are sensitive to quadruplex groove structure. Targeting the quadruplex grooves can provide an enhanced ability to distinguish between quadruplex and duplex conformations as well as among different quadruplex structures (35). Wilson also described, along with posters from Bruce Armitage and coworkers (Carnegie Mellon University), hybridization methods that use the same PNA sequence to target both the G- and C-rich strands of oncogene promoters. Such homologous and complementary targeting of both strands is obviously an advantage for opening the duplex and for target selectivity (36).
The meeting ended with overview comments by Chaires and Hurley and a discussion of possible dates and locations for the Second International Meeting on Quadruplex DNA. Given the success of the first meeting and the dramatic current pace of quadruplex research, great enthusiasm abounded for holding a second meeting soon.
Acknowledgment
The authors gratefully acknowledge support for nucleic acid research in their laboratories from NIH (GM 61587 and AI064200 to W.D.W.) and from the Ministry of Education, Science, Sports and Culture, Japan (to H. S.). We would especially like to thank Brad Chaires and Laurence Hurley for organizing the meeting and for their comments and suggestions, which were a great help in writing this review.
REFERENCES
- 1.Sun D, Thompson B, Cathers BE, Salazar M, Kerwin SM, Trent JO, Jenkins TC, Neidle S, Hurley LH. Inhibition of human telomerase by a G-quadruplex-interactive compound. J. Med. Chem. 1997;40:2113–2116. doi: 10.1021/jm970199z. [DOI] [PubMed] [Google Scholar]
- 2.Phan AT, Luu KN, Patel DJ. Different loop arrangements of intramolecular human telomeric (3+1) G-quadruplexes in K+ solution. Nucleic Acids Res. 2006;34:5715–5719. doi: 10.1093/nar/gkl726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Luu KN, Phan AT, Kuryavyi V, Lacroix L, Patel DJ. Structure of the human telomere in K+ solution: an intramolecular (3 + 1) G-quadruplex scaffold. J. Am. Chem. Soc. 2006;128:9963–9970. doi: 10.1021/ja062791w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Phan AT, Kuryavyi V, Burge S, Neidle S, Patel DJ. Structure of an unprecedented G-quadruplex scaffold in the human c-kit promoter. J. Am. Chem. Soc. 2007;129:4386–4392. doi: 10.1021/ja068739h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xu Y, Noguchi Y, Sugiyama H. The new models of the human telomere d[AGGG(TTAGGG)3] in K+ solution. Bioorg. Med. Chem. 2006;14:5584–5591. doi: 10.1016/j.bmc.2006.04.033. [DOI] [PubMed] [Google Scholar]
- 6.Matsugami A, Xu Y, Noguchi Y, Sugiyama H, Katahira M. Structure of a human telomeric DNA sequence stabilized by 8-bromoguanosine substitutions, as determined by NMR in a K(+) solution. FEBS J. 2007;274:3545–3556. doi: 10.1111/j.1742-4658.2007.05881.x. [DOI] [PubMed] [Google Scholar]
- 7.Ambrus A, Chen D, Dai J, Bialis T, Jones RA, Yang D. Human telomeric sequence forms a hybrid-type intramolecular G-quadruplex structure with mixed parallel/antiparallel strands in potassium solution. Nucleic Acids Res. 2006;34:2723–2735. doi: 10.1093/nar/gkl348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dai J, Punchihewa C, Ambrus A, Chen D, Jones RA, Yang D. Structure of the intramolecular human telomeric G-quadruplex in potassium solution: a novel adenine triple formation. Nucleic Acids Res. 2007;35:2440–2450. doi: 10.1093/nar/gkm009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dai J, Ambrus A, Punchihewa C, Jones RA, Yang D. Structure of the hybrid-2 type intramolecular human telomeric G-quadruplex in K+ solution: insights into structure polymorphism of the human telomeric sequence. Nucleic Acids Res. 2007;35 doi: 10.1093/nar/gkm522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Qi J, Shafer RH. Human telomere quadruplex: refolding and selection of individual conformers via RNA/DNA chimeric editing. Biochemistry. 2007;46:7599–7606. doi: 10.1021/bi602392u. [DOI] [PubMed] [Google Scholar]
- 11.De Cian A, Mergny JL. Quadruplex ligands act as molecular chaperones for tetramolecular quadruplex formation. Nucleic Acids Res. 2007;35:2483–2493. doi: 10.1093/nar/gkm098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gros J, Rosu F, Amrane S, De Cian A, Lacroix L, Mergny JL. Guanines are a quartet's best friend: impact of base substitutions on the kinetics and stability of tetramolecular quadruplexes. Nucleic Acids Res. 2007;35:3064–3075. doi: 10.1093/nar/gkm111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fadrna E, Spackova N, Stefl R, Koca J, Cheatham TE, Sponer J. Molecular dynamics simulations of guanine quadruplex loops: advances and force field limitations. Biophys. J. 2004;87:227–242. doi: 10.1529/biophysj.103.034751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Perez A, Marchan I, Svozil D, Sponer J, Cheatham TE, Laughton CA, Orozco M. Refinenement of the AMBER force field for nucleic acids: improving the description of alpha/gamma conformers. Biophys. J. 2007;92:3817–3829. doi: 10.1529/biophysj.106.097782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Siddiqui-Jain A, Grand CL, Bearss DJ, Hurley LH. Direct evidence for a G-quadruplex in a promoter region and its targeting with a small molecule to repress c-MYC transcription. Proc. Natl. Acad. Sci., U.S.A. 2002;99:11593–11598. doi: 10.1073/pnas.182256799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang D, Hurley LH. Structure of the biologically relevant G-quadruplex in the c-MYC promoter. Nucleosides, Nucleotides Nucleic Acids. 2006;25:951–968. doi: 10.1080/15257770600809913. [DOI] [PubMed] [Google Scholar]
- 17.Huppert JL, Balasubramanian S. Prevalence of quadruplexes in the human genome. Nucleic Acids Res. 2005;33:2908–2916. doi: 10.1093/nar/gki609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huppert JL, Balasubramanian S. G-quadruplexes in promoters throughout the human genome. Nucleic Acids Res. 2007;35:406–413. doi: 10.1093/nar/gkl1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Davis JT, Spada GP. Supramolecular architectures generated by self-assembly of guanosine derivatives. Chem. Soc. Rev. 2007;36:296–313. doi: 10.1039/b600282j. [DOI] [PubMed] [Google Scholar]
- 20.Miyoshi D, Karimata H, Wang ZM, Koumoto K, Sugimoto N. Artificial G-wire switch with 2,2′-bipyridine units responsive to divalent metal ions. J. Am. Chem. Soc. 2007;129:5919–5925. doi: 10.1021/ja068707u. [DOI] [PubMed] [Google Scholar]
- 21.Miyoshi D, Inoue M, Sugimoto N. DNA logic gates based on structural polymorphism of telomere DNA molecules responding to chemical input signals. Angew. Chem. Int. Ed. 2006;45:7716–7719. doi: 10.1002/anie.200602404. [DOI] [PubMed] [Google Scholar]
- 22.Cuesta J, Read MA, Neidle S. The design of G-quadruplex ligands as telomerase inhibitors. Mini-Rev. Med. Chem. 2003;3:11–21. doi: 10.2174/1389557033405502. [DOI] [PubMed] [Google Scholar]
- 23.Gunaratnam M, Greciano O, Martins C, Reszka AP, Schultes CM, Morjani H, Riou JF, Neidle S. Mechanism of acridine-based telomerase inhibition and telomere shortening. Biochem. Pharmacol. 2007;74:679–689. doi: 10.1016/j.bcp.2007.06.011. [DOI] [PubMed] [Google Scholar]
- 24.Gomez D, O’Donohue MF, Wenner T, Douarre C, Macadre J, Koebel P, Giraud-Panis MJ, Kaplan H, Kolkes A, Shin-ya K, Riou JF. The G-quadruplex ligand telomestatin inhibits POT1 binding to telomeric sequences in vitro and induces GFPPOT1 dissociation from telomeres in human cells. Cancer Res. 2006;66:6908–6912. doi: 10.1158/0008-5472.CAN-06-1581. [DOI] [PubMed] [Google Scholar]
- 25.Duquette ML, Handa P, Vincent J, Taylor AF, Maizels N. Intracellular transcription of G-rich DNAs induces formation of G-loops, novel structures containing G4 DNA. Genes Dev. 2004;18:1618–1629. doi: 10.1101/gad.1200804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Duquette ML, Pham P, Goodman MF, Maizels N. AID binds to transcription-induced structures in c-MYC that map to regions associated with translocation and hypermutation. Oncogene. 2005;24:5791–5798. doi: 10.1038/sj.onc.1208746. [DOI] [PubMed] [Google Scholar]
- 27.Eddy J, Maizels N. Gene function correlates with potential for G4 DNA formation in the human genome. Nucleic Acids Res. 2006;34:3887–3896. doi: 10.1093/nar/gkl529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Duquette ML, Huber MD, Maizels N. G-rich proto-oncogenes are targeted for genomic instability in B cell lymphomas. Cancer Res. 2007;67:2586–2594. doi: 10.1158/0008-5472.CAN-06-2419. [DOI] [PubMed] [Google Scholar]
- 29.Schaffitzel C, Berger I, Postberg J, Hanes J, Lipps HJ, Pluckthun A. In vitro generated antibodies specific for telomeric guanine-quadruplex DNA react with stylonychia lemnae macronuclei. Proc. Natl. Acad. Sci. U.S.A. 2001;98:8572–8577. doi: 10.1073/pnas.141229498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Paeschke K, Simonsson T, Postberg J, Rhodes D, Lipps HJ. Telomere end-binding proteins control the formation of G-quadruplex DNA structures in vivo. Nat. Struct. Mol. Biol. 2005;10:847–854. doi: 10.1038/nsmb982. [DOI] [PubMed] [Google Scholar]
- 31.Girvan AC, Teng Y, Casson LK, Thomas SD, Juliger S, Ball MW, Klein JB, Pierce WM, Jr, Barve SS, Bates PJ. AGRO100 inhibits activation of nuclear factor-kappaB (NF-kappaB) by forming a complex with NF-kappaB essential modulator (NEMO) and nucleolin. Mol. Cancer Ther. 2006;71:1790–1799. doi: 10.1158/1535-7163.MCT-05-0361. [DOI] [PubMed] [Google Scholar]
- 32.Mi Y, Thomas SD, Xu X, Casson LK, Miller DM, Bates PJ. Apoptosis in leukemia cells is accompanied by alterations in the levels and localization of nucleolin. J. Biol. Chem. 2003;278:8572–8579. doi: 10.1074/jbc.M207637200. [DOI] [PubMed] [Google Scholar]
- 33.Khateb S, Weisman-Shomer P, Ludwig A, Hershko-Shani I, Fry M. Nucleic Acids Res. 2007;35 doi: 10.1093/nar/gkm636. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Freyer MW, Buscaglia R, Kaplan K, Cashman D, Hurley LH, Lewis EA. Biophysical studies of the c-MYC NHE III1 promoter: model quadruplex interactions with a cationic porphyrin. Biophys. J. 2007;92:1–9. doi: 10.1529/biophysj.106.097246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.White EW, Tanious F, Ismail MA, Reszka AP, Neidle S, Boykin DW, Wilson WD. Structure-specific recognition of quadruplex DNA by organic cations: influence of shape, substituents and charge. Biophys. Chem. 2007;126:140–153. doi: 10.1016/j.bpc.2006.06.006. [DOI] [PubMed] [Google Scholar]
- 36.Marin VL, Armitage BA. Hybridization of complementary and homologous peptide nucleic acid oligomers to a guanine quadruplex-forming RNA. Biochemistry. 2006;45:1745–1754. doi: 10.1021/bi051831q. [DOI] [PMC free article] [PubMed] [Google Scholar]