Abstract
The role of glycoprotein membrane-spanning domains in the process of membrane fusion is poorly understood. It has been demonstrated that replacing all or part of the membrane-spanning domain of a viral fusion protein with sequences that encode signals for glycosylphosphatidylinositol linkage attachment abrogates membrane fusion activity. It has been suggested, however, that the actual amino acid sequence of the membrane-spanning domain is not critical for the activity of viral fusion proteins. We have examined the function of Moloney murine leukemia virus envelope proteins with substitutions in the membrane-spanning domain. Envelope proteins bearing substitutions for proline 617 are processed and incorporated into virus particles normally and bind to the viral receptor. However, they possess greatly reduced or undetectable capacities for the promotion of membrane fusion and infectious virus particle formation. Our results imply a direct role for the residues in the membrane-spanning domain of the murine leukemia virus envelope protein in membrane fusion and its regulation. They also support the thesis that membrane-spanning domains possess a sequence-dependent function in other protein-mediated membrane fusion events.
INTRODUCTION
Protein-mediated membrane fusion is a central process in the biology of a cell, and membrane fusion events also occur between cells during conception and development. Recently, insight has been gained into the identities of many of the components of the cellular membrane fusion machinery, but an understanding of their biochemical contributions to the membrane fusion event itself has proven to be more elusive. Investigations of viral membrane proteins have supplied material with which this lacuna can be filled. Besides greatly enhancing our insight into the entry of viruses into cells, studies of these proteins continue to play critical roles in the development of our understanding of such processes as transmembrane protein assembly, processing, subcellular targeting, and association with intracellular molecules. It is possible, however, that the greatest contribution to biology of research into the function of these viral proteins may lie in the ongoing unraveling of the mechanism of protein-mediated membrane fusion.
An enveloped virus possesses a lipid bilayer that it acquires while budding from an infected cell. To initiate an infection the virus must fuse its membrane envelope with that of a cell. This membrane fusion is effected by a protein or proteins that are inserted in the viral bilayer and is generally thought to commence with the close juxtaposition of the viral and cellular membranes (Hernandez et al., 1996; Melikyan and Chernomordik, 1997). The proximity of the membranes is achieved through the binding of one or more viral proteins to cell surface components. This association may result in fusion of the viral and cellular membrane at the cell surface or in the uptake of the virus into the cell through endocytosis, with the subsequent triggering of membrane fusion by means of protein conformational changes induced by the reduction in the vesicular pH. The physical event that is believed to promote membrane fusion is the insertion of a “fusion peptide” from one of the viral proteins into or, at least, the establishment of contact with the cellular membrane. These fusion peptides are generally considered uncharged regions of the protein that are buried within until the moment for the promotion of membrane fusion arrives and they are “exposed.” The insertion of these fusion peptides is postulated to disrupt the normal organization of the lipids in their vicinity. Studies of viral membrane proteins with amino acid residue substitutions in these fusion peptides and of the membrane fusion activity in model systems of synthesized fusion peptides lend support for these concepts. Recent data bolster certain proposals for the mechanism of fusion peptide exposure and presentation to the cellular membrane (Carr and Kim, 1993; Bullough et al., 1994). More detailed hypotheses that incorporate the foregoing ideas and that concern the number of viral fusion proteins required to cooperate to promote a membrane fusion event, the oligomeric state of those proteins, the physical state or phases of the membrane lipid components during fusion, and the mixing of the lipid layers have been expounded (Melikyan and Chernomordik, 1997), but there is less of a consensus about their validity (Durrer et al., 1996).
One aspect of virus protein-mediated membrane fusion that had been largely overlooked until recently is the effect of the viral protein on the virus membrane. It is not clear why fusion peptide insertion into the cellular membrane should be sufficient to produce membrane fusion rather than just a hole in the cellular membrane. In particular, the role of the membrane-spanning domain of the viral transmembrane proteins has only recently been examined. Experiments in a number of laboratories have examined the effects of replacing all or part of the membrane-spanning domain of a viral fusion protein with sequences that encode signals for glycosylphosphatidylinositol (GPI) linkage attachment (Weiss and White, 1993; Kemble et al., 1994; Ragheb and Anderson, 1994a; Melikyan et al., 1995). In each case a defect in membrane fusion was observed. Similarly, engineered viral fusion proteins that are not membrane bound do not fuse membranes. These findings suggest that a protein membrane-spanning domain is critical for the function of the fusion proteins. Alternatively, it is possible that the GPI linkage somehow inhibits membrane fusion.
One major issue raised by these data is whether it is a protein membrane-spanning domain per se that is required for membrane fusion or whether the amino acid sequence of the membrane-spanning domain is critical. In support of the former hypothesis is the great variety in the sequences of the membrane-spanning domains in viral fusion proteins. In addition, it has been reported that fusion-competent proteins can be constructed from viral proteins whose membrane-spanning domains have been replaced with those of proteins that are not believed to participate in membrane fusion (Wilk et al., 1996; Odell et al., 1997). On the other hand, the membrane-spanning domains of some homologous viral fusion proteins, such as those of the murine leukemia viruses, are well conserved, an unexpected result if the sequence is unimportant to function. It is evident that the membrane-spanning domain may be playing a role in viral assembly, such as in proper folding of the viral fusion protein or in its selective incorporation in virions, but it is no clearer how heterologous membrane-spanning domains would execute these functions when they replace the normal viral protein membrane-spanning domain. Additional support for the proposition that the sequence of membrane-spanning domains is critical for the promotion of membrane fusion is provided by the findings that charged residues in the putative membrane-spanning domain of the human immunodeficiency virus type 1 (HIV-1) envelope glycoprotein (Helseth et al., 1990; Owens et al., 1994) and glycine residues in the membrane-spanning domain of vesicular stomatitis virus (VSV) G protein (Cleverley and Lenard, 1998) appear to play important roles in the membrane fusion mediated by those viral proteins, although interpretation of the data is complicated by potential inconsistencies between publications (Helseth et al., 1990; Owens et al., 1994; Odell et al., 1997) and incomplete characterization of the mutant glycoproteins (Cleverley and Lenard, 1998).
In retroviruses, it is the envelope protein that is responsible for the initial events in the infection of a cell, that is, the binding of the virus to a receptor on the cell surface and the subsequent fusion of viral and cellular membranes (Hunter and Swanstrom, 1990). This membrane fusion results in the release of the contents of the viral particle into the cell, where viral uncoating and replication can occur.
The Moloney murine leukemia virus (Mo-MuLV) envelope (Env) protein is encoded as a 665-amino acid residue polypeptide that has a cleaved signal sequence (Shinnick et al., 1981; Henderson et al., 1984). During its progress through the secretory system the protein forms a trimer (Kamps et al., 1991) and is subsequently proteolytically processed into two subunits (Witte et al., 1977; Henderson et al., 1984), gp70 (amino acids 34–468) and p15E (amino acids 470–665), that are linked through a labile disulfide bond (Pinter and Fleissner, 1977; Pinter et al., 1978, 1997). Once the envelope protein has reached the cell surface, it is incorporated into a budding C-type retroviral particle. The gp70 subunit (conventionally referred to as SU) is on the outside of the particle, whereas the p15E transmembrane protein (TM) possesses an extraparticle domain, a transmembrane domain, and a 35-amino acid domain that resides within the particle (Pinter and Honnen, 1983). During a late stage of viral particle maturation the carboxyl-terminal 16 amino acid residues of the p15E protein are removed by the retroviral gag–pol-encoded protease resulting in a 12-kDa envelope transmembrane protein (Sutcliffe et al., 1980; Shinnick et al., 1981; Green et al., 1981; Henderson et al., 1984; Crawford and Goff, 1985; Katoh et al., 1985; Schultz and Rein, 1985). The Env protein present on the membrane of an infected cell is therefore structurally different from that found in a mature particle. Removal of the carboxyl-terminal region activates the Env so that it is capable of fusing membranes in a receptor-dependent manner (Ragheb and Anderson, 1994b; Rein et al., 1994). The Mo-MuLV Env is a member of a family of homologous murine, feline, and ape leukemia virus Envs (Shinnick et al., 1981; Guilhot et al., 1987; Delassus et al., 1989; Ott et al., 1990). The conservation of amino acid sequence in these proteins is greatest in the TM proteins.
In the present study we have explored the roles of conserved amino acid residues in the amino-terminal half of the presumptive membrane-spanning domain of the Mo-MuLV Env and have found that substitutions can result in dramatic and specific alterations in the capacity of the envelope protein to promote membrane fusion and in the regulation of fusion activity.
MATERIALS AND METHODS
Construction of Plasmids Encoding Mutant Mo-MuLV Envelope Proteins
Mutagenesis of Mo-MuLV was performed on env-containing subclones of pMOVΨ− (Mann et al., 1983) in pTZ18U using the Bio-Rad (Hercules, CA) Muta-Gene phagemid mutagenesis kit; regions of all plasmids derived from synthesized oligonucleotides were sequenced to confirm their structure. Amino acid numbering is from the initiation methionine codon of Env; nucleotide numbering is from the complete genomic sequence (Shinnick et al., 1981). For mutations in the membrane-spanning domain, a unique BglII site was created by introducing a silent mutation at Mo-MuLV Env codon 603. A degenerate double-stranded oligonucleotide encoding substitutions of alanine, glycine, or valine for proline 617 was inserted between the introduced BglII site and the natural Mo-MuLV env ClaI site. The P617A substitution created a unique Ecl136II site; the resulting plasmid, pMoenvP617A, was used in all subsequent cloning. Plasmid pMoenvP617A was cut with BglII and Ecl136II and ligated with the oligonucleotides corresponding to all of the other mutations (W606A, W606G, W606E, W606S, W606I, W606F, F607A, F607G, F607V, F607W, F607Y, G616A, G616V, and G616P).
The Mo-MuLV envelope proteins were expressed from pLTRSDSA, a vector that contains the Mo-MuLV long terminal repeat (LTR) enhancer and promoter, and encodes the Mo-MuLV splice donor and acceptor sequences, and the SV-40 polyadenylation addition signal. pLTRSDSA was derived from the Mo-MuLV vector pCRIPgag-2 (Danos and Mulligan, 1988) by first replacing sequences between the XbaI site 5′ to env and the NheI site in the 3′ LTR of Mo-MuLV (nucleotides 5766–7846) with the sequence 5′-TCTAGCCCTCGAGGCTAGC-3′. This removed the XbaI site but retained the NheI site and inserted a unique XhoI site. The fragment from the NheI site in the 5′ LTR through the EcoRI site at the end of the SV40 polyadenylation sequence was cloned between the XbaI and EcoRI sites of pUC19, and the internal HindIII fragment was removed, deleting most of the gag and pol. The penv1min (wild-type env) and penv1Δ650–665 plasmids were constructed. For construction of penv1Δ650–665 the coding oligonucleotide was 5′-CGATTAAGTCCAATTTGTTAAAGACAGGATATCAGTGGTTCA AGGCCTTGTGACGTCG-3′. The complementary oligonucleotide possessed a 5′-CTAG overhang and left a 5′-CG overhang on the coding strand. For the wild-type envelope-encoding vector (penv1min) a double-stranded oligonucleotide (coding oligonucleotide, 5′-CCTTGGTTTTGACTCAACAATATCACCAGCTTAAGC CTATAGAGTTACGAGCCCTAGGT-3′) was cloned into the fragment of penv1Δ650–665 from which a small StuI–NheI fragment had been removed. The complementary oligonucleotide possessed a 5′-CTAG overhang. The mutant envelope proteins were moved from the cloning vector into both of the expression vectors, penv1min and penv1Δ650–665, by cutting the cloning and expression plasmids with PflMI and ClaI cleaving at the natural Mo-MuLV env sites and ligating the 1000-bp fragment from the cloning vector with the 6-kb fragment of the expression vector.
Syncytia Formation Assays
NIH 3T3 cells were transiently transfected with the mutant Env-encoding plasmids using LipofectAMINE (Life Technologies, Gaithersburg, MD) and Opti-MEM media (Life Technologies). NIH 3T3 cells were plated at 5 × 105 cells per 60-mm plate 24 h before transfection. The cells were washed and incubated for 30 min at 37°C with 2 ml of Opti-MEM media. The DNA-LipofectAMINE-Opti-MEM mixture (4 μg of DNA, 24 μl of LipofectAMINE, and 300 μl of Opti-MEM media) was incubated for 30 min at 25°C. After the 30-min incubations, 2.4 ml of Opti-MEM media were added to the DNA-LipofectAMINE mixture; the resulting solution was layered onto the NIH 3T3 cells. Eight hours later the transfection mixture was removed, and the cells were incubated with Dulbecco’s modified Eagle’s media (Sigma, St. Louis, MO) containing 10% calf serum (Life Technologies), 0.1 mg/ml streptomycin (Sigma), and 10 U/ml penicillin (Sigma) (DMEM CS/PS) for 32 h before counting syncytia and the number of nuclei in syncytia in 20 different fields. There were ∼1200 cells per field.
When syncytia formation promotion by Mo-MuLV envelope proteins expressed in 293T cells was examined, 2 × 106 293T cells were transfected with 4 μg of DNA using LipofectAMINE as above. NIH 3T3 cells and the transfected 293T cells were trypsinized and mixed by plating 1 × 106 NIH 3T3 cells and 1 × 106 transfected 293T cells on a 60-mm tissue culture dish. After 24 h of growth in DMEM with fetal calf serum, 0.1 mg/ml streptomycin (Sigma), and 10 U/ml penicillin (Sigma) the syncytia were counted as above.
Establishment of Stable Cell Lines Expressing Mutant Envelope Proteins
NIH 3T3 cells were stably transfected with each mutant that was cloned into the expression vector encoding the full-length cytoplasmic domain penv1min. NIH 3T3 cells were plated at 5 × 105 cells per 60-mm plate 24 h before transfection. The cells were washed and incubated for 30 min at 37°C with 2 ml Opti-MEM media. The DNA-LipofectAMINE-Opti-MEM mixture (8 μg of mutant DNA, 0.4 μg of pJ6Ωpuro [Morgenstern and Land, 1990], 48 μl of LipofectAMINE, and 300 μl of Opti-MEM media) was incubated for 30 min at 25°C. After the 30-min incubations, 2.4 ml of Opti-MEM media were added to the DNA-LipofectAMINE mixture; the resulting solution was layered onto the NIH 3T3 cells. Eight hours later the transfection mixture was removed, and the cells were incubated with DMEM CS/PS for 32 h before transferring the cells to 10-cm plates at three different dilutions (1:5, 1:10, and 1:20). The following day the media were changed to DMEM CS/PS containing 5 μg/ml puromycin (Sigma). Colonies appeared after 2 wk and were picked for screening by immunoprecipitation and interference assays.
Immunoprecipitation Assays
The stable cell lines were plated at 1 × 106 cells on 60-mm plates 24 h before labeling. For metabolic labeling, the cells were incubated with 2 ml of DMEM lacking cysteine and methionine (Life Technologies) and containing 10% dialyzed calf serum (Life Technologies-BRL) and 50 μCi/ml 35S-cysteine/[35S]methionine (Amersham, Arlington Heights, IL) for 4 h at 37°C. After the 4-h incubation, the media were removed, and the cells were washed and lysed with 1× radioimmunoprecipitation (RIP) buffer (20 mM Tris, pH 7.4, 500 mM NaCl, and 0.5% NP-40). Cell debris was removed by centrifugation at 14,000 rpm in a microfuge, and the samples were precleared by incubation with 1:100 vol goat serum (Sigma) at 37°C for 30 min and 1:10 vol prepared Pansorbin (Staphylococcus aureus) cells (Calbiochem, La Jolla, CA) for 10 min at room temperature. The Pansorbin cells were removed by centrifugation, and the supernatant liquid was incubated with 1:75 diluted goat anti-Rauscher leukemia virus gp70 antiserum (lot 80S-019; Quality Biotech, Camden, NJ) overnight at 4°C. Pansorbin cells were added for 10 min at 25°C. After centrifugation the Pansorbin cells and immunoprecipitated protein were washed and suspended in SDS-PAGE sample buffer. The samples were then analyzed by SDS-PAGE (8% SDS-polyacrylamide gel) and fluorography.
For metabolic labeling of the 293T or gpGFP cells (described below) that were transiently transfected with the mutant envs and used in the syncytia formation assays described above or transduction experiments described below, the cells were incubated with 5 ml of DMEM lacking cysteine and methionine (Life Technologies) and containing 10% dialyzed calf serum (Life Technologies) and 50 μCi/ml 35S-cysteine/[35S]methionine (Amersham) for 16 h at 37°C. The cell supernatant medium was filtered through a 0.45-μm filter and spun through a 30% sucrose cushion in a Beckman (Palo Alto, CA) 50.2Ti rotor at 25,000 rpm and 4°C for 2.5 h. The viral pellet was suspended in 1 ml of 1× RIP buffer and immunoprecipitated following the protocol above. The gpGFP cells were lysed and immunoprecipitated as described previously.
Viral Interference Assays
The stable cell lines established previously were plated out at 5 × 105 cells per 60-mm plate 24 h before infection with recombinant β-galactosidase-conveying Mo-MuLV (E86nlslacZ cell supernatant medium). The E86nlslacZ cells were developed in our laboratory by cotransfecting GP+E-86 cells (Markowitz et al., 1988) with MFG.S-nlslacZ, a retroviral vector that encodes a nuclear-localized β-galactosidase (Ory et al., 1996) and pJ6Ωpuro. Single colonies were selected and screened for β-galactosidase production. The supernatant medium from E86nlslacZ cells was collected and filtered through a 0.45-μm filter. A 1:100 dilution of the virus supernatant medium was mixed with 8 μg/ml polybrene and incubated with the stable cells for 4 h. The recombinant virus-containing medium was then replaced with DMEM CS/PS medium. Forty-eight hours after the infection, the cells were fixed with 0.5% glutaraldehyde (Sigma) and then incubated with 1 mg/ml β-galactosidase detection reagent 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-gal; Fisher Scientific, Pittsburgh, PA) in the presence of 1 mM MgCl2, 50 mM K3Fe(CN)6, and 50 mM K4Fe(CN)6 for 3 h before the determination of the proportion of blue cells (Sanes et al., 1986). The total number of cells as well as the number of blue cells were counted for 4 fields per plate. Interference was calculated by comparing the reduction of virus titer determined on cells expressing mutant Envs and those expressing the wild-type Env.
Viral Transduction Assays
The mutant envelope proteins with the full-length cytoplasmic domain were transiently transfected into gpGFP cells following the protocol above for syncytia assays, except that DMEM with fetal calf serum was used to culture the cells. The gpGFP cells were developed in our laboratory by cotransfecting MFG.S-GFP-S65T, a retroviral vector encoding the Aequorea victoria GFP S65T mutant (a kind gift from Dirk Lindemann, Institut für Virologie und Immunbiologie Universität Würzburg, Würzburg, Germany), and pJ6Ωpuro into ΦNX cells, a second-generation 293T-based retroviral packaging cell line (Pear et al., 1993; Grignani et al., 1998). Single colonies were selected and screened for production of high-titer replication-incompetent virus resulting from transient transfection with penv1min. Medium from the transiently transfected gpGFP cells was removed 32 h after transfection and filtered through a 0.45-μm filter. It was then incubated with 1 × 106 NIH 3T3 cells in the presence of 8 μg/ml polybrene for 4 h. The recombinant virus-containing medium was then replaced with DMEM CS/PS. Forty-eight hours later the cells were removed from the plate, suspended in 1× PBS containing 1 mM EDTA, and analyzed by flow cytometry with a Coulter Electronics (Hialeah, FL) XL-MCL flow cytometer using a 525-nm bandpass filter and a 488-nm air-cooled argon laser.
RESULTS
The amino acid sequence of the membrane-spanning domain and surrounding residues of the Mo-MuLV envelope TM protein is conserved between murine and feline leukemia viruses (Figure 1). It is noteworthy that the amino-terminal half of the membrane-spanning domain is rich in serine and threonine residues, whereas the carboxyl-terminal half contains almost exclusively aliphatic residues. The effects of substitutions in tryptophan 606 and phenylalanine 607 were analyzed, because of the likelihood that these two residues lay at the interface between the virus membrane and the extracellular domain and of substitutions in the glycine–proline residue pair (amino acids 616 and 617), which is unusual in the center of a membrane-spanning domain (Brandl and Deber, 1986).
Figure 1.
Amino acid sequence of the ecotropic Mo-MuLV membrane-spanning domain and surrounding residues of TM compared with that of other murine, feline, and gibbon ape leukemia viruses (GeneBank accession numbers 74692, M33469, K02730, 74707, 74702, and M26927, respectively). The absolutely conserved residues in the putative membrane-spanning domain are represented by the bold type, whereas the specific residues that were mutated are indicated by an asterisk.
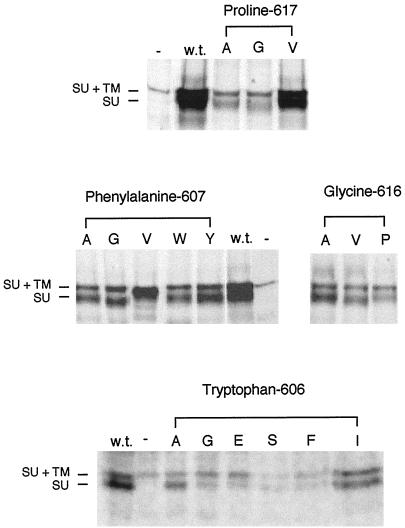
Mutant Envelope Proteins Are Processed Normally Except for F607V
Stable cell lines of each mutant were established by cotransfecting the plasmids encoding the mutant envelope proteins with the full-length cytoplasmic domain and a selection plasmid (pJ6Ωpuro; Morgenstern and Land, 1990) into NIH 3T3 cells. The stable cell lines were then used in immunoprecipitation assays to ensure that the various point mutations did not have a significant affect on the processing of the envelope proteins. Each stable cell line was labeled with 35S-cysteine/[35S]methionine for 4 h before lysing the cells and immunoprecipitating the envelope protein with anti-SU antibody. The presence of both the 85-kDa Mo-MuLV uncleaved N-glycosylated Env and the 70-kDa glycosylated SU subunit indicates that all of the proteins are properly expressed and processed, with the exception of F607V, which appears to be underglycosylated and not cleaved into SU and TM (Figure 2). It should be noted that a stable cell line expressing barely detectable levels of the W606S Env (Figure 2) was obtained only after several experiments. It was, indeed, not possible to obtain cell lines expressing normal levels of several of the mutant envelope proteins bearing substitutions for tryptophan 606.
Figure 2.
The mutant envelope proteins are processed normally in NIH 3T3 cells, with the exception of F607V. Cells (5 × 105) that were stably expressing the various mutant envelope proteins were labeled with 50 μCi/ml 35S-cysteine/[35S]methionine for 4 h before lysis with 1× RIP buffer. The cell debris was removed before two rounds of immunoprecipitation of the envelope proteins with antibody against SU. The immunoprecipitated envelope proteins were analyzed on an 8% SDS-PAGE gel and exposed to autoradiography film for 1 wk. Some variation in expression level from cells with different sites of stable env gene integration is the expected result. The amino acid substitutions in the mutant envelope proteins expressed in the lysates are indicated by their representations in the single-letter amino acid code beneath the headings noting the altered residues. GP+E-86 cells were used as the positive control for expression of wild-type Mo-MuLV envelope protein (w.t.), whereas NIH 3T3 cells were used as the negative control (−). At left are indicated the positions of wild-type uncleaved SU+TM (85 kDa) and SU (70 kDa). There is a cross-reactive protein in the cell lysate that migrates slightly slower than SU+TM (85 kDa).
Envelope Proteins Bearing Substitutions for Proline 617 Are Unable to Transduce Cells despite Being Incorporated at Normal Levels into Virus Particles
To determine whether the mutant envelope proteins are functional in a virus particle, infectivity assays were performed. gpGFP cells are a packaging cell line that has been stably transfected with the Mo-MuLV gag and pol genes and a retroviral vector encoding the Aequorea victoria GFP S65T mutant that are able to produce virus particles containing the GFP RNA. Because these cells do not express an envelope protein, they are not able to produce infectious virus particles unless a plasmid encoding an envelope protein is transfected into the cells. Plasmids encoding the mutant Mo-MuLV envelope proteins with full-length cytoplasmic domains were transiently transfected into the gpGFP cells. The media from these cells were removed, filtered, and placed on uninfected NIH 3T3 cells, which are susceptible to infection by Mo-MuLV. If the mutant envelope protein is able to promote infectious virion formation, then the infected cells will express GFP plasmid and fluoresce. Infected cells are detected by flow cytometry. Viruses produced by cells expressing the P617A, P617G, and F607V Envs were unable to transduce cells (Table 1). Those bearing the P617V Env had a 1000-fold decrease in transduction capacity compared with those bearing the wild-type Env, whereas W606E Env virions had a 100-fold decrease. All of the other mutant envelope proteins were able to transduce cells at levels comparable with that produced by virions bearing the wild-type envelope protein (Table 1). There are two conceivable sources for the complete inability or decreased ability of some of the mutant envelope proteins to transduce cells—a defect in incorporation of the envelope proteins into the virus particle or a failure by the mutant envelope protein to promote membrane fusion.
Table 1.
Transduction of NIH 3T3 cells by virions bearing mutant Mo-MuLV envelope proteins
| Mutant envelope protein | Virus titer (TU/ml) |
|---|---|
| W606A | 1.03 ± 0.03 × 105 |
| W606G | 6.52 ± 0.32 × 104 |
| W606E | 8.4 ± 2.7 × 103 |
| W606S | 3.14 ± 0.58 × 104 |
| W606I | 5.11 ± 0.71 × 104 |
| W606F | 4.68 ± 0.52 × 104 |
| F607A | 1.04 ± 0.09 × 105 |
| F607G | 9.77 ± 0.61 × 104 |
| F607V | <3 |
| F607W | 1.05 ± 0.06 × 105 |
| F607Y | 1.04 ± 0.04 × 105 |
| G616A | 6.95 ± 0.70 × 104 |
| G616V | 4.84 ± 1.14 × 104 |
| G616P | 7.65 ± 0.79 × 104 |
| P617A | <3 |
| P617G | <3 |
| P617V | 0.7 ± 1.2 × 102 |
| Cells not expressing an envelope protein | <3 |
| Cells expressing the wild-type envelope protein | 1.00 × 105 |
gpGFP cells were transiently transfected with the plasmids encoding the various mutant envelope proteins. After 40 h the supernatant medium was removed and used for infection of the NIH 3T3 cells. Cells that were infected express GFP and were detected by flow cytometry. Virus titer is given as transducing units per ml (TU/ml).
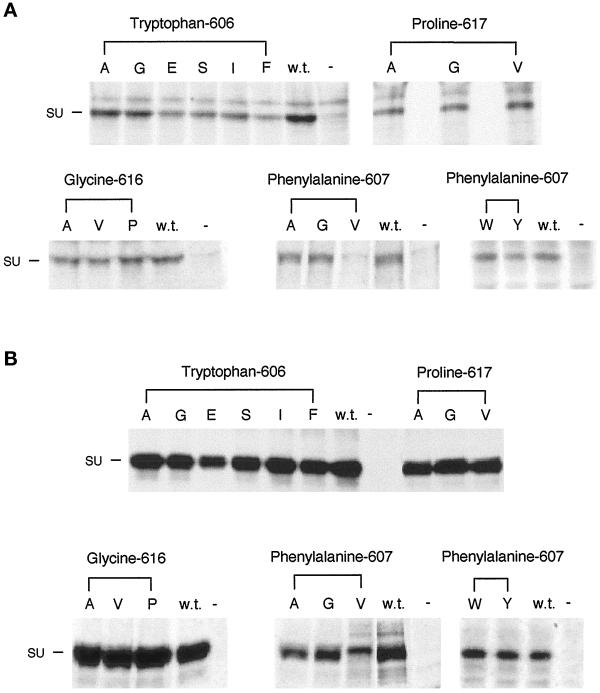
To ensure that the mutant envelope proteins were incorporated into virus particles, the supernatant medium from the transiently transfected gpGFP cells used in the infection assays was analyzed. The cells were incubated with 35S-cysteine/[35S]methionine, and the supernatant medium was removed after 16 h and passed through a 0.45-μm filter, and the virus was collected by ultracentrifugation through a sucrose cushion to remove any SU that may have been shed into the medium. The envelope proteins of the suspended virions were immunoprecipitated with anti-SU antibody (Figure 3A). Immunoprecipitations were also carried out on the cell lysate to examine expression levels and processing of the envelope proteins (Figure 3B). Wild-type virus produced from cells transfected with the wild-type env gene bears the 70-kDa SU subunit and not the 85-kDa SU-TM precursor. The predominant immunoreactive env gene product in the lysate of the human 293T-derived gpGFP cell lysate is also SU. All of the mutant envelope proteins, including those bearing substitutions for proline 617, are incorporated into virus particles at normal levels, with the exception of the W606E and F607V mutant Envs, which are incorporated into virus particles less efficiently or not at all, respectively. The normal level of incorporation of Envs possessing substitutions for proline 617 suggests that their inability to transduce cells is a result of a defect either in receptor binding or membrane fusion.
Figure 3.
Analysis of the incorporation into virus particles (A) and processing (B) of the various mutant Mo-MuLV envelope proteins in gpGFP cells. gpGFP cells (5 × 105) were transiently transfected with plasmids encoding the mutant and wild-type envelope proteins 48 h before labeling with 35S-cysteine/[35S]methionine for 16 h. The amino acid substitutions in the mutant envelope proteins expressed in the transfected cells are indicated by their representations in the single-letter amino acid code beneath the headings noting the altered residues. Analyses of cell lysates and virus particles budded from cells transfected with the plasmid encoding the wild-type Mo-MuLV envelope protein (w.t.) and from cells that were mock transfected (−) are also presented. At left is indicated the position of SU (70 kDa). (A) Immunoprecipitation of the cell supernatant medium was carried out by filtering it through a 0.45-μm filter and spinning it through a 30% sucrose cushion for 2 h at 25,000 rpm and 4°C. The envelope proteins were immunoprecipitated with antibody against SU, and they were analyzed as above. There is a cross-reactive protein that migrates slightly slower than SU (70 kDa). (B) Immunoprecipitation of the cell lysate was carried out by lysing the cells with 1× RIP buffer. The cell debris was removed before two rounds of immunoprecipitation of the envelope proteins with antibody against SU. The immunoprecipitated envelope proteins were analyzed on an 8% SDS-PAGE gel and exposed to autoradiography film for 1 wk.
Envelope Proteins Bearing Substitutions for Proline 617 Are Not Able to Promote Normal Levels of Receptor-mediated Membrane Fusion
To examine the ability of the various mutant envelope proteins to promote receptor-mediated-membrane fusion, syncytia assays were carried out. NIH 3T3 cells, which possess the Mo-MuLV receptor and are susceptible to infection by Mo-MuLV, were transiently transfected with the various plasmids encoding the mutant envelope proteins lacking the last 16 amino acid residues in the cytoplasmic domain (R peptide). The last 16 amino acid residues in the cytoplasmic domain of TM must be removed for fusion to occur (Ragheb and Anderson, 1994b; Rein et al., 1994). Normally this cleavage is mediated by the retroviral protease after the envelope protein has been incorporated into a virus particle (Sutcliffe et al., 1980; Green et al., 1981; Shinnick et al., 1981; Henderson et al., 1984; Crawford and Goff, 1985; Katoh et al., 1985; Schultz and Rein, 1985), so that only the envelope protein in virus particles is capable of mediating membrane fusion, whereas the wild-type envelope protein expressed on the surface of a cell is not active. Cells that are expressing the truncated envelope proteins can bind to the receptor of neighboring NIH 3T3 cells and potentially fuse and form syncytia or multinucleated cells. The transiently transfected cells are allowed to grow for 36 h before syncytium formation is analyzed (Table 2).
Table 2.
Syncytia formation promoted by mutant Mo-MuLV envelope proteins
| Mutant envelope protein | Syncytia formationa |
|---|---|
| W606A | 124 ± 28 |
| W606G | 156 ± 45 |
| W606E | 90 ± 20 |
| W606S | 76 ± 14 |
| W606I | 104 ± 20 |
| W606F | 102 ± 28 |
| W606S (full-length cytoplasmic domain) | 8 ± 3 |
| F607A | 72 ± 33 |
| F607G | 104 ± 60 |
| F607V | <0.25 |
| F607W | 120 ± 28 |
| F607Y | 111 ± 32 |
| G616A | 61 ± 19 |
| G616V | 70 ± 5 |
| G616P | 121 ± 48 |
| P617A | 2 ± 1 |
| P617G | <0.25 |
| P617V | 12 ± 6 |
| Wild-type with full-length cytoplasmic domain | <0.25 |
| Wild-type with truncated cytoplasmic domain | 100 |
NIH 3T3 cells were transiently transfected with plasmids encoding the mutant envelope proteins lacking the last 16 cytoplasmic amino acids. The number of syncytia and the number of nuclei in syncytia were counted 36 h after transfection.
The numbers are normalized to the percentage of nuclei in syncytia formed as a result of expression of the wild-type envelope protein with a truncated cytoplasmic domain (8.3%).
The P617G and F607V envelope proteins were unable to promote receptor-mediated membrane fusion, whereas the fusion ability of the P617A and P617V envelope proteins was greatly reduced. The G616A, G616V, W606S, and F607A envelope proteins possess moderately reduced membrane fusion promotion capacities (Table 2). All of the other mutant envelope proteins were able to induce syncytia formation at levels comparable with that of the wild-type Env.
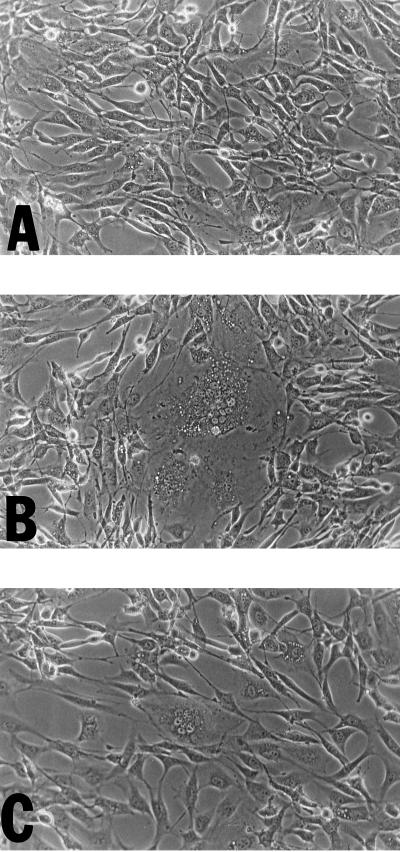
To further our understanding of the basis of the regulation of membrane fusion by the cytoplasmic domain of TM, the syncytia formation capacities of cells expressing mutant envelope proteins possessing full-length cytoplasmic domains were determined. Whereas the wild-type envelope protein with a full-length cytoplasmic domain is unable to promote syncytia formation (Figure 4A), the envelope protein bearing a deletion of the last 16 amino acid residues promotes extensive syncytia formation (Figure 4B). It was discovered that the W606S envelope protein with the full-length cytoplasmic domain was able to promote syncytia formation, although at a greatly reduced level (Figure 4C and Table 2).
Figure 4.
Analysis of the ability of the W606S mutant envelope protein to undergo receptor-mediated-membrane fusion in the presence of the full-length cytoplasmic domain. Four micrograms of plasmids (A) penv1min, (B) penvΔ650–665, and (C) penvW606S were transiently transfected into 5 × 105 NIH 3T3 cells. The transfected cells were allowed to grow for 36 h before syncytia were recorded. Syncytia can be observed in B and C but not in A.
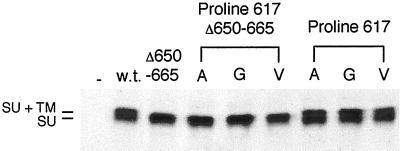
It was important to establish the level of expression and processing of the Mo-MuLV envelope proteins bearing substitutions of P617 in the cells capable of syncytia formation. During transient transfection of the NIH 3T3 cells only a low percentage (3–5%) of the cells express the envelope protein, which is consequently difficult to detect through immunological methods. We therefore examined syncytia formation between 293T cells (50–80% of which express transfected genes and which do not bear the Mo-MuLV receptor) expressing the truncated mutant or wild-type envelope proteins and NIH 3T3 cells (Rein et al., 1994) and investigated envelope protein expression and processing. As was found upon expression of the mutant envelope proteins in NIH 3T3 cells, the P617G envelope protein was unable to promote syncytia formation, whereas the fusion ability of the P617A and P617V envelope proteins was greatly reduced (Table 3). Expression and processing of the truncated Mo-MuLV envelope proteins bearing substitutions of P617 were similar to those of the truncated wild-type protein in the 293T cells (Figure 5). Expression and processing of the full-length Mo-MuLV envelope proteins bearing substitutions of P617 were also found to be similar to those of the full-length wild-type protein (Figure 5). The facts that the MuLV envelope proteins are processed in the late Golgi (Bedgood and Stallcup, 1992) and that the Mo-MuLV envelope proteins bearing P617 substitutions are processed normally and also incorporated at wild-type levels into virus particles (Figure 3A) indicate that the mutant envelope proteins are present in mature form at the cell surface at wild-type levels.
Table 3.
Syncytia formation promoted by Mo-MuLV envelope proteins expressed in 293T cells
| Mutant envelope proteins | Syncytia formationa |
|---|---|
| P617A | 3 ± 1 |
| P617G | <0.25 |
| P617V | 6 ± 1 |
| Wild-type with full-length cytoplasmic domain | <0.25 |
| Wild-type with truncated cytoplasmic domain | 100a |
293T cells were transiently transfected with plasmids encoding the mutant envelope proteins lacking the last 16 cytoplasmic amino acids and cocultivated with NIH 3T3 cells. The number of syncytia and the number of nuclei in syncytia were counted 48 h after transfection.
The numbers are normalized to the percentage of nuclei in syncytia formed as a result of expression of the wild-type envelope protein with a truncated cytoplasmic domain (7%).
Figure 5.
Analysis of the processing of the proline 617 mutants with and without a full-length cytoplasmic domain in 293T cells. 293T cells (1 × 106) were transiently transfected with plasmids encoding the mutant and wild-type envelope proteins with or without the full-length cytoplasmic domain 48 h before labeling with 35S-cysteine/[35S]methionine for 4 h. The amino acid substitutions in the mutant envelope proteins expressed in the transfected cells are indicated by their representations in the single-letter amino acid code beneath the heading noting the altered residue. Analysis of the lysates from cells transfected with the plasmids encoding the wild-type Mo-MuLV envelope protein with the full-length cytoplasmic domain (w.t.) or without the full-length cytoplasmic domain (Δ650–665) and from cells that were mock transfected (−) are also presented. At left is indicated the positions of SU+TM (85 kDa) and SU (70 kDa). Immunoprecipitation and analysis by gel electrophoresis and autoradiography were conducted as described in Figure 3B. The mobility of the SU+TM of proteins with the cytoplasmic domain truncation is increased, whereas that of the mature SU is unaffected.
Mutant Envelope Proteins Are Able to Bind to the Cellular Receptor
To test the receptor-binding ability of the mutant envelope proteins, interference assays were performed using the stable clones of NIH 3T3 cells expressing the mutant env genes analyzed before (Figure 2). Retroviral interference or resistance to superinfection is the phenomenon whereby a cell infected by a retrovirus is resistant to infection by other viruses that use the same cellular receptor (Steck and Rubin, 1966a,b). This resistance to superinfection occurs when the endogenous envelope protein in the infected cell binds the cellular receptor and thus prevents exogenous virus from binding to that receptor.
Media from cells that produce replication-defective Mo-MuLV retrovirus that conveys nuclear-localized β-galactosidase expression on infected cells were incubated with the stable NIH 3T3 Env-expressing clones. The susceptibility of these cells to superinfection was measured by staining with X-gal, a synthetic substrate of β-galactosidase. A reduction in the number of X-gal-stained cells indicates that the expressed mutant Env is able to bind to the cellular receptor. The results suggest that the mutants are able to bind to the cellular receptor and prevent superinfection to varying degrees (Table 4). The extent of interference correlates with the level of envelope protein expression except for the F607V Env (Figure 2), indicating that none of the mutant Envs (besides F607V Env) has major defects in receptor binding. In particular, expression of the proline 617 substitution mutants leads to interference, indicating that they are not defective in receptor-binding activity. The low level of interference displayed by the W606S Env is consistent with its poor expression (Figure 2); higher expression would probably have led to syncytia formation (Figure 4C). It can be concluded that the Envs bearing substitutions for proline 617 have a specific defect in the promotion of the membrane fusion step of Mo-MuLV entry.
Table 4.
Cellular receptor binding by mutant Mo-MuLV envelope proteins
| Mutant envelope proteins | Interference |
|---|---|
| W606A | +++ |
| W606G | + |
| W606E | + |
| W606S | + |
| W606I | ++ |
| W606F | ++ |
| F607A | ++ |
| F607G | +++ |
| F607V | + |
| F607W | +++ |
| F607Y | +++ |
| G616A | +++ |
| G616V | +++ |
| G616P | ++ |
| P617A | +++ |
| P617G | +++ |
| P617V | +++ |
| NIH 3T3 cells not expressing envelope protein | − |
| Cells expressing wild-type envelope protein | +++ |
NIH 3T3 cells that were stably transfected with plasmids encoding the mutant envelope proteins were infected with virus produced by E86nlslacZ cells. Forty-eight hours after infection the cells were stained with X-gal, and the percentage of blue cells was calculated. The cells expressing the wild-type envelope protein were transduced at a level 13-fold lower than that of the NIH 3T3 cells.
DISCUSSION
The entry of the Mo-MuLV into a cell is conceptually constituted of a number of steps, each of which is likely to correspond to a stage occurring in cellular membrane fusion events. It is notable that the various phases of the process involve discrete domains of the envelope protein The binding of the virus to the cell, mediated by the amino-terminal domain of the SU protein (Heard and Danos, 1991; Battini et al., 1992; Ott and Rein, 1992), induces conformational changes, probably promoted by an SU-TM linkage-dissolving thiol-disulfide exchange reaction (Pinter et al., 1997) that lead to exposure of fusion peptides at the amino termini of α-helices of a trimeric coiled coil (Fass and Kim, 1995; Fass et al., 1996). Interaction of the fusion peptide with the cellular membrane leads to membrane fusion. The data presented here support the hypothesis that the membrane-spanning domain of the TM protein must be included as an important component in the membrane fusion event and that the role it plays is highly dependent on its amino acid sequence.
The function of conserved amino acid residues at the amino-terminal edge and at the center of the putative Mo-MuLV TM membrane-spanning domain has been examined. Envelope proteins bearing substitutions for glycine 616 have been demonstrated to possess close to wild-type capacities in all assays. This is contrary to a prediction made on the basis of studies of the role of glycine residues in the membrane-spanning domain of the VSV-Indiana G protein (Cleverley and Lenard, 1998). Substitutions of both of the glycine residues in the VSV G protein membrane-spanning domain with either alanine or leucine residues greatly reduced membrane fusion activity, although single substitutions had only small effects (Cleverley and Lenard, 1998), similar to those found with the Mo-MuLV TM mutants bearing substitutions for glycine 616.
Envelope proteins bearing substitutions for phenylalanine 607 possess essentially wild-type transduction and membrane fusion capacities with the exception of the F607V mutant. Whereas the F607V Env retained some capacity to bind to the receptor intracellularly, as measured in the interference assay, it appeared to be processed abnormally and thereby incapable of promoting membrane fusion or viral entry. These data suggest that, although a phenylalanine residue is not required at amino acid position 607, and smaller or similarly shaped residues can be accommodated there, the β-branched residue valine cannot. It is possible, therefore, that the phenylalanine is normally found within a narrow pocket into which the branched valine residue cannot fit, and that exposure of the valine leads to misfolding.
Substitutions for Tryptophan 606 and the Mechanism of Membrane Fusion Regulation
The role of tryptophan 606, which we predict is at the interface between the exterior domain of TM and the viral envelope, appears to be more complex. Envelope proteins bearing substitutions of alanine, glycine, isoleucine, or phenylalanine for tryptophan 606 possess essentially wild-type transduction and syncytia formation promotion capacities. Although the W606E Env exhibits nearly normal membrane fusion capabilities in the assay performed in the NIH 3T3 cells, it appears to be expressed at lower levels in the human 293T-derived cells and to be incorporated at decreased levels into virions with consequent reduced transduction titers (Figure 2). Perhaps the presence of a charged residue diminishes the stability of the W606E Env or its progress through the secretory system of the 293T-derived cells.
Most remarkable are the effects of the substitution of serine for tryptophan 606 on Env protein function. The W606S Env possesses moderately reduced capacity for transduction and displays slightly diminished syncytia formation promotion potential when assayed using a construct that deletes the carboxyl-terminal 16 amino acid residues of TM, resulting in the expression of a glycoprotein that resembles the mature Env protein in virus particles. It is of greatest interest, however, that expression of the W606S Env in NIH 3T3 cells, which possess the Mo-MuLV receptor, in a form containing the full-length wild-type cytoplasmic domain results in cell–cell fusion, although at reduced levels compared with those produced by the Env lacking the last 16 amino acid residues of TM (Table 2). This substitution is the first outside of the TM cytoplasmic domain (TMCD) that has been found to render the Mo-MuLV Env capable of causing syncytia formation when expressed in NIH 3T3 cells in the presence of a full-length cytoplasmic domain. None of the other mutant Envs described in this article displayed this capacity. It is noteworthy that substitutions in the extracellular domain of the human T-cell leukemia virus type 1 TM glycoprotein near the membrane-spanning domain have also been reported to enhance envelope protein-mediated membrane fusion, although extensive syncytia formation is observed even with expression of the wild-type human T-cell leukemia virus type 1, and the mutant Envs had reduced or abolished capacities to participate in infectious virus-particle formation (Rosenberg et al., 1997).
Various models of the means by which the cytoplasmic domain of the Mo-MuLV TM regulates fusion can be conceived. We propose that a structure in the TMCD regulates the stability of the oligomeric state of the envelope protein and thereby its capacity for the promotion of membrane fusion. In the presence of an intact TMCD the Env trimer is stabilized so that exposure of the fusion peptide upon receptor binding is disfavored. When the TMCD is disrupted through cleavage, truncation, or mutation, the trimer is destabilized so that the fusion peptide can be exposed upon receptor binding. In a sense, the regulation of fusion would involve the propagation of a conformational change across a membrane. The results obtained with the W606S Env are consistent with such a proposal. The existence of such a mutant Env with a substitution outside of the TMCD that promotes syncytia formation is predicted by our hypothesis. Perhaps the substitution counteracts the presence of the cytoplasmic domain by reducing trimer stability. One alternative conjecture is that the modified regulation of membrane fusion occurs through alterations of the conformation of the membrane-spanning domain, which itself may participate in intermonomer interactions and directly in membrane fusion.
Membrane-spanning Domain Sequences and the Promotion of Membrane Fusion
Proline 617, which is at the center of the membrane-spanning domain of the Mo-MuLV TM protein, plays an essential role in the promotion of membrane fusion. Substitutions of alanine, glycine, or valine residues for proline 617 either totally eliminate or reduce by 3 orders of magnitude transduction by viruses bearing the mutant Envs and also dramatically diminish syncytia formation by cells bearing the mutant Mo-MuLV Envs. Our other assays of function indicate that there is a specific defect in membrane fusion promotion. The membrane fusion deficiencies caused by substitutions for membrane-spanning domain proline residues seem to be restricted to those replacing proline 617, because substitution of a leucine residue for proline 629, which lies at the interface between the membrane and the cytoplasm (cysteine 630 is palmitoylated; Yang and Compans, 1996) has no major effect on Mo-MuLV Env-mediated transduction and syncytia formation (our unpublished results).
It is possible that substitutions for proline 617 affect the conformation of the TM membrane-spanning domain and thereby alter the interaction of the domain with the membrane. For example, the wild-type TM membrane-spanning domain may influence the curvature of the lipid bilayer and thereby promote membrane fusion. A consideration of the roles that proline residues play in membrane-spanning domains suggests an additional hypothesis.
Although proline residues are comparatively rarely found in the membrane-spanning domains of proteins that do not transport molecules across the membrane, they are commonly found in such domains in G-protein-coupled receptors, where they appear to play critical roles in the regulation of signal transduction (Konopka et al., 1996), and in transport proteins (Brandl and Deber, 1986). Proline residues introduce kinks and free backbone carbonyl oxygen atoms into membrane-integrating helices (Fox and Richards, 1982; Terwilliger et al., 1982) and have been predicted to be found predominantly on the hydrophilic side of pore-forming helices in ion channels (Woolfson et al., 1991). Substitution of an alanine residue for proline 14 of the lytic bee venom peptide melittin greatly reduces its capacity to induce voltage-dependent ion conductance in planar bilayers (Dempsey et al., 1991). Studies of analogues of the ion channel-forming fungal antibacterial peptide alamethicin indicate that the proline at position 14 is required for optimal channel activity (Kaduk et al., 1997) and is essential for hemolytic activity (Dathe et al., 1998).
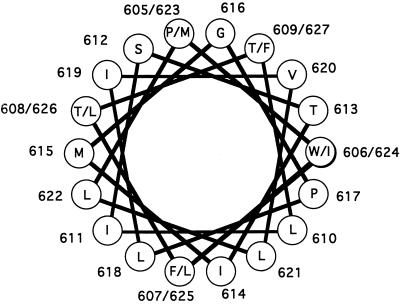
The Mo-MuLV TM membrane-spanning domain could form a kinked amphipathic α-helix (Figure 6) similar to those formed by melittin and alamethicin and presumably formed by ion channels. The hydoxyl amino acids would lie on one side of the helix, and the proline would be at the juncture of the polar and less polar faces of the helix. It is probable that the polar face participates in the association of monomers within the membrane, whereas the more hydrophobic face interacts with the membrane lipids (Dieckmann and DeGrado, 1997). An inference from a synthesis of all the foregoing data suggests that the role of proline 617 of the Mo-MuLV Env is to participate in the formation of the aqueous fusion pore that arises during membrane fusion.
Figure 6.
Helical wheel representation of the proposed α-helical membrane-spanning domain of Mo-MuLV. The helical wheel is a depiction of an ideal α-helix with 3.6 residues per turn and a repeating unit of 18 residues or five complete turns. The residues whose role was examined in this article are indicated by the outlined numbers and letters. Glycine 616 is on the polar face of the helix, whereas proline 617 is at the interface between the polar face and apolar face.
This concept is consistent with the results obtained in a particularly informative set of experiments in which it was demonstrated that when cells expressing an influenza virus hemagglutinin protein possessing a GPI linkage were mixed with cells containing sialic acid (the viral receptor) on their surface glycoproteins, mixing of the outer leaflets of the lipid bilayers was detectable at lowered pH, whereas the mixing of cellular contents was not achieved (Kemble et al., 1994; Melikyan et al., 1995). Mixing of both the outer leaflets and cellular contents results when the hemagglutinin possesses the wild-type membrane-spanning domain. These results would be expected if such mixing of the outer leaflets, referred to as “hemifusion,” were an intermediate in the process of protein-mediated membrane fusion, as has been proposed, and if the GPI-linked protein were competent for only part of the fusion process. It is plausible in this context that the P617A and P617G Mo-MuLV Envs promote hemifusion but do not promote completion of the process of membrane fusion.
Previously the issue of whether there were particular sequence requirements for the promotion of complete membrane fusion by a membrane-spanning domain or whether any peptide that crossed the membrane would suffice was incompletely resolved. The conservation of membrane-spanning domains within some families of homologous viral fusion proteins provides support for the concept that the actual sequences are important. The putative membrane-spanning domain of the HIV-1 Env possesses two conserved positively charged residues. Substitution of the first residue, lysine 683, with an isoleucine residue results in a major reduction in HIV-1 Env-mediated syncytia formation and the elimination of infectious virus formation without affecting envelope protein processing or incorporation into virions (Helseth et al., 1990). The substitution of the second residue, arginine 696, with a serine residue (Helseth et al., 1990) or an isoleucine residue (Odell et al., 1997) appears to have little effect on HIV-1 Env function, whereas substitution with a leucine residue (Owens et al., 1994) was reported to eliminate HIV-1 Env-mediated syncytia formation. It should be noted that the putative membrane-spanning domain encompassing both lysine 683 and arginine 696 could extend across 41 residues, whereas one initiating at the residue following lysine 683 could possess 23 residues (residues 707 and 709 are arginines), probably a sufficient length to span the membrane. As noted above, substitutions of both of the glycine residues in the VSV G protein membrane-spanning domain greatly reduced membrane fusion activity, although single substitutions had only small effects (Cleverley and Lenard, 1998). It was suggested that the mutant VSV G proteins bearing doubly substituted membrane-spanning domains were capable of promoting hemifusion but not complete membrane fusion (Cleverley and Lenard, 1998).
On the other hand, replacement of the VSV G protein or HIV-1 membrane-spanning domains with those of the cellular CD4 or CD22 protein, respectively, led to the retention of membrane fusion capacities (Wilk et al., 1996; Odell et al., 1997). In the HIV-1 Env-CD22 chimeric proteins the 22-amino acid-residue region beginning after lysine 683 was replaced with a 19-residue CD22 membrane-spanning domain (Wilk et al., 1996). Our data, in contrast to the foregoing, provide the first evidence that a hydrophobic amino acid residue within the membrane-spanning domain of a viral fusion protein plays a role directly in the process of membrane fusion.
A resolution to the paradox is possible. The large variation in the sequences of the membrane-spanning domains of viral fusion proteins from unrelated viruses indicates that many such sequences are compatible with the promotion of membrane fusion. Indeed, the membrane-spanning domains of some cellular proteins may fortuitously possess such sequences and membrane fusion-promoting properties but are inactive in their normal contexts. However, there are membrane-spanning sequences, such as the Mo-MuLV TM proteins bearing substitutions for proline 617, that are incompatible with the promotion of membrane fusion. It seems evident that during the course of evolution the viruses have specifically retained sequences that abet membrane fusion. This proposal does not exclude the possibility that a viral fusion protein membrane-spanning domain may have additional functions. It is also possible that viral fusion proteins may differ in their requirements for specific membrane-spanning domain sequences.
In summary, the present studies indicate that residues in or near the Mo-MuLV TM protein membrane-spanning domain play important roles in the promotion and regulation of membrane fusion. A conserved proline residue in the center of a putative kinked amphipathic membrane-spanning α-helix appears to play a critical role in the process of MoMuLV envelope protein-mediated membrane fusion. Future experiments should uncover the structural and biochemical bases for the function of the TM membrane-spanning domain and the applicability of the results to cellular and other viral fusion proteins.
ACKNOWLEDGMENTS
We thank Watjana Lilaonitkul for work in the construction of some of the plasmids used in this research and Yi Gao for development of the gpGFP cells. This work was supported by National Institutes of Health biophysics training grant GM08296, the Purdue Research Foundation, and the Leukemia Research Foundation. This article is dedicated to the memory of Marian E. Koshland.
Abbreviations used:
- DMEM CS/PS
Dulbecco’s modified Eagle’s media containing 10% calf serum, 0.1 mg/ml streptomycin, and 10 U/ml penicillin
- Env
envelope protein
- GFP
Aequorea victoria green fluorescent protein
- GPI
glycosylphosphatidylinositol
- HIV-1
human immunodeficiency virus type 1
- LTR
long terminal repeat
- Mo-MuLV
Moloney murine leukemia virus
- RIP
radioimmunoprecipitation
- SU
surface protein
- TM
transmembrane protein
- TMCD
transmembrane protein cytoplasmic domain
- VSV
vesicular stomatitis virus
- X-gal
5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside
REFERENCES
- Battini JL, Heard JM, Danos O. Receptor choice determinants in the envelope glycoproteins of amphotropic, xenotropic, and polytropic murine leukemia viruses. J Virol. 1992;66:1468–1475. doi: 10.1128/jvi.66.3.1468-1475.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedgood RM, Stallcup MR. A novel intermediate in processing of murine leukemia virus envelope glycoproteins. Proteolytic cleavage in the late Golgi region. J Biol Chem. 1992;267:7060–7065. [PubMed] [Google Scholar]
- Brandl CJ, Deber CM. Hypothesis about the function of membrane-buried proline residues in transport proteins. Proc Natl Acad Sci USA. 1986;83:917–921. doi: 10.1073/pnas.83.4.917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bullough PA, Hughson FM, Skehel JJ, Wiley DC. Structure of influenza hemagglutinin at the pH of membrane fusion. Nature. 1994;371:37–43. doi: 10.1038/371037a0. [DOI] [PubMed] [Google Scholar]
- Carr CM, Kim PS. A spring-loaded mechanism for the conformational change of influenza hemagglutinin. Cell. 1993;73:823–832. doi: 10.1016/0092-8674(93)90260-w. [DOI] [PubMed] [Google Scholar]
- Cleverley DZ, Lenard J. The transmembrane domain in viral fusion: essential role for a conserved glycine residue in vesicular stomatitis virus G protein. Proc Natl Acad Sci USA. 1998;95:3425–3430. doi: 10.1073/pnas.95.7.3425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford S, Goff SP. A deletion mutation in the 5′ part of the pol gene of murine leukemia virus blocks proteolytic processing of the gag and pol polyproteins. J Virol. 1985;53:899–907. doi: 10.1128/jvi.53.3.899-907.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danos O, Mulligan RC. Safe and efficient generation of recombinant retroviruses with amphotropic and ecotropic host ranges. Proc Natl Acad Sci USA. 1988;85:6460–6464. doi: 10.1073/pnas.85.17.6460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dathe M, Kaduk C, Tachikawa E, Melzig MF, Wenschuh H, Bienert M. Proline at position 14 of alamethicin is essential for hemolytic activity, catecholamine secretion from chromaffin cells and enhanced metabolic activity in endothelial cells. Biochim Biophys Acta. 1998;1370:175–183. doi: 10.1016/s0005-2736(97)00260-5. [DOI] [PubMed] [Google Scholar]
- Delassus S, Sonigo P, Wain HS. Genetic organization of gibbon ape leukemia virus. Virology. 1989;173:205–213. doi: 10.1016/0042-6822(89)90236-5. [DOI] [PubMed] [Google Scholar]
- Dempsey CE, Bazzo R, Harvey TS, Syperek I, Boheim G, Campbell ID. Contribution of proline 14 to the structure and actions of melittin. FEBS Lett. 1991;281:240–244. doi: 10.1016/0014-5793(91)80402-o. [DOI] [PubMed] [Google Scholar]
- Dieckmann GR, DeGrado WF. Modeling transmembrane helical oligomers. Curr Opin Struct Biol. 1997;7:486–494. doi: 10.1016/s0959-440x(97)80111-x. [DOI] [PubMed] [Google Scholar]
- Durrer P, Galli C, Hoenke S, Corti C, Gluck R, Vorherr T, Brunner J. H+-induced membrane insertion of influenza virus hemagglutinin involves the HA2 amino-terminal fusion peptide but not the coiled coil region. J Biol Chem. 1996;271:13417–13421. doi: 10.1074/jbc.271.23.13417. [DOI] [PubMed] [Google Scholar]
- Fass D, Harrison SC, Kim PS. Retrovirus envelope domain at 1.7 angstrom resolution. Nat Struct Biol. 1996;3:465–469. doi: 10.1038/nsb0596-465. [DOI] [PubMed] [Google Scholar]
- Fass D, Kim PS. Dissection of a retrovirus envelope protein reveals structural similarity to influenza hemagglutinin. Curr Biol. 1995;5:1377–1383. doi: 10.1016/s0960-9822(95)00275-2. [DOI] [PubMed] [Google Scholar]
- Fox RO, Jr, Richards FM. A voltage-gated ion channel model inferred from the crystal structure of alamethicin at 1.5-Å resolution. Nature. 1982;300:325–330. doi: 10.1038/300325a0. [DOI] [PubMed] [Google Scholar]
- Green N, Shinnick TM, Witte O, Ponticelli A, Sutcliffe JG, Lerner RA. Sequence-specific antibodies show that maturation of Moloney leukemia virus envelope polyprotein involves removal of a COOH-terminal peptide. Proc Natl Acad Sci USA. 1981;78:6023–6027. doi: 10.1073/pnas.78.10.6023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grignani F, Kinsella T, Mencarelli A, Valtieri M, Riganelli D, Grignani F, Lanfrancone L, Peschle C, Nolan GP, Pelicci PG. High-efficiency gene transfer and selection of human hematopoietic progenitor cells with a hybrid EBV/retroviral vector expressing the green fluorescence protein. Cancer Res. 1998;58:14–19. [PubMed] [Google Scholar]
- Guilhot S, Hampe A, D’Auriol L, Galibert F. Nucleotide sequence analysis of the LTRs and env genes of SM-FeSV and GA-FeSV. Virology. 1987;161:252–258. doi: 10.1016/0042-6822(87)90194-2. [DOI] [PubMed] [Google Scholar]
- Heard JM, Danos O. An amino-terminal fragment of the Friend murine leukemia virus envelope glycoprotein binds the ecotropic receptor. J Virol. 1991;65:4026–4032. doi: 10.1128/jvi.65.8.4026-4032.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helseth E, Olshevsky U, Gabuzda D, Ardman B, Haseltine W, Sodroski J. Changes in the transmembrane region of the human immunodeficiency virus type 1 gp41 envelope glycoprotein affect membrane fusion. J Virol. 1990;64:6314–6318. doi: 10.1128/jvi.64.12.6314-6318.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson LE, Sowder R, Copeland TD, Smythers G, Oroszlan S. Quantitative separation of murine leukemia virus proteins by reversed-phase high-pressure liquid chromatography reveals newly described gag and env cleavage products. J Virol. 1984;52:492–500. doi: 10.1128/jvi.52.2.492-500.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez LD, Hoffman LR, Wolfsberg TG, White JM. Virus-cell and cell-cell fusion. Annu Rev Cell Dev Biol. 1996;12:627–661. doi: 10.1146/annurev.cellbio.12.1.627. [DOI] [PubMed] [Google Scholar]
- Hunter E, Swanstrom R. Retrovirus envelope glycoproteins. Curr Top Microbiol Immunol. 1990;157:187–253. doi: 10.1007/978-3-642-75218-6_7. [DOI] [PubMed] [Google Scholar]
- Kaduk C, Duclohier H, Dathe M, Wenschuh H, Beyermann M, Molle G, Bienert M. Influence of proline position upon the ion channel activity of alamethicin. Biophys J. 1997;72:2151–2159. doi: 10.1016/S0006-3495(97)78858-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamps CA, Lin YC, Wong PK. Oligomerization and transport of the envelope protein of Moloney murine leukemia virus-TB and of ts1, a neurovirulent temperature-sensitive mutant of MoMuLV-TB. Virology. 1991;184:687–694. doi: 10.1016/0042-6822(91)90438-h. [DOI] [PubMed] [Google Scholar]
- Katoh I, Yoshinaka Y, Rein A, Shibuya M, Odaka T, Oroszlan S. Murine leukemia virus maturation: protease region required for conversion from “immature” to “mature” core form and for virus infectivity. Virology. 1985;145:280–292. doi: 10.1016/0042-6822(85)90161-8. [DOI] [PubMed] [Google Scholar]
- Kemble GW, Danieli T, White JM. Lipid-anchored influenza hemagglutinin promotes hemifusion, not complete fusion. Cell. 1994;76:383–391. doi: 10.1016/0092-8674(94)90344-1. [DOI] [PubMed] [Google Scholar]
- Konopka JB, Margarit SM, Dube P. Mutation of Pro-258 in transmembrane domain 6 constitutively activates the G protein-coupled alpha-factor receptor. Proc Natl Acad Sci USA. 1996;93:6764–6769. doi: 10.1073/pnas.93.13.6764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann R, Mulligan RC, Baltimore D. Construction of a retrovirus packaging mutant and its use to produce helper-free defective retrovirus. Cell. 1983;33:153–159. doi: 10.1016/0092-8674(83)90344-6. [DOI] [PubMed] [Google Scholar]
- Markowitz D, Goff S, Bank A. A safe packaging line for gene transfer: separating viral genes on two different plasmids. J Virol. 1988;62:1120–1124. doi: 10.1128/jvi.62.4.1120-1124.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melikyan GB, Chernomordik LV. Membrane rearrangements in fusion mediated by viral proteins. Trends Microbiol. 1997;5:349–355. doi: 10.1016/S0966-842X(97)01107-4. [DOI] [PubMed] [Google Scholar]
- Melikyan GB, White JM, Cohen FS. GPI-anchored influenza hemagglutinin induces hemifusion to both red blood cell and planar bilayer membranes. J Cell Biol. 1995;131:679–691. doi: 10.1083/jcb.131.3.679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgenstern JP, Land H. A series of mammalian expression vectors and characterization of their expression of a reporter gene in stably and transiently transfected cells. Nucleic Acids Res. 1990;18:1068. doi: 10.1093/nar/18.4.1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odell D, Wanas E, Yan J, Ghosh HP. Influence of membrane anchoring and cytoplasmic domains on the fusogenic activity of vesicular stomatitis virus glycoprotein G. J Virol. 1997;71:7996–8000. doi: 10.1128/jvi.71.10.7996-8000.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ory DS, Neugeboren BA, Mulligan RC. A stable human-derived packaging cell line for production of high titer retrovirus/vesicular stomatitis virus G pseudotypes. Proc Natl Acad Sci USA. 1996;93:11400–11406. doi: 10.1073/pnas.93.21.11400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ott D, Friedrich R, Rein A. Sequence analysis of amphotropic and 10A1 murine leukemia viruses: close relationship to mink cell focus-inducing viruses. J Virol. 1990;64:757–766. doi: 10.1128/jvi.64.2.757-766.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ott D, Rein A. Basis for receptor specificity of nonecotropic murine leukemia virus surface glycoprotein gp70SU. J Virol. 1992;66:4632–4638. doi: 10.1128/jvi.66.8.4632-4638.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owens RJ, Burke C, Rose JK. Mutations in the membrane-spanning domain of the human immunodeficiency virus envelope glycoprotein that affect fusion activity. J Virol. 1994;68:570–574. doi: 10.1128/jvi.68.1.570-574.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pear WS, Nolan GP, Scott ML, Baltimore D. Production of high-titer helper-free retroviruses by transient transfection. Proc Natl Acad Sci USA. 1993;90:8392–8396. doi: 10.1073/pnas.90.18.8392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinter A, Fleissner E. The presence of disulfide-linked gp70–p15(E) complexes in AKR murine leukemia virus. Virology. 1977;83:417–422. doi: 10.1016/0042-6822(77)90187-8. [DOI] [PubMed] [Google Scholar]
- Pinter A, Honnen WJ. Topography of murine leukemia virus envelope proteins: characterization of transmembrane components. J Virol. 1983;46:1056–1060. doi: 10.1128/jvi.46.3.1056-1060.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinter A, Kopelman R, Li Z, Kayman SC, Sanders DA. Localization of the labile disulfide bond between SU and TM of the murine leukemia virus envelope protein complex to a highly conserved CWLC motif in SU that resembles the active site sequence of thiol-disulfide exchange enzymes. J Virol. 1997;71:8073–8077. doi: 10.1128/jvi.71.10.8073-8077.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinter A, Lieman-Hurwitz J, Fleissner E. The nature of the association between the murine leukemia virus envelope proteins. Virology. 1978;91:345–351. doi: 10.1016/0042-6822(78)90382-3. [DOI] [PubMed] [Google Scholar]
- Ragheb JA, Anderson WF. Uncoupled expression of Moloney murine leukemia virus envelope polypeptides SU and TM: a functional analysis of the role of TM domains in viral entry. J Virol. 1994a;68:3207–3219. doi: 10.1128/jvi.68.5.3207-3219.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ragheb JA, Anderson WF. pH-independent murine leukemia virus ecotropic envelope-mediated cell fusion: implications for the role of the R peptide and p12E TM in viral entry. J Virol. 1994b;68:3220–3231. doi: 10.1128/jvi.68.5.3220-3231.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rein A, Mirro J, Haynes JG, Ernst SM, Nagashima K. Function of the cytoplasmic domain of a retrovrial transmembrane protein: p15E-p2E cleavage activates the membrane fusion capability of the murine leukemia virus Env protein. J Virol. 1994;68:1773–1781. doi: 10.1128/jvi.68.3.1773-1781.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg AR, Delamarre L, Pique C, Pham D, Dokhelar MC. The ectodomain of the human T-cell leukemia virus type 1 TM glycoprotein is involved in postfusion events. J Virol. 1997;71:7180–7186. doi: 10.1128/jvi.71.10.7180-7186.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanes JR, Rubenstein JL, Nicolas JF. Use of a recombinant retrovirus to study postimplantation cell lineage in mouse embryos. EMBO J. 1986;5:3133–3142. doi: 10.1002/j.1460-2075.1986.tb04620.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz A, Rein A. Maturation of murine leukemia virus env proteins in the absence of other viral proteins. Virology. 1985;145:335–339. doi: 10.1016/0042-6822(85)90168-0. [DOI] [PubMed] [Google Scholar]
- Shinnick TM, Lerner RA, Sutcliffe JG. Nucleotide sequence of Moloney murine leukemia virus. Nature. 1981;293:543–548. doi: 10.1038/293543a0. [DOI] [PubMed] [Google Scholar]
- Steck FT, Rubin H. The mechanism of interference between an avian leukosis virus and Rous sarcoma virus. I. Establishment of interference. Virology. 1966a;29:628–641. doi: 10.1016/0042-6822(66)90287-x. [DOI] [PubMed] [Google Scholar]
- Steck FT, Rubin H. The mechanism of interference between an avian leukosis virus and Rous sarcoma virus. II. Early steps of infection by RSV of cells under conditions of interference. Virology. 1966b;29:642–653. doi: 10.1016/0042-6822(66)90288-1. [DOI] [PubMed] [Google Scholar]
- Sutcliffe JG, Shinnick TM, Green N, Liu FT, Niman HL, Lerner RA. Chemical synthesis of a polypeptide predicted from nucleotide sequence allows detection of a new retroviral gene product. Nature. 1980;287:801–805. doi: 10.1038/287801a0. [DOI] [PubMed] [Google Scholar]
- Terwilliger TC, Weissman L, Eisenberg D. The structure of melittin in the form I crystals and its implication for melittin’s lytic and surface activities. Biophys J. 1982;37:353–361. doi: 10.1016/S0006-3495(82)84683-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss CD, White JM. Characterization of stable Chinese hamster ovary cells expressing wild-type, secreted, and glycosylphosphatidylinositol-anchored human immunodeficiency virus type 1 envelope glycoprotein. J Virol. 1993;67:7060–7066. doi: 10.1128/jvi.67.12.7060-7066.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilk T, Pfeiffer T, Bukovsky A, Moldenhauer G, Bosch V. Glycoprotein incorporation and HIV-1 infectivity despite exchange of the gp160 membrane-spanning domain. Virology. 1996;218:269–274. doi: 10.1006/viro.1996.0190. [DOI] [PubMed] [Google Scholar]
- Witte ON, Tsukamoto AA, Weissman IL. Cellular maturation of oncornavirus glycoproteins: topological arrangement of precursor and product forms in cellular membranes. Virology. 1977;76:539–553. doi: 10.1016/0042-6822(77)90236-7. [DOI] [PubMed] [Google Scholar]
- Woolfson DN, Mortishire-Smith RJ, Williams DH. Conserved positioning of proline residues in membrane-spanning helices of ion-channel proteins. Biochem Biophys Res Commun. 1991;175:733–737. doi: 10.1016/0006-291x(91)91627-o. [DOI] [PubMed] [Google Scholar]
- Yang C, Compans RW. Palmitoylation of the murine leukemia virus envelope glycoprotein transmembrane subunits. Virology. 1996;221:87–97. doi: 10.1006/viro.1996.0355. [DOI] [PubMed] [Google Scholar]