Abstract
We have identified a novel Ras-interacting protein from Dictyostelium, RIP3, whose function is required for both chemotaxis and the synthesis and relay of the cyclic AMP (cAMP) chemoattractant signal. rip3 null cells are unable to aggregate and lack receptor activation of adenylyl cyclase but are able, in response to cAMP, to induce aggregation-stage, postaggregative, and cell-type-specific gene expression in suspension culture. In addition, rip3 null cells are unable to properly polarize in a cAMP gradient and chemotaxis is highly impaired. We demonstrate that cAMP stimulation of guanylyl cyclase, which is required for chemotaxis, is reduced ∼60% in rip3 null cells. This reduced activation of guanylyl cyclase may account, in part, for the defect in chemotaxis. When cells are pulsed with cAMP for 5 h to mimic the endogenous cAMP oscillations that occur in wild-type strains, the cells will form aggregates, most of which, however, arrest at the mound stage. Unlike the response seen in wild-type strains, the rip3 null cell aggregates that form under these experimental conditions are very small, which is probably due to the rip3 null cell chemotaxis defect. Many of the phenotypes of the rip3 null cell, including the inability to activate adenylyl cyclase in response to cAMP and defects in chemotaxis, are very similar to those of strains carrying a disruption of the gene encoding the putative Ras exchange factor AleA. We demonstrate that aleA null cells also exhibit a defect in cAMP-mediated activation of guanylyl cyclase similar to that of rip3 null cells. A double-knockout mutant (rip3/aleA null cells) exhibits a further reduction in receptor activation of guanylyl cyclase, and these cells display almost no cell polarization or movement in cAMP gradients. As RIP3 preferentially interacts with an activated form of the Dictyostelium Ras protein RasG, which itself is important for cell movement, we propose that RIP3 and AleA are components of a Ras-regulated pathway involved in integrating chemotaxis and signal relay pathways that are essential for aggregation.
INTRODUCTION
Dictyostelium is an excellent experimental system in which to examine the signaling pathways that control chemotaxis because of the availability of genetic and biochemical approaches as well as physiological assays to study mutant phenotypes (Firtel, 1995; Van Haastert, 1995; Chen et al., 1996; Parent and Devreotes, 1996; Chung and Firtel, 1999). During the early phase of development, up to 105 cells chemotactically aggregate to form a multicellular organism in response to the chemoattractant cyclic AMP (cAMP) emitted from cells. This process requires the coordinated regulation of pathways that control the activation of adenylyl cyclase and the relay of the cAMP signal, chemotaxis toward cAMP, the activation of guanylyl cyclase and the Dictyostelium homologue of mammalian Akt/PKB required for chemotaxis, and the activation of the expression of genes required for this process (including the receptor, Gα subunit, and cell adhesion molecules) (Firtel, 1995; Van Haastert, 1995; Chen et al., 1996; Parent and Devreotes, 1996; Meili et al., 1999). Activation of Dictyostelium Akt/PKB requires the function of the PI3 kinases PI3K1 and PI3K2, which are related to mammalian p110α PI3K (Zhou et al., 1995; Meili et al., 1999). All of these pathways are regulated through the cell surface serpentine cAMP receptor cAR1 and the coupled G protein containing the Gα2 subunit. Disruption of genes encoding the cAMP receptor, the sole Gβ subunit, or the Gα2 subunit results in complete abrogation of all four pathways (Firtel, 1995; Van Haastert, 1995; Chen et al., 1996; Parent and Devreotes, 1996; Meili et al., 1999).
Activation of the cAMP receptor by extracellular cAMP results in stimulation of the activity of the adenylyl cyclase ACA, which has a molecular architecture similar to that of mammalian adenylyl cyclases (Pitt et al., 1992). The activation is mediated by the Gβγ subunit and requires cytosolic proteins including CRAC, a pleckstrin homology domain containing protein that translocates to the plasma membrane in response to receptor activation, and Pianissimo (Lilly et al., 1993; Insall et al., 1994b; Lilly and Devreotes, 1995; Chen et al., 1997). In addition, activation of adenylyl cyclase and cAMP accumulation requires the function of the putative Ras exchange factor (GEF) Aimless (AleA) and the MAPK ERK2 (Segall et al., 1995; Insall et al., 1996). The control of chemotaxis is even more complex; it requires the coordinated regulation of the actin cytoskeleton, the function of myosin II, and the unconventional myosins IB and IC (Schleicher and Noegel, 1992; Peterson et al., 1995; Chen et al., 1996; Wessels et al., 1996; Uyeda and Titus, 1997; Zigmond et al., 1997). One of the second messengers required for chemotaxis is cyclic GMP (cGMP), which is thought to function, in part, through a cGMP-dependent protein kinase to activate myosin II kinase (Dembinsky et al., 1996; Van Haastert and Kuwayama, 1997). Guanylyl cyclase activity is very rapidly and transiently stimulated in response to chemoattractants and requires a MAPK pathway distinct from that containing the MAPK ERK2 (Van Haastert and Van Lookeren Campagne, 1984; Ma et al., 1997; Van Haastert and Kuwayama, 1997). MEK1, the cloned component of this pathway, is also required for the reorganization of the actin cytoskeleton, which may be dependent on MEK1’s role in regulating guanylyl cyclase activation (H. Ma and R.A. Firtel, unpublished data). Another signaling pathway needed for proper chemotaxis involves the Dictyostelium homologue of mammalian Akt/PKB that requires the function of the PI3 kinases PI3K1 and PI3K2, which are related to mammalian p110α PI3K (Meili et al., 1999).
In addition to its control of the activation of adenylyl cyclase, the putative Ras GEF AleA is required for proper chemotaxis (Insall et al., 1996). aleA null cells are unable to chemotax effectively or properly polarize in a chemoattractant gradient. The cells produce pseudopodia along the perimeter of the cell, in contrast to wild-type cells, in which a single, predominant pseudopod is extended at the leading edge in the direction of the chemoattractant signal. Dictyostelium has five known Ras proteins, two of which (RasD and RasG) are most closely related to their metazoan counterparts (Reymond et al., 1984; Pawson et al., 1985; Esch et al., 1993). RasG is preferentially expressed during early growth and development, whereas RasD is maximally expressed during the multicellular stages. Disruption of RasD does not exhibit growth or aggregation-stage defects (R. Insall, unpublished data). Cells expressing constitutively active RasD exhibit aberrant morphogenesis after the multicellular aggregate is formed and no abnormal phenotypes during aggregation (Reymond et al., 1986). rasG null cells, on the other hand, exhibit defects in cytokinesis and general cell movement (Tuxworth et al., 1997). In contrast to the cell movement defect of aleA null cells, which is observed only during chemotaxis, the rasG null cell movement defect is observed for randomly moving cells.
To identify proteins that may interact with different Dictyostelium Ras proteins, we undertook a two-hybrid screen using the activated form of mammalian Ha-Ras (Ha-RasG12V) as the bait. The logic was that using mammalian Ha-Ras rather than each individual Dictyostelium Ras protein might identify a broad spectrum of interacting proteins whose function may regulate pathways similar to those regulated by mammalian Ras proteins. The initial two-hybrid screen identified three Ha-Ras–interacting proteins, RIP1, RIP2, and RIP3 (Lee et al., 1997). RIP2 (RasGAP1) is related to mammalian IQGAPs and is required for proper cytokinesis during vegetative growth and for morphogenesis during multicellular development. This paper describes the analysis of RIP3, which we show preferentially interacts with the Dictyostelium RasG protein in a two-hybrid system. rip3 null cells have phenotypes very similar to those of aleA null cells; they are unable to activate adenylyl cyclase in response to a cAMP signal and have an impaired ability to chemotax. A rip3/aleA double-knockout strain exhibits a complete impairment in the ability to chemotax in a cAMP gradient, suggesting that both genes may regulate Ras-dependent pathways essential for chemotaxis. Our results suggest that RIP3 is a component of the Ras regulatory network that is required for signal relay and proper chemotaxis. Our findings suggest that RIP3 functions with AleA and possibly RasG in coordinating two essential functions during aggregation: chemotaxis and signal relay.
MATERIALS AND METHODS
Cloning of RIP3 Genomic Clone
Part of the RIP3 genomic clone encoding the amino-terminal two-thirds of the RIP3 open reading frame (ORF) and ∼2.0 kilobase (kb) of the 5′ regulatory sequence were cloned by construction of a mini genomic library. This domain of the RIP3 gene was mapped to ∼5 kb of an NdeI/BglII fragment. Dictyostelium DNA was digested with these two restriction enzymes and run on an agarose gel. The region between 4 and 6 kb was excised and cloned into a Bluescript II vector constructed to carry NdeI and BamHI restriction sites. The ligated DNA was transformed into Escherichia coli, and colonies were screened using a PCR-amplified hybridization probe from nucleotides 1880–2210 of the RIP3 ORF. Positive clones were picked and analyzed by DNA sequencing to ensure that the 3′ portion of the RIP3 gene was contained in the plasmids.
Construction of the rip3 Null Strain
To construct a rip3 null strain, the Bsr selectable marker cassette was cloned into the BamHI restriction site at nucleotide 1492 of the ORF. The RIP3 vector carrying the Bsr cassette was digested with SpeI and EcoRV restriction endonucleases, and the DNA was electroporated into wild-type KAx-3 cells selecting for Bsr-resistant clones after selection for 5 d in Bsr-containing medium. Rapid transformants were plated clonally, and random clones were picked and screened by PCR and Southern blot hybridization to identify Dictyostelium clones in which the RIP3 gene was disrupted with the Bsr cassette. There is a one-to-one correlation between the rip3 null aggregation-deficient phenotype and those clones carrying a rip3 gene disruption. Several independently derived clones were analyzed for various developmental properties. After demonstrating that all have a similar developmental phenotype and the inability to chemotax, indicating that expression of the RIP3 gene complements the null phenotype, a single clone was used for all subsequent experiments.
Activation of Adenylyl and Guanylyl Cyclases
Adenylyl cyclase assays were performed as previously described by Devreotes et al. (1987). Briefly, cells were starved with pulses of cAMP for 5 h and treated with caffeine and vigorous shaking for 30 min at room temperature. Caffeine inhibits adenylyl cyclase and is used to bring the cells to a basal level of enzyme activity. Cells were washed twice and resuspended at 8 × 107 cells/ml in 5 mM Na2HPO4, 5 mM NaH2PO4, pH 6.2, and 2 mM MgSO4. Receptor-mediated activation was performed by stimulating cells with 10 μM cAMP. At specific time points, cells were lysed and assayed for 2 min at room temperature. For guanidine thiotriphosphate (GTPγS)-mediated activation, the cells were lysed in the presence or absence of 40 μM GTPγS and 1 μM cAMP, incubated on ice for 4 min, and assayed for 2 min.
To measure cGMP production in response to cAMP stimulation, cells were prepared, stimulated, and assayed as described previously (Van Haastert and Van der Heijden, 1983; Ma et al., 1997). Samples (100 μl) were taken at appropriate intervals and processed using the cGMP 3H assay system (Amersham, Arlington Heights, IL) following the manufacturer’s instructions. The assay of each strain was independently repeated at least three times. All mutant strains were assayed along with wild-type cells as a direct comparison and as a control that the activation response was normal. Results of a representative experiment are shown.
Video Imaging and Chemotaxis Assays
The video imaging and chemotaxis assays were performed as previously described (Ma et al., 1997; Meili et al., 1999). Briefly, log-phase vegetative cells were washed three times with Na/K phosphate buffer and resuspended at a density of 2 × 106 to 3 × 106 cells/ml in Na/K phosphate buffer and pulsed with 30 nM cAMP for 5 h at 6-min intervals. Cells were washed and resuspended in Na/KPO4 buffer. Cells were plated in Na/K phosphate buffer at a density of 6 × 104 cells/cm2 onto a plate with a hole covered by a 0.17-mm glass coverslip and allowed to adhere to the surface for ∼30 min. With an Eppendorf Patchman micromanipulator, a glass capillary needle (Eppendorf Femtotip) filled with a 150 μM cAMP solution was brought into the field of view of an inverted microscope. The response was recorded using time-lapse video and NIH Image software, and the images were recorded directly on a computer hard drive.
For phase-contrast video microscopy, log-phase cells were washed and plated on a 60-mm Petri dish containing a thin agar layer. Cells were recorded using a 4× phase objective. The movies were recorded using a S-VHS time-lapse videotape recorded with a CCD72S video camera (DAGE MTI, Michigan City, IN) using a Nikon (Garden City, NY) Optiphot-2 microscope and a 4× phase contrast lens. Individual frames were captured into an image-processing program (NIH Image) with the help of a SCION frame grabber board.
Visible light images of chemotaxing cells were taken with a Nikon Eclipse TE 300 inverted microscope equipped for differential interference contrast imaging with a Plan Fluor ELWD 20×/0.45 or a Plan Fluor ELWD 40×/0.60 lens. Individual frames were captured from a CCD72S video camera into an image-processing program (NIH Image) with the help of a SCION frame grabber board as described by Meili et al. (1999).
RESULTS
Identification of RIP3
RIP3 was identified in a two-hybrid screen using an activated form of mammalian Ha-Ras (Ha-RasG12V) as a bait (Lee et al., 1997). Two inserts were identified of 1111 and 746 base pairs derived from the 3′ end of the RIP3 gene (Figure 1A). (One insert was identified three times, the other once.) The insert from the yeast two-hybrid clone was used to screen a Dictyostelium cDNA library that yielded two partial cDNAs. The longest cDNA was used as a probe in a Southern blot analysis to map RIP3 to genomic DNA fragments to identify restriction sites to make a mini Dictyostelium genomic library. These results were then used to clone the remainder of the ORF and an ∼2.0-kb upstream sequence containing the RIP3 promoter (see MATERIALS AND METHODS). Visual examination of the open reading frame derived from the genomic and cDNA clones shows polyglutamine stretches and other regions with highly reiterated polyamino acid sequences (Figure 1A). (The accession number for RIP3 is AF159241.) Although RIP3 has a higher fraction of such sequences than most Dictyostelium genes, such regions have been observed in numerous Dictyostelium genes and most are not thought to play a role in the function of the protein (Burki et al., 1991; Mann and Firtel, 1991; Pitt et al., 1992). A BLAST search did not suggest that RIP3 is highly homologous to known proteins. It did, however, identify a 36-amino acid nonsimple sequence region of RIP3 with homology to a region of a mammalian protein (clone JC310, GenBank number C38637) that was identified in a screen for mammalian proteins that suppress activated Ras function in the yeast Saccharomyces cerevisiae (Figure 1B; Colicelli et al., 1991). As the two two-hybrid clones do not contain this region of homology, it cannot be the region that is responsible for the interaction with Ha-Ras in the two-hybrid screen.
Figure 1.
Sequence of RIP3. (A) The derived amino acid sequence of RIP3. The shaded areas contain a repetitive amino acid sequence that is often found in Dictyostelium ORFs and that is not thought to have an important function in the protein. RIP3 has a higher fraction of this sequence than other identified proteins. The domain that is homologous to the mammalian protein cloned, JC310, which was identified in a screen for mammalian genes that inhibit activated Ras function in yeast, is underlined. The two arrows mark the beginning of the ORFs of two different two-hybrid clones that were identified in our screen using activated mammalian Ha-Ras as bait. (B) Comparison of the amino acid homology between RIP3 and mammalian ORF JC310.
RIP3 Is a RasG-interacting Protein
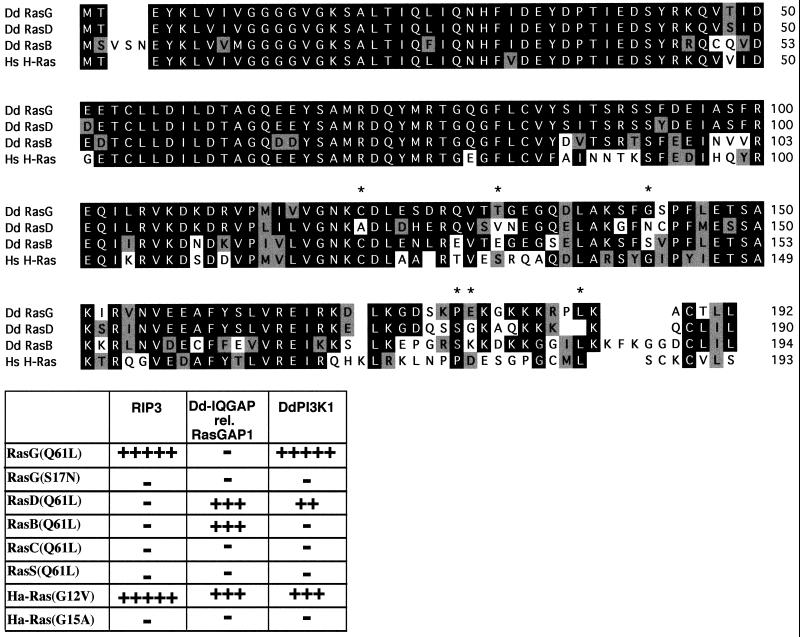
To examine whether RIP3 interacts with Dictyostelium Ras proteins, we performed a two-hybrid assay using wild-type and activated forms of the five known Dictyostelium Ras proteins and activated Ha-Ras. The Dictyostelium RasG and RasD proteins are very homologous to each other, having only three conserved amino acid sequence changes in the first 110 amino acids (S/T, D/E, Y/F) and the Ras proteins most homologous to human Ha-Ras (Figure 2A; Dictyostelium RasB is included in the comparison) (Reymond et al., 1984; Pawson et al., 1985; Esch et al., 1993). Dictyostelium RasG and RasD and human Ha-Ras show weaker homology in the last 40% of the protein. As shown in Figure 2B, RIP3 preferentially interacts with the Dictyostelium Ras protein RasGQ61L and only very weakly interacts with RasDQ61L. RIP3 exhibits strong interaction with activated human Ha-Ras (Ha-RasG12V), as expected from its isolation in a two-hybrid screen using activated Ha-RasG12V as the bait. RIP3 does not interact with the dominant negative form of Ha-Ras (Ha-RasG15A) or RasG (RasGS17N). In contrast, the domain of the Dictyostelium IQGAP-related gene DdRasGAP1 that interacts with Ha-RasG12V, RasBQ61L, and RasDQ61L showed no interaction with RasGQ61L (S. Lee and R.A. Firtel, unpublished data). The Dictyostelium PI3 kinase PI3K1 (Zhou et al., 1995) has a Ras-interacting domain that is related to the Ras-interacting domain of the mammalian family of p110 PI3 kinases (C. Ellsworth, S. Lee, T.B.K. Reddy, and R.A. Firtel, unpublished data). This domain interacts strongly with RasGQ61L, only weakly with RasDQ61L, and not at all with RasBQ61L. These control experiments indicate that our observation that RIP3 interacts with RasG and not any of the other four Dictyostelium Ras proteins tested in these experiments is presumably not an expression artifact of the yeast two-hybrid system or the inability of the yeast two-hybrid system to demonstrate interaction of RasB and RasD with other Dictyostelium proteins. The difference in interaction between RasG and Ha-Ras with RIP3 compared with the lack of interaction with RasD was unexpected considering the level of homology of the proteins. Although we do not know which residues are responsible for these interaction differences in the two-hybrid system, there are six residues (marked with asterisks in Figure 2A) in the terminal ∼40% of the protein that are highly conserved between Ha-Ras and RasG but not RasD.
Figure 2.
Interaction of Dictyostelium RIP3 with different Ras proteins. (A) Amino acid sequence comparison of Dictyostelium RasG, RasD, and RasB and human Ha-Ras. The asterisks indicate positions of conservation between RasG and Ha-Ras but not RasD. (B) The carboxyl-terminal region of RIP3 that was identified and cloned in two-hybrid screens using mammalian Ha-RasG12V as bait was used in two-hybrid assays to examine interaction of this domain with the activated form of the five identified Dictyostelium Ras proteins (RasG, RasD, RasB, RasC, and RasS). In addition, the dominant negative or nonactivatable form of RasG (RasGS17N) and interaction with Ha-RasG12V and Ha-RasG15A is shown. As controls, the interactions of the previously identified Dictyostelium IQGAP and the related gene RasGAPa as well as the Ras-interacting domain of Dictyostelium P110-related PI3 kinase DdPI3K1 are shown. The level of two-hybrid interactions was quantitated by the level of β-galactosidase production and the intensity of blue staining of yeast colonies.
RIP3 Is Developmentally Regulated and Required for Aggregation
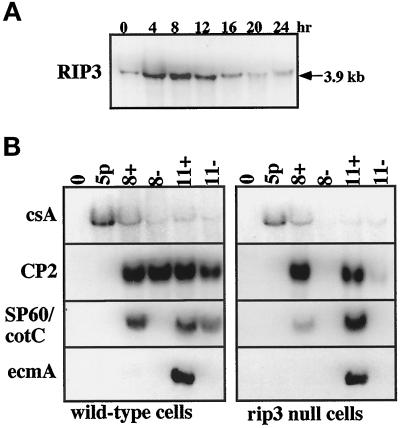
To examine the expression pattern of RIP3, a developmental RNA blot was made using RNA isolated from various stages in Dictyostelium development. As shown in Figure 3A, RIP3 is expressed at low levels in vegetative cells. The expression pattern is maximal during aggregation (4–8 h) and decreases thereafter.
Figure 3.
Developmental kinetics of RIP3 gene expression and the requirement of RIP3 for expression of aggregation and postaggregative genes. (A) RNA blot analysis of the expression pattern of RIP3 using RNA isolated at different stages of Dictyostelium development. Mound formation occurs at ∼8 h and culmination initiates at ∼18–20 h. (B) Expression of aggregation-stage, postaggregative, and cell-type-specific genes in wild-type and rip3 null cells in suspension culture in response to cAMP. Cells were pulsed for 5 h with 30 nM cAMP (5p). Cells were given cAMP under slow-shake suspension conditions, which allow cell-cell interactions (120 rpm), to 300 μM and were supplemented to 300 μM every 2 h. 8+ and 11+ represent time points (hours) after the initiation of the experiment. The plus sign represents the addition of exogenous cAMP. Minus cultures (8− and 11−) did not receive exogenous cAMP. Cultures were split after 5 h of cAMP pulsing. csA (Contact Sites A) is a marker for aggregation-stage gene expression; CP2 is a marker for postaggregative gene expression. SP60/CotC is a prespore-specific gene; ecmA is a prestalk-specific gene.
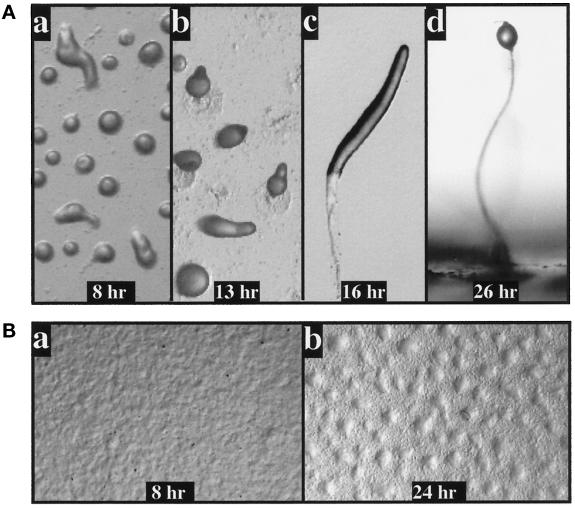
To examine the function of RIP3, the gene was disrupted by homologous recombination as described in MATERIALS AND METHODS. Clones were picked randomly, and disruption of the RIP3 gene was confirmed by Southern blot analysis. There was a one-to-one correlation between the rip3 null phenotype and disruption of the RIP3 gene by Southern blot analysis (our unpublished results). Wild-type cells form aggregates by 8 h, and by 13 h a tip forms (Figure 4A, a and b). The tip elongates to form a standing finger, which falls over, becoming a migrating slug or pseudoplasmodium (Figure 4Ac). As shown in Figure 4B, rip3 null cells are unable to aggregate, producing only some rippling at 8 h (Figure 4Ba). At 24 h, there is some accumulation of cells into very loose, diffuse mounds (Figure 4Bb); however, the majority of the cells exhibit little sign of aggregation, and development does not proceed further. This phenotype is similar to that observed in aleA null cells (Insall et al., 1996; our unpublished results). Expression of the RIP3 ORF from the cloned RIP3 promoter complements the rip3 null phenotype (our unpublished results). Expression of the construct in wild-type cells, which leads to a high overexpression of RIP3 transcripts, does not cause an observable aggregation or developmental phenotype (our unpublished results).
Figure 4.
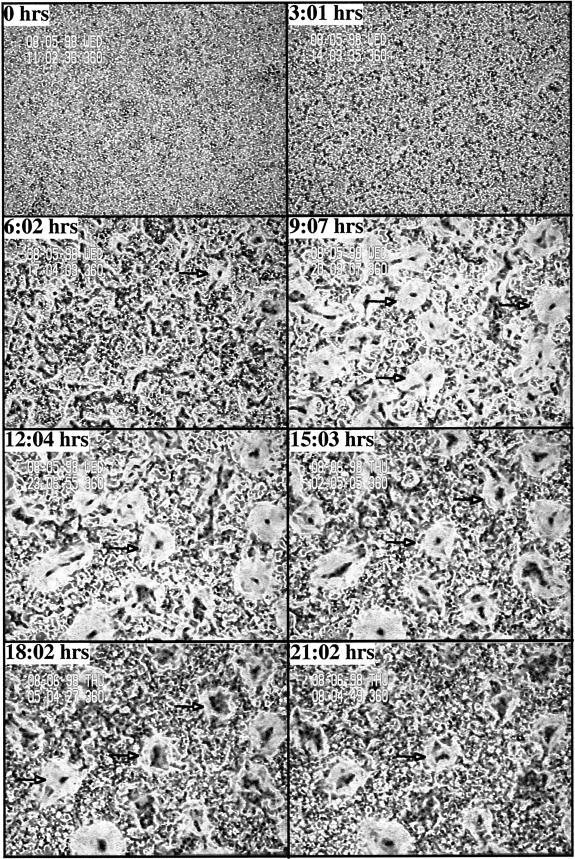
Developmental phenotypes of wild-type and rip3 null cells. (A) The developmental phenotypes of wild-type KAx-3 cells. Mound formation is visible at 8 h. At 13 h, most of the aggregates have formed a tip, and at 16 h, the cells have formed a migrating slug or pseudoplasmodium. Cells have culminated by 24–26 h. (B) Developmental phenotype of rip3 null cells. At 8 h, the cells exhibit a small amount of rippling. At 24 h, very loose cellular associations are observed. Half or less of the cells are associated with the aggregates. Most cells show no sign of participation in aggregate formation.
During aggregation, Dictyostelium cells respond to nanomolar oscillatory pulses of cAMP to induce the expression of genes required for this process (Gerisch, 1968; Noegel et al., 1986; Mann and Firtel, 1987; Firtel, 1995). These genes include the cAMP receptor cAR1, the coupled Gα subunit Gα2, and the cell adhesion molecule Contact Sites A (csA) (Noegel et al., 1986; Mann et al., 1988; Kumagai et al., 1989; Saxe et al., 1991). These genes are induced in wild-type strains during aggregation, with expression peaking at 4–8 h of development or in shaking culture in response to cAMP pulsing (Firtel, 1995; Ginsburg et al., 1995). When rip3 null cells are plated for development, no csA expression is detected (our unpublished results). We examined whether the inability of rip3 null cells to aggregate might be caused by an inability of the cells to respond to cAMP and activate aggregation-stage gene expression or whether the cells can be induced if pulsed with exogenous cAMP. As shown in Figure 3B, csA mRNA is normally expressed in rip3 null cells in response to cAMP signaling, suggesting that the aggregation defect is not due to an inability to induce aggregation-stage gene expression.
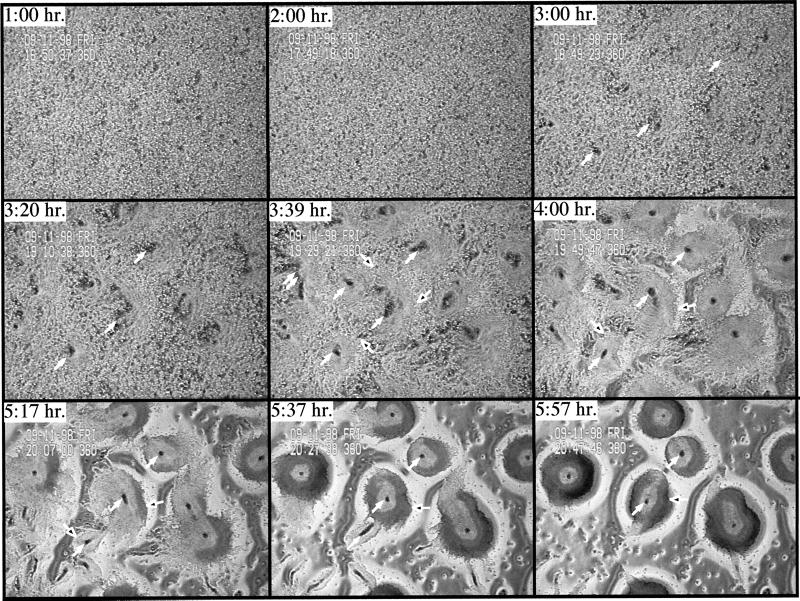
The aggregation defects of rip3 null cells were examined in more detail using time-lapse video microscopy (Ma et al., 1997). In wild-type cells, aggregation centers and waves of cAMP are visualized as changes in the conformation of cells, resulting in lighter and darker regions within the field of cells. For wild-type cells, these patterns are first seen by 3 h, 40 min after plating (Figure 5A). Aggregation domains become defined shortly thereafter (Figure 5A). The initial stages of chemotaxis, as seen by the movement of cells toward the centers, are visible by 4 h, 20 min. Aggregates are formed by 6 h. The phase contrast patterns observed for rip3 null cells are quite different. Wave patterns within the field of rip3 null cells observed on the videotapes are very limited (our unpublished results), and the initial formation of aggregation centers is delayed until 6 h (Figure 5B). By 9 h, multiple aggregation centers are visible. In contrast to wild-type cells, the rip3 null cell aggregation domains are quite small, suggesting an impairment in the ability to activate and relay the cAMP signaling pathway and/or to demonstrate cell shape changes in response to the cAMP. In addition, half of the rip3 null cell aggregation domains form and then disperse after a few hours, and the same domains are not observed when the time lapse recordings are examined (our unpublished results). In addition, the cells do not aggregate and no true mounds are formed, even after 15 h. The domains are still observed at 21 h, although they are fewer and less organized. These data suggest that the inability to chemotax to form a mound could be caused by defects in chemotaxis and/or the response to cAMP signaling.
Figure 5.
Phase contrast video microscopic analysis of aggregate formation in wild-type and rip3 null cells. (A) Phase contrast video microscopy of wild-type cells is depicted as described previously. Briefly, cells are plated as a monolayer on nonnutrient agar and examined by phase contrast video microscopy using a 4× objective. The solid white arrows point to some of the aggregation centers. The open white arrows point to the outer regions of individual aggregation domains. Only a few of these are marked. (B) rip3 null cells. A similar analysis was performed on rip3 null cells.
rip3 Null Cells Form Aggregates in Response to cAMP Signaling and Induce Cell-Type-Specific Gene Expression
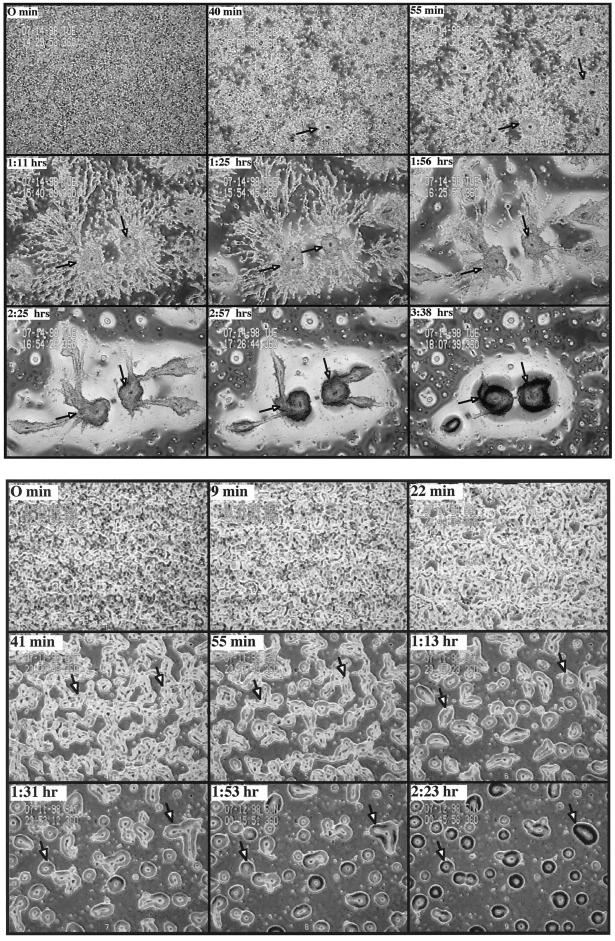
Some aggregation-deficient strains that are impaired in the ability to aggregate are able to form multicellular organisms after the cells have been pulsed with 30 nM cAMP for 5 h to mimic the normal oscillatory pulses of cAMP that occur during aggregation (Devreotes et al., 1987; Insall et al., 1994b, 1996; Ma et al., 1997). Figure 6B demonstrates that when rip3 null cells are pulsed with cAMP and plated on a nonnutrient agar surface they form mounds, although mound formation is delayed compared with wild-type cells, which form mounds in 1.5 h under these conditions (Figure 6A). Wild-type cells proceed through development and form migrating slugs by 9 h and mature fruiting bodies by 15 h (Figure 6A; our unpublished results). The majority of rip3 null mounds, however, arrest for 3–10 h before tip formation. By 30 h, less than half of these mounds have formed fruiting bodies (our unpublished results). Protein kinase (PKA) is required for multiple aggregation-stage pathways (Mann and Firtel, 1991; Schaap et al., 1995; Mann et al., 1997), and constitutive, high levels of expression of the catalytic subunit of cAMP-dependent PKA can bypass the inability of the aggregation-deficient null mutations in the MAPK ERK2 and the aggregation-stage adenylyl cyclase ACA to form aggregates and undergo morphogenesis (Aubry et al., 1997; Wang and Kuspa, 1997). rip3 null cells constitutively expressing the catalytic subunit of cAMP-dependent PKA do not form aggregates when plated on nonnutrient agar under standard conditions, indicating that PKA is unable to suppress the rip3 null phenotype (our unpublished results).
Figure 6.
Phase contrast video microscopic analysis of wild-type and rip3 null cells aggregating after being pulsed for 5 h with cAMP. Wild-type (A) or rip3 null (B) cells were pulsed for 5 h with 30 nM cAMP and plated on nonnutrient agar as described in Figure 5.
We investigated, by time-lapse video microscopy (as described in Figure 5), whether the aggregates formed by wild-type and rip3 null cells after cAMP pulsing occur through the chemotactic aggregation of cells. Figure 6A shows that wild-type cells form aggregation domains, which are larger than those formed by the same strain when plated for development with previous exogenous pulsing with cAMP (compare with Figure 5A). These domains are observed within 40 min of plating and start to chemotax and form aggregates by 55 min. Large aggregates form in ∼3 h. In contrast, rip3 null cells form numerous microcenters similar to those observed for mek1 null cells (Ma et al., 1997), which leads to the formation of many small aggregates (Figure 6B). Closer analysis of the time-lapse videotapes reveals that the rip3 null cells, like mek1 null cells, coalesce to form the aggregates rather than chemotaxing like wild-type cells (our unpublished results). The coalescence may be facilitated by cell-cell interactions mediated through cell adhesion molecules such as csA that are induced in response to cAMP pulses given before the cells are plated for development. These data suggest that rip3 null cells exhibit a chemotaxis defect.
We examined whether rip3 null cells express postaggregative genes, which are induced at mound formation in wild-type cells, and cell-type-specific genes in response to cAMP signaling (Firtel, 1995). Postaggregative genes such as CP2 are expressed at the mound stage or in cells in suspension that have been previously pulsed for 5 h with 30 nM cAMP to express aggregation-stage genes and then induced with continuous micromolar levels of cAMP (Pears et al., 1985; Datta et al., 1986). This leads to the induction of cell-type-specific (e.g., ecmA [prestalk-specific] and SP60/CotC [prespore-specific]) genes, which requires cell-cell contact in addition to exogenous, high, continuous levels of cAMP (Mehdy and Firtel, 1985; Jermyn et al., 1987). The morphogen differentiation-inducing factor is also required for maximal prestalk gene expression and is supplied endogenously (Jermyn et al., 1987; Williams et al., 1987). CP2 and the cell-type-specific genes ecmA (prestalk-specific) and SP60/CotC (prespore-specific) are not induced in rip3 null cells plated for development (our unpublished results), as might be expected for cells with an aggregation defect. In suspension culture, CP2 is induced in wild-type cells with or without exogenous cAMP (Figure 3B). Under assay conditions (slow-shake culture) that permit cell-cell contacts to form, there is sufficient endogenous cAMP produced for CP2 gene expression in wild-type cells. However, rip3 null cells induce CP2 in suspension culture only in the presence of exogenous cAMP (Figure 3B). The absence of the expression of CP2 in rip3 null cells under these assay conditions suggests that rip3 null cells may be defective in cAMP production. The cell-type-specific genes ecmA and SP60/CotC are induced by both rip3 null and wild-type cells and require exogenous cAMP, as previously demonstrated for wild-type cells (Mehdy and Firtel, 1985; Jermyn et al., 1987; Dynes et al., 1994). The combined results indicate that rip3 null cells are not defective in cAMP-induced gene expression but are defective in cAMP production.
rip3 Null Cells Are Defective in Receptor Activation of Adenylyl and Guanylyl Cyclases
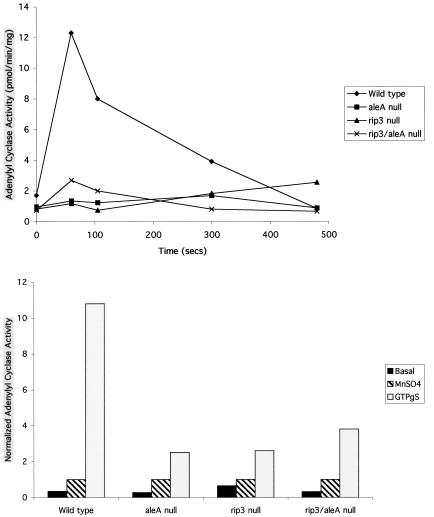
The developmental defects of rip3 null cells suggest that they are impaired in the ability to activate adenylyl cyclase and relay the cAMP signal. To examine this possibility, receptor- and G protein-mediated activation of adenylyl cyclase were measured in wild-type, rip3, and aleA null cells. aleA null cells exhibit phenotypes (Insall et al., 1996) similar to those of the rip3 null cells described here. Cells were pulsed with cAMP for 5 h to maximally express receptors and G proteins. The pulsed cells were stimulated in vivo with cAMP, lysed at specific time points, and assayed for adenylyl cyclase activity, as described previously (Devreotes et al., 1987). As depicted in Figure 7A, wild-type cells exhibit a 7.4-fold stimulation of adenylyl cyclase activity, which peaks at the 60-s time point, consistent with previous results (Roos and Gerisch, 1976). In agreement with previously published results, aleA null cells exhibit very little or no activation (Insall et al., 1996). Similarly, rip3 null cells display very low levels of activity.
Figure 7.
Adenylyl cyclase activation in wild-type and mutant cell lines. (A) Receptor-mediated stimulation of adenylyl cyclase activity. Differentiated cells were stimulated with 10 μM cAMP, lysed at specific time points, and immediately assayed as described in MATERIALS AND METHODS. (B) GTPγS-mediated stimulation of adenylyl cyclase activity. Differentiated cells were lysed with or without GTPγS, or in the presence MnSO4 as a measure of unregulated enzyme activity, incubated on ice for 4 min, and assayed as described in MATERIALS AND METHODS. The results are expressed as a ratio of the adenylyl cyclase activity to MnSO4 activity. The absolute MnSO4 activities are 4.0, 3.9, 2.5, and 2.5 pmol·min−1·mg−1 for wild-type, aleA null, rip3 null, and rip3/aleA null cells, respectively. The results presented are representative of at least two independent experiments.
Activation of adenylyl cyclase is mediated by the Gβγ subunits and can be activated in wild-type cells that are lysed in the presence of GTPγS (Wu et al., 1995). This activation does not require the coupled Gα2 subunit, as GTPγS activation occurs in gα2 null cells, presumably because lysis of cells in the presence of GTPγS releases Gβγ from other heterotrimeric G proteins. Cells pulsed with cAMP for 5 h were lysed in the absence or presence of GTPγS and assayed for adenylyl cyclase activity. In wild-type cells, GTPγS induces the activity of adenylyl cyclase 31.5-fold compared with basal activity (Figure 7B). As we observed with the in vivo stimulation, rip3 and aleA null cells exhibit greatly reduced adenylyl cyclase activity in the presence of GTPγS (Figure 7B). The inefficiency of GTPγS stimulation of adenylyl cyclase activity in rip3 null cells is unexpected, as it suggests that RIP3 function is required for function of the G protein (see DISCUSSION). The activity of adenylyl cyclase can be stimulated by MnSO4, which gives a measurement of the unregulated activity of the enzyme. Wild-type, rip3 null, and aleA null cells show a similar level of MnSO4-stimulated activity, suggesting that the total adenylyl cyclase enzymatic activity is similar in the three strains (Figure 7B). These results were confirmed by Western blot analysis (our unpublished results).
Many of the developmental phenotypes of rip3 null cells are similar to those of aleA null cells (Insall et al., 1996). To examine this similarity in more detail, we created a rip3/aleA double-knockout strain. Adenylyl cyclase assays in this strain were done in parallel with those in the wild-type, rip3, and aleA null strains. Interestingly, the rip3/aleA double-knockout strain shows a slight adenylyl cyclase activity in response to cAMP stimulation (Figure 7B) and a slightly higher GTPγS-stimulated activity than either null strain (Figure 7B).
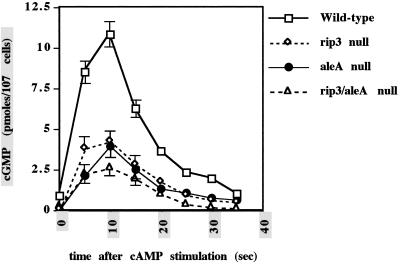
Receptor-mediated activation of guanylyl cyclase is required for chemotaxis (Van Haastert and Kuwayama, 1997). As described previously (Van Haastert and Van der Heijden, 1983), cAMP stimulation of wild-type cells pulsed for 5 h (aggregation-competent cells) results in a rapid, transient activation of guanylyl cyclase (Figure 8). In contrast, rip3 and aleA null cells show a significantly reduced cAMP-induced activation of guanylyl cyclase activation response that was ∼40% that of the wild-type response in three separate experiments. When the rip3/aleA null strain was examined, the level of stimulation was further reduced to ∼25% that of wild-type cells in three separate experiments. These results indicate that rip3 and aleA null cells are defective in the chemoattractant-mediated activation of guanylyl and adenylyl cyclases.
Figure 8.
cAMP stimulation of guanylyl cyclase activity in wild-type, rip3 null, aleA null, and rip3/aleA null strains. Cells were pulsed for 5 h with 30 nM cAMP and stimulated with cAMP, and measurements were taken for the quantitation of cGMP as described in MATERIALS AND METHODS. Unregulated activity of the enzyme was measured in the presence of 5 mM MnSO4. Each strain was analyzed a minimum of three times in association with wild-type cells. The results of representative experiments are shown. The differences between rip3 and aleA null cells, wild-type cells and rip3/aleA null cells, and rip3 and aleA null strains are reproducible.
RIP3 Is Required for Proper Chemotaxis
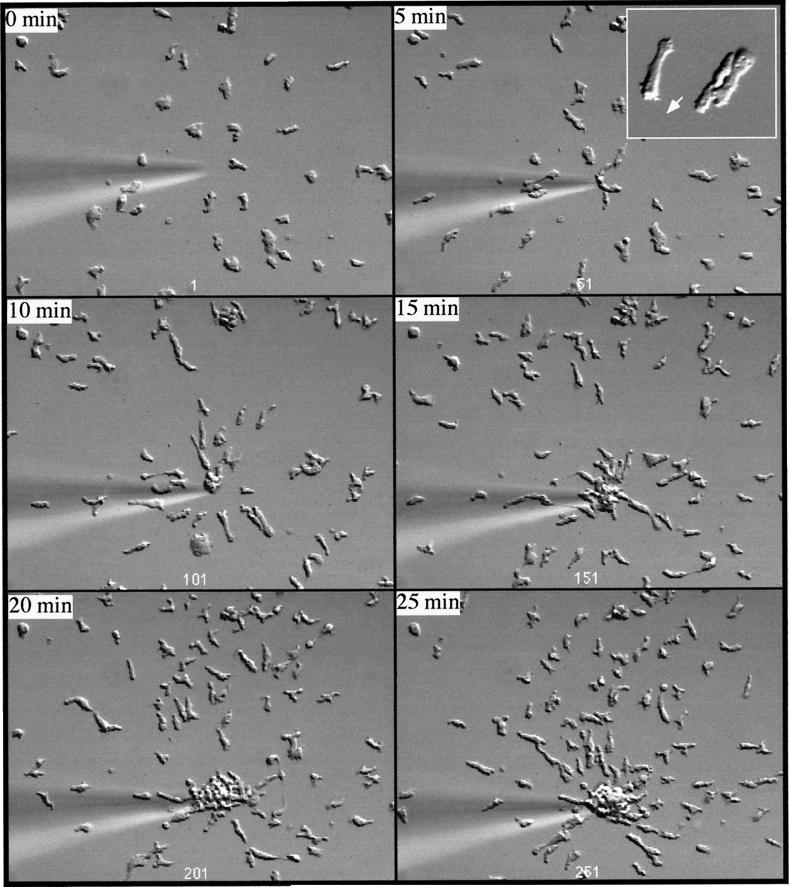
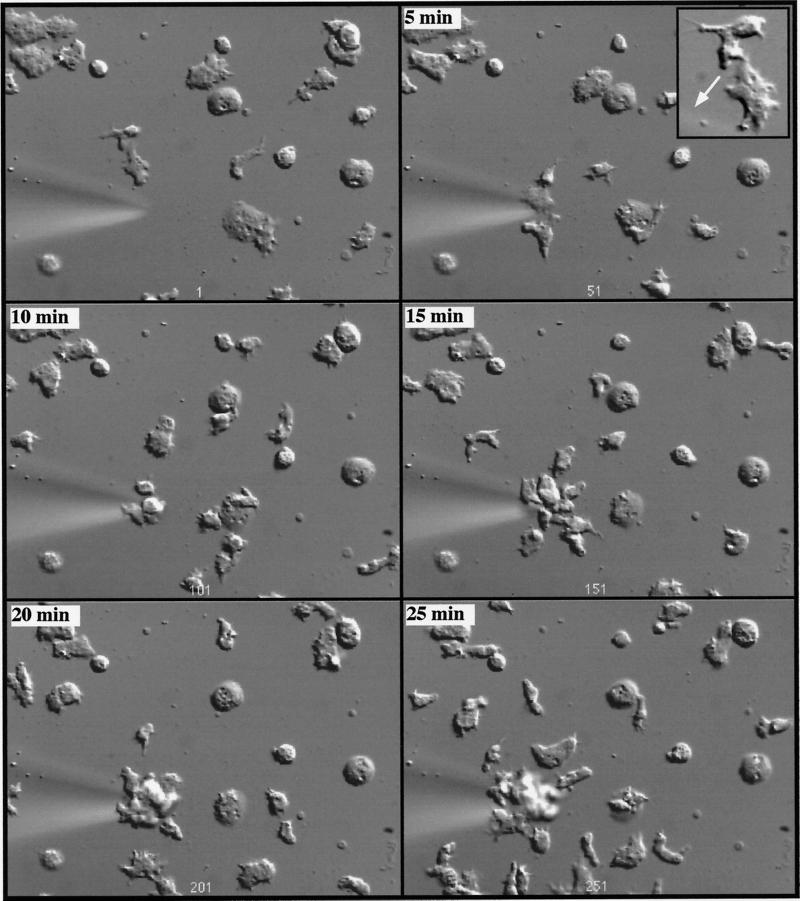
To examine whether rip3 null cells are impaired in the ability to chemotax toward cAMP, we used an assay in which cells chemotax on the glass coverslip toward cAMP that is emitted from a micropipet (Gerisch et al., 1975; Meili et al., 1999). As depicted in Figure 9A, wild-type cells become highly polarized and move toward the micropipet by extending pseudopodia in the direction of the micropipet (up the cAMP concentration gradient). Time-lapse video microscopy reveals that the vast majority of wild-type cells exhibit this highly polarized cell shape and produce very few pseudopodia in directions that are either perpendicular or oblique to the axis of the cAMP gradient (Figure 9A, inset). In contrast, rip3 null cells are less polarized and migrate more slowly than wild-type cells (Figure 9B). Detailed analysis of the time-lapse video indicates that the cells produce numerous pseudopodia and filopodia at right or oblique angles to the direction of the cAMP gradient (Figure 9B, inset). These results indicate that rip3 null cells are impaired in their ability to chemotax up a concentration gradient of chemoattractants, which is consistent with the aggregation-defective phenotypes described above.
Figure 9.
Chemotaxis of wild-type, rip3 null, and rip3/aleA null strains. Cells were pulsed for 5 h with 30 nM cAMP, plated on coverslips as described previously, and examined by differential interference contrast microscopy. The micropipet shown contains 150 μM cAMP. Cells chemotax toward the cAMP gradient produced by diffusion of the cAMP from the micropipet. Insets in A and B depict the shapes of wild-type and rip3 null cells, respectively. (A) Analysis of wild-type cells was done with a 20× objective, whereas that of rip3/aleA null cells was done with a 40× objective. (A) Wild-type cells. (B) rip3 null cells. (C) rip3/aleA null cells.
aleA null cells exhibit aberrant chemotaxis toward cAMP (Insall et al., 1996). To further examine potential genetic interactions of RIP3 and AleA in controlling cell movement, we performed a similar chemotaxis assay using the double-knockout cells. As shown in Figure 9C, these cells exhibit very little movement or polarization toward cAMP. When the rip3/aleA double-knockout cells are pulsed and plated for development, only a small fraction of the cells coalesce to form mound-like structures by 24 h (our unpublished results), and development does not proceed further.
DISCUSSION
Chemotaxis requires coordinated changes in the actin and myosin cytoskeletons (see INTRODUCTION for references). In the case of Dictyostelium, these changes are mediated by chemoattractants such as cAMP and folic acid that function through discrete cell surface serpentine receptors coupled to heterotrimeric G proteins containing different Gα protein subunits. Aggregation in Dictyostelium requires the integrated regulation of pathways controlling chemotaxis, activation of adenylyl and guanylyl cyclases, and relay of cAMP. These processes are mediated through the cAMP receptor and downstream signal transduction pathways that require the heterotrimeric G protein containing the Gα2 subunit.
Our analysis demonstrates that rip3 null cells are impaired in the signal relay pathway and chemotaxis. rip3 null cells show only a minimal level of cAMP-mediated activation of adenylyl cyclase in cells lysed at various times after stimulation. When cells are lysed in the presence or absence of GTPγS to quantitate GTP-dependent stimulation of adenylyl cyclase activity, rip3 and aleA null cells exhibit very little adenylyl cyclase activity compared with wild-type cells, although MnSO4-stimulated activity is similar in all strains. The most simple genetic interpretation of these biochemical results suggests that RIP3 and AleA are required for Gβγ stimulation of adenylyl cyclase. One possible function of RIP3 is to control the function of other components of the pathway that are required for Gβγ-stimulated adenylyl cyclase activity. As the function of the pleckstrin homology domain-containing protein CRAC is essential for activation of adenylyl cyclase (Insall et al., 1994a; Lilly and Devreotes, 1995; Parent et al., 1998), one possible role of RIP3 is to potentiate one of these processes. Previous results and the results presented here indicate that aimless and rip3 null cells exhibit similar defects in adenylyl cyclase activation. Moreover, of the numerous genes that are required for proper aggregation (Firtel, 1995; Chen et al., 1996; Parent and Devreotes, 1996), only RIP3 and AleA are required for both chemotaxis and signal relay. It is possible that RIP3 and AleA function in a similar pathway, especially considering that RIP3 interacts in vitro with RasG and AleA is a putative Ras GEF.
rip3 null cells do not express aggregation-stage or postaggregative and cell-type-specific genes when the cells are plated for development. These defects are probably due to the defect in the activation of cAMP and signal relay, as rip3 null cells induce all three classes of genes in response to exogenous cAMP. PKA activity is required for multiple developmental pathways, including aggregation (Mann and Firtel, 1991; Reymond et al., 1995; Schulkes and Schaap, 1995; Mann et al., 1997). We demonstrate that constitutive expression of the PKA catalytic subunit does not complement the rip3 null phenotype, indicating that rip3 null cells have sufficient levels of endogenous PKA activity for these processes. This suggests that there is probably a low level of activation of adenylyl cyclase in vivo. Consistent with this possibility, we observe a low level of GTPγS-stimulated activity in vitro and the formation of rudimentary aggregation domains, which requires a low level of cAMP relay, when rip3 null cells are plated for development as visualized in the phase contrast time-lapse video imaging.
It is very striking that rip3 null cells have a defect in chemotaxis. The cells are less polarized than wild-type cells in a cAMP gradient, being unable to properly elongate. Instead, rip3 null cells extend pseudopodia laterally as well as in the direction of the cAMP signal. This defect is observed whether the micropipet is close to or farther away from cells, indicating that the defect occurs in response to different concentration gradients of cAMP. As with the regulation of adenylyl cyclase, aleA and rip3 null strains exhibit similar chemotaxis defects. We have shown that the activation of guanylyl cyclase is reduced in rip3 null cells. However, it is not clear that the reduced activation of guanylyl cyclase is sufficient to cause the chemotaxis defects we observe. Although we have no direct evidence, our view is that RIP3 controls other pathways that are also required for chemotaxis and that it is the defect in these pathways or a combined defect in multiple pathways that leads to the chemotaxis defects. Our analysis of the rip3/aleA double knockout indicates that these cells exhibit an even greater impairment in the ability to chemotax and do not respond to cAMP gradients produced by cAMP diffusing from a micropipet. The stronger phenotype of the double knockout has two interpretations. The first is that the proteins function in two direct, parallel pathways leading to chemotaxis and that the combined partial impairment of both pathways results in a sufficient loss of function to render the cells unable to chemotax. The second interpretation is that RIP3 and AleA function on the same pathway but both null phenotypes are leaky with respect to their requirement for the pathway. The double knockout would result in more efficient blocking of the pathway. As AleA is a putative Ras exchange factor (GEF) and RIP3 has specific interactions with RasG, which is known to be required for proper cell movement, it is reasonable to postulate that AleA and RIP3 control a common pathway by either regulating or being regulated by the Ras protein, presumably RasG. However, as the rasG null phenotype is not the same as that of rip3 null cells during aggregation, it is possible that RIP3 interacts genetically with another, not-yet-identified Ras protein or has a function that does not require interaction with RasG or another Ras protein. Although we expect Ras to be involved, we cannot distinguish between functions of AleA and RIP3 that do or do not require Ras and the phenotypes we observe.
One of our most striking observations is that this pathway that we expect involved Ras-mediated pathways is important for the activation of adenylyl cyclase and chemotaxis. It is not unexpected that such a coordination would be important to produce an integrated biological response. Although this coordination was expected to be mediated, at least partially, through chemoattractant activation of a common heterotrimeric G protein, the direct coupling of these pathways through pathways containing components that interact with Ras was unexpected. The phenotypes of AleA and RIP3 during chemotaxis are more severe than those of rasG null cells. We cannot exclude the possibility that another yet-to-be-identified Ras may be the key component of this pathway or that RIP3 may exert functions independent of possible interactions with RasG. It is possible that RIP3 could function as a scaffolding for various components of the chemotaxis and signal relay pathways and that RasG may coordinate or activate solely the chemotaxis component of the aggregation-stage pathways. RIP3 exhibits a restricted homology to a mammalian gene that was identified as a protein that represses the phenotype of activated Ras in yeast (Colicelli et al., 1991). Unfortunately, this restricted homology does not shed any light on the function of either the mammalian gene or RIP3. It is possible that this homology is more an indicator of a Ras-interacting or other regulatory domain than the biological function of either gene. Identification of interacting proteins or second-site suppressers should further elucidate the pathway that requires RIP3 function.
ACKNOWLEDGMENTS
We thank members of the Firtel and Devreotes laboratories for helpful suggestions. This work was funded in part by Wellcome grants to R.I. and U.S. Public Health Service grants to C.A.P. and R.A.F.
REFERENCES
- Aubry L, Maeda M, Insall R, Devreotes PN, Firtel RA. The Dictyostelium mitogen-activated protein kinase ERK2 is regulated by Ras and cAMP-dependent protein kinase (PKA) and mediates PKA function. J Biol Chem. 1997;272:3883–3886. doi: 10.1074/jbc.272.7.3883. [DOI] [PubMed] [Google Scholar]
- Burki E, Anjard C, Scholder JC, Reymond CD. Isolation of two genes encoding putative protein kinases regulated during Dictyostelium discoideum development. Gene. 1991;102:57–65. doi: 10.1016/0378-1119(91)90538-m. [DOI] [PubMed] [Google Scholar]
- Chen MY, Insall RH, Devreotes PN. Signaling through chemoattractant receptors in Dictyostelium. Trends Genet. 1996;12:52–57. doi: 10.1016/0168-9525(96)81400-4. [DOI] [PubMed] [Google Scholar]
- Chen MY, Long Y, Devreotes PN. A novel cytosolic regulator, pianissimo, is required for chemoattractant receptor and G protein-mediated activation of the 12 transmembrane domain adenylyl cyclase in Dictyostelium. Genes Dev. 1997;11:3218–3231. doi: 10.1101/gad.11.23.3218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung CY, Firtel RA. Dictyostelium: a model experimental system for elucidating the pathways and mechanisms controlling chemotaxis. In: Conn PM, Means A, editors. Molecular Regulation. Totowa, NJ: Humana Press; 1999. (in press). [Google Scholar]
- Colicelli J, Nicolette C, Birchmeier C, Rodgers L, Riggs M, Wigler M. Expression of three mammalian cDNAs that interfere with RAS function in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1991;88:2913–2917. doi: 10.1073/pnas.88.7.2913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta S, Gomer RH, Firtel RA. Spatial and temporal regulation of a foreign gene by a prestalk-specific promotor in transformed Dictyostelium discoideum. Mol Cell Biol. 1986;6:811–820. doi: 10.1128/mcb.6.3.811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dembinsky A, Rubin H, Ravid S. Chemoattractant-mediated increases in cGMP induce changes in Dictyostelium myosin II heavy chain-specific protein kinase C activities. J Cell Biol. 1996;134:911–921. doi: 10.1083/jcb.134.4.911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devreotes P, Fontana D, Klein P, Sherring J, Theibert A. Transmembrane signaling in Dictyostelium. Methods Cell Biol. 1987;28:299–331. doi: 10.1016/s0091-679x(08)61653-2. [DOI] [PubMed] [Google Scholar]
- Dynes JL, Clark AM, Shaulsky G, Kuspa A, Loomis WF, Firtel RA. LagC is required for cell-cell interactions that are essential for cell-type differentiation in Dictyostelium. Genes Dev. 1994;8:948–958. doi: 10.1101/gad.8.8.948. [DOI] [PubMed] [Google Scholar]
- Esch RK, Weeks G, Firtel RA. Regulation and function of two developmentally regulated ras genes in Dictyostelium. In: Lacal JC, McCormick F, editors. The Ras Superfamily of GTPases. New York: CRC Press; 1993. pp. 173–186. [Google Scholar]
- Firtel RA. Integration of signaling information in controlling cell-fate decisions in Dictyostelium. Genes Dev. 1995;9:1427–1444. doi: 10.1101/gad.9.12.1427. [DOI] [PubMed] [Google Scholar]
- Gerisch G. Cell aggregation and differentiation in Dictyostelium. In: Moscona AA, Monroy A, editors. Current Topics in Developmental Biology. New York: Academic Press; 1968. pp. 157–197. [DOI] [PubMed] [Google Scholar]
- Gerisch G, Hulser D, Malchow D, Wick U. Cell communication by periodic cyclic-AMP pulses. Philos Trans R Soc Lond B Biol Sci. 1975;272:181–192. doi: 10.1098/rstb.1975.0080. [DOI] [PubMed] [Google Scholar]
- Ginsburg GT, Gollop R, Yu YM, Louis JM, Saxe CL, Kimmel AR. The regulation of Dictyostelium development by transmembrane signaling. J Euk Microbiol. 1995;42:200–205. doi: 10.1111/j.1550-7408.1995.tb01565.x. [DOI] [PubMed] [Google Scholar]
- Insall R, Kuspa A, Lilly PJ, Shaulsky G, Levin LR, Loomis WF, Devreotes P. CRAC, a cytosolic protein containing a pleckstrin homology domain, is required for receptor and G protein-mediated activation of adenylyl cyclase in Dictyostelium. J Cell Biol. 1994a;126:1537–1545. doi: 10.1083/jcb.126.6.1537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Insall RH, Borleis J, Devreotes PN. The aimless RasGEF is required for processing of chemotactic signals through G-protein-coupled receptors in Dictyostelium. Curr Biol. 1996;6:719–729. doi: 10.1016/s0960-9822(09)00453-9. [DOI] [PubMed] [Google Scholar]
- Insall RH, Soede RDM, Schaap P, Devreotes PN. Two cAMP receptors activate common signaling pathways in Dictyostelium. Mol Biol Cell. 1994b;5:703–711. doi: 10.1091/mbc.5.6.703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jermyn KA, Berks M, Kay RR, Williams JG. Two distinct classes of prestalk-enriched mRNA sequences in Dictyostelium discoideum. Development. 1987;100:745–755. doi: 10.1242/dev.100.4.745. [DOI] [PubMed] [Google Scholar]
- Kumagai A, Pupillo M, Gundersen R, Miake-Lye R, Devreotes PN, Firtel RA. Regulation and function of Gα protein subunits in Dictyostelium. Cell. 1989;57:265–275. doi: 10.1016/0092-8674(89)90964-1. [DOI] [PubMed] [Google Scholar]
- Lee S, Escalante R, Firtel RA. A Ras GAP is essential for cytokinesis and spatial patterning in Dictyostelium. Development. 1997;124:983–996. doi: 10.1242/dev.124.5.983. [DOI] [PubMed] [Google Scholar]
- Lilly P, Wu LJ, Welker DL, Devreotes PN. A G-protein beta-subunit is essential for Dictyostelium development. Genes Dev. 1993;7:986–995. doi: 10.1101/gad.7.6.986. [DOI] [PubMed] [Google Scholar]
- Lilly PJ, Devreotes PN. Chemoattractant and GTP gamma S-mediated stimulation of adenylyl cyclase in Dictyostelium requires translocation of CRAC to membranes. J Cell Biol. 1995;129:1659–1665. doi: 10.1083/jcb.129.6.1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma H, Gamper M, Parent C, Firtel RA. The Dictyostelium MAP kinase kinase DdMEK1 regulates chemotaxis and is essential for chemoattractant-mediated activation of guanylyl cyclase. EMBO J. 1997;16:4317–4332. doi: 10.1093/emboj/16.14.4317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann SKO, Brown JM, Briscoe C, Parent C, Pitt G, Devreotes PN, Firtel RA. Role of cAMP-dependent protein kinase in controlling aggregation and postaggregative development in Dictyostelium. Dev Biol. 1997;183:208–221. doi: 10.1006/dbio.1996.8499. [DOI] [PubMed] [Google Scholar]
- Mann SKO, Firtel RA. Cyclic AMP regulation of early gene expression in Dictyostelium discoideum: mediation via the cell surface cyclic AMP receptor. Mol Cell Biol. 1987;7:458–469. doi: 10.1128/mcb.7.1.458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann SKO, Firtel RA. A developmentally regulated, putative serine/threonine protein kinase is essential for development in Dictyostelium. Mech Dev. 1991;35:89–101. doi: 10.1016/0925-4773(91)90060-j. [DOI] [PubMed] [Google Scholar]
- Mann SKO, Pinko C, Firtel RA. Regulation of Dictyostelium early gene expression in cAMP bypass mutants. Dev Biol. 1988;130:406–410. doi: 10.1016/0012-1606(88)90447-2. [DOI] [PubMed] [Google Scholar]
- Mehdy MC, Firtel RA. A secreted factor and cyclic AMP jointly regulate cell-type-specific gene expression in Dictyostelium discoideum. Mol Cell Biol. 1985;5:705–713. doi: 10.1128/mcb.5.4.705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meili R, Ellsworth C, Lee S, Reddy TBK, Ma H, Firtel RA. Chemoattractant-mediated transient activation and membrane localization of Akt/PKB is required for efficient chemotaxis to cAMP in Dictyostelium. EMBO J. 1999;18:2092–2105. doi: 10.1093/emboj/18.8.2092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noegel A, Gerisch G, Stadler J, Westphal M. Complete sequence and transcript regulation of a cell adhesion protein from aggregating Dictyostelium cells. EMBO J. 1986;5:1473–1476. doi: 10.1002/j.1460-2075.1986.tb04384.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parent CA, Blacklock BJ, Froehlich WM, Murphy DB, Devreotes PN. G protein signaling events are activated at the leading edge of chemotactic cells. Cell. 1998;95:81–91. doi: 10.1016/s0092-8674(00)81784-5. [DOI] [PubMed] [Google Scholar]
- Parent CA, Devreotes PN. Molecular genetics of signal transduction in Dictyostelium. Annu Rev Biochem. 1996;65:411–440. doi: 10.1146/annurev.bi.65.070196.002211. [DOI] [PubMed] [Google Scholar]
- Pawson T, Amiel T, Hinze E, Auersperg N, Neave N, Sobolewski A, Weeks G. Regulation of a Ras-related protein during development of Dictyostelium discoideum. Mol Cell Biol. 1985;5:33–39. doi: 10.1128/mcb.5.1.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pears CJ, Mahbubani HM, Williams JG. Characterization of two highly diverged but developmentally coregulated cysteine proteinases in Dictyostelium discoideum. Nucleic Acids Res. 1985;13:8853–8866. doi: 10.1093/nar/13.24.8853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson MD, Novak KD, Reedy MC, Ruman JI, Titus MA. Molecular genetic analysis of myoC, a Dictyostelium myosin I. J Cell Sci. 1995;108:1093–1103. doi: 10.1242/jcs.108.3.1093. [DOI] [PubMed] [Google Scholar]
- Pitt GS, Milona N, Borleis J, Lin KC, Reed RR, Devreotes PN. Structurally distinct and stage-specific adenylyl cyclase genes play different roles in Dictyostelium development. Cell. 1992;69:305–315. doi: 10.1016/0092-8674(92)90411-5. [DOI] [PubMed] [Google Scholar]
- Reymond CD, Gomer RH, Mehdy MC, Firtel RA. Developmental regulation of a Dictyostelium gene encoding a protein homologous to mammalian Ras protein. Cell. 1984;39:141–148. doi: 10.1016/0092-8674(84)90199-5. [DOI] [PubMed] [Google Scholar]
- Reymond CD, Gomer RH, Nellen W, Theibert A, Devreotes P, Firtel RA. Phenotypic changes induced by a mutated Ras gene during the development of Dictyostelium transformants. Nature. 1986;323:340–343. doi: 10.1038/323340a0. [DOI] [PubMed] [Google Scholar]
- Reymond CD, Schaap P, Veron M, Williams JG. Dual role of cAMP during Dictyostelium development. Experientia. 1995;51:1166–1174. doi: 10.1007/BF01944734. [DOI] [PubMed] [Google Scholar]
- Roos W, Gerisch G. Receptor-mediated adenylate cyclase activation in Dictyostelium discoideum. FEBS Lett. 1976;68:170–172. doi: 10.1016/0014-5793(76)80429-2. [DOI] [PubMed] [Google Scholar]
- Saxe CL, III, Johnson RL, Devreotes PN, Kimmel AR. Expression of a cAMP receptor gene of Dictyostelium and evidence for a multigene family. Genes Dev. 1991;5:1–8. doi: 10.1101/gad.5.1.1. [DOI] [PubMed] [Google Scholar]
- Schaap P, Brandt R, Van Es S. Regulation of Dictyostelium adenylyl cyclases by morphogen-induced modulation of cytosolic pH or Ca++ levels. Dev Biol. 1995;167:179–188. doi: 10.1006/dbio.1995.1070. [DOI] [PubMed] [Google Scholar]
- Schleicher M, Noegel AA. Dynamics of the Dictyostelium cytoskeleton during chemotaxis. New Biol. 1992;4:461–472. [PubMed] [Google Scholar]
- Schulkes C, Schaap P. cAMP-dependent protein kinase activity is essential for preaggregative gene expression in Dictyostelium. FEBS Lett. 1995;368:381–384. doi: 10.1016/0014-5793(95)00676-z. [DOI] [PubMed] [Google Scholar]
- Segall JE, Kuspa A, Shaulsky G, Ecke M, Maeda M, Gaskins C, Firtel RA. A MAP kinase necessary for receptor-mediated activation of adenylyl cyclase in Dictyostelium. J Cell Biol. 1995;128:405–413. doi: 10.1083/jcb.128.3.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuxworth RI, Cheetham JL, Machesky LM, Spiegelmann GB, Weeks G, Insall RH. Dictyostelium RasG is required for normal motility and cytokinesis, but not growth. J Cell Biol. 1997;138:605–614. doi: 10.1083/jcb.138.3.605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uyeda TQ, Titus MA. The myosins of Dictyostelium. In: Maeda Y, Inouye K, Takeuchi I, editors. Dictyostelium: A Model System for Cell and Developmental Biology. Tokyo: Universal Academy Press; 1997. pp. 43–64. [Google Scholar]
- Van Haastert PJM. Transduction of the chemotactic cAMP signal across the plasma membrane of Dictyostelium cells. Experientia. 1995;51:1144–1154. doi: 10.1007/BF01944732. [DOI] [PubMed] [Google Scholar]
- Van Haastert PJM, Kuwayama H. cGMP as second messenger during Dictyostelium chemotaxis. FEBS Lett. 1997;410:25–28. doi: 10.1016/s0014-5793(97)00416-x. [DOI] [PubMed] [Google Scholar]
- Van Haastert PJM, Van der Heijden PR. Excitation adaptation and deadaptation of the cAMP mediated cGMP response in Dictyostelium discoideum. J Cell Biol. 1983;96:347–353. doi: 10.1083/jcb.96.2.347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Haastert PJM, Van Lookeren Campagne MM. Transient kinetics of a cGMP-dependent cGMP-specific phosphodiesterase from Dictyostelium discoideum. J Cell Biol. 1984;98:709–716. doi: 10.1083/jcb.98.2.709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B, Kuspa A. Dictyostelium development in the absence of cAMP. Science. 1997;277:251–254. doi: 10.1126/science.277.5323.251. [DOI] [PubMed] [Google Scholar]
- Wessels D, Titus M, Soll DR. A Dictyostelium myosin I plays a crucial role in regulating the frequency of pseudopods formed on the substratum. Cell Motil Cytoskeleton. 1996;33:64–79. doi: 10.1002/(SICI)1097-0169(1996)33:1<64::AID-CM7>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- Williams JG, Ceccarelli A, McRobbie S, Mahbubani H, Kay RR, Farly A, Berks M, Jermyn KA. Direct induction of Dictyostelium prestalk gene expression by D1F provides evidence that D1F is a morphogen. Cell. 1987;49:185–192. doi: 10.1016/0092-8674(87)90559-9. [DOI] [PubMed] [Google Scholar]
- Wu LJ, Valkema R, Van Haastert PJM, Devreotes PN. The G protein beta subunit is essential for multiple responses to chemoattractants in Dictyostelium. J Cell Biol. 1995;129:1667–1675. doi: 10.1083/jcb.129.6.1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou KM, Takegawa K, Emr SD, Firtel RA. A phosphatidylinositol (PI) kinase gene family in Dictyostelium discoideum: biological roles of putative mammalian p110 and yeast Vps34p PI 3-kinase homologs during growth and development. Mol Cell Biol. 1995;15:5645–5656. doi: 10.1128/mcb.15.10.5645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zigmond SH, Joyce M, Borleis J, Bokoch GM, Devreotes PN. Regulation of actin polymerization in cell-free systems by GTPγS and Cdc42. J Cell Biol. 1997;138:363–374. doi: 10.1083/jcb.138.2.363. [DOI] [PMC free article] [PubMed] [Google Scholar]