Abstract
The neutral form of the chromophore in wild-type green fluorescent protein (wtGFP) undergoes excited state proton transfer (ESPT) upon excitation, resulting in characteristic green (508 nm) fluorescence. This ESPT reaction involves a proton relay from the phenol hydroxyl of the chromophore to the ionized side chain of E222, and results in formation of the anionic chromophore in a protein environment optimized for the neutral species (the I* state). Reorientation or replacement of E222, as occurs in the S65T and E222Q GFP mutants, disables the ESPT reaction and results in loss of green emission following excitation of the neutral chromophore. Previously, it has been shown that the introduction of a second mutation (H148D) into S65T GFP allows the recovery of green emission, implying that ESPT is again possible. A similar recovery of green fluorescence is also observed for the E222Q/H148D mutant, suggesting that D148 is the proton acceptor for the ESPT reaction in both double mutants. The mechanism of fluorescence emission following excitation of the neutral chromophore in S65T/H148D and E222Q/H148D has been explored through the use of steady state and ultrafast time-resolved fluorescence and vibrational spectroscopy. The data are contrasted with the single mutant S65T GFP. Time-resolved fluorescence studies indicate very rapid (< 1 ps) formation of I* in the double mutants, followed by vibrational cooling on the picosecond time scale. The time resolved IR difference spectra are markedly different to those of wtGFP or its anionic mutants. In particular, no spectral signatures are apparent in the picosecond IR difference spectra that would correspond to alteration in the ionization state of D148, leading to the proposal that a low barrier hydrogen bond (LBHB) is present between the phenol hydroxyl of the chromophore and the side chain of D148, with different potential energy surfaces for the ground and excited states. This model is consistent with recent high resolution structural data in which the distance between the donor and acceptor oxygen atoms is ≤ 2.4Å. Importantly, these studies indicate that the hydrogen bond network in wtGFP can be replaced by a single residue, an observation which, when fully explored, will add to our understanding of the various requirements for proton transfer reactions within proteins.
Introduction
The transfer of a proton between donor and acceptor groups is a fundamental chemical reaction that occurs throughout chemistry and biology. Many enzymes catalyze proton transfer reactions,1–3 while in respiratory complexes the coupling of proton and electron transfer results in movement of protons across a membrane, and the establishment of a proton gradient that can then be used for the synthesis of ATP.4, 5 The rate of proton transfer is governed by the energy required to bring the reactants together, the magnitude of the barrier that must be crossed (or potentially tunnelled through) during the actual proton transfer process, and the energy required to separate the products of the reaction.6 Once the reactants are positioned, the ability of the proton to be transferred is modulated by the factors that influence the nature of hydrogen bonding interactions,7 such as the dielectric of the local environment, the distance between donor and acceptor groups, and their relative pKa values. These factors can conspire to reduce or eliminate the barrier between donor and acceptor groups, and there has been extensive discussion of the role of low barrier hydrogen bonds in, for example, enzyme catalysis.8, 9
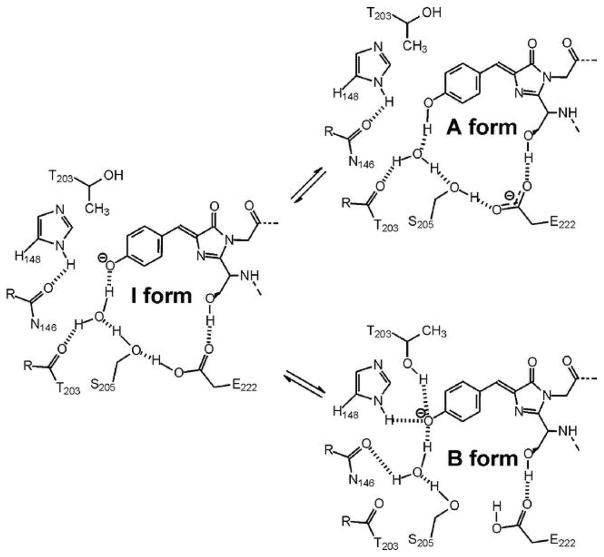
Biological systems in which proton transfer is initiated by light absorption are particularly attractive as model systems for understanding the detailed physical and chemical aspects of these reactions. One such system is the green fluorescent protein (GFP), a commonly used tool in cellular and molecular imaging owing to the presence of an intrinsically fluorescent chromophore formed autocatalytically from residues S65, Y66 and G67.10–13 Alterations in the structure of residues in and around the chromophore perturb the photophysics of GFP allowing a detailed correlation to be established between chromophore structure and fluorescence. In wild-type GFP (wtGFP), excitation into either the neutral (λmax 395 nm, A) or anionic (λmax 475 nm, B) forms of the chromophore results in very efficient green (λem 508 and 504 nm, respectively) fluorescence.14–16 The 508 nm emission following excitation of A arises from an anionic species formed in the excited state (I*) by deprotonation of the chromophore and concomitant protonation of the E222 carboxylate group (Figure 1). The directly excited B* and indirectly excited I* states differ in that the former has an environment optimized for the anionic form while the latter retains the unrelaxed neutral environment. Only very weak short lived emission is detected from the neutral form following excitation at 395 nm.17 The A* to I* excited state proton transfer (ESPT) reaction has been studied in detail and occurs via a hydrogen bond network comprised of a water molecule and the side chain of S205, that results in protonation of the E222 carboxylate side chain.18, 19 Replacement of residues directly involved in proton transfer such as E222 or that modulate the orientation of these residues such as S65 disables ESPT and results in loss of green emission from I*.
Figure 1.
Model for the interconversion of the A and B forms of the chromophore via the I form. Adapted from Brejc et al.18
Many mutants of GFP have been generated in order to expand the range of applications of fluorescent proteins. One common change involves the replacement of S65, a residue directly involved in chromophore formation, with a threonine (S65T).18, 20 This mutation leaves the conjugated part of the chromophore, where the excitation is localized, unchanged, but modifies the side chain. Unlike wtGFP, the chromophore in S65T is sensitive to pH, and conversion between A and B forms of the chromophore occurs with a pKa of 6.21, 22 Thus, S65T GFP acts as a pH sensor. In addition, at physiological pH the anionic B form predominates, thereby increasing the fluorescent signal resulting from direct excitation of this form of the chromophore. The mutation of S65 to a threonine alters contacts between the side chain of this residue and E222, causing the side chain of the latter to move so that it can no longer participate in ESPT.18, 21 Thus, for S65T, the I* state cannot form, and excitation into A results in only weak fluorescence directly from A*.
S65T has been used as the foundation for the generation of additional fluorescent proteins. Intriguingly, Remington and colleagues demonstrated that replacement of H148 with an aspartate results in the recovery of green fluorescence following excitation of the A state.21 H148 is situated close to the chromophore’s phenolic hydroxyl residue, leading to the suggestion that the carboxylate of D148 is the proton acceptor for ESPT in this mutant. In the current work we have explored the excited state dynamics of S65T/H148D GFP using steady state and ultrafast time resolved fluorescence (TRF) and ultrafast TR infrared (TRIR) spectroscopies, and contrasted them with the single mutant S65T GFP. These studies indicate that excitation of the A state in the double mutant leads to very rapid formation of the anionic chromophore (cro) in the excited state (I*). However, unlike wtGFP, there is no evidence for a change in ionization state of a carboxylate residue upon formation of I*, leading to the hypothesis that a low barrier hydrogen bond is present between the phenol hydroxyl and the carboxylate of D148 in the ground state, and that electronic excitation results in rapid translational motion of the proton along the cro…H…D148 potential energy surface. This hypothesis is supported by additional experiments which show that green fluorescence following A state excitation can also be recovered in E222Q GFP by introduction of the H148D mutation. This interpretation of the dynamics and spectroscopy of the H148D mutants is in accord with the recent high resolution structural data from Remington and co-workers, demonstrating the presence of a very short (≤2.4Å) hydrogen bond between the chromophore phenol oxygen and D148 in S65T/H148D.23 The consequences of the H148D mutation for radiationless decay of the I* state are described.
Experimental
Plasmids containing the gene for wtGFP and S65T GFP in the pRSETb vector were obtained from Prof. Rebekka Wachter (Arizona State University). The mutations E222Q and H148D were incorporated using the QuikChange mutagenesis kit (Stratagene). The following primers were used with the mutation in bold:
E222Q:
| Forward: | 5′ CAACTATAACTCAGACAATGTATACATCATGG 3′ |
| Reverse: | 5′ CCATGATGTATACATTGTCTGAGTTATAGTTG 3′ |
H148D:
| Forward: | 5′ CCACATGGTCCTTCTTCAGTTTGTAACAGCTGC 3′ |
| Reverse: | 5′ GCAGCTGTTACAAACTGAAGAAGGACCATGTGG 3′ |
Following transformation of BL21-DE3/pLysS cells (Stratagene) with the appropriate plasmid, protein was expressed overnight at 25°C in the presence of 0.8 mM IPTG (Fisher). Subsequently, cells were lysed using a French press, centrifuged for 45 min at 33,000 rpm, and the protein purified by metal-affinity chromatography using Ni-NTA resin (Novagen). Fractions containing pure GFP were combined and dialyzed against 20 mM potassium phosphate pH 7.5 buffer containing 300 mM sodium chloride, and then concentrated using Microcon YM10 centrifuge filters (Millipore). An overnight, room temperature chymotrypsin digest (1 mg per 50 mg GFP) was used to cleave the affinity tag, which was then removed by passing the reaction mixture through fresh Ni-NTA resin. The extent of chromophore maturation in the mutants was comparable to that observed for wtGFP. TRIR and TRF experiments were typically conducted at pH 6.5 to 7.5 in order to minimize problems associated with protein aggregation that occurred at lower pH. For experiments conducted in D2O, proteins were exhaustively exchanged with solvent deuterium by repetitive cycles of lyophilization and reconstitution with D2O. Due to instability of the GFP mutants at low pH, TRIR experiments were conducted using protein samples in pH 7.5 buffer.
Absorbance spectra were acquired with a Cary 100 Bio UV/visible spectrophotometer (Varian) while steady state fluorescence spectra were acquired using a Fluorolog-3 fluorimeter (Jobin Yvon-Spex). Quantum yields (Φf) were estimated using wtGFP (Φf = 0.8) as a standard.13 The ability of the GFP mutants to photoconvert, for example via irreversible decarboxylation of an acidic residue, was studied using both an unfiltered Hg lamp as well as selected wavelengths generated by an argon-ion laser. The off-resonance Raman instrument and analysis methods have been described previously.22
The two instruments employed for ultrafast spectroscopy are based in the Ultrafast Spectroscopy Laboratory of the UK Central Laser Facility, and have been described in detail elsewhere.19, 24 The TRIR spectrometer measures IR difference spectra as a function of time after excitation with a sub 200 fs 400 nm pump pulse. The pump pulse energy was < 1 μJ focused to a spot size > 200 μm, and a broadband difference frequency IR pulse generated from signal and idler of a BBO (β-Barium Borate) based optical parametric amplifier was used as the probe. The time resolution was about 400 fs and the spectral resolution 3 cm−1. Typical sample concentrations were 1 mM and the sample was rastered in the beam to eliminate photodamage. The IR spectrum of water vapor was used to calibrate the TRIR data.
In most measurements the relative angle between the pump and probe beams was set to the magic angle (54.7°) to suppress effect of orientational relaxation. However, additional data may be obtained from polarization resolved measurements. The angle between the electronic and vibrational transition moments (θ) can be determined by exciting the samples with the 400 nm pump radiation polarized parallel or perpendicular to the linearly polarized IR probe. In either case the measured pump-on minus pump-off IR data were combined to generate the transient IR difference spectra at each delay time. For parallel and perpendicular polarization resolved data θ is obtained from the anisotropy, D, defined as the ratio of parallel to perpendicular intensity in the TRIR difference spectra25
via the relation
The assumption is that the protein itself does not rotate significantly between excitation and probe pulses (which is reasonable on the picosecond time scale of the measurements reported here). Thus, for angles θ varying from 0 to 90° D may take values between 0.5 and 3. The calculation cannot distinguish between θ and θ + 90° so two solutions exist. The application of the anisotropy method in assigning the observed vibrational modes has been discussed elsewhere.25
The time resolved fluorescence spectra were measured with the ultrafast Kerr gate method, which required even weaker excitation pulse intensities and sample concentrations of only a few μM. The spectral resolution was 4 nm and the temporal resolution about 4 ps (determined by the polarizability anisotropy relaxation time of the CS2 gate). Thus the Kerr gate method has a lower time resolution than the up-conversion method, but yields better spectral resolution.
Results
Absorption and fluorescence spectra
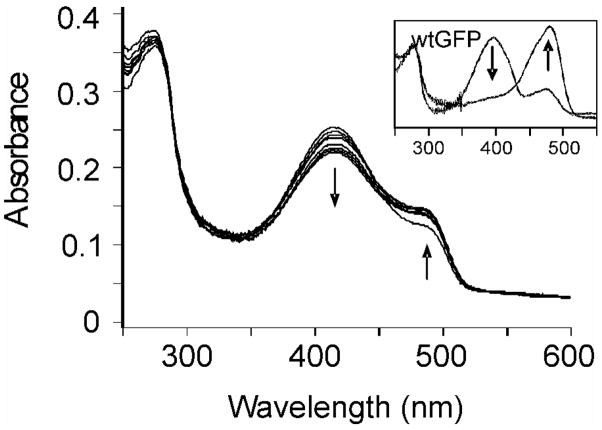
Absorbance and fluorescence of the GFP mutant, S65T/H148D, have been characterized and are in good agreement with earlier data.21 The absorption spectrum of the double mutant shows two bands at 411 and 488 nm, which are assigned to the neutral and anionic ground states, respectively (Figure 2). As observed for S65T, the absorption spectrum of S65T/H148D is sensitive to pH. However, introduction of D148 has caused the pKa observed for S65T (pKa = 6.0) to increase to 7.8,21 so that at pH 7.5 a far greater fraction of S65T/H148D is in the neutral form compared to S65T alone. Significantly the introduction of D148 has also caused the λmax of the neutral chromophore to red shift to 411 nm, a shift of 16 nm compared to wtGFP (395 nm) and S65T GFP (394 nm). In contrast, the λmax for the anionic chromophore in S65T/H148D is 488 nm, comparable to that of S65T (489 nm), but red-shifted relative to wtGFP (475 nm). The absorbance spectrum and pH sensitivity of E222Q/H148D are very similar to those of S65T/H148D, showing the red shift of the dominant neutral peak to 410 nm and the appearance of an anionic shoulder at 485 nm at higher pH.26
Figure 2.

Absorbance and excitation spectra in D2O of S65T and S65T/H148D at pD 7.1 (normalized at 280 nm).
Fluorescence emission spectra from the two double mutants are also virtually identical. Each shows strong green fluorescence with a maximum at 508 nm. Thus the emission spectrum following excitation of the neutral form is very similar to that of the wtGFP. The Φf of the two mutants varies with pH, ranging from 0.05 at pH 5.5 to 0.15 at pH 7.5. This pH dependence of Φf is similar to that reported by Remington and co-workers for S65T/H148D.21 In deuterated buffer (pD 7.1), the 508 nm excitation spectrum of S65T/H148D shows two distinct maxima corresponding to the presence of both neutral and anionic chromophore. In contrast to the absorption spectrum, where the B form is the minor component, the excitation spectrum has almost equal intensity for excitation of A and B forms (Figure 2). The excitation spectrum of E222Q/H148D is similar, showing both the neutral and anionic chromophore, and with a higher intensity of the B form than expected from the absorption spectrum. This disagreement between the excitation and absorption spectra suggests distinct fates for the two excitation pathways leading to the green emission.
Under steady state conditions, the two double mutants show no A* emission upon neutral excitation, even when fully exchanged into deuterated buffer (which enhances the A* emission in wtGFP due to the slower proton transfer). This is in contrast to wt and S65T GFP. Despite the absence of a distinct A* emission, some underlying heterogeneity of the spectrum is suggested by its observed dependence on excitation wavelength. Specifically, as the excitation wavelength increases from 410 to 484 nm, the emission spectra of both S65T/H148D and E222Q/H148D narrow slightly on the blue edge (shown for S65T/H148D in Figure 3). This leaves open the possibility that a weak red shifted A* emission is hidden underneath the dominant I* emission.
Figure 3.

Normalized steady state fluorescence emission of S65T/H148D (pH 7.5) with 410 nm and 484 nm excitation.
To further investigate the excited state chemistry of S65T/H148D and E222Q/H148D, we have recorded the time dependent emission spectra of the proteins using the ultrafast Kerr gated method, and compared them with those for S65T GFP (Figure 4). For S65T the early time spectra are dominated by a rapidly decaying blue shifted A* emission. The emission is broad as was observed for the corresponding A* emission in wtGFP.17 At longer times a narrow B* emission is also resolved which can be assigned to residual population of B form at pD 5.1. For the double mutants there is no evidence in the early time gated spectra for two distinct emission maxima arising from the decay of the A* and the concomitant formation of the I* states. In contrast to wtGFP, a single band is observed with a spectrum very similar to that of the I* emission of the wild-type which decays as a function of time after excitation. Detailed inspection of the normalized time gated spectra shows that the spectrum also narrows as a function of time after excitation (Figure 5). The narrowing is most noticeable as a collapse of the fluorescence on the blue edge on the timescale of tens of picoseconds. No isoemissive point was observed. The time dependence displayed in Figure 4 is reminiscent of the excitation wavelength dependence reported in Figure 3.
Figure 4.
TRF of S65T/H148D, E222Q/H148D and S65T at pD 5.1 in D2O with times delays noted. The short rise times seen in the first 6 ps for S65T/H148D and S65T simply reflect the convolution of the 4 ps instrument response with the emission. The apparent vibronic structure in the S65T emission may reflect interference effects in the collection optics.
Figure 5.

Time-resolved fluorescence emission (normalized at 508 nm) of wt GFP pD 7.1, S65T pD 5.1, S65T/H148D pD 6.1 and E222Q/H148D pD 6.1 in D2O.
The wavelength resolved time dependent fluorescence intensity is similar in the two double mutant proteins (Figure 6). The fluorescence measured at the red edge of the peak wavelength (515 nm) rises instantaneously within the 4 ps time resolution of the experiment and relaxes exponentially with a lifetime of approximately (300 ± 100) ps (Table 1). In contrast, the decay at 490 nm is non-single exponential over the first 50 ps with a dominant fast component of (10 ± 3) ps and a slow relaxation component of (160 ± 40) ps which is similar to that observed at 515 nm. Critically, the time dependence of the fluorescence has been measured in both H2O and D2O equilibrated samples and these are, within experimental limits, the same. This is in sharp contrast to the case in wtGFP, where deuteration dramatically slows the proton transfer rate.15, 16, 19, 27 These data suggest that there is a negligible barrier to proton transfer in the excited electronic state. The fast relaxation component reflects the dynamics of the spectral narrowing observed in the gated spectra (Figure 5). We assign this component to vibrational cooling of the rapidly formed I* state (discussed below).
Figure 6.

Fluorescence decay at 515 and 490 nm of S65T/H148D and E222Q/H148D in both D2O (pD 6.1) and H2O (pH 6.5).
Table 1.
Fluorescence Decay Rates for S65T/H148D and E222Q/H148D
| Decay rate constants | TRF 490 nm | TRF 490 nm | TRF 515 nm | TRF 515 nm | TRIR phenol | TRIR C=O |
|---|---|---|---|---|---|---|
| S65T/H148D | (H2O) | (D2O) | (H2O) | (D2O) | ||
| t1 | 11 ± 7 ps | 7 ± 3 ps | 300 ± 40 ps | 300 ± 100 ps | 7 ± 4 ps | 71 ± 19 ps |
| t2 | 160 ± 40 ps | 160 ± 35 ps | - | - | 113 ± 40 ps | 1.5 ± 0.3 ns |
| E222Q/H148D | (H2O) | (D2O) | (H2O) | (D2O) | ||
| t1 | 16 ± 3 ps | 12 ± 2 ps | 190 ± 20 ps | 190 ± 17 ps | 5 ± 2 ps | poor fit |
| t2 | 165 ± 30 ps | 150 ± 20 ps | - | - | 225 ± 355 ps |
Vibrational spectroscopy
Steady state Raman spectra of the neutral forms of both double mutant proteins (pH 5.5) show chromophore vibrational modes similar to wtGFP and S65T,22 with a dominant delocalized C=N stretch mode at 1554 cm−1 and C=C stretch at 1642 cm−1 (data not shown). For S65T these modes appear at 1560 and 1646 cm−1 respectively. Thus, the 21 nm red shift in λmax for A* excitation caused by introduction of D148 (394 to 415 nm) has had a remarkably small effect on the ground state structure of the chromophore. This correlation is in line with previous observations where it was noted that while solvent and environmental effects caused alterations in λmax for the neutral chromophore, they had little impact on positions of the normal modes.22 For example, while λmax for the neutral model chromophore 4′-hydroxybenzylidene-2,3-dimethylimidazolinone (HBDI) is at 368 nm, 26 nm to the blue of the corresponding value for S65T, the C=C stretch mode for this compound is at 1641 cm−1. In contrast, for the anionic form of the chromophore, there is a strong correlation between λmax and the position of normal modes in the vibrational spectrum, thus establishing a direct link between ground and excited state effects.22 This correlation was previously rationalized by hypothesizing that the protein had evolved to accommodate the anionic form of the chromophore, since this is the fluorescent state. Thus, the present data suggest that the neutral ground state structure of the double mutant is not markedly different to that found in S65T GFP; the red shift in λmax must result primarily from selective stabilization of the excited state by the D148 mutation.
The time-resolved emission spectra of both double mutants are consistent with deprotonation of the neutral chromophore in the excited state (A*) to form the fluorescent I* state. This process mirrors the photochemistry of wtGFP, in which the abstracted proton is transferred through a H-bond network with E222 as the final proton acceptor. This H-bond network is disrupted in the S65T and E222Q mutant proteins, and recovery of ESPT in the S65T/H148D and E222Q/H148D double mutants is hypothesized to result from D148 acting as the proton acceptor. To shed further light on this possibility, TRIR spectra following 400 nm excitation have been collected at time delays between 1 ps and 2 ns after excitation for S65T GFP and the two double mutant proteins, and are compared with TRIR data for wtGFP (Figure 7). In wtGFP a temporal evolution of the TRIR spectrum is observed which can be assigned to formation of E222-COOH (1710 cm−1) and disappearance of the corresponding carboxylate (1565 cm−1). The rest of the spectrum is dominated by bleach modes which can be assigned to the chromophore and some shifts of protein modes in response to electronic excitation.25, 27 In sharp contrast, the TRIR for the three mutants show only bleach and decay with no evolution in spectral profile. This is in agreement with the TRF (Figure 5) which show only an instantaneous (within the time resolution) formation and decay of the green emission. Compared with wtGFP, these mutants show a more rapid return to the ground state (e.g. approximately 300 ps bleach recovery for S65T/H148D versus 3 ns for wtGFP) and no long-lived intermediates. This is in reasonable agreement with the excited state lifetimes (Table 1).
Figure 7.

Time-resolved difference IR spectra of a) wt GFP, b) S65T pD 5.1, c) S65T/H148D pD 7.1, and d) E222Q/H148D pD 7.1 following excitation at 400 nm.
Importantly, in the double mutant TRIR spectrum there is no evidence for formation of the neutral carbonyl of D148 or loss of the associated D148 carboxylate absorption, although the fluorescence spectra clearly show emission from the I* state. As noted above, in wtGFP these modes appear at 1710 and 1565 cm−1, respectively. Similar frequencies are expected for D148, although a strengthening of H-bonding interactions involving this residue would cause a shift in υCOOH to lower frequency and a corresponding increase in frequency of the antisymmetric COO−,28–30 and plausibly to a situation where these modes are obscured by other transients in the TRIR spectrum. However, this possibility is not supported by the polarization-resolved TRIR spectra (see below).
A further remarkable difference with wtGFP is the lack of a transient absorption around 1560–1580 cm−1 in either of the double mutants. This band has multiple origins in GFP mutants, but all are associated with the formation of the A* state. Isotope labeling was previously used to assign an instantaneous transient absorption in this region of the spectrum to the cro A* C=O in wtGFP.25 Interestingly, S65T also shows the immediate formation of an absorption at 1560 cm−1 that could similarly be assigned to the A* C=O. Thus, there is substantial shift of the C=O stretch mode to lower wavenumber upon excitation, reflecting a weakening of the bond in response to photoexcitation-induced electronic rearrangement.25 However, in S65T there is also a significant new bleach observed in a complex band around 1640 cm−1, which is also observed in other mutants in which proton relay has been blocked.31 The 1560–80 cm−1 transient is also observed in those mutants, so protein modes shifted from 1640 cm−1 by interaction with A* may also contribute to absorption in this region of the spectrum. In contrast, for S65T/H148D and E222Q/H148D, the formation of I* from A* is so rapid that transients arising from A*, or protein modes perturbed by it, cannot be observed within the 400 fs time resolution of the TRIR experiment. The absence of a transient chromophore mode associated with I* in the two double mutants is consistent with previous TRIR studies on T203V/E222Q,19, 25 a GFP variant proposed to exist exclusively in the I ground state,32 and in which no strong excited state absorptions were observed in the TRIR spectra.
An intense promptly formed transient absorption can be observed at 1660 cm−1 in the spectra of both S65T/H148D and E222Q/H148D double mutants. This band, which relaxes on the hundreds of picoseconds time scale does not appear in the wtGFP magic angle spectrum (Figure 7a). However, a mode at this frequency does appear with a sigmoidal shape in the parallel polarized transient absorption of wtGFP, and was assigned as a protein mode.25 Thus, the 1660cm−1 mode observed here (Figure 7b–d) may be of the same, protein, origin. A plausible alternative assignment is to an excited state mode of the chromophore itself. The measured anisotropy is similar to that for the ground state C=O localized mode (Figure 8), indicating that the transient has its vibrational transition moment pointing in the same direction. This in turn suggests an assignment to the C=O mode of I*, formed during a prompt proton transfer. However, no such intense mode appears in wtGFP as I* is formed, or in mutants where I* is directly excited (e.g. T203V/E222Q10). The definite assignment of this mode as protein or chromophore localized requires isotopic substitution studies on the double mutants.25 However, whatever the final assignment the distinction between the double mutants and wtGFP TRIR spectra is once again very clear.
Figure 8.
Polarization-resolved TRIR data for S65T/H148D pD 7.1 at 100 ps after excitation.
Finally, the bleach at 1600 cm−1 in wtGFP is reliably assigned to a stretching mode of the chromophore phenyl group, and is narrow in most GFP mutants.25, 31 In S65T/H148D this band is much broader than in wt or S65T GFP. However, the high value of the anisotropy (Figure 8) is identical to that for the narrower wtGFP mode; such a high value of the anisotropy is characteristic of the phenyl mode, confirming that this is the same mode in the double mutant. This in turn suggests that the phenyl mode has been significantly broadened in the double mutant, presumably by its interaction with D148.
A possible complication in TRIR is the potential for irreversible photoconversion of the sample to an anionic form of the chromophore. Absorption spectra before and after the experimental runs were examined and no conversion was noted under these conditions. Further study with S65T/H148D using off-resonance Raman indicated only limited photoconversion with much higher levels of exposure of UV light (a combination of 333, 351 and 363.8 nm at 30 mW and using a spot size of 1 cm2) and requiring longer (>30 minutes) exposure (Figure 9). A two hour recovery time resulted in negligible return to the neutral form implying this limited conversion is irreversible. Exposure to these wavelengths for greater than two hours resulted in photobleaching of the sample. Importantly, these studies confirm that no appreciable photoconversion is expected under the irradiation conditions employed in the TRIR experiments.
Figure 9.
Photoconversion of S65T/H148D using a combination of wavelengths (333, 351, and 364 nm) followed for 2 hours. WtGFP photoconversion with unfiltered Hg lamp is shown in inset.
Discussion
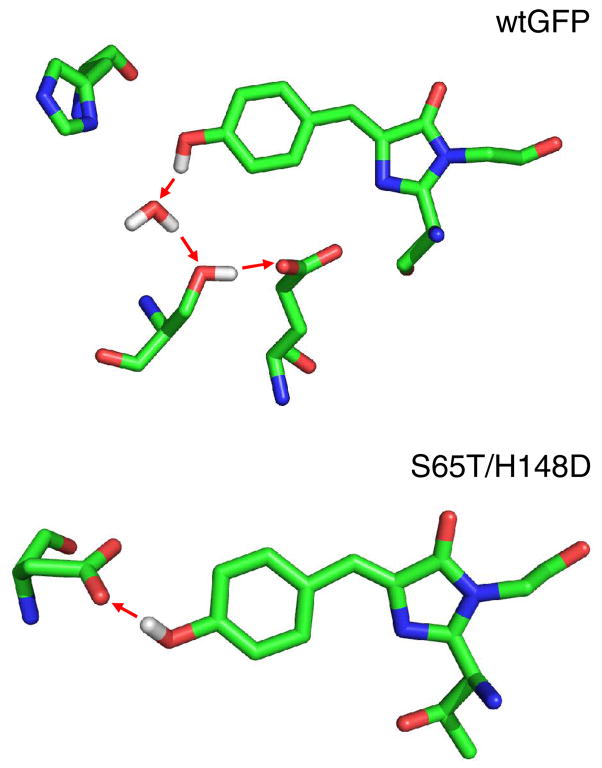
The spectroscopic results for the two double mutants reveal a mechanism for ESPT which differs greatly from that of wtGFP. Specifically the ground state A → A* spectrum is perturbed, the proton transfer is ultrafast (probably sub-picosecond, Figure 6–7) and insensitive to deuteration (Figure 6) and no distinct carbonyl mode can be assigned to the formation of the protonated acceptor. Rather than proton transfer through a network to a distant receptor, as occurs in the wild type protein, the present results suggest an interpretation based on the existence of a low barrier H-bond between D148 and the chromophore in which the proton participates in a polar covalent bond with both oxygens (Figure 10).33 This arrangement allows barrierless ESPT upon photoexcitation within a strongly coupled chromophore D148 complex, as illustrated in Figure 11 and discussed further below. Recent work by Remington and co-workers on the structure of S65T/H148D GFP provides concrete evidence of the presence of such a low barrier H-bond in these mutants. This group has resolved the structure to 1.5 Å, revealing an extremely short (≤ 2.4Å) H-bond connecting the chromophore to D148 (S.J. Remington, personal communication).
Figure 10.
Comparison of the proposed H+ transfer routes in wtGFP and S65T/H148D upon excitation.
Figure 11.
A representation of the proposed potential energy surfaces for wtGFP (A) and S65T/H148D GFP (B). In wtGFP the phenolic OH bond is shorter and the acceptor remote. In S65T/H148D the donor and acceptor are close yielding the low barrier and flat potential surface. On excitation, wtGFP forms A* which converts to I* via a barrier on the picosecond time scale. In S65T/H148D the transfer is barrierless and occurs in < 1 ps. The I* state so formed is vibrationally hot and cools to form the emissive state. The hot state may also access a nonradiative part of the potential energy surface.
The interaction between the chromophore and D148 is evident in the TRIR spectrum, where the formation of a new H-bond is suggested by the perturbation of the phenol mode at 1600 cm−1 (Figure 7). We interpret the observed broadening as indicative of either static or dynamic disorder, due to small changes in geometry around the shared proton, leading to significant shifts in the frequency of the phenol localized mode. The chromophore D148 interaction is also evident in the red shifted electronic absorption spectrum of the A → A* transition. Raman and TRIR data suggest that ground state modes not associated with the phenol ring are unperturbed by D148. Thus the spectral shift mainly arises from stabilization of the chromophore excited electronic state. This is interpreted as arising from the formation of an extended H-bond leading to a localization of charge on the phenolic oxygen, such that the phenol develops a more quinoidal structure than is found in S65T or wtGFP.
Another major difference in the TRIR spectra between S65T/H148D and wtGFP is the absence of an excited state absorption at 1560 cm−1 in the former. A band is observed at this position in S65T, as well as in wtGFP, and is assigned to the C=O absorption of A* and protein modes perturbed by interaction with it in both S65T and wtGFP. The absence of this band in S65T/H148D is consistent with the rapid formation of I* from A* within the time resolution of the TRIR measurements.
In both H148D mutants a temporal evolution in the protonation state of D148 was anticipated in TRIR, based on previous observations in wtGFP.19 However, this is not observed. No transient C=O stretch mode due to the formation of aspartic acid was seen (expected above 1716 cm−1)34; losses of the asymmetric (1576 cm−1) and symmetric (1410 cm−1) stretch modes of the ionized aspartate were also not observed, even though I* is formed promptly and exists for several hundred picoseconds. The absence of any change assignable to the protonation state of the carbonyl of D148 is unexpected (unlike the weak or undetectable contribution of I* which has been noted for other mutants10). It suggests that the spectra of the initial ionic and final neutral states of D148 are not very different. We propose that the presence of a low barrier H-bond in the ground state can explain the lack of evolution in our TRIR data. The polar covalent nature of the short H-bond between the phenolic proton and D148 implies the presence in the ground state of the neutral-like aspartic acid. Upon excitation, D148 does not change protonation state, but merely binds the proton more tightly. Thus no large shifts are observed in D148 vibrational modes. The proton-phenol oxygen bond however does change upon excitation, converting from a polar covalent to a hydrogen bond in an ultrafast barrierless process (Figure 11). This allows for the instantaneous appearance of the anionic chromophore and green fluorescence. This is very different from the situation in wtGFP, where the proton acceptor, E222, converts from the ionized to the acid form upon chromophore excitation, with the conversion observed in the TRIR spectra.
An important question with regard to the mechanism is whether or not A* fluoresces prior to proton transfer. While the narrowing at the blue edge of the emission spectrum might indicate the presence of short-lived A* fluorescence, the absence of an isoemissive point argues against a simple two state model. In addition the lack of an observable isotope effects argues against a distinct A* state feeding the anionic form in an activated process. Finally there is no corresponding 10 ps rise-time of the 508 nm emission, unlike that seen for wtGFP.16, 17 We propose therefore, on the basis of both the undetectable A* emission in gated fluorescence with 4ps time resolution and the absence of A* related modes in TRIR with 400 fs time resolution, that A* does not fluoresce directly but is quenched on a sub picosecond time scale with a major fate being ESPT. A consequence of the ultrafast proton transfer is the formation of a vibrationally hot excited state population in the initial I* state. This is proposed to be the source of the higher energy emission, which relaxes as the excited state cools on the picosecond timescale through vibrational energy transfer to the protein matrix. A time scale of 10 ps is typical for vibrational cooling.
The distinction between absorption and excitation spectra (Figure 2) suggests a further level of complication in the photophysics of S65T/H148D GFP. The enhanced emission from the directly excited B state is explicable either because A* has an additional decay route competing with ultrafast ESPT or because the vibrationally hot I* state can transform into a nonradiative conformation which is not accessible from B*. In either case it suggests a new degree of control over the fluorescence emission of GFP mutants.
These ideas are summarized in a representation of the reaction coordinate associated with the proton transfer reaction shown in Figure 11. To a certain extent the picture used here is borrowed from extensive investigations of excited state intramolecular proton transfer, which has been studied in the condensed and gas phase for a number of systems.35–37 These ideas can be extended to intermolecular proton transfer in the case of proteins because the structure of the protein holds the reaction partners in place, whereas in solution they would be free to diffuse apart. Significantly, it is this feature of S65T/H148D GFP photophysics which makes it suitable as a model system to study low barrier or barrierless proton transfer reactions in proteins. In intramolecular systems the proton transfer may occur on a sub-picosecond timescale and be insensitive to hydrogen/deuterium exchange,38 a situation mirrored for the intermolecular (in the sense of chromophore to D148) proton transfer reactions that occur in the GFP double mutants.
In Figure 11 the proposed reaction coordinates for wtGFP and double mutant proton transfer are contrasted. For wtGFP the multi-step proton relay reaction is represented by a single effective barrier between the chromophore donor and the E222 acceptor, since the precise shape of the reaction coordinate is not known. The key factor is that there is a significant barrier to proton transfer in the ground state such that the A form is the most stable. In S65T/H148D the distance between the O atoms in the donor chromophore and the acceptor is far shorter than any of the distances between O atoms along the H-bond network in wtGFP. Even if the donor acceptor proton affinity are unchanged this has the effect of producing a lower barrier, stretching both the OH bond of the chromophore and probably modifying the C=O of the acceptor. The lower barrier will in turn imply a greater range of distances are accessible to the OH bond at a given temperature, possibly contributing to the observed broadening of the chromophore phenyl mode. In the figure the pKa for the donor and acceptor are assumed unchanged, so the proton nevertheless remains localized on the chromophore, such that it remains in the A state. It should be emphasized that Figure 11 is a simplified one dimensional representation of what is in reality a multidimensional surface. Indeed theory and experiment for the intramolecular case show that the probability of proton transfer is likely to be modified by vibrational modes which modulate the distance between the O atoms involved.35
Upon electronic excitation the pKa of the chromophore is reduced.39, 40 In the case of wtGFP the effect is to make the anionic I* form more stable. However, the proton transfer proceeds from A* to I* through a barrier, leading to the observed picosecond lifetime of the A* and its sensitivity to isotopic exchange. The effect of the same pKa change for the S65T/H148D is to make the proton transfer barrierless, such that the I* state is reached on a sub-picosecond time scale after excitation of A*, by essentially a translation of the proton between the two O atoms. This mechanism is consistent with the absence of deuterium isotope effect and the prompt rise of the 508 nm emission observed. It is our proposal that the similarity of the initial and final structures, suggested by the absence of a product state contribution in the TRIR spectra, is also consistent with the strong low barrier H bond formed in the ground electronic state.
The sub-picosecond proton transfer means that reaction occurs faster than the excess energy can be dissipated, so the initial I* state is formed vibrationally hot. The hot initial state may either transform to a non-radiative state (accounting for the relative inefficiency of emission from A* excitation compared to direct B* excitation) or cool on a 10 ps timescale. Vibrational cooling results in the narrowing of the emission spectrum with time and the observation of a fast component on the blue edge of the time resolved fluorescence spectra.
Although somewhat speculative this model is consistent with the range of experimental observations reported. Of course the potential energy surfaces displayed need to be supported by detailed high level quantum chemical calculations. Even if such surfaces become available (in itself a challenging objective) the subsequent calculation of the ultrafast rate coefficients will require non equilibrium dynamics calculations.
Conclusions
The observed recovery of ESPT in S65T and E222Q GFP through the incorporation of a proximal proton acceptor indicates that multiple pathways of proton transfer in GFP proteins are possible. While proton transfer in wtGFP occurs along a hydrogen bond network, culminating with the protonation of E222,19 proton transfer in S65T/H148D and E222Q/H148D instead occurs directly from the chromophore to D148. This is made possible by the pre-existence of a low barrier or barrierless H-bond between the phenol group of the chromophore and the side chain of aspartate. These results demonstrate the capability of control over the pathway of proton transfer in GFP through mutagenesis. Importantly this sets the scene for using GFP mutants as a model system for understanding the molecular basis for proton transfer in biological systems.
Acknowledgments
We are grateful to the Science & Technology Facilities Council for access to the facilities. SRM is grateful to EPSRC for financial support, and JN thanks the Leverhulme trust for a fellowship. This work was supported by NIH grant GM66818 to PJT. We also think Prof. Remington for sharing data from X-ray structural studies prior to publication.
References
- 1.Bell AF, Feng Y, Hofstein HA, Parikh S, Wu J, Rudolph MJ, Kisker C, Whitty A, Tonge PJ. Stereoselectivity of Enoyl-CoA Hydratase Results from Preferential Activation of One of Two Bound Substrate Conformers. Chem Biol. 2002;9(11):1247–55. doi: 10.1016/s1074-5521(02)00263-6. [DOI] [PubMed] [Google Scholar]
- 2.Reece SY, Hodgkiss JM, Stubbe J, Nocera DG. Proton-coupled electron transfer: the mechanistic underpinning for radical transport and catalysis in biology. Philos Trans R Soc Lond B Biol Sci. 2006;361(1472):1351–64. doi: 10.1098/rstb.2006.1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Richard JP, Amyes TL. Proton transfer at carbon. Curr Opin Chem Biol. 2001;5(6):626–33. doi: 10.1016/s1367-5931(01)00258-7. [DOI] [PubMed] [Google Scholar]
- 4.Fillingame RH. Coupling H+ transport and ATP synthesis ln F1F0-ATP synthases: Glimpses of interacting parts in a dynamic molecular machine. J Exp Biol. 1997;200(2):217–224. doi: 10.1242/jeb.200.2.217. [DOI] [PubMed] [Google Scholar]
- 5.Hosler JP, Ferguson-Miller S, Mills DA. Energy transduction: proton transfer through the respiratory complexes. Annu Rev Biochem. 2006;75:165–87. doi: 10.1146/annurev.biochem.75.062003.101730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kresge AJ. What Makes Proton-Transfer Fast. Acc Chem Res. 1975;8(10):354–360. [Google Scholar]
- 7.Jeffrey G. An Introduction to Hydrogen Bonding. Oxford University Press; New York: 1997. [Google Scholar]
- 8.Cleland WW, Frey PA, Gerlt JA. The low barrier hydrogen bond in enzymatic catalysis. J Biol Chem. 1998;273(40):25529–32. doi: 10.1074/jbc.273.40.25529. [DOI] [PubMed] [Google Scholar]
- 9.Shan SO, Herschlag D. The change in hydrogen bond strength accompanying charge rearrangement: Implications for enzymatic catalysis. Proc Natl Acad Sci U S A. 1996;93(25):14474–14479. doi: 10.1073/pnas.93.25.14474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shimomura O. Structure of the Chromophore of Aequorea Green Fluorescent Protein. FEBS Lett. 1979;104(2):220–222. [Google Scholar]
- 11.Cody CW, Prasher DC, Westler WM, Prendergast FG, Ward WW. Chemical Structure of the Hexapeptide Chromophore of the Aequorea Green Fluorescent Protein. Biochemistry. 1993;32(5):1212–1218. doi: 10.1021/bi00056a003. [DOI] [PubMed] [Google Scholar]
- 12.Tsien RY. The green fluorescent protein. Annu Rev Biochem. 1998;67:509–544. doi: 10.1146/annurev.biochem.67.1.509. [DOI] [PubMed] [Google Scholar]
- 13.Palm GJ, Wlodawer A. Spectral variants of green fluorescent protein. Methods Enzymol. 1999;302:378–94. doi: 10.1016/s0076-6879(99)02034-0. [DOI] [PubMed] [Google Scholar]
- 14.Ward WW, Prentice HJ, Roth AF, Cody CW, Reeves SC. Spectral Perturbations of the Aequorea Green Fluorescent Protein. Photochem Photobiol. 1982;35(6):803–808. [Google Scholar]
- 15.Lossau H, Kummer A, Heinecke R, PollingerDammer F, Kompa C, Bieser G, Jonsson T, Silva CM, Yang MM, Youvan DC, Michel-Beyerle ME. Time-resolved spectroscopy of wild-type and mutant Green Fluorescent Proteins reveals excited state deprotonation consistent with fluorophore-protein interactions. Chem Phys. 1996;213(1–3):1–16. [Google Scholar]
- 16.Chattoraj M, King BA, Bublitz GU, Boxer SG. Ultra-fast excited state dynamics in green fluorescent protein: Multiple states and proton transfer. Proc Natl Acad Sci U S A. 1996;93(16):8362–8367. doi: 10.1073/pnas.93.16.8362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jaye AA, Stoner-Ma D, Matousek P, Towrie M, Tonge PJ, Meech SR. Time-resolved emission spectra of green fluorescent protein. Photochem Photobiol. 2006;82(2):373–379. [Google Scholar]
- 18.Brejc K, Sixma TK, Kitts PA, Kain SR, Tsien RY, Ormo M, Remington SJ. Structural basis for dual excitation and photoisomerization of the Aequorea victoria green fluorescent protein. Proc Natl Acad Sci U S A. 1997;94(6):2306–2311. doi: 10.1073/pnas.94.6.2306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stoner-Ma D, Jaye AA, Matousek P, Towrie M, Meech SR, Tonge PJ. Observation of excited-state proton transfer in green fluorescent protein using ultrafast vibrational spectroscopy. J Am Chem Soc. 2005;127(9):2864–2865. doi: 10.1021/ja042466d. [DOI] [PubMed] [Google Scholar]
- 20.Heim R, Cubitt AB, Tsien RY. Improved Green Fluorescence. Nature. 1995;373(6516):663–664. doi: 10.1038/373663b0. [DOI] [PubMed] [Google Scholar]
- 21.Elsliger MA, Wachter RM, Hanson GT, Kallio K, Remington SJ. Structural and spectral response of green fluorescent protein variants to changes in pH. Biochemistry. 1999;38(17):5296–301. doi: 10.1021/bi9902182. [DOI] [PubMed] [Google Scholar]
- 22.Bell AF, He X, Wachter RM, Tonge PJ. Probing the ground state structure of the Green Fluorescent Protein chromophore using Raman spectroscopy. Biochemistry. 2000;39(15):4423–4431. doi: 10.1021/bi992675o. [DOI] [PubMed] [Google Scholar]
- 23.Shu X, Kallio K, Shi X, Abbyad P, Kanchanawong P, Childs W, Boxer SG, Remington SJ. Ultrafast Excited-State Dynamics in the Green Fluorescent Protein Variant S65T/H148D. 1. Mutagenesis and Structural Studies. Biochemistry. 2007 doi: 10.1021/bi7009037. Epub Oct 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kwok WM, Ma C, Matousek P, Parker AW, Phillips D, Toner WT, Towrie M, Umapathy S. A determination of the structure of the intramolecular charge transfer state of 4-dimethylaminobenzonitrile (DMABN) by time-resolved resonance Raman spectroscopy. J Phys Chem. 2001;A105(6):984–990. [Google Scholar]
- 25.Stoner-Ma D, Melief EH, Nappa J, Ronayne KL, Tonge PJ, Meech SR. Proton relay reaction in green fluorescent protein (GFP): Polarization-resolved ultrafast vibrational spectroscopy of isotopically edited GFP. J Phys Chem. 2006;B110(43):22009–22018. doi: 10.1021/jp065326u. [DOI] [PubMed] [Google Scholar]
- 26.Stoner-Ma D. PhD Thesis. Stony Brook University; Stony Brook: 2006. Spectroscopic studies of green fluorescent protein. [Google Scholar]
- 27.van Thor JJ, Zanetti G, Ronayne KL, Towrie M. Structural events in the photocycle of green fluorescent protein. J Phys Chem B. 2005;109(33):16099–16108. doi: 10.1021/jp051315+. [DOI] [PubMed] [Google Scholar]
- 28.Tonge PJ, Moore GR, Wharton CW. Fourier-transform infrared studies of the alkaline isomerization of mitochondrial cytochrome c and the ionization of carboxylic acids. Biochem J. 1989;258:599–605. doi: 10.1042/bj2580599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cassidy CS, Reinhardt LA, Cleland WW, Frey PA. Hydrogen bonding in complexes of carboxylic acids with 1-alkylimidazoles: steric and isotopic effects on low barrier hydrogen bonding. J Chem Soc Perkin Trans. 1999;2(3):635–641. [Google Scholar]
- 30.Tobin JB, Whitt SA, Cassidy CS, Frey PA. Low-Barrier Hydrogen-Bonding in Molecular-Complexes Analogous to Histidine and Aspartate in the Catalytic Triad of Serine Proteases. Biochemistry. 1995;34(21):6919–6924. doi: 10.1021/bi00021a002. [DOI] [PubMed] [Google Scholar]
- 31.Stoner-Ma D, Jaye AA, Ronayne KL, Nappa J, Tonge PJ, Meech SR. Ultrafast Electronic and Vibrational Dynamics of Stabilized A State Mutants of the Green Fluorescent Protein (GFP): Snipping the Proton Wire. Chem Phys. 2007 doi: 10.1016/j.chemphys.2008.02.021. submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wiehler J, Jung G, Seebacher C, Zumbusch A, Steipe B. Mutagenic stabilization of the photocycle intermediate of green fluorescent protein (GFP) ChemBioChem. 2003;4:1164–1171. doi: 10.1002/cbic.200300595. [DOI] [PubMed] [Google Scholar]
- 33.Schiott B, Iversen BB, Madsen GKH, Larsen FK, Bruice TC. On the electronic nature of low-barrier hydrogen bonds in enzymatic reactions. Proc Nat Acad Sci U S A. 1998;95(22):12799–12802. doi: 10.1073/pnas.95.22.12799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Barth A. The infrared absorption of amino acid side chains. Prog Biophys Mol Biol. 2000;74(3–5):141–173. doi: 10.1016/s0079-6107(00)00021-3. [DOI] [PubMed] [Google Scholar]
- 35.Guallar V, Batista VS, Miller WH. Semiclassical molecular dynamics simulations of intramolecular proton transfer in photoexcited 2-(2 ′-hydroxyphenyl)-oxazole. J Chem Phys. 2000;113(21):9510–9522. [Google Scholar]
- 36.Sobolewski AL, Domcke W. Ab initio study of excited-state intramolecular proton dislocation in salicylic acid. Chem Phys. 1998;232(3):257–265. [Google Scholar]
- 37.Mitra S, Das R, Bhattacharyya SP, Mukherjee S. Energetic and dynamic aspects of intramolecular proton transfer in 4-methyl-2,6-diformylphenol: A detailed analysis with AM1 potential energy surfaces. J Phys Chem A. 1997;101(3):293–298. [Google Scholar]
- 38.Rodriguez-Cordoba W, Zugazagoitia JS, Collado-Fregoso E, Peon J. Excited state intramolecular proton transfer in schiff bases. Decay of the locally excited enol state observed by femtosecond resolved fluorescence. J Phys Chem A. 2007;111(28):6241–6247. doi: 10.1021/jp072415d. [DOI] [PubMed] [Google Scholar]
- 39.Scharnagl C, Raupp-Kossmann RA. Solution pK(a) values of the green fluorescent protein chromophore from hybrid quantum-classical calculations. J Phys Chem B. 2004;108(1):477–489. [Google Scholar]
- 40.Wehry EL, Rogers LB. Application of Linear Free Energy Relations to Electronically Excited States of Monosubstituted Phenols. J Am Chem Soc. 1965;87(19):4234. [Google Scholar]