Abstract
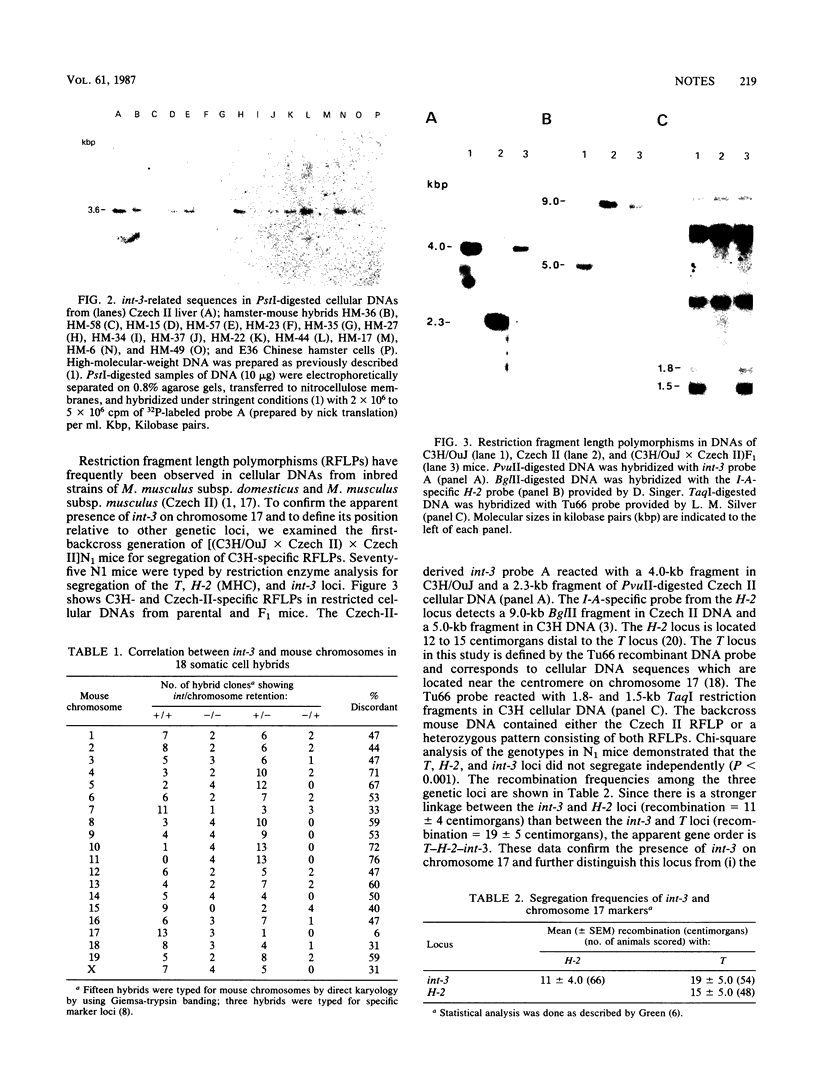
Mus musculus subsp. musculus (Czech II) mammary tumor DNA frequently contains an integrated proviral genome of the mouse mammary tumor virus (MMTV) within a specific 0.5-kilobase-pair region of the cellular genome (designated int-3). Viral integration at this site results in activation of expression of an adjacent cellular gene. We mapped int-3 to mouse chromosome 17 by analysis of PstI-restricted cellular DNAs from mouse-hamster somatic cell hybrids. Restriction analysis of cellular DNA from (C3H/OuJ X Czech II) X Czech II backcross mice established the gene order T-H-2-int-3. These results demonstrated that the int-3 locus is distinct from two other common integration regions for mouse mammary tumor virus (designated int-1 and int-2) in mammary tumor DNA and suggest that several cellular genes may be at risk for virally induced activation during mammary tumor development.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Callahan R., Drohan W., Gallahan D., D'Hoostelaere L., Potter M. Novel class of mouse mammary tumor virus-related DNA sequences found in all species of Mus, including mice lacking the virus proviral genome. Proc Natl Acad Sci U S A. 1982 Jul;79(13):4113–4117. doi: 10.1073/pnas.79.13.4113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen J. C., Murphey-Corb M. Targeted integration of baboon endogenous virus in the BEVI locus on human chromosome 6. Nature. 1983 Jan 13;301(5896):129–132. doi: 10.1038/301129a0. [DOI] [PubMed] [Google Scholar]
- Davis M. M., Cohen D. I., Nielsen E. A., Steinmetz M., Paul W. E., Hood L. Cell-type-specific cDNA probes and the murine I region: the localization and orientation of Ad alpha. Proc Natl Acad Sci U S A. 1984 Apr;81(7):2194–2198. doi: 10.1073/pnas.81.7.2194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickson C., Smith R., Brookes S., Peters G. Tumorigenesis by mouse mammary tumor virus: proviral activation of a cellular gene in the common integration region int-2. Cell. 1984 Jun;37(2):529–536. doi: 10.1016/0092-8674(84)90383-0. [DOI] [PubMed] [Google Scholar]
- Escot C., Hogg E., Callahan R. Mammary tumorigenesis in feral Mus cervicolor popaeus. J Virol. 1986 May;58(2):619–625. doi: 10.1128/jvi.58.2.619-625.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallahan D., Callahan R. Mammary tumorigenesis in feral mice: identification of a new int locus in mouse mammary tumor virus (Czech II)-induced mammary tumors. J Virol. 1987 Jan;61(1):66–74. doi: 10.1128/jvi.61.1.66-74.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallahan D., Escot C., Hogg E., Callahan R. Mammary tumorigenesis in feral species of the genus Mus. Curr Top Microbiol Immunol. 1986;127:354–361. doi: 10.1007/978-3-642-71304-0_43. [DOI] [PubMed] [Google Scholar]
- Kozak C. A. Genetic mapping of a mouse chromosomal locus required for mink cell focus-forming virus replication. J Virol. 1983 Oct;48(1):300–303. doi: 10.1128/jvi.48.1.300-303.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak C. A. Retroviruses as chromosomal genes in the mouse. Adv Cancer Res. 1985;44:295–336. doi: 10.1016/s0065-230x(08)60030-5. [DOI] [PubMed] [Google Scholar]
- Lemons R. S., Nash W. G., O'Brien S. J., Benveniste R. E., Sherr C. J. A gene (Bevi) on human chromosome 6 is an integration site for baboon type C DNA provirus in human cells. Cell. 1978 Aug;14(4):995–1005. doi: 10.1016/0092-8674(78)90353-7. [DOI] [PubMed] [Google Scholar]
- Lemons R. S., O'Brien S. J., Sherr C. J. A new genetic locus, Bevi, on human chromosome 6 which controls the replication of baboon type C virus in human cells. Cell. 1977 Sep;12(1):251–262. doi: 10.1016/0092-8674(77)90203-3. [DOI] [PubMed] [Google Scholar]
- Nusse R., Varmus H. E. Many tumors induced by the mouse mammary tumor virus contain a provirus integrated in the same region of the host genome. Cell. 1982 Nov;31(1):99–109. doi: 10.1016/0092-8674(82)90409-3. [DOI] [PubMed] [Google Scholar]
- Nusse R., van Ooyen A., Cox D., Fung Y. K., Varmus H. Mode of proviral activation of a putative mammary oncogene (int-1) on mouse chromosome 15. Nature. 1984 Jan 12;307(5947):131–136. doi: 10.1038/307131a0. [DOI] [PubMed] [Google Scholar]
- Peters G., Brookes S., Smith R., Dickson C. Tumorigenesis by mouse mammary tumor virus: evidence for a common region for provirus integration in mammary tumors. Cell. 1983 Jun;33(2):369–377. doi: 10.1016/0092-8674(83)90418-x. [DOI] [PubMed] [Google Scholar]
- Peters G., Kozak C., Dickson C. Mouse mammary tumor virus integration regions int-1 and int-2 map on different mouse chromosomes. Mol Cell Biol. 1984 Feb;4(2):375–378. doi: 10.1128/mcb.4.2.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters G., Lee A. E., Dickson C. Activation of cellular gene by mouse mammary tumour virus may occur early in mammary tumour development. Nature. 1984 May 17;309(5965):273–275. doi: 10.1038/309273a0. [DOI] [PubMed] [Google Scholar]
- Robbins J. M., Gallahan D., Hogg E., Kozak C., Callahan R. An endogenous mouse mammary tumor virus genome common in inbred mouse strains is located on chromosome 6. J Virol. 1986 Feb;57(2):709–713. doi: 10.1128/jvi.57.2.709-713.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Röhme D., Fox H., Herrmann B., Frischauf A. M., Edström J. E., Mains P., Silver L. M., Lehrach H. Molecular clones of the mouse t complex derived from microdissected metaphase chromosomes. Cell. 1984 Mar;36(3):783–788. doi: 10.1016/0092-8674(84)90358-1. [DOI] [PubMed] [Google Scholar]
- Silver L. M. Genetic organization of the mouse t complex. Cell. 1981 Dec;27(2 Pt 1):239–240. doi: 10.1016/0092-8674(81)90406-2. [DOI] [PubMed] [Google Scholar]