Abstract
Background
Inflammatory bowel disease (IBD) is a clinically, and likely genetically, heterogeneous group of disorders. A recent report suggests that genetic variations in the TNFSF15 gene contribute to the susceptibility of IBD in both Japanese and caucasian populations.
Aims
To confirm the association between TNFSF15 high and low risk haplotypes and IBD in a caucasian population.
Methods
Five SNPs that comprise the two common haplotypes were genotyped in 599 caucasian patients with Crohn’s disease (CD), 382 caucasian patients with Ulcerative Colitis (UC) and 230 ethnically-matched healthy controls, including both Jews and non-Jews.
Results
The previously reported ‘risk’ haplotype was not associated with CD or UC (88.2% in CD cases vs. 88.3% in controls, p=0.96; 88.1% in UC cases vs. 88.3% in controls, p=0.78). We did however observe an increased frequency of the “protective” haplotype in Non-Jewish controls for both CD and UC (38.8% CD cases vs. 50% controls, p=0.01; 37.3% UC cases vs. 50% controls, p=0.01) with no such effect observed in the Jewish samples. There was an interactive effect between ethnicity and the protective haplotype in CD (p=0.04).
Conclusions
We observed a protective haplotype, consisting of the minor alleles for all five markers, to have a higher frequency in the non-Jewish controls than in CD and UC. Of further interest, the haplotype frequency was in the opposite direction, in our Jewish case-control panels (both CD and UC), leading us to conclude (1) TNFSF15 is indeed an IBD susceptibility gene, and (2) the disease susceptibility is ethnic specific.
Keywords: Inflammatory bowel disease, Crohn’s disease, Ulcerative Colitis, genotype, genetics
Introduction
Inflammatory bowel disease (IBD) is a group of complex idiopathic disorders, resulting from gene-environment interactions. The major forms of IBD are Crohn’s disease (CD) and ulcerative colitis (UC). A major distinction between CD and UC is the distribution of the inflammation. Inflammation in CD can affect any portion of the gastrointestinal tract, affecting most commonly the terminal ileum of the small intestine. It is discontinuous, asymmetric and can involve all layers of the bowel wall, with aggressive disease including intestinal perforations, strictures, and fistulas to adjacent tissues. UC is confined to the colon and is continuous, symmetric, and affects only the innermost layers of the bowel wall. In North America, the incidence rates for UC range from 2.2 to 14.3 cases per 100,000 persons-years and for CD it ranges 3.1 to 14.6 cases per 100,000 persons 1. The prevalence of UC ranges from 37 to 246 cases/100,000 people and 26 to 199/100,000 persons for CD.
Factors such as ethnicity and the presence of affected first-degree relatives have been repeatedly shown to be significant in the frequency of disease expression. Relatives of persons with either CD or UC have an increased risk to develop either form of IBD 2. The relative risk to first-degree relatives of a proband with IBD is 4–10 fold 3. There have been several familial aggregation studies that indicate UC and CD are genetically complex traits that involve incomplete penetrance due to genetically multigenic and/or heterogeneous defects 4. Linkage studies, identifying seven different IBD linkage regions, paved the path to potential candidate genes5–14. Several candidate gene studies have since emerged, revealing association to IBD or one of its clinical subphenotypes, including MDR1 15, 16, IBD5 (inclusive of OCTN) 14, 18, DLG5 17, 18, ICAM-1 19 and the most notable association, predisposing in ~30% of CD patients, the NOD2/CARD15 association in the IBD1 region 5, 6. These findings confirmed the role genetics has in IBD pathogenesis and stimulated interest in identifying further susceptibility genes. Our group has recently focused efforts on the candidate gene TNFSF15 (also known as TL1A) reported by Yamazaki and colleagues in 2005. Yamazaki 20 found a core haplotype of TNFSF15 showing association with increased risk for CD in a Japanese case control and in a caucasian family study population. They also identified a common haplotype that was found to be associated with a lowered risk of disease susceptibility in both their family-based and in case-control samples.
TNFSF15 was first cloned in 2002 at Human Genome Sciences 21. It is thought to be involved in the inflammatory process of both rheumatoid arthritis and atherosclerosis 22. However, TNFSF15 seems to exert its effects most profoundly in the gut 23–27. In vitro studies have shown that TNFSF15 is produced by human umbilical vein endothelial cells (HUVEC), as well as by T-cells, monocytes/macrophages, and dendritic cells 23, 25, 28–30. TNFSF15 enhances the induction of interferon-gamma (IFN-γ) expression in human T cells, and natural killer cells when combined with IL-12/18 23–25. TNFSF15 is a very potent enhancer of IFN-γ production in that as little as 100pg/ml is capable of inducing a 5–10 fold increase in IFN-γ levels from IL-12/IL-18 activated human T-cells 24. In a mouse model of Listeria monocytogenes, Hsu et al demonstrated a central role for this molecule in providing protection against dissemination of this pathogen 30. Thus, TNFSF15 is central for optimizing Th1 responses for protection of mucosal pathogens, yet can be critical for generation of enhanced Th1 responses in abnormal mucosal inflammation such as seen with Crohn’s disease. Therefore, defining genetic variations in the TNFSF15 gene that could alter expression and/or function of this molecule may allow a better understanding of the role of this molecule in gut homeostasis and abnormal inflammation.
Our group sought to replicate the CD-TNFSF15 association that Yamazaki et al 2005 observed in our own caucasian case control cohort. Given the importance of ethnicity in genetics, especially that of IBD, we analyzed our data overall and within each ethnic group, Jewish and non-Jewish.
Materials and Methods
Patients
We ascertained 1211 caucasian subjects seen at Cedars-Sinai Medical Center. The cohort consisted of 599 CD patients, 382 ulcerative colitis patients and 230 controls. Part of this cohort has been studied previously 31–33. The Institutional Review Board at Cedars-Sinai Medical Center approved the study protocol, and informed consent was obtained from all study subjects. The diagnosis of Crohn’s disease, ulcerative colitis and clinical sub-phenotypes were based on standard criteria including clinical, endoscopic, radiographic, and histopathological criteria 31, 32, 34–36. Patients with indeterminate colitis were excluded. All of the controls were geographically and ethnically selected to be maximally similar to our patients, while having no family history of IBD or autoimmune disorders. Also, we evaluated our cohort on ethnicity, Jewish or non-Jewish. We defined Jewish as those individuals with at least one of four grandparents of Jewish origin 37, 38 and more specifically, our Jewish population is primarily of Ashkenazi descent.
Genotyping
DNA was extracted from the peripheral whole blood collected using PureGene DNA isolation kit (Gentra Systems; Minneapolis, MN.) Five single nucleotide polymorphisms (SNP) previously found to be associated with CD in a Japanese cohort (rs3810936, rs6478108, rs6478109, rs7848647, rs7869487) were tested 20. These SNPs were used to construct 2 common haplotypes previously seen in both a Japanese and a caucasian population, as shown in Figure 1 20. Probes and primers were designed for the TaqMan MGB allelic discrimination method (Applied Biosystems, Foster City, CA.) The fluorescent amplifications were measured using the ABI Prism 7000 Sequence Detection System.
Figure 1.
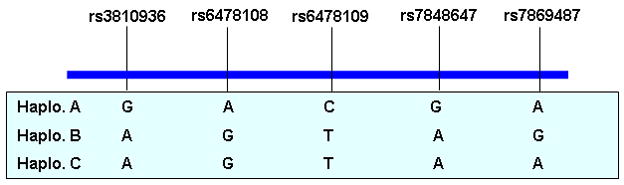
Main haplotype structure of TNFSF15 gene
Statistical Analysis
None of the markers exhibited significant deviation from the Hardy-Weinberg equilibrium in our sample set (p=0.05 level). Genotype distributions of individual markers were compared between cases and controls. Individual haplotypes were estimated by using Bayesian statistical method as implemented in the PHASE software (v2.0) 39, 40. For each haplotype, a dominant model was assumed, i.e., carriers of the particular haplotype versus non-carriers were compared. Chi-square (χ2) tests were used to test associations between the haplotypes and disease status and disease phenotype characteristics. A one-tailed p-value of less than 0.05 under the χ2 distribution was considered statistically significant. All analyses were done in total sample first, and then separated in Jewish and non-Jewish population. Mutiple logistic regression was used to test the interactive effect between ethnicity and haplotype in the total sample. Statistical analysis was conducted by SAS software (SAS Institute; Cary, NC.).
Considering the modest sample size in our Jewish control population, we also did a power calculation in the analysis. Two types of power analyses were conducted. First, we calculated the power for testing association between TNFSF15 and disease under the current sample size in the Jewish population by using the odds ratio reported for the caucasian population. Second, we computed the power for detecting interaction between ethnicity and TNFSF15 under different estimates of joint gene-ethnicity effect, using the reported odds ratio for estimate of marginal gene effect and ethnicity effect. The alternative hypothesis was 1-sided and type I error rate was 0.05. The program QUANTO was used for the power calculation.
Results
The five genotyped SNPs formed three common haplotypes in our population (A, B and C), and all were observed in the prior work by Yamazaki 20. The haplotypes carrier frequencies are shown in Tables 1–3. We were unable to confirm the association Yamazaki found between Haplotype A, the risk haplotype, and their Japanese case control population. In our IBD population the Haplotype A carrier frequency was 88.5% in cases vs. 88.3% in controls, in CD it was 88.2% in cases vs. 88.3% in controls and in UC it was 89.1% in cases and 88.3% in controls, with p-values 0.93, 0.96, and 0.78 respectively. Also, when stratified by Jewish ethnicity, Haplotype A was not found to be associated with CD, UC or IBD.
Table 1.
TNFSF15 haplotype carrier associations with CD
| Haplotype | Overall Haplotype Carrier Frequency | Non-Jew Haplotype Carrier Frequency | Jew Haplotype Carrier Frequency | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Cases(n=599) | Controls(n=230) | P | OR(95% CI) | Cases(n=345) | Controls(n=176) | P | OR(95% CI) | Cases(n=254) | Controls(n=54) | P | OR(95% CI) | |
| A | 88.2% | 88.3% | 0.96 | 0.99(0.62, 1.59) | 87.8% | 88.1% | 0.94 | 0.98(0.56, 1.71) | 88.6% | 88.9% | 0.95 | 0.97(0.38, 2.46) |
| B | 36.1% | 44.4% | 0.03 | 0.71 (0.53, 0.96) | 38.8% | 50.0% | 0.01 | 0.64 (0.44, 0.92) | 32.3% | 25.9% | 0.36 | 1.4 (0.7, 2.6) |
| C | 5.5% | 7.0% | 0.43 | 0.78 (0.42, 1.45) | 5.2% | 6.3% | 0.63 | 0.83 (0.38, 1.79) | 5.9% | 9.3% | 0.36 | 0.62 (0.2, 2.27) |
Table 3.
TNFSF15 haplotype carrier associations with IBD (CD+UC)
| Haplotype | Overall Haplotype Carrier Frequency | Non-Jews Haplotype Carrier Frequency | Jews Haplotype Carrier Frequency | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Cases (n=981) | Controls (n=230) | P | OR (95% CI) | Cases (n=581) | Controls (n=176) | P | OR (95% CI) | Cases (n=400) | Controls (n=54) | P | OR (95% CI) | |
| A | 88.5% | 88.3% | 0.93 | 1.02 (0.65, 1.6) | 88.6% | 88.1% | 0.83 | 1.1 (0.63, 1.78) | 88.3% | 88.9% | 0.81 | 0.94(0.38, 2.31) |
| B | 34.9% | 44.4% | 0.007 | 0.67(0.5, 0.9) | 38.2% | 50.0% | 0.005 | 0.62 (0.44, 0.87) | 30.0% | 25.9% | 0.54 | 1.22 (0.64, 2.33) |
| C | 5.0% | 7.0% | 0.23 | 0.7(0.39, 1.26) | 4.5% | 6.3% | 0.34 | 0.7(0.34, 1.45) | 5.8% | 9.3% | 0.36 | 0.6(0.21, 2.11) |
Overall, Haplotype A had a carrier frequency in controls of 88.3%, Haplotype B 44.4%, and Haplotype C 7.0%. However, the overall result masked distinct ethnic differences. In non-Jews, the haplotype carrier frequencies in controls were 88.1%, 50.0% and 6.3% respectively, while in Jews they were 88.9%, 25.9% and 9.3%. When the results are separated by ethnicity into non-Jewish and Jewish subgroups, Haplotype B carriers, consisting of the rare allele for each marker, were found to be protective against CD, but specifically only in non-Jews (38.8% in CD vs 50.0% in controls, OR: 0.64 CI: 0.44–0.92, p=0.01.) These results were similar to the caucasian population of the Yamazaki et al. In contrast, Haplotype B frequencies had an opposite trend in our Jewish population, with 32.3% in the CD cases versus 25.9% in controls; this difference was not significant (p=0.36). The ethnic distinction was further verified through logistic regression, in which an interactive effect between ethnicity and Haplotype B in CD was detected (p=0.04).
The same haplotype analysis was performed on the UC cohort. The combined UC cohort (including both Jewish and non-Jewish panels) did show an association between Haplotype B and UC as shown in Table 2 (p=0.005, OR: 0.62 CI: 0.44–0.86.) However, the association was clarified when results were separated by ethnicity, with the non-Jewish population having a significant association with Haplotype B (37.3% vs 50.0%, OR: 0.59 CI: 0.40–0.88, p=0.01.)In the Jewish UC cohort, there was no difference in Haplotype B carrier frequency (26.0% in cases and 25.9% in controls.
Table 2.
TNFSF15 haplotype carrier associations with UC
| Haplotype | Overall Haplotype Carrier Frequency | Non-Jew Haplotype Carrier Frequency | Jew Haplotype Carrier Frequency | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Cases(n=382) | Controls(n=230) | P | OR(95% CI) | Cases(n=236) | Controls n=176)( | P | OR(95% CI) | Cases(n=146) | Controls(n=54) | P | OR (95% CI) | |
| A | 89.1% | 88.3% | 0.78 | 1.1 (0.64, 1.8) | 89.8% | 88.1% | 0.57 | 1.2(0.642.22) | 87.7% | 88.9% | 0.81 | 0.89 (0.33, 2.37) |
| B | 33.0% | 44.4% | 0.005 | 0.62 (0.44, 0.86) | 37.3% | 50.0% | 0.01 | 0.59 (0.4,0.88) | 26.0% | 25.9% | 0.99 | 1 (0.49, 2.05) |
| C | 4.2% | 7.0% | 0.14 | 0.58 (0.29, 1.19) | 3.4% | 6.3% | 0.18 | 0.53 (0.21, 1.34) | 5.5% | 9.3% | 0.34 | 0.57 (0.16, 2.32) |
The CD cohort was further stratified by clinical phenotypes: uc-like, fibrostenotic, internal perforating, perianal penetrating and small bowel surgery to investigate anymore detailed associations. No association was found between clinical phenotypes and TNFSF15 haplotypes in the non-Jewish population (data not shown). However, in the Jewish population, patients undergoing small bowel surgery appeared to have a higher frequency of Haplotype B (p=0.005, OR 2.2 CI 1.3, 3.8) and lower frequency of Haplotype A (p=0.009, OR 0.33, 95%CI: 0.14, 0.76) than those without small bowel surgery. Since previous research studies have shown age and gender effects in IBD susceptibility genes, namely DLG541, 42, we analyzed for these variables with the TNFSF15 haplotypes. We found the distribution of the major haplotype is similar in early-onset vs. late-onset CD/UC and male vs. female (Supplementary materials, Tables 1–4.) Finally, since a similar effect was seen in both CD and UC, we combined both disease groups and the result was that the protective effect achieved even greater statistical significance (Table 3.)
Table 4.
Power for detecting interaction between different joint gene-ethnicity effects
| OR (gene-Jewish ethnicity) | * Power (1-beta) |
|---|---|
| 1.9 | 0.61 |
| 2 | 0.67 |
| 2.1 | 0.72 |
| 2.2 | 0.76 |
| 2.3 | 0.8 |
| 2.4 | 0.83 |
| 2.5 | 0.86 |
assuming type I error=0.05 (one-sided)
Discussion
We sought to replicate the conclusions drawn from Yamazaki’s caucasian panel results, identifying both a high and low-risk haplotype association with CD. While we were unable to confirm association with the high-risk Haplotype A, we did see similar results Yamazaki found in their caucasian case control group. Neither group showed association between Haplotype A and CD, UC or IBD in this population. The protective Haplotype B was observed in both Yamazaki’s and our caucasian panels. Our results demonstrated an association between the low-risk Haplotype (B) and CD, UC and IBD in the non-Jewish caucasian population. We identified a distinct ethnic relationship, in that the result was confined to non-Jews. This confirms and clarifies the Yamazaki et al 2005 20 finding that Haplotype B is protective against IBD in a caucasian population, but it is specifically in a non-Jewish caucasian population. The frequency trend of Haplotype B was in fact in the opposite direction in our Jewish CD and UC panels compared with the direction seen in our non-Jewish panels and Yamazaki’s caucasian cohorts. One possible explanation for this difference in frequency between our overall study cohort and Yamazaki’s is that latter study consisted mainly of patients from non-Jewish descent. This protective haplotype association with both UC and with CD and IBD (CD+UC) in our non-Jewish cohort, demonstrates that this gene is not CD or UC specific, but is in fact an IBD gene.
When analyzing TNFSF15 haplotype association with the various CD phenotypes, we observed an association between Haplotype B and small bowel surgery in Jewish patients only p=0.006, OR 2.2 (CI 1.25, 3.75.) This further supports our hypothesis that there are ethnic differences in the susceptibility, powered by the gene.
One limitation of our study is that the sample size for the Jewish controls is relatively modest. We agree that the sample size may not be large enough for identifying the association in Jewish case-control if we assume the same gene effect as non-Jews (power: 0.55–0.72). However, since we observed an opposite trend of effect of Haplotype B in Jews compared with non-Jews, this finding justified us performing an interaction test. Table 4 shows the power to detect interaction under the different estimates for joint gene-ethnicity effect in our current sample size, assuming the odds ratios for marginal gene effect and marginal Jewish ethnicity effect are 0.7 (from the reference) and 2.5, respectively. From the table, we can see that when the odds ratio for the joint effect is greater than 2.0, there will be appropriate power to identify the interaction (>0.7). The results of our study support the interactive effect between ethnicity and TNFSF15 gene in CD, which indicated that the effects of gene may depend on the ethnic group. To confirm the finding, the results should be repeated in other samples.
Only some of the genes providing genetic susceptibility genes for IBD have been identified. Some of the genes that have been implicated in IBD susceptibility, such as NOD2/CARD15, IBD5, interleukin 1 receptor antagonist, HLA class II and herein TNFSF15, demonstrate different gene frequencies in different ethnic groups43–47. Thus, lack of an association in one ethnic group cannot be used to exclude a relationship with disease status in other ethnic groups. In fact, it emphasizes the importance of defining haplotype frequencies and gene marker associations for different ethnic groups.
It is worthwhile to make distinctions between populations’ ethnicities, as it is well recognized that there is a 2–4 fold increased frequency of Crohn’s disease in Jews as compared with non-Jewish groups in the same geographic area 48, 49. Genes predisposing Jewish people to CD may be different than those in a non-Jewish caucasian population due to the population’s unique evolutionary history 49. Various studies have focused their efforts on understanding the apparent frequency discrepancy of IBD in Jews vs. non-Jews dating back to the 1960s 50–52 and most recently a study of potential risk factors for IBD in a Canadian population found that being Jewish was significantly associated with CD and UC (OR=4.32, 95% CI 1.10–16.9 and OR=7.46, 95% CI, 2.33–23.89 respectively) 53. This finding helps illustrate that it would be expected that there will be some difference in genetic predisposition amongst Jewish and non-Jewish populations.
Acknowledgments
Supported by the National Institute of Diabetes & Digestive & Kidney Diseases Program Project Grant (DK46763), IBD TL1A and Crohn’s Disease Mucosal Inflammation (DK56328), and the Cedars-Sinai Board of Governors’Chair in Medical Genetics.
Bibliography
- 1.Loftus EV., Jr Clinical epidemiology of inflammatory bowel disease: Incidence, prevalence, and environmental influences. Gastroenterology. 2004;126:1504–17. doi: 10.1053/j.gastro.2004.01.063. [DOI] [PubMed] [Google Scholar]
- 2.Bonen DK, Cho JH. The genetics of inflammatory bowel disease. Gastroenterology. 2003;124:521–36. doi: 10.1053/gast.2003.50045. [DOI] [PubMed] [Google Scholar]
- 3.Orholm M, Munkholm P, Langholz E, Nielsen OH, Sorensen TI, Binder V. Familial occurrence of inflammatory bowel disease. N Engl J Med. 1991;324:84–8. doi: 10.1056/NEJM199101103240203. [DOI] [PubMed] [Google Scholar]
- 4.Yang H, Rotter JI. The genetic background of inflammatory bowel disease. Hepatogastroenterology. 2000;47:5–14. [PubMed] [Google Scholar]
- 5.Hugot JP, Chamaillard M, Zouali H, Lesage S, Cezard JP, Belaiche J, Almer S, Tysk C, O’Morain CA, Gassull M, Binder V, Finkel Y, Cortot A, Modigliani R, Laurent-Puig P, Gower-Rousseau C, Macry J, Colombel JF, Sahbatou M, Thomas G. Association of NOD2 leucine-rich repeat variants with susceptibility to Crohn’s disease. Nature. 2001;411:599–603. doi: 10.1038/35079107. [DOI] [PubMed] [Google Scholar]
- 6.Ogura Y, Bonen DK, Inohara N, Nicolae DL, Chen FF, Ramos R, Britton H, Moran T, Karaliuskas R, Duerr RH, Achkar JP, Brant SR, Bayless TM, Kirschner BS, Hanauer SB, Nunez G, Cho JH. A frameshift mutation in NOD2 associated with susceptibility to Crohn’s disease. Nature. 2001;411:603–6. doi: 10.1038/35079114. [DOI] [PubMed] [Google Scholar]
- 7.Williams CN, Kocher K, Lander ES, Daly MJ, Rioux JD. Using a genome-wide scan and meta-analysis to identify a novel IBD locus and confirm previously identified IBD loci. Inflamm Bowel Dis. 2002;8:375–81. doi: 10.1097/00054725-200211000-00001. [DOI] [PubMed] [Google Scholar]
- 8.Cho JH, Nicolae DL, Gold LH, Fields CT, LaBuda MC, Rohal PM, Pickles MR, Qin L, Fu Y, Mann JS, Kirschner BS, Jabs EW, Weber J, Hanauer SB, Bayless TM, Brant SR. Identification of novel susceptibility loci for inflammatory bowel disease on chromosomes 1p, 3q, and 4q: evidence for epistasis between 1p and IBD1. Proc Natl Acad Sci U S A. 1998;95:7502–7. doi: 10.1073/pnas.95.13.7502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rioux JD, Silverberg MS, Daly MJ, Steinhart AH, McLeod RS, Griffiths AM, Green T, Brettin TS, Stone V, Bull SB, Bitton A, Williams CN, Greenberg GR, Cohen Z, Lander ES, Hudson TJ, Siminovitch KA. Genomewide search in Canadian families with inflammatory bowel disease reveals two novel susceptibility loci. Am J Hum Genet. 2000;66:1863–70. doi: 10.1086/302913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hampe J, Shaw SH, Saiz R, Leysens N, Lantermann A, Mascheretti S, Lynch NJ, MacPherson AJ, Bridger S, van Deventer S, Stokkers P, Morin P, Mirza MM, Forbes A, Lennard-Jones JE, Mathew CG, Curran ME, Schreiber S. Linkage of inflammatory bowel disease to human chromosome 6p. Am J Hum Genet. 1999;65:1647–55. doi: 10.1086/302677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ma Y, Ohmen JD, Li Z, Bentley LG, McElree C, Pressman S, Targan SR, Fischel-Ghodsian N, Rotter JI, Yang H. A genome-wide search identifies potential new susceptibility loci for Crohn’s disease. Inflamm Bowel Dis. 1999;5:271–8. doi: 10.1097/00054725-199911000-00005. [DOI] [PubMed] [Google Scholar]
- 12.Duerr RH, Barmada MM, Zhang L, Pfutzer R, Weeks DE. High-density genome scan in Crohn disease shows confirmed linkage to chromosome 14q11–12. Am J Hum Genet. 2000;66:1857–62. doi: 10.1086/302947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.van Heel DA, Dechairo BM, Dawson G, McGovern DP, Negoro K, Carey AH, Cardon LR, Mackay I, Jewell DP, Lench NJ. The IBD6 Crohn’s disease locus demonstrates complex interactions with CARD15 and IBD5 disease-associated variants. Hum Mol Genet. 2003;12:2569–75. doi: 10.1093/hmg/ddg281. [DOI] [PubMed] [Google Scholar]
- 14.Rioux JD, Daly MJ, Silverberg MS, Lindblad K, Steinhart H, Cohen Z, Delmonte T, Kocher K, Miller K, Guschwan S, Kulbokas EJ, O’Leary S, Winchester E, Dewar K, Green T, Stone V, Chow C, Cohen A, Langelier D, Lapointe G, Gaudet D, Faith J, Branco N, Bull SB, McLeod RS, Griffiths AM, Bitton A, Greenberg GR, Lander ES, Siminovitch KA, Hudson TJ. Genetic variation in the 5q31 cytokine gene cluster confers susceptibility to Crohn disease. Nat Genet. 2001;29:223–8. doi: 10.1038/ng1001-223. [DOI] [PubMed] [Google Scholar]
- 15.Brant SR, Panhuysen CI, Nicolae D, Reddy DM, Bonen DK, Karaliukas R, Zhang L, Swanson E, Datta LW, Moran T, Ravenhill G, Duerr RH, Achkar JP, Karban AS, Cho JH. MDR1 Ala893 polymorphism is associated with inflammatory bowel disease. Am J Hum Genet. 2003;73:1282–92. doi: 10.1086/379927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Potocnik U, Ferkolj I, Glavac D, Dean M. Polymorphisms in multidrug resistance 1 (MDR1) gene are associated with refractory Crohn disease and ulcerative colitis. Genes Immun. 2004;5:530–9. doi: 10.1038/sj.gene.6364123. [DOI] [PubMed] [Google Scholar]
- 17.Stoll M, Corneliussen B, Costello CM, Waetzig GH, Mellgard B, Koch WA, Rosenstiel P, Albrecht M, Croucher PJ, Seegert D, Nikolaus S, Hampe J, Lengauer T, Pierrou S, Foelsch UR, Mathew CG, Lagerstrom-Fermer M, Schreiber S. Genetic variation in DLG5 is associated with inflammatory bowel disease. Nat Genet. 2004;36:476–80. doi: 10.1038/ng1345. [DOI] [PubMed] [Google Scholar]
- 18.Peltekova VD, Wintle RF, Rubin LA, Amos CI, Huang Q, Gu X, Newman B, Van Oene M, Cescon D, Greenberg G, Griffiths AM, St George-Hyslop PH, Siminovitch KA. Functional variants of OCTN cation transporter genes are associated with Crohn disease. Nat Genet. 2004;36:471–5. doi: 10.1038/ng1339. [DOI] [PubMed] [Google Scholar]
- 19.Yang H, Vora DK, Targan SR, Toyoda H, Beaudet AL, Rotter JI. Intercellular adhesion molecule 1 gene associations with immunologic subsets of inflammatory bowel disease. Gastroenterology. 1995;109:440–8. doi: 10.1016/0016-5085(95)90331-3. [DOI] [PubMed] [Google Scholar]
- 20.Yamazaki K, McGovern D, Ragoussis J, Paolucci M, Butler H, Jewell D, Cardon L, Takazoe M, Tanaka T, Ichimori T, Saito S, Sekine A, Iida A, Takahashi A, Tsunoda T, Lathrop M, Nakamura Y. Single nucleotide polymorphisms in TNFSF15 confer susceptibility to Crohn’s disease. Hum Mol Genet. 2005;14:3499–506. doi: 10.1093/hmg/ddi379. [DOI] [PubMed] [Google Scholar]
- 21.Migone TS, Zhang J, Luo X, Zhuang L, Chen C, Hu B, Hong JS, Perry JW, Chen SF, Zhou JX, Cho YH, Ullrich S, Kanakaraj P, Carrell J, Boyd E, Olsen HS, Hu G, Pukac L, Liu D, Ni J, Kim S, Gentz R, Feng P, Moore PA, Ruben SM, Wei P. TL1A is a TNF-like ligand for DR3 and TR6/DcR3 and functions as a T cell costimulator. Immunity. 2002;16:479–92. doi: 10.1016/s1074-7613(02)00283-2. [DOI] [PubMed] [Google Scholar]
- 22.Kang YJ, Kim WJ, Bae HU, Kim DI, Park YB, Park JE, Kwon BS, Lee WH. Involvement of TL1A and DR3 in induction of pro-inflammatory cytokines and matrix metalloproteinase-9 in atherogenesis. Cytokine. 2005;29:229–35. doi: 10.1016/j.cyto.2004.12.001. [DOI] [PubMed] [Google Scholar]
- 23.Prehn JL, Mehdizadeh S, Landers CJ, Luo X, Cha SC, Wei P, Targan SR. Potential role for TL1A, the new TNF-family member and potent costimulator of IFN-gamma, in mucosal inflammation. Clin Immunol. 2004;112:66–77. doi: 10.1016/j.clim.2004.02.007. [DOI] [PubMed] [Google Scholar]
- 24.Papadakis KA, Zhu D, Prehn JL, Landers C, Avanesyan A, Lafkas G, Targan SR. Dominant role for TL1A/DR3 pathway in IL-12 plus IL-18-induced IFN-gamma production by peripheral blood and mucosal CCR9+ T lymphocytes. J Immunol. 2005;174:4985–90. doi: 10.4049/jimmunol.174.8.4985. [DOI] [PubMed] [Google Scholar]
- 25.Bamias G, Martin C, 3rd, Marini M, Hoang S, Mishina M, Ross WG, Sachedina MA, Friel CM, Mize J, Bickston SJ, Pizarro TT, Wei P, Cominelli F. Expression, localization, and functional activity of TL1A, a novel Th1-polarizing cytokine in inflammatory bowel disease. J Immunol. 2003;171:4868–74. doi: 10.4049/jimmunol.171.9.4868. [DOI] [PubMed] [Google Scholar]
- 26.Bamias G, Mishina M, Nyce M, Ross WG, Kollias G, Rivera-Nieves J, Pizarro TT, Cominelli F. Role of TL1A and its receptor DR3 in two models of chronic murine ileitis. Proc Natl Acad Sci U S A. 2006;103:8441–6. doi: 10.1073/pnas.0510903103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Young HA, Tovey MG. TL1A: a mediator of gut inflammation. Proc Natl Acad Sci U S A. 2006;103:8303–4. doi: 10.1073/pnas.0602655103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim S, Zhang L. Identification of naturally secreted soluble form of TL1A, a TNF-like cytokine. J Immunol Methods. 2005;298:1–8. doi: 10.1016/j.jim.2004.12.019. [DOI] [PubMed] [Google Scholar]
- 29.Yang CR, Hsieh SL, Teng CM, Ho FM, Su WL, Lin WW. Soluble decoy receptor 3 induces angiogenesis by neutralization of TL1A, a cytokine belonging to tumor necrosis factor superfamily and exhibiting angiostatic action. Cancer Res. 2004;64:1122–9. doi: 10.1158/0008-5472.can-03-0609. [DOI] [PubMed] [Google Scholar]
- 30.Hsu MJ, Lin WW, Tsao WC, Chang YC, Hsu TL, Chiu AW, Chio CC, Hsieh SL. Enhanced adhesion of monocytes via reverse signaling triggered by decoy receptor 3. Exp Cell Res. 2004;292:241–51. doi: 10.1016/j.yexcr.2003.09.019. [DOI] [PubMed] [Google Scholar]
- 31.Vasiliauskas EA, Kam LY, Karp LC, Gaiennie J, Yang H, Targan SR. Marker antibody expression stratifies Crohn’s disease into immunologically homogeneous subgroups with distinct clinical characteristics. Gut. 2000;47:487–96. doi: 10.1136/gut.47.4.487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Abreu MT, Taylor KD, Lin YC, Hang T, Gaiennie J, Landers CJ, Vasiliauskas EA, Kam LY, Rojany M, Papadakis KA, Rotter JI, Targan SR, Yang H. Mutations in NOD2 are associated with fibrostenosing disease in patients with Crohn’s disease. Gastroenterology. 2002;123:679–88. doi: 10.1053/gast.2002.35393. [DOI] [PubMed] [Google Scholar]
- 33.Mow WS, Vasiliauskas EA, Lin YC, Fleshner PR, Papadakis KA, Taylor KD, Landers CJ, Abreu-Martin MT, Rotter JI, Yang H, Targan SR. Association of antibody responses to microbial antigens and complications of small bowel Crohn’s disease. Gastroenterology. 2004;126:414–24. doi: 10.1053/j.gastro.2003.11.015. [DOI] [PubMed] [Google Scholar]
- 34.Greenstein AJ, Lachman P, Sachar DB, Springhorn J, Heimann T, Janowitz HD, Aufses AH., Jr Perforating and non-perforating indications for repeated operations in Crohn’s disease: evidence for two clinical forms. Gut. 1988;29:588–92. doi: 10.1136/gut.29.5.588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vasiliauskas EA, Plevy SE, Landers CJ, Binder SW, Ferguson DM, Yang H, Rotter JI, Vidrich A, Targan SR. Perinuclear antineutrophil cytoplasmic antibodies in patients with Crohn’s disease define a clinical subgroup. Gastroenterology. 1996;110:1810–9. doi: 10.1053/gast.1996.v110.pm8964407. [DOI] [PubMed] [Google Scholar]
- 36.Gasche C, Schober E, Turetschek K. Small bowel barium studies in Crohn’s disease. Gastroenterology. 1998;114:1349. doi: 10.1016/s0016-5085(98)70454-8. [DOI] [PubMed] [Google Scholar]
- 37.Roth MP, Petersen GM, McElree C, Feldman E, Rotter JI. Geographic origins of Jewish patients with inflammatory bowel disease. Gastroenterology. 1989;97:900–4. doi: 10.1016/0016-5085(89)91495-9. [DOI] [PubMed] [Google Scholar]
- 38.Yang H, McElree C, Roth MP, Shanahan F, Targan SR, Rotter JI. Familial empirical risks for inflammatory bowel disease: differences between Jews and non-Jews. Gut. 1993;34:517–24. doi: 10.1136/gut.34.4.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stephens M, Smith NJ, Donnelly P. A new statistical method for haplotype reconstruction from population data. Am J Hum Genet. 2001;68:978–89. doi: 10.1086/319501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stephens M, Donnelly P. A comparison of bayesian methods for haplotype reconstruction from population genotype data. Am J Hum Genet. 2003;73:1162–9. doi: 10.1086/379378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Friedrichs F, Brescianini S, Annese V, Latiano A, Berger K, Kugathasan S, Broeckel U, Nikolaus S, Daly MJ, Schreiber S, Rioux JD, Stoll M. Evidence of transmission ratio distortion of DLG5 R30Q variant in general and implication of an association with Crohn disease in men. Hum Genet. 2006;119:305–11. doi: 10.1007/s00439-006-0133-1. [DOI] [PubMed] [Google Scholar]
- 42.Biank V, Friedrichs F, Babusukumar U, Wang T, Stoll M, Broeckel U, Kugathasan S. DLG5 R30Q variant is a female-specific protective factor in pediatric onset Crohn’s disease. Am J Gastroenterol. 2007;102:391–8. doi: 10.1111/j.1572-0241.2006.01011.x. [DOI] [PubMed] [Google Scholar]
- 43.Inoue N, Tamura K, Kinouchi Y, Fukuda Y, Takahashi S, Ogura Y, Inohara N, Nunez G, Kishi Y, Koike Y, Shimosegawa T, Shimoyama T, Hibi T. Lack of common NOD2 variants in Japanese patients with Crohn’s disease. Gastroenterology. 2002;123:86–91. doi: 10.1053/gast.2002.34155. [DOI] [PubMed] [Google Scholar]
- 44.Negoro K, McGovern DP, Kinouchi Y, Takahashi S, Lench NJ, Shimosegawa T, Carey A, Cardon LR, Jewell DP, van Heel DA. Analysis of the IBD5 locus and potential gene-gene interactions in Crohn’s disease. Gut. 2003;52:541–6. doi: 10.1136/gut.52.4.541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tountas NA, Casini-Raggi V, Yang H, Di Giovine FS, Vecchi M, Kam L, Melani L, Pizarro TT, Rotter JI, Cominelli F. Functional and ethnic association of allele 2 of the interleukin-1 receptor antagonist gene in ulcerative colitis. Gastroenterology. 1999;117:806–13. doi: 10.1016/s0016-5085(99)70338-0. [DOI] [PubMed] [Google Scholar]
- 46.Cavanaugh J. International collaboration provides convincing linkage replication in complex disease through analysis of a large pooled data set: Crohn disease and chromosome 16. Am J Hum Genet. 2001;68:1165–71. doi: 10.1086/320119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Trachtenberg EA, Yang H, Hayes E, Vinson M, Lin C, Targan SR, Tyan D, Erlich H, Rotter JI. HLA class II haplotype associations with inflammatory bowel disease in Jewish (Ashkenazi) and non-Jewish caucasian populations. Hum Immunol. 2000;61:326–33. doi: 10.1016/s0198-8859(99)00134-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yang HYTK, Rotter JI. Inflammatory Bowel Disease. In: King RARJ, Motulsky AG, editors. The Genetic Basis of Common Diseases. 2. New York: Oxford University Press; 2002. pp. 266–297. [Google Scholar]
- 49.Diamond J, Rotter JI. The evolution of human genetic diseases. In: RA King JR, Motulsky AG, editors. Genetic Basis of Common Diseases. 2. New York: Oxford University Press; 2002. pp. 50–64. [Google Scholar]
- 50.Acheson ED. The distribution of ulcerative colitis and regional enteritis in United States veterans with particular reference to the Jewish religion. Gut. 1960;1:291–3. doi: 10.1136/gut.1.4.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Weiner HA, Lewis CM. Some notes on the epidemiology of nonspecific ulcerative colitis. An apparent increase in incidence in Jews. Am J Dig Dis. 1960;5:406–18. doi: 10.1007/BF02232626. [DOI] [PubMed] [Google Scholar]
- 52.Korelitz B. International Workshop on Epidemiology and Genetics of IBD. Liverpool, U. K: 1983. Observation of the familial and ethnic aspects of Crohn’s disease and ulcerative colitis in New York City. [Google Scholar]
- 53.Bernstein CN, Wajda A, Svenson LW, MacKenzie A, Koehoorn M, Jackson M, Fedorak R, Israel D, Blanchard JF. The epidemiology of inflammatory bowel disease in Canada: a population-based study. Am J Gastroenterol. 2006;101:1559–68. doi: 10.1111/j.1572-0241.2006.00603.x. [DOI] [PubMed] [Google Scholar]


