Abstract
The Tim23 protein is an essential inner membrane (IM) component of the yeast mitochondrial protein import pathway. Tim23p does not carry an amino-terminal presequence; therefore, the targeting information resides within the mature protein. Tim23p is anchored in the IM via four transmembrane segments and has two positively charged loops facing the matrix. To identify the import signal for Tim23p, we have constructed several altered versions of the Tim23 protein and examined their function and import in yeast cells, as well as their import into isolated mitochondria. We replaced the positively charged amino acids in one or both loops with alanine residues and found that the positive charges are not required for import into mitochondria, but at least one positively charged loop is required for insertion into the IM. Furthermore, we find that the signal to target Tim23p to mitochondria is carried in at least two of the hydrophobic transmembrane segments. Our results suggest that Tim23p contains separate import signals: hydrophobic segments for targeting Tim23p to mitochondria, and positively charged loops for insertion into the IM. We therefore propose that Tim23p is imported into mitochondria in at least two distinct steps.
INTRODUCTION
Eukaryotic membrane proteins face many problems during their biogenesis. For example, membrane proteins must be targeted to the correct organelle within the cell. They also must be inserted into the lipid bilayer in the correct topological arrangement. In addition, since organelles such as mitochondria are encompassed by two membranes, proteins destined for the inner membrane (IM) must first cross the outer membrane (OM). At present, little is known about the mechanisms by which eukaryotic proteins are targeted to specific membranes and inserted in their correct conformation.
Most mitochondrial proteins are synthesized in the cytosol and imported into the organelle via a multistep pathway that includes interaction with cytosolic chaperones, binding to receptors on the OM surface, and translocation across one or both of the mitochondrial membranes (for review see Schatz and Dobberstein, 1996; Stuart and Neupert, 1996; Stuart et al., 1996; Jensen and Kinnally, 1997; Pfanner and Meijer, 1997). Cytosolic chaperones bind precursors to prevent premature folding or aggregation, and one chaperone MSF also plays a role in targeting the precursor to the mitochondria (Hachiya et al., 1994, 1995; Komiya et al., 1996). On the mitochondrial surface, precursors encounter several proteins proposed to act as receptors, including Tom70p, Tom37p, Tom22p, and Tom20p (Hines et al., 1990; Söllner et al., 1990, 1992; Schlossmann et al., 1994; Gratzer et al., 1995; Mayer et al., 1995). The outer membrane receptors, along with Tom40p, Tom6p, Tom7p, and Tom8p, make up the TOM complex, which translocates precursors across the mitochondrial outer membrane (Kiebler et al., 1990, 1993; Moczko et al., 1992; Söllner et al., 1992).
Translocation of precursors across the IM is mediated by the TIM complex, which includes Tim44p, Tim23p, Tim17p, and a matrix-localized Hsp70 protein, called mt-Hsp70 (Kang et al., 1990; Maarse et al., 1992, 1994; Emtage and Jensen, 1993). Tim23p and Tim17p are proposed to form a protein-translocating channel in the IM (Emtage and Jensen, 1993; Maarse et al., 1994; Ryan et al., 1994; Lohret et al., 1997). Tim44p and mt-Hsp70 are thought to “pull” precursors through the channel (Pfanner et al., 1994; Stuart et al., 1994; Glick, 1995; von Ahsen et al., 1995) by a process that requires matrix ATP (Chen and Douglas, 1987; Eilers et al., 1987, 1988; Pfanner and Neupert, 1987; Pfanner et al., 1987; Stuart et al., 1994; Wachter et al., 1994) and a electrochemical potential across the IM (Schleyer et al., 1982; Pfanner and Neupert, 1985, 1987; Chen and Douglas, 1987; Eilers et al., 1987). Recently, a new IM complex, containing Tim54p and Tim22p, has been shown to mediate the insertion of at least some polytopic proteins into the IM (Sirrenberg et al., 1996; Kerscher et al., 1997). Two intermembrane space proteins, Tim12p and Tim10p, appear to be part of this new complex (Koehler et al., 1998; Sirrenberg et al., 1998).
Most imported mitochondrial proteins are synthesized with an amino-terminal targeting signal called a presequence. Presequences vary in length and primary amino acid sequence, yet share a common motif consisting of a number of positively charged amino acids, a lack of acidic residues, no long stretches of hydrophobic residues, and the ability to form an amphipathic structure (Allison and Schatz, 1986; Roise et al., 1986, 1988; Roise, 1992). Once in the matrix, the presequence is removed by a two-subunit–processing protease, called MPP (McAda and Douglas, 1982; Yaffe et al., 1985; Jensen and Yaffe, 1988; Pollock et al., 1988; Witte et al., 1988; Yang et al., 1988). Some proteins destined for the mitochondrial IM carry a cleavable presequence followed by one or more hydrophobic membrane-spanning segments (Stuart and Neupert, 1996). The transmembrane segments are proposed to either function as stop-transfer sequences in the IM (Miller and Cumsky, 1991, 1993), or to facilitate the insertion of the polypeptide into the IM after its complete import into the matrix (Mahlke et al., 1990; Herrmann et al., 1997).
Some imported mitochondrial proteins do not carry cleavable, amino-terminal presequences. The import information therefore resides within the mature part of the protein. The targeting signal for Bcs1p, an IM protein without an amino-terminal presequence, has recently been identified (Fölsch et al., 1996). Bcs1p has a positively charged stretch of amino acids immediately adjacent to its single transmembrane-spanning segment. This positively charged segment has the capability to form an amphipathic α-helix, and exposing this region of Bcs1p by deletion of the N terminus and transmembrane domain resulted in the mislocalization of the truncated Bcs1 protein to the matrix. Fölsch et al. (1996) proposed that the positively charged stretch functions as an internal targeting signal functionally analogous to amino-terminal presequences.
Other proteins without presequences that are localized to the mitochondrial IM include the yeast ADP/ATP carrier proteins (Aac1p, Aac2p, Aac3p; Lawson and Douglas, 1988), the mammalian uncoupling protein (UCP; Aquila et al., 1985; Liu et al., 1988), and the yeast phosphate carrier (PiC; Zara et al., 1991). The PiC, UCP, and the Aac proteins belong to the mitochondrial carrier family and contain six transmembrane segments and three matrix-facing, positively charged loops between the transmembrane segments (Aquila et al., 1985; Runswick et al., 1987; Gawaz et al., 1990; Lawson et al., 1990; Palmieri et al., 1993). The positive charges in the matrix loops have been proposed to function as internal targeting signals similar to that in Bcs1p (Fölsch et al., 1996). In addition, mitochondrial carrier family proteins are thought to be composed of a threefold repeat structure of two transmembrane segments with an intervening loop (Runswick et al., 1987). Consistent with this idea, redundant targeting information has been found in UCP and the Aac1 proteins (Pfanner et al., 1987; Liu et al., 1988, 1990; Smagula and Douglas, 1988a,b).
Tim23p, Tim17p, and Tim22p are three homologous proteins of the IM import machinery and are also synthesized without an amino-terminal presequence (Dekker et al., 1993; Emtage and Jensen, 1993; Maarse et al., 1994; Ryan et al., 1994). The Tim23 protein appears to have four transmembrane domains and is inserted in the IM with both its amino and carboxyl termini facing the intermembrane space (Bauer et al., 1996; Ryan et al., 1998; Emtage, Kerscher, and Jensen, unpublished data). This proposed topology places two positively charged segments of Tim23p in the matrix. To test the possibility that the matrix-facing, positively charged loops of Tim23p mediate import into the mitochondrial IM, we replaced the positively charged amino acids in one or both loops with alanine residues. We find that the positive charges are not required for import into mitochondria, but at least one positively charged loop is required for insertion into the IM. We find that the signal to target Tim23p to mitochondria is carried in at least two of the hydrophobic transmembrane segments, but these segments are not sufficient to insert Tim23p into the IM. Our results suggest that Tim23p contains separate and distinct import signals: hydrophobic segments for targeting Tim23p to mitochondria, and positively charged loops for insertion into the IM. The import information for Tim23p thus differs from that of other IM proteins, such as the Bcs1 protein, and Tim23p appears to contain novel import signals that have not been previously described. We propose that Tim23p is imported into mitochondria in at least two distinct steps.
MATERIALS AND METHODS
Yeast Strains and Genetic Methods
The haploid tim23::URA3 ura3 trp1 leu2 strain KRR146 was obtained by crossing the MATα ura3 trp1 strain BY134 (Brachmann et al., 1997) with strain KRR123 (Ryan et al., 1998). Strain KRR146 also carries plasmid pKR1, a TIM23-LEU2-CYH2 plasmid (Ryan et al., 1998). wt Strain D273–10b has been described (Sherman, 1964). Yeast transformations were performed as described (Schiestl and Gietz, 1989). Standard yeast media and genetic techniques were used (Kaiser et al., 1994).
Plasmid Constructions
Tim23p-HA construct. pAD91, a CEN-LEU2 plasmid containing the Tim23 protein with an insertion of the hemagglutinin (HA) epitope in the middle of loop L2, was constructed as follows. First, a SacI/NotI fragment containing amino acids 1–168 of Tim23p was isolated from plasmid pKR34 (see below). The SacI/NotI fragment was inserted into SacI/NotI-digested pKR31, which encodes amino acids 173–222 of Tim23p (Ryan, unpublished data). The resulting plasmid pAD90 encodes Tim23 with a NotI site in the middle of loop L2. A NotI fragment encoding the triple HA epitope (Field et al., 1988) was cloned into the NotI site of pAD90, forming pAD91. pAD91 encodes amino acids 1–168 of Tim23p, followed by amino acids GGR, the triple HA epitope, residues GGR, and then amino acids 173–222 of Tim23p. pJE8, a CEN-LEU2 plasmid encoding Tim23p with the triple-HA epitope inserted at its carboxyl terminus, has been described (Emtage, Kerscher, and Jensen, unpublished data).
L1Neut, L3Neut and L1L3Neut Constructs.
pAD62, a LEU2 plasmid that expresses a L1Neut, a Tim23 protein with the positively charged residues in the first loop changed to neutral alanine residues (K131A, K143A and R144A), was created as follows. First, lys131 was changed to ala131 using the PCR (Saiki et al., 1985) using oligo 176 (5′-GTTCAATTGCAATGCTCCGGGACTATTC-3′), oligo 20 (5′-AATACGACTCACTATAG-3′), and plasmid pKR1 (Ryan and Jensen, 1993) as a template. The PCR fragment was digested with XbaI and MunI. In a second PCR reaction, Lys143 and Arg144 were changed to alanines using oligo 185 (5′-TTGCAATTGAACACCGTCCTGAATCACATTACTGCGGCAGGTCCCTTCTTAG-3′), oligo 21 (5′-ATTAACCCTCACTAAAG-3′), and pKR1. The PCR fragment was digested with MunI and BamHI, added to the first PCR fragment, and ligated into XbaI-BamHI–digested pJE50, a LEU2-TIM23 plasmid (Emtage, unpublished data), forming pAD62. pAD66, which carries the L1Neut-coding sequences downstream of the SP6 promoter, was formed by inserting a SalI and BamHI fragment from pAD62 into the SalI–BamHI sites of SP6-TIM23 plasmid pJE29 (Ryan et al., 1998).
pAD47, which expresses the L3Neut protein behind the SP6 promoter, was constructed as follows. Positively charged residues in the third loop were changed to alanines (K190A, K193A, and K196A) using PCR amplification from a pJE29 template, oligo 166 (5′-TAACCCATGGGTGCCAAACCTGCTGAAGACGCGAACAAAGCGCC-3′) and oligo 11 (5′-CGATTTAGGTGACACTATAG-3′). The PCR fragment was digested with NcoI and EcoRI and ligated into NcoI-EcoRI–digested pJE29. pAD58, a LEU2 plasmid that expresses a L3Neut was constructed by inserting the SacII–HindIII fragment from pAD47 into the SacII–HindIII sites of the LEU2-TIM23 plasmid pJE7 (Emtage and Jensen, 1993).
pAD64, a LEU2 plasmid that expresses L1L3Neut, a Tim23 protein with the positively charged residues in both the first loop and the third loop changed to alanines, was constructed as follows. A 1-kilobase pair (kbp) SacI–SacII fragment from pAD62 containing the mutated residues in loop 1 was inserted into SacI-SacII–digested pAD58. pAD67, which expresses L1L3Neut behind the SP6 promoter, was formed by inserting a SalI and BamHI fragment from pAD64 into the SalI–BamHI sites of pJE29.
Tim23Np and Tim23Cp Constructs.
pKR14, an SP6-containing plasmid that expresses Tim23Np, and pKR15, which carries Tim23Cp, have been described previously (Ryan et al., 1998). Tim23Np consists of amino acids 1–96 of Tim23p, and Tim23Cp contains residues 95–222 of Tim23p.
Tim23p Deletion Constructs.
A series of either N-terminal or C-terminal deletions of TIM23 were constructed using specific oligonucleotides and PCR. Constructs were subcloned into either a CEN6-LEU2 plasmid (pRS315, Sikorski and Hieter, 1989) for expression in yeast, or into the SP6-containing plasmids, pSP64 or pSP65 (Promega, Madison, WI), for in vitro synthesis. All Tim23p constructs for expression in yeast carry 560 base pairs (bp) of promoter sequences upstream of the coding region and 950 bp downstream of coding sequences (Emtage and Jensen, 1993). All SP6 constructs carry 77 bp of upstream sequences (Ryan et al., 1998). Deletion junctions were engineered to contain a NotI site, which adds three extra amino acids (GGR).
pKR34 carries the Δ3Δ4 construct, which contains amino acids 1–168 of Tim23p followed by residues GGR, inserted into pRS315. pKR41 contains Δ3Δ4 inserted in pSP64. pAD20 carries the Δ1Δ2 construct, which contains residues 1–96 of Tim23p, GGR introduced by the NotI site, followed by residues 173–222 of Tim23p, inserted into pSP64. pAD57 carries the Δ1Δ4 construct, which contains residues 1–96 of Tim23p, GGR, amino acids 173–191 of Tim23p, and ends in GGR. pAD73 carries the Δ2Δ3 construct, which contains amino acids 1–132 of Tim23p, followed by GGR, and then residues 195–222 of Tim23p. Δ2Δ3 carries a chimeric loop (KLGGRLK) between TM1 and TM4 of Tim23p, consisting of the first two amino acids of loop L1, amino acids GGR created by the cloning procedure, and the last two amino acids of loop L3.
pAD29 carries Tim23p lacking TM4, which contains the first 196 amino acids of Tim23p, followed by GGR, inserted into pSP64. pAD75 is a pRS315-based version of the same protein. pKR2, a pRS315-based plasmid containing Tim23p lacking TM4 and loop L3 (residues 1–191 followed by GGR), has been described (Ryan and Jensen, 1993). pKR42 is an SP6-based version of the same Tim23p construct.
pAD32 is an SP6-based plasmid, which carries Tim23p lacking TM1, TM2, loop L3, and TM4 (residues 1–96 of Tim23p, followed by GGR, followed by 173–191 of Tim23p, followed by GGR). pAD33 is an SP6-based plasmid, which expresses Tim23p lacking TM1, TM3, and TM4 (residues 1–96 of Tim23p, GGR, followed by residues 173–196 of Tim23p, followed by GGR). pAD68 is an SP6-based plasmid carrying Tim23p lacking TM1, TM2, and TM3 (1–96 amino acids of Tim23p, GGR, followed by amino acids 195–222 of Tim23p).
pKR40, a pRS315-based plasmid, contains Tim23p lacking TM2, TM3, and TM4 and contains residues 1–132 of Tim23p followed by GGR. pKR33 is an SP6-based version of the same protein. pAD72 is an SP6-based construct that expresses Tim23p lacking TM1, TM3, and TM4 (residues 1–96 of Tim23p, followed by GGR, followed by amino acids 143–168, followed by GGR).
Imports into Isolated Mitochondria
Mitochondria were isolated from wt strain D273–10b as described (Sherman, 1964), except that SEH buffer (250 mM sucrose, 1 mM EDTA, 20 mM HEPES-KOH, pH 7.4) was used in place of breaking buffer. Radiolabeled proteins were made from SP6-containing plasmids using 1.5 mCi/ml [35S]-methionine (1000 Ci/mmol, Amersham, Arlington Heights, IL) in a coupled transcription/translation system (SP6 TNT System, Promega, Madison, WI) according to the manufacturer’s instructions. For import reactions, mitochondria were suspended in import buffer (Scherer et al., 1992) to a final concentration of 1 mg/ml protein. Mitochondria (200 μg) and 10 μl of lysate containing the radiolabeled protein were used per reaction. Import reactions were incubated at 30°C for 30 min and were stopped by placing the samples on ice and the addition of carbonyl cyanide m-chlorophenyl hydrazone (Sigma, St. Louis, MO) to a final concentration of 30 μM. Samples were treated with the indicated amounts of trypsin (Sigma) or proteinase K (Calbiochem, San Diego, CA) for 20 min on ice, followed by the addition of either 1 mg/ml soybean trypsin inhibitor (Sigma) or 1 mM phenylmethylsulfonyl fluoride (Sigma). Disrupting of the OM (forming mitoplasts) was performed by diluting mitochondria with 9 volumes of 20 mM HEPES, pH 7.4, followed by incubation on ice for 30 min. After imports and protease treatment, mitochondria or mitoplasts were reisolated by centrifugation at 12,500 × g for 10 min through a 1-ml sucrose cushion (0.625 M sucrose, 20 mM HEPES-KOH, pH 7.4). For analysis, pellets were resuspended in 1× sample buffer (125 mM Tris, pH 6.8, 2% SDS, 20% glycerol) containing 4% β-mercaptoethanol and subjected to SDS-PAGE (Laemmli, 1970). Radiolabeled proteins were visualized by fluorography (Bonner and Laskey, 1974).
Cellular Fractionation
tim23::URA3 trp1 leu2 cyh2 strain KRR146 containing plasmids expressing Tim23p (pKR50), L1Neut (pAD62), L3Neut (pAD58), or L1L3Neut (pAD64) were grown to an OD600 of 1.5 in YEP medium containing 2% sodium lactate, pH 5.5. Cells were converted to spheroplasts, homogenized, and separated into a 9,600 × g mitochondrial pellet and a postmitochondrial supernatant as described (Daum et al., 1982), except that SEH buffer was used in place of breaking buffer. Proteins from the cell fractions were separated by SDS-PAGE and transferred (Laemmli, 1970; Haid and Suissa, 1983) to Immobilon filters (Millipore, Bedford, MA). Filters were probed with 1:10,000 dilution of antiserum to the β subunit of the F1-ATPase (F1β) (a gift from M. Yaffe, University of California, San Diego), hexokinase (a gift from M. Yaffe), or against Tim23p (Emtage and Jensen, 1993). Immune complexes were visualized using a 1:10,000 dilution of HRP-conjugated secondary antibody (Amersham) followed by chemiluminescence (Supersignal, Pierce Chemical, Rockford, IL).
Miscellaneous
Quantitation of import reactions was done using Molecular Dynamics ImageQuant software version 1.1 (Molecular Dynamics, Sunnyvale, CA). Gels were exposed to a Molecular Dynamics Phosphor screen overnight and scanned using a Molecular Dynamics Storm 860 phosphorimager (Molecular Dynamics). Alternatively, fluorographs were scanned using a UMAX VistaScan flatbed scanner, and the results were quantitated with ImageQuant. Antibodies to the HA epitope (Niman et al., 1983), Tom70p (a gift from G. Schatz, Biocenter, Basel, Switzerland), and α-MPP (Jensen and Yaffe, 1988) were used to decorate immune blots.
RESULTS
One of Two Sets of Positively Charged Segments within Tim23p Is Required for Function, but Not for Targeting to Mitochondria
The Tim23 protein has four predicted transmembrane segments and is proposed to be inserted in the IM with both its amino and carboxyl termini facing the intermembrane space (Figure 1A; Bauer et al., 1996; Ryan et al., 1998; Emtage, Kerscher, and Jensen, unpublished data). This topology places two positively charged segments of Tim23p in the matrix. One segment, called loop L1, is located between the first and second transmembrane regions, and the other segment, called loop L3, lies between the third and fourth transmembrane stretches (Figure 1A). Loop L1, which is 14 amino acids in length (KLQLNTVLNHITKR), and loop L3, which is 7 amino acids long (KSSKGLK), both contain three positively charged residues and no acidic amino acids. In contrast, loop L2, which is proposed to face the intermembrane space (IMS), contains 8 amino acids (DALRGKHD), 2 of which are negatively charged and 2 are positively charged.
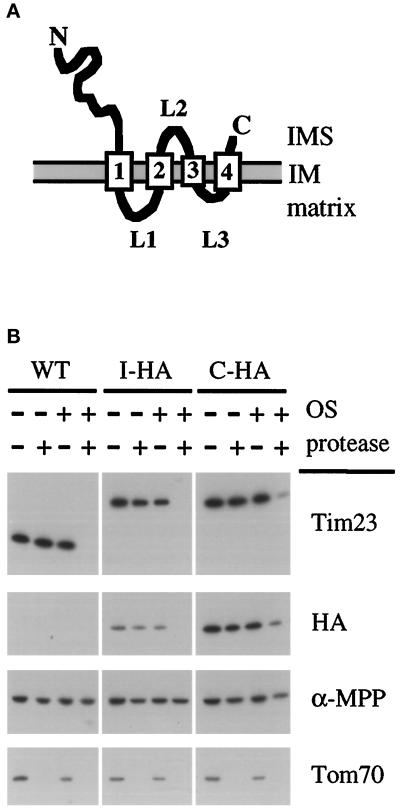
Figure 1.
A working model for the topology of the Tim23 protein in the mitochondrial inner membrane (IM). (A) The Tim23p contains four predicted TM segments and its amino and carboxyl termini have been shown to face the intermembrane space (IMS). TM segments are indicated by the numbered rectangles, and loops L1 and L3 are proposed to face the matrix as indicated. (B) An HA epitope inserted into loop L2 of Tim23p faces the IMS. Mitochondria were isolated yeast cells expressing Tim23p with the HA epitope in loop L2 from plasmid pAD91 (called I-HA). As controls mitochondria were isolated from wt cells (WT), and cells expressing Tim23p with the HA epitope inserted at its C terminus from plasmid pJE8 (called C-HA). Intact mitochondria were digested with 100 μg/ml proteinase K for 30 min at 0°C, and mitochondria whose OM was disrupted by OS were digested with 50 μg/ml proteinase K. Mitochondrial proteins were immune blotted with antibodies to the amino-terminal domain of Tim23p, the HA epitope, the matrix protein α-MPP, or the OM protein, Tom70p.
To further support the model for the configuration of Tim23p in the IM, we inserted an epitope tag into loop L2 of Tim23p and asked whether this tag faced the mitochondrial IMS. We inserted the influenza HA epitope (Field et al., 1988) between residues 168 and 173 of Tim23p. Surprisingly, we found that the Tim23p-HA fusion protein was functional since it rescued the lethality of a tim23::URA3 disruption. Mitochondria were isolated from cells expressing Tim23p with the HA tag in loop L2 (called I-HA), as well as from wt cells, or cells expressing Tim23p with the HA tag at its carboxyl terminus (called C-HA; Emtage, Kerscher, and Jensen, unpublished data). As shown in Figure 1B, immune blotting showed that both the internal HA tag (I-HA) and the carboxyl-terminal HA tag (C-HA) were protected from protease digestion in intact mitochondria, but that both tags were digested when the mitochondrial OM was disrupted by osmotic shock (OS). Control blots showed that the matrix marker α-MPP was not accessible to protease digestion even when the OM was disrupted. Our results thus support the model that Tim23p has four transmembrane segments with two matrix-facing loops (loops L1 and L3) and one loop facing the IMS (loop L2).
Tim23p is a member of a set of proteins that are imported into mitochondria without an amino-terminal, cleavable presequence. The import signal for one of these proteins Bcs1p was recently shown to be an internal, positively charged segment facing the matrix that had many of the properties of a mitochondrial presequence (Fölsch et al., 1996). We therefore tested the possibility that the matrix-facing, positively charged loops of Tim23p mediate its import into mitochondria. As diagrammed in Figure 2A, we made three mutant versions of Tim23p: L1Neut, in which we replaced the two lysines and one arginine in loop L1 with alanines; L3Neut, where we substituted alanines for the three lysines in loop L3; and L1L3Neut, in which we replaced the six positively charged amino acids in both loop L1 and L3 with alanines. We first examined the ability of these constructs to provide Tim23p function in yeast cells. LEU2-containing plasmids expressing either Tim23p, L1Neut, L3Neut, or L1L3Neut were transformed into tim23::URA3 disruption strain KRR146, which also contains the TIM23-CEN-CYH2 plasmid pKR1 (Figure 2B). Leu+ transformants were patched onto medium lacking leucine (SD−Leu). Since CYH2-containing cells are unable to grow in the presence of cycloheximide (Sikorski and Boeke, 1991), we tested our transformants for their ability to lose the TIM23-CYH2 plasmid by replica plating them onto medium containing cycloheximide (YEPD + CYH). Tim23p is essential for cell viability (Emtage and Jensen, 1993); therefore, only transformants carrying a second copy of functional TIM23 will be able to grow on cycloheximide-containing medium. We found that both L1Neut and L3Neut provided wt Tim23p activity when grown at 24°C, 30°C and 37°C on both fermentable and nonfermentable medium. In contrast, the L1L3Neut construct did not grow on media with cycloheximide at any temperature, and thus did not provide Tim23p function. Our results suggest that only one of the two positively charged matrix segments within Tim23p is required for its function. Full Tim23p activity is observed when the lysine and arginine residues are replaced by alanine in either loop L1 or L3, but activity is lost when the positive charges are removed from both the matrix-facing loops.
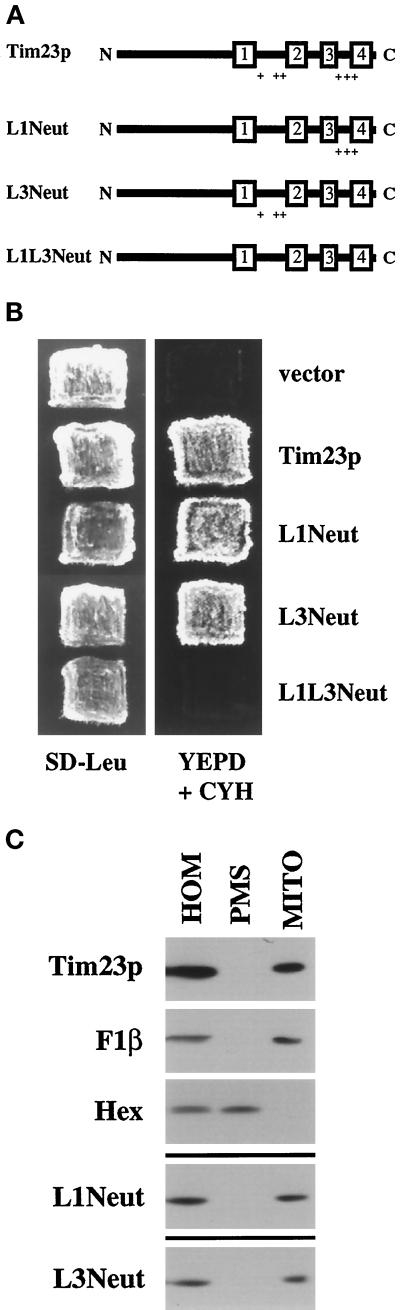
Figure 2.
One of two sets of positively charged segments within Tim23p is required for function, but not for targeting to mitochondria in vivo. (A) Schematic representation of the Tim23 protein and mutant Tim23p constructs used in this study. The numbered rectangles represent the four hydrophobic regions corresponding to proposed membrane-spanning segments. The + signs denote positively charged amino acids within the loops (L1 or L3) between the TM segments. (B) Positively charged amino acids in either segment L1 or L3 are needed for Tim23p function. tim23::URA3 trp1 leu2 cyh2 strain KRR146 carrying TIM23-TRP1-CYH2 plasmid pKR1 was transformed with either the LEU2 vector pRS315 or pRS315 carrying DNA inserts encoding Tim23p (pJE50), L1Neut (pAD62), L3Neut (pAD58), or L1L3Neut (pAD64). Leu+ colonies were patched out onto synthetic complete medium lacking leucine (SD−Leu). Patches were grown at 30°C for 2 d, and then replica plated onto YEPD medium containing 10 mg/L cycloheximide (YEPD + CYH). Yeast cells that contain a functional Tim23 protein are able to lose the TIM23-TRP1-CYH2 plasmid and grow in the presence of cycloheximide. (C) Tim23p lacking positive charges in segment L1 or L3 are still targeted to mitochondria in vivo. tim23::URA3 trp1 leu2 cyh2 strain KRR146 containing plasmids expressing Tim23p (pJE50), L1Neut (pAD62), or L3Neut (pAD58) were homogenized (HOM) and separated into a mitochondrial pellet (MITO) and a postmitochondrial supernatant (PMS). Proteins from the cell fractions representing equivalent cell amounts were analyzed by SDS-PAGE and immune blotting with antisera to Tim23p, the β-subunit of the F1-ATPase (F1β, a mitochondrial marker), or hexokinase (Hex, a cytoplasmic marker). Immune blots of fractions from cells expressing L1Neut and L3Neut decorated with antiserum to F1β and hexokinase were identical to those shown for Tim23p cells and are not shown.
To examine the level and the location of the different Tim23p constructs in yeast, we grew tim23::URA3 cells expressing either Tim23p, L1Neut, or L3Neut. Cells were homogenized (HOM) and separated into a mitochondrial fraction (MITO) and a postmitochondrial supernatant (PMS) by centrifugation. When we analyzed our fractions by immune blotting, we found that all the Tim23 proteins cofractionated with F1β, a mitochondrial protein (Figure 2C). No Tim23p, L1Neut, or L3Neut was found in the supernatant with the cytosolic hexokinase (Hex) protein. While the L1Neut and L3Neut proteins are targeted to mitochondria, their steady-state levels appear to be reduced when compared with wt Tim23p. When the level of Tim23p, L1Neut, and L3Neut were standardized to the amount of F1β in each cell fractionation, we found that L1Neut and L3Neut were reduced two- to threefold compared with wt Tim23p. We also examined the level of the L1L3Neut construct, which did not complement the tim23 disruption. Immune blotting of yeast cells showed that the amount of L1L3Neut is reduced at least 100-fold as compared with the level of wt Tim23p. We propose that the altered Tim23p constructs are more rapidly turned over in cells since they are not efficiently inserted into the mitochondrial IM (see below).
Internal Positively Charged Segments Mediate the Insertion of Tim23p into the IM, but Are Not Required for Import into Mitochondria
To directly examine the role of the positively charged loops in Tim23p, we examined the import of the different constructs into isolated mitochondria. Radiolabeled Tim23, L1Neut, L3Neut, and L1L3Neut proteins were made by in vitro transcription and translation and were then incubated with isolated mitochondria (Figure 3A). After the import reaction, samples were divided into aliquots. One aliquot was treated with trypsin to digest proteins that were not imported into the mitochondria. Mitochondrial proteins were isolated by centrifugation and separated by SDS-PAGE, and the radiolabeled proteins were visualized by fluorography. We found that Tim23p, L1Neut, L3Neut, and L1L3Neut were all imported into mitochondria and protected from protease digestion to the same extent (Figure 3A, mitos + protease). While the majority of Tim23p, L1Neut, L3Neut, and L1L3Neut molecules required an IM potential for their import, a small amount of all four proteins were protected from protease digestion after import into mitochondria treated with valinomycin (−Δψ). Whether this small amount of protein represented potential-independent import or protease-resistant material is not clear. Nonetheless, we conclude that the positively charged loops L1 and L3 are not required for the efficient import of Tim23p into mitochondria. We next examined whether the different Tim23p constructs were correctly inserted into the mitochondrial IM. wt Tim23p resides within the IM with a 9-kDa amino-terminal hydrophilic domain facing the IMS (Bauer et al., 1996; Lohret et al., 1997; Ryan et al., 1998; Emtage, Kerscher, and Jensen, unpublished data). When the OM of mitochondria is disrupted (forming mitoplasts), the amino-terminal domain can be digested by protease yielding a characteristic 14-kDa fragment. This fragment represents the carboxyl-terminal domain of Tim23p that is embedded in the IM. We imported Tim23p, L1Neut, L3Neut, and L1L3Neut into mitochondria, disrupted the mitochondrial OM by OS, and then digested the mitoplasts with proteinase K (Figure 3A, mitoplasts + protease). We found that the L1Neut and L3Neut constructs were inserted in the IM, but not to the same extent as wt Tim23p. Reduced amounts of the 14-kDa protease-protected fragment were seen after import of L1Neut and L3Neut as compared with Tim23p. In contrast, virtually no 14-kDa fragment was seen after import of the L1L3Neut construct.
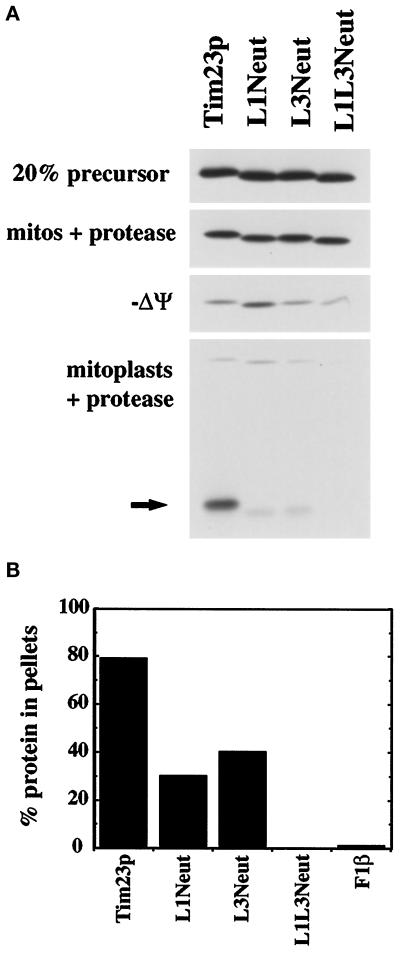
Figure 3.
Internal positively charged segments mediate the insertion of Tim23p into the IM but are not required for import into mitochondria. (A) The Tim23, L1Neut, L3Neut, and L1L3Neut proteins were synthesized in the presence of 35S-methionine and imported into isolated mitochondria as described in MATERIALS AND METHODS. To dissipate the IM potential (−Δψ), mitochondria were preincubated with 250 mM KCl and 40 μM valinomycin. After import, mitochondria were treated with 200 μg/ml trypsin, split into aliquots, and reisolated by centrifugation. Two sets of samples were resuspended in SDS-sample buffer (mitos + protease; −Δψ mitos + protease). In the other samples, the OM was disrupted by OS, and the resulting mitoplasts were treated with 50 μg/ml proteinase K. Mitoplasts were isolated by centrifugation and resuspended in SDS-sample buffer (mitoplasts + protease). Proteins were separated by SDS-PAGE, and radiolabeled proteins were visualized by fluorography; 20% of the translation product used in the import reactions is also shown. The arrow indicates the 14-kDa fragment of Tim23p protected from protease digestion in mitoplasts, indicative of the proper insertion of Tim23p into the IM. (B) Radiolabeled Tim23, L1Neut, L3Neut, L1L3Neut, and F1β proteins were imported into mitochondria and treated with 200 μg/ml proteinase K. Mitochondria were isolated by centrifugation, and the pellets were resuspended in 0.1 M sodium carbonate, pH 11.4. After incubation on ice for 30 min, samples were spun at 100,000 × g for 30 min at 4°C. Pellets and supernatants were subjected to SDS-PAGE, fluorography, and densitometry. The amount of the Tim23, L1Neut, L3Neut, L1L3Neut, and F1β proteins found in the pellet fraction is indicated.
To further determine whether the altered Tim23p constructs were inserted into the IM, we asked whether the proteins could be extracted from mitochondria after treatment with alkali (Figure 3B). Tim23p, L1Neut, L3Neut, L1L3Neut, and the peripheral membrane protein F1β were imported into mitochondria and then treated with protease to remove any proteins that were not imported into the organelle. Mitochondrial pellets were resuspended in 0.1 M sodium carbonate and separated into a membrane pellet and supernatant fraction by centrifugation. We found that 80% of the imported Tim23p protein remained with the mitochondrial membranes after alkali treatment, whereas virtually all the F1β protein was removed. Compared with Tim23p, a lesser amount of L1Neut and L3Neut remained membrane associated (∼25% and 40%, respectively). In contrast, virtually all of the L1L3Neut protein was removed from the membranes. Our results suggests that while the positively charged loops of Tim23p are not required for import into mitochondria, they play an important role in the insertion of Tim23p into the IM. One set of positive charges, carried in either loop L1 or L3, are sufficient for the partial insertion of Tim23p into the IM, whereas complete insertion requires both sets of positively charged loops.
The Hydrophobic Carboxyl Terminus of Tim23p Carries Redundant Targeting Information
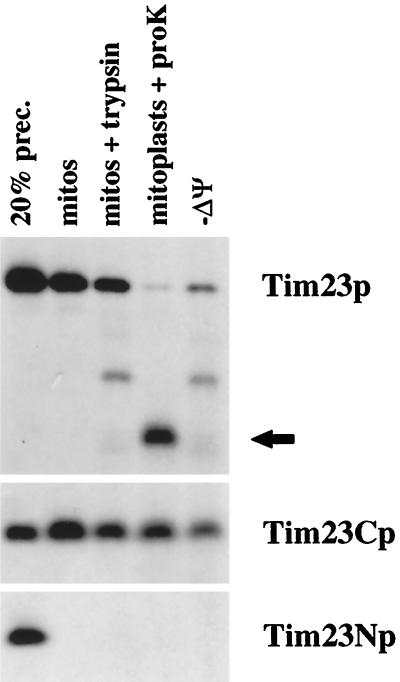
We found that the hydrophilic amino-terminal domain of Tim23p does not carry targeting information. A Tim23p construct lacking its first 9 kDa lacks function (Ryan et al., 1998), but it is efficiently imported into mitochondria and inserted into the IM (Figure 4). We synthesized Tim23p, along with Tim23Np, which contains the amino-terminal portion (amino acids 1–96) of Tim23p, and Tim23Cp, which contains the carboxyl-terminal domain (residues 95–222) of Tim23p, and incubated the three proteins with isolated mitochondria. In the presence of energized mitochondria, wt Tim23p was imported into mitochondria (Figure 4, mitos) and was protected from exogenously added protease after the import reaction (Figure 4, mito + trypsin). When the OM was disrupted by OS after import of Tim23p, proteinase K digestion produced the 14-kDa fragment indicative of IM insertion (Figure 4, mitoplasts + protease). In the absence of membrane potential (−Δψ), the amount of Tim23p imported into mitochondria was reduced, and virtually no Tim23p was inserted into the IM. The carboxyl-terminal domain of Tim23p, Tim23Cp, was also efficiently imported into mitochondria (Figure 4, mitos + protease; mitoplasts + protease). Surprisingly, a significant amount of Tim23Cp was imported into valinomycin-treated mitochondria (Figure 4, −Δψ). In contrast to Tim23p and Tim23Cp, the amino-terminal portion of Tim23p, Tim23Np, was not imported into energized mitochondria. Tim23Np did not even bind to mitochondria and failed to pellet with the organelles after the import reaction (Figure 4, mitos). These results support our previous studies indicating that the import signal within Tim23p resides within the carboxyl-terminal half of the molecule (Ryan et al., 1998).
Figure 4.
The hydrophobic carboxyl terminus of Tim23p carries targeting information. Tim23Np, which consists of amino acids 1–96 of Tim23p, Tim23Cp, which contains residues 95–222, and wt Tim23p were synthesized in the presence of 35S-methionine and imported into isolated mitochondria in the presence or absence (−Δψ) of membrane potential. After import, mitochondria were treated with 200 μg/ml trypsin, split into aliquots, and reisolated by centrifugation. Two sets of samples were resuspended in SDS-sample buffer (mitos + protease; −Δψ mitos + protease). In the other samples, the OM was disrupted by OS, and the resulting mitoplasts were treated with 50 μg/ml proteinase K. Mitoplasts were isolated by centrifugation and resuspended in SDS-sample buffer (mitoplasts + protease). Proteins were separated by SDS-PAGE, and radiolabeled proteins were visualized by fluorography. Twenty percent of the translation product used in the import reactions is also shown. The arrow indicates the 14-kDa fragment of Tim23p protected from protease digestion in mitoplasts, indicative of the proper insertion of Tim23p into the IM.
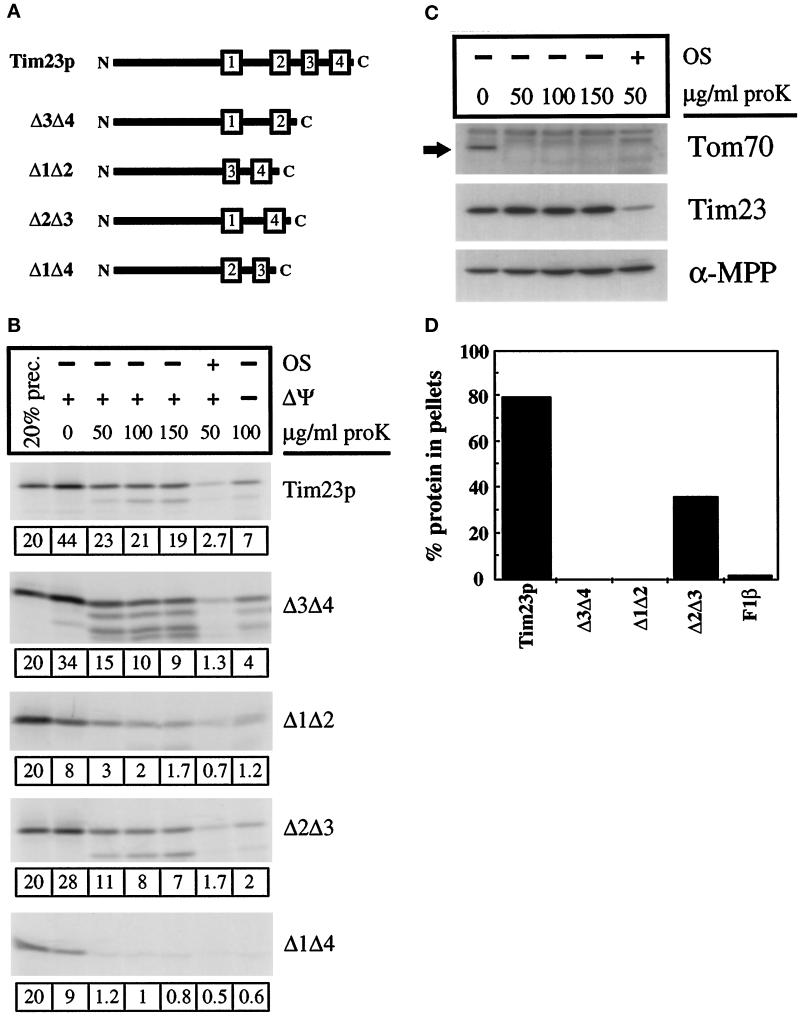
As described above, we found that the positively charged loops of Tim23p are required for IM insertion, but not for import into the organelle. To localize the mitochondrial import signal within the carboxyl-terminal region of Tim23p, we have created constructs lacking one or more of the hydrophobic transmembrane (TM) segments. As shown in Figure 5A, we generated a protein lacking the third and fourth TM segments, called Δ3Δ4, a protein lacking TM segments 1 and 2, called Δ1Δ2, a protein lacking TM segments 2 and 3, called Δ2Δ3, and a protein lacking TM segments 1 and 4, called Δ1Δ4. When expressed in yeast cells, none of these constructs provides Tim23p function.
Figure 5.
The hydrophobic carboxyl terminus of Tim23p carries redundant targeting information. (A) Schematic representation of Tim23p deletion constructs used in this study. The numbered rectangles represent the hydrophobic regions corresponding to proposed membrane-spanning segments within Tim23p. All constructs carry the 96-amino acid amino-terminal domain of Tim23p. Δ3Δ4, Δ1Δ2, and Δ1Δ4 all retain the normal loops between the TM segments, whereas Δ2Δ3 contains a hybrid loop (see MATERIALS AND METHODS). (B) Tim23, Δ1Δ2, Δ3Δ4, Δ2Δ3, and Δ1Δ4 imports. Radiolabeled proteins were imported into isolated mitochondria. For one set of samples, the IM potential was dissipated with valinomycin before import (−ΔΨ). After import, samples were split into aliquots and treated with the indicated amounts of proteinase K. In another set of imports, mitochondria were converted to mitoplasts by osmotic shock (OS), followed by protease treatment. Mitochondrial pellets were analyzed by SDS-PAGE and fluorography. Twenty percent of the translation product used in the import reactions is also shown. The amount of the imported protein, calculated as a percentage of the total material added to the import reaction, is shown below each gel. (C) Immune blots. Mitochondria were subjected to mock import conditions and subsequent protease treatment or OS and protease treatment, concurrently with import samples in Figure 5B. Mitochondria or mitoplasts were pelleted and proteins were analyzed by SDS-PAGE and immune blotting with antisera against Tim23p, α-MPP (a matrix marker), and Tom70p (an OM marker). (D) Radiolabeled Tim23, Δ3Δ4, Δ1Δ2, Δ2Δ3, and F1β proteins were imported into mitochondria and treated with 200 μg/ml proteinase K. Mitochondria were isolated by centrifugation, and the pellets were resuspended in 0.1 M sodium carbonate (pH 11.4). After incubation on ice for 30 min, samples were spun at 100,000 × g for 30 min at 4°C. Pellets and supernatants were subjected to SDS-PAGE, fluorography, and densitometry. The amount of the Tim23, Δ3Δ4, Δ1Δ2, Δ2Δ3, and F1β proteins found in the pellet fraction is indicated.
We synthesized Tim23p, along with the different deletion constructs, and asked whether they could be imported into isolated mitochondria. As shown in Figure 5B, Tim23p and the Δ3Δ4, Δ1Δ2, and Δ2Δ3 proteins were all imported into energized mitochondria, but import for all of the proteins was reduced in the absence of membrane potential (−Δψ). Δ1Δ4 differed from the other constructs and was not imported into mitochondria to a protease-protected location (Figure 5E). Since Δ3Δ4 and Δ1Δ2 are both imported into mitochondria, our results suggest that the Tim23p carboxyl terminus carries two targeting signals. Furthermore, since Δ3Δ4, Δ1Δ2, and Δ2Δ3 were capable of import but Δ1Δ4 was not suggests that the targeting information is located in or near TM segments 1 and 4.
Quantitation of the imports of Tim23p, Δ3Δ4, Δ1Δ2, and Δ2Δ3 indicated that while similar amounts of the altered constructs pelleted with mitochondria as compared with wt Tim23p, none were protected from protease digestion to the same extent as Tim23p. It is likely that Δ3Δ4, Δ1Δ2, and Δ2Δ3 were more sensitive than Tim23p to digestion after import because they were not completely imported into the organelle. Demonstrating that the mitochondrial OM remained intact in our studies with mitochondria, we found that the amino-terminal domain of the endogenous Tim23 protein (which faces the IMS) was protected from protease digestion (Figure 5C). In contrast, the N-terminal domain of Tim23p was readily digested when the mitochondrial OM was disrupted by OS.
We suggest that altered Tim23 constructs were arrested at an early step in the import pathway, and much of the proteins were incompletely translocated across the OM. Consistent with their incomplete import, we found that Δ3Δ4 and Δ1Δ2 were not inserted into the IM and could be extracted from mitochondrial membranes by carbonate treatment (Figure 5D). While 80% of the imported Tim23p protein remained with the mitochondrial membranes after carbonate treatment, almost all the Δ3Δ4, Δ1Δ2, and F1β proteins were removed. Also indicating that Δ3Δ4 and Δ1Δ2 were not inserted into the IM, we failed to detect any protease-resistant fragment in mitoplasts after import of Δ3Δ4 and Δ1Δ2. All of the Δ3Δ4 and Δ1Δ2 proteins were completely digested when the OM was disrupted. Although the Δ3Δ4 and Δ1Δ2 proteins were not inserted into the IM, we found that both proteins were membrane associated after their import. When the mitochondrial OM was disrupted, Δ3Δ4 and Δ1Δ2 were not released with soluble IMS proteins and instead pelleted with the mitoplast fraction. We suggest that Δ3Δ4 and Δ1Δ2 are stuck in the OM import machinery at an early step in the import pathway.
Import of either the Δ3Δ4 or Δ1Δ2 proteins into mitochondria did not require the positively charged residues in the matrix-facing loops. A Δ3Δ4 construct, in which the two lysines and one arginine in loop L1 were replaced by alanines, and a Δ1Δ2 construct, in which the three lysines were replaced by alanine, were imported to the same extent as the Δ3Δ4 or Δ1Δ2 construct containing the positively charged loop. These results support our conclusion that the import signalfor Tim23p is separate from the signal required for insertion into the IM.
In contrast to Δ3Δ4 and Δ1Δ2, ∼40% of the Δ2Δ3 protein remained with the membrane fraction after carbonate treatment (Figure 5D). Our results suggest that a significant amount of the Δ2Δ3 protein was inserted in the IM. Δ2Δ3 contains the first and fourth TM segments of Tim23p and, as described above, may carry two sets of Tim23p-targeting information. Therefore, the observation that Δ2Δ3 was imported more completely than either Δ3Δ4 or Δ1Δ2 may not be surprising. Δ2Δ3 also carries a chimeric loop consisting of the first two amino acids of loop L1, amino acids GGR created by the cloning procedure, and the last two amino acids of loop L3. This hybrid loop (KLGGRLK), which has three positively charged residues and no acidic residues, appears to function as an effective IM insertion signal. Our results suggest that positively charged amino acids may play a more critical role in IM insertion than a specific amino acid sequence or secondary structure.
Efficient Import of Tim23p Requires a Pair of Hydrophobic Segments
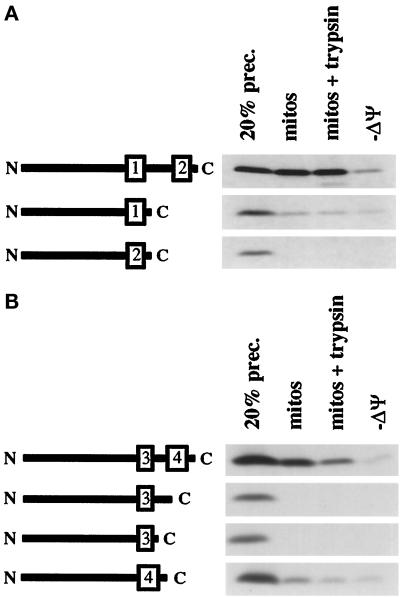
Our results above suggest that Tim23p carries redundant targeting information in TM segments 1 and 4. To test whether either TM segment 1 or 4 is sufficient for targeting, we created Tim23p constructs that contain only a single TM segment. As shown in Figure 6A, starting with a Tim23p construct that lacks the first two TM segments (Δ1Δ2), we removed TM segment 4. Similarly we removed both loop L3 and the fourth TM segment from Δ1Δ2, and we also made a construct that lacks loop L3 and the third TM segment. We found that while Δ1Δ2 was imported into mitochondria to a protease-protected location, constructs lacking TM segment 4 failed to be imported (Figure 6A). A Tim23p construct that contains only TM segment 4 is imported into mitochondria, but ∼10-fold less efficiently than the Δ1Δ2 construct, which contains both TM3 and TM4. A construct that contains only TM3 is not imported and fails to even bind to mitochondria. Our results suggest that TM segment 4 functions as a more effective targeting signal when paired with TM segment 3.
Figure 6.
Efficient import of Tim23p requires a pair of hydrophobic segments. (A) Further deletions of the Δ1Δ2 protein inhibit import. The Δ1Δ2 construct contains the third and fourth TM segments of Tim23p, as well as the intervening loop L3. Constructs lacking the fourth TM segment, lacking loop L3 and the fourth TM segment, or lacking loop L3 and the third TM segment were generated as described in MATERIALS AND METHODS. Schematic representations of these constructs are shown on the left side of the gel. The four proteins were synthesized and imported into isolated mitochondria either in the presence or absence (−ΔΨ) of a membrane potential. After import, samples were either not treated (mitos) or treated with 200 μg/ml trypsin (mitos + trypsin; −ΔΨ). Mitochondrial pellets were analyzed by SDS-PAGE and fluorography. (B) Further deletions of the Δ3Δ4 protein inhibit import. The Δ3Δ4 construct contains the first and second TM segments of Tim23p, as well as the intervening loop L1. A construct lacking the second TM segment and loop L1, or a construct lacking the first TM segment and loop L1, was produced. Diagrams illustrating the constructs are shown to the left of the gel. The three proteins were synthesized and imported either in the presence or absence (−ΔΨ) of a mitochondrial membrane potential. After import, samples were either not treated (mitos) or treated with 200 μg/ml trypsin (mitos + trypsin; −ΔΨ) before analysis.
We similarly found that the targeting activity of TM segment 1 is increased in combination with TM segment 2, as compared with TM segment 1 alone. Starting with a construct that lacks TM segments 3 and 4 (Δ3Δ4), w deleted TM segment 2 (Figure 6B). We also created a construct that carries only TM segment 2. While Δ3Δ4 was imported into mitochondria, very little of the protein containing only TM1 was imported into mitochondria. A construct containing only TM2 was not imported. Our results suggest that the import information of Tim23p is carried in TM segments 1 and 4, and both segments need the cooperation of adjacent hydrophobic segments to be recognized by the import machinery. This conclusion is also supported by our observation that a Tim23p construct that carries only TM segments 1 and 4 (Δ2Δ3) is imported into mitochondria (and inserted into the IM) almost as efficiently as the wt Tim23 protein (Figure 4B).
Tim23p Lacking the Fourth TM Segment Is Not Efficiently Imported into Mitochondria
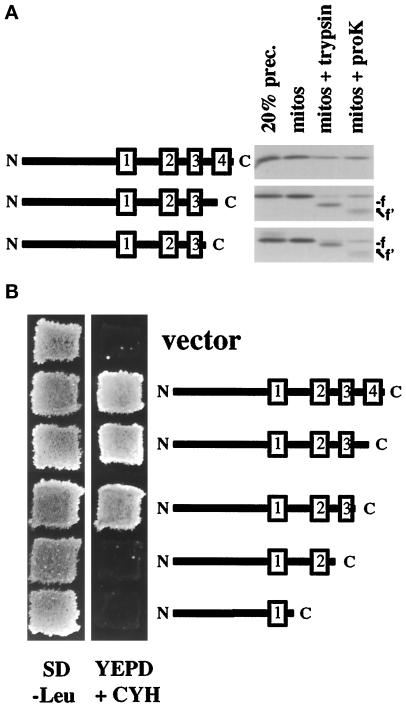
Our results, suggesting that the Tim23p TM segments need to cooperate to promote efficient import into mitochondria, raise the possibility that a specific secondary structure, such as paired TM segments, is recognized by the import machinery. Supporting this idea, we found that Tim23p constructs lacking TM segment 4 were incompletely imported into mitochondria. Tim23p lacking TM segment 4 or a construct lacking both loop L3 and TM segment 4 were incubated with isolated mitochondria along with the wt Tim23 protein (Figure 7A). When mitochondria were treated with protease after the import reaction, we found that most of Tim23p was inside the mitochondria and protected from digestion. In contrast, ∼80% of both constructs lacking TM 4 were digested to a smaller form by trypsin digestion (Figure 7A, mitos + trypsin, labeled f) or by proteinase K digestion (Figure 7A, mitos + proK, labeled f′). Both of the constructs lacking TM 4 appeared to get stuck in transit across the OM at the same point since protease treatment generated fragments of identical size from both proteins. The estimated mass of the proteinase K fragment (∼17.5 kDa) represents a Tim23 protein lacking TM 3, TM 4, and loop L3. We conclude from these results that after recognition and binding of the paired TM-targeting signals, the wt Tim23 protein is imported into mitochondria in an N-to-C direction. Constructs that lack TM segment 4 cannot form a correctly paired structure. Therefore, TM segment 3, in the absence of TM 4, is not efficiently recognized by the import machinery, and the carboxyl-terminal region of Tim23p remains outside the OM accessible to protease digestion.
Figure 7.
Tim23p lacking the fourth TM segment is not efficiently imported into mitochondria. (A) Tim23p lacking the fourth TM segment is incompletely imported into mitochondria. Constructs of Tim23p lacking the fourth TM segment (Δ4) or lacking loop L3 and the fourth TM segment (ΔL3Δ4) were generated. Schematic representations of the constructs are shown to the left of the gel. The three proteins were synthesized and incubated with mitochondria. After the import reaction, samples were either analyzed directly (mitos) or after treatment with 200 μg/ml trypsin (mitos + trypsin) or 200 μg/ml proteinase K (mitos + proK). Mitochondrial pellets were analyzed by SDS-PAGE and fluorography. (B) The fourth TM segment and loop L3 are not required for Tim23p function. tim23::URA3 trp1 leu2 cyh2 strain KRR146 carrying TIM23-TRP1-CYH2 plasmid pKR1 was transformed with six different plasmids: LEU2-containing plasmid pJE50, which expresses wt Tim23p; pAD75, which expresses Tim23p lacking the fourth TM segment (TM4); pKR2, which lacks loop L3 and TM4; pKR34, which lacks the TM3 and TM4; pKR40, which lacks TM2, TM3, and TM4; and the empty vector pRS315. Leu+ colonies were patched out onto synthetic complete medium lacking leucine (SD−Leu). Patches were grown at 30°C for 2 d, and then replica-plated onto YEPD medium containing 10 mg/L cycloheximide (YEPD + CYH). Yeast cells that contain a functional Tim23 protein are able to lose the TIM23-TRP1-CYH2 plasmid and grow in the presence of cycloheximide.
While the majority of the molecules lacking TM 4 got stuck during import into mitochondria, a small number of proteins were completely imported. As shown in Figure 7A, ∼10–20% of the construct without TM 4 was protected from protease digestion after import. Supporting this conclusion, we found that constructs lacking TM 4, or both loop L3 and TM4, provide functional Tim23p activity in yeast cells (Figure 7B). Since both constructs can rescue the lethality of a tim23::URA3 disruption, some fraction of these proteins must be imported into mitochondria and inserted into the IM. Surprisingly, while TM segment 4 and loop L3 appear to play an important role in Tim23p import, these sequences do not seem critical for Tim23p function. Constructs lacking TM 4 and loop L3, however, are not fully functional, as they cannot rescue tim23::URA3 strains at elevated temperatures (Ryan and Jensen, 1993).
DISCUSSION
Tim23p, along with several other proteins of the mitochondrial IM, do not carry amino-terminal presequences. The most likely topology for Tim23p places the protein in the IM with four TM segments, with its hydrophilic amino-terminal domain facing the matrix, and with two positively charged loops facing the matrix. We replaced the positively charged amino acids in one or both loops with alanine residues and found that the positive charges are not required for import into mitochondria, but at least one positively charged loop is required for insertion into the IM. We found that the signal to import Tim23p across the OM and into mitochondria is carried in the first and fourth hydrophobic TM segments. These TM segments can mediate the import of Tim23p into mitochondria, but they are not sufficient to insert Tim23p into the IM. These hydrophobic segments represent novel mitochondrial targeting information and differ dramatically from the positively charged import signals carried on most matrix-targeted precursor proteins. Our results suggest that Tim23p contains separate and distinct targeting signals: hydrophobic signals for import into the organelle and positively charged loops for IM insertion. We therefore propose that Tim23p is imported into mitochondria in at least two independent steps using machinery different than that used by presequence-containing proteins.
The import of Tim23p appears to differ from another IM protein Bcs1p whose targeting signal has been recently characterized (Fölsch et al., 1996). Bcs1p, like Tim23p, does not carry an amino-terminal presequence and its targeting signal has been shown to be a positively charged stretch of amino acids immediately adjacent to a single TM-spanning segment. This positively charged region, which has the capacity to form an amphipathic helix, is proposed to function in a manner analogous to presequences. The TM segment of Bcs1p is thought to be a stop-transfer sequence preventing complete translocation of Bcs1p into the matrix. In contrast to Bcs1p, we find that the positively charged loops in Tim23p do not function as import signals. Tim23p constructs lacking the positive charges in loops L1 or L3 are still imported into the organelle.
Our results suggest that the positively charged loops of Tim23p mediate insertion into the IM. The mitochondrial IM has two separate import complexes, the Tim54p–Tim22p complex and the Tim23p–Tim17p complex (Sirrenberg et al., 1996, 1998; Kerscher et al., 1997; Koehler et al., 1998). We have recently shown that Tim23p is inserted into the IM via the Tim54p/Tim22p machinery (Kerscher et al., 1997). In contrast, Bcs1p appears to use the Tim23p/Tim17p pathway (Fölsch et al., 1996, Kerscher and Jensen, unpublished data). Furthermore, matrix-destined precursor proteins with amino-terminal presequences appear to be translocated across the IM by the Tim23p/Tim17p machinery (Sirrenberg et al., 1996; Kerscher et al., 1997; Emtage, Kerscher, and Jensen, unpublished data; Kerscher and Jensen, unpublished data). Tim23p must therefore carry a different signal directing it to the Tim54p–Tim22p complex. We propose that proteins that carry either an amino-terminal presequence, or an internal segment capable of forming an amphipathic helix, are recognized by the Tim23p–Tim17p complex, while the positively charged loops of Tim23p (which are not amphipathic) are recognized by the Tim54p–Tim22p complex. In addition to Tim23p, several other polytopic proteins, including Tim22p, Tim17p, Aac1p, and PiC, are inserted into the IM via the Tim54p/Tim22p machinery (Sirrenberg et al., 1996; Kerscher et al., 1997). We predict that the insertion of these IM proteins is mediated by positively charged, matrix-facing loops similar to those in Tim23p.
While the positively charged loops are required for the IM insertion of Tim23p, these loops do not mediate the import of Tim23p into mitochondria. Instead, hydrophobic sequences in TM segments 1 and 4 appear to meditate the import of Tim23p into the organelle. Supporting our hypothesis that the import signal for this class of proteins is hydrophobic, Kübrich et al. (1998) have recently identified a translocation intermediate of Aac1 during its transfer across the OM. The majority of the Aac1p intermediate is exposed to the IMS, but remains stuck in the OM with its carboxyl terminus exposed to the matrix. This intermediate of Aac1p is strikingly similar to many of our Tim23p constructs that are unable to insert into the IM. Interestingly, the Aac1p intermediate cannot be removed from the mitochondrial membranes by high-salt treatment, suggesting that its association with the OM import machinery is via hydrophobic interactions.
Although the L1L3Neut version of Tim23p does not carry positively charged residues in loops L1 or L3, do basic residues located in other parts of the molecule, in particular in the amino-terminal domain or at the C terminus, contribute to the potential-dependent import of L1L3Neut into mitochondria? In preliminary studies, we have mutated the basic residues in the C terminus of Tim23p and find that the altered protein rescues the tim23::URA3 disruption strain and is efficiently imported into isolated mitochondria (Davis and Jensen, unpublished observations). We also find that Tim23 proteins lacking the amino-terminal domain are efficiently imported into mitochondria in the absence of positively charges in either loop L1 or L3 (Davis and Jensen, unpublished observations). We therefore argue that the potential-dependent import of L1L3Neut is independent of basic residues. Experiments to determine whether the TM segments of Tim23p are sufficient for import of a passenger protein into mitochondria are in progress.
Aac1p, an ATP/ADP carrier, and PiC, the phosphate carrier, are members of the mitochondrial carrier family. Carrier family members contain six TM segments and are composed of a threefold repeat structure of two TM segments with an intervening loop. Studies with Aac1p indicate that it carries import information in the first one-third of the protein and in the carboxyl-terminal two-thirds (Adrian et al., 1986; Pfanner et al., 1987; Smagula and Douglas, 1988a,b). Similarly, another member of the carrier family UCP, the mammalian brown fat-UCP, has at least two internal targeting signals (Liu et al., 1988, 1990). Like Aac1p and UCP, we find that Tim23p has redundant import information, with targeting signals in the first and fourth TM segments of Tim23p.
Although TM segment 1 and TM segment 4 of Tim23p promote import, they do not function efficiently when present as the sole TM domain. The targeting activity of TM1 is much more effective when present with TM2, and TM4 works better in concert with TM3. It is therefore possible that some sort of secondary structure, such as paired TM segments are important for import across the OM. Supporting this possibility, we find that Tim23p lacking its fourth TM segment gets stuck in the OM during its import into mitochondria. Protease digestion indicates most of the Tim23 protein is inside the OM with TM3 extending outside the organelle. Our results argue that Tim23p is imported into mitochondria in an N-to-C direction, and that unpaired TM segments are not efficiently recognized by the import machinery. Similar observations suggesting that coordination between TM segments or that specific secondary structures are important determinants for import have been noted in studies with other IM proteins (Liu et al., 1988, 1990).
Recently, Káldi et al. (1998) analyzed the import pathway of Tim23p and identified two import signals within Tim23p. One import signal was reported to be located in the first 62 amino acid residues of Tim23p and mediated the translocation of the Tim23 protein across the OM in the presence or absence of membrane potential. We find no evidence for an import signal in the amino-terminal region of Tim23p. Tim23Np, which contains the first 96 residues of Tim23p, is not imported into isolated mitochondria, even in the presence of a membrane potential (Ryan et al., 1998; Figure 4). Quantitation of gels indicates that <1% of the Tim23Np protein added to the import reaction is protected from protease digestion. Whether this protected material is a small amount of Tim23 protein actually imported into the organelle or represents incompletely digested protein is unclear. Nonetheless, in our hands the Tim23p amino-terminal domain does not appear to contain a significant import signal in experiments with isolated mitochondria. Supporting this view, we find that constructs carrying the Tim23p amino-terminal region with either TM2 or TM3 are also not imported (Figure 6).
Káldi et al. (1998) identified a second import signal in Tim23p, the positively charged loop L3, and proposed that L3 functioned as an internal import signal. When L3 was placed at the amino terminus of a passenger protein, the authors observed that the chimeric protein (called IS23-DHFR) was imported into the matrix. We find no evidence that L3 functions as an import signal, but instead find that L3 mediates the insertion of Tim23p into the IM after it has crossed the OM. We find that Tim23p constructs lacking the positively charged residues in loop L3 are still imported into mitochondria. In addition, when we placed loop L3 of Tim23p in front of the DHFR protein, the L3-DHFR construct was not imported and did not even bind to mitochondria (Davis and Jensen, unpublished observations). We speculate that the passenger protein, called d2–20 (Klaus et al., 1996), used by Káldi et al. to construct IS23-DHFR, contains additional basic residues that, when combined with the positive charges in L3, generate a functional presequence. Supporting this view, Káldi et al. found the import of IS23-DHFR, like other presequence-containing proteins, was dependent upon Tim23p function. In contrast, import of authentic Tim23p is dependent upon Tim54p/Tim22p and does not require Tim23p function.
If the import signal for Tim23p is truly a hydrophobic TM segment, then several questions remain. For example, what mitochondrial import machinery specifically recognizes this signal? In vitro studies suggest that proteins with presequences prefer the OM Tom20p/Tom22p receptors for their import, while proteins with internal targeting information, such as Aac1p, utilize Tom70p (Söllner et al., 1989, 1990). Consistent with this idea, we find that Tim23p uses the Tom70p receptor for its import (Emtage and Jensen, unpublished observations). Our studies suggest that Tom70p may specifically recognize the hydrophobic import signals within proteins like Tim23p, whereas the Tom22p/Tom20p receptors appear to interact with positively charged presequences. Recent studies, however, suggest that Tom70p may also recognize presequences (Brix et al., 1997; Komiya et al., 1997), raising the possibility that Tom70p interacts with both types of import signals. Experiments are currently underway to identify mitochondrial proteins that directly recognize the hydrophobic import signals within Tim23p.
Another unanswered question is why Tim23p is targeted to the mitochondria and not to other cellular organelles, such as the endoplasmic reticulum. Since the Tim23p hydrophobic segments have no apparent distinguishing features, why aren’t they recognized by the signal recognition particle or other endoplasmic reticulum translocation machinery? Whether cells contain a cytosolic factor that recognizes the signals within Tim23p, and targets it specifically to mitochondria, awaits further studies.
ACKNOWLEDGMENTS
We thank Carolyn Machamer, Dan Isaac, Mike Maceyka, Hiromi Sesaki, Jason Holder, and Oliver Kerscher for critical comments on the manuscript. We also thank Mike Yaffe for the F1β and hexokinase antisera and Jeff Schatz for antiserum to Tom70p. This work was supported by grant R01-GM-46803 from the United States Public Health Service to R.E.J.; Medical Scientist Training Program grant GM-07309 to K.R.R; and a National Institute of Health Predoctoral Training grant 5T32GN07445 to A.J.D.
REFERENCES
- Adrian GS, McCammon MT, Montgomery DL, Douglas MG. Sequences required for delivery and localization of the ADP/ATP translocator to the mitochondrial inner membrane. Mol Cell Biol. 1986;6:626–634. doi: 10.1128/mcb.6.2.626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allison DS, Schatz G. Artificial mitochondrial presequences. Proc Natl Acad Sci USA. 1986;83:9011–9015. doi: 10.1073/pnas.83.23.9011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aquila H, Link TA, Klingenberg M. The uncoupling protein from brown fat mitochondria is related to the mitochondrial ADP/ATP carrier. Analysis of sequence homologies and of folding of the protein in the membrane. EMBO J. 1985;4:2369–2376. doi: 10.1002/j.1460-2075.1985.tb03941.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer M, Sirrenberg C, Neupert W, Brunner M. Role of Tim23 as voltage sensor and presequence receptor in protein import into mitochondria. Cell. 1996;87:33–41. doi: 10.1016/s0092-8674(00)81320-3. [DOI] [PubMed] [Google Scholar]
- Bonner WM, Laskey RA. A film detection method for tritium-labeled proteins and nucleic acids in polyacrylamide gels. Eur J Biochem. 1974;46:83–88. doi: 10.1111/j.1432-1033.1974.tb03599.x. [DOI] [PubMed] [Google Scholar]
- Brachmann CB, Davies A, Cost GJ, Caputo E, Li J, Hieter P, Boeke JD. Designer deletion strains from Saccharomyces cerevisiae 288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast. 1998;14:115–132. doi: 10.1002/(SICI)1097-0061(19980130)14:2<115::AID-YEA204>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- Brix J, Dietmeier K, Pfanner N. Differential recognition of preproteins by the purified cytosolic domains of the mitochondrial import receptors Tom20, Tom22, and Tom70. J Biol Chem. 1997;272:20730–20735. doi: 10.1074/jbc.272.33.20730. [DOI] [PubMed] [Google Scholar]
- Chen WJ, Douglas MG. Phosphodiester bond cleavage outside mitochondria is required for the completion of protein import into the mitochondrial matrix. Cell. 1987;49:651–658. doi: 10.1016/0092-8674(87)90541-1. [DOI] [PubMed] [Google Scholar]
- Daum G, Böhni PC, Schatz G. Import of proteins into mitochondria. Cytochrome b2 and cytochrome c peroxidase are located in the intermembrane space of yeast mitochondria. J Cell Biol. 1982;257:13028–13033. [PubMed] [Google Scholar]
- Dekker P, Keil P, Rassow J, Maarse AC, Pfanner N, Meijer M. Identification of MIM23, a putative component of the protein import machinery of the mitochondrial inner membrane. FEBS Lett. 1993;330:66–70. doi: 10.1016/0014-5793(93)80921-g. [DOI] [PubMed] [Google Scholar]
- Eilers M, Hwang S, Schatz G. Unfolding and refolding of a purified precursor protein during import into isolated mitochondria. EMBO J. 1988;7:1139–1145. doi: 10.1002/j.1460-2075.1988.tb02923.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eilers M, Oppliger W, Schatz G. Both ATP and an energized inner membrane are required to import a purified precursor protein into mitochondria. EMBO J. 1987;6:1073–1077. doi: 10.1002/j.1460-2075.1987.tb04860.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emtage JLT, Jensen RE. MAS6 encodes an essential inner membrane component of the yeast mitochondrial import pathway. J Cell Biol. 1993;122:1003–1012. doi: 10.1083/jcb.122.5.1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Field J, Nikawa J, Broek D, MacDonald B, Rodgers L, Wilson IA, Lerner RA, Wigler M. Purification of a RAS-responsive adenylyl cyclase complex from Saccharomyces cerevisiae by use of an epitope addition method. Mol Cell Biol. 1988;8:2159–2165. doi: 10.1128/mcb.8.5.2159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fölsch H, Guiard B, Neupert W, Stuart RA. Internal targeting signal of the BCS1 protein: a novel mechanism of import into mitochondria. EMBO J. 1996;15:479–487. [PMC free article] [PubMed] [Google Scholar]
- Gawaz M, Douglas MG, Klingenberg M. Structure-function studies of adenine nucleotide transport in mitochondria. II. Biochemical analysis of distinct AAC1 and AAC2 proteins in yeast. J Biol Chem. 1990;265:14202–14208. [PubMed] [Google Scholar]
- Glick BS. Can Hsp70 proteins act as force-generating motors? Cell. 1995;80:11–14. doi: 10.1016/0092-8674(95)90444-1. [DOI] [PubMed] [Google Scholar]
- Gratzer S, Lithgow T, Bauer RE, Lamping E, Paltauf F, Kohlwein SD, Haucke V, Junne T, Schatz G, Horst M. Mas37p, a novel receptor subunit for protein import into mitochondria. J Cell Biol. 1995;129:25–34. doi: 10.1083/jcb.129.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hachiya N, Komiya T, Alam R, Iwahashi J, Sakaguchi M, Omura T, Mihara K. MSF, a novel cytoplasmic chaperone which functions in precursor targeting to mitochondria. EMBO J. 1994;13:5146–5154. doi: 10.1002/j.1460-2075.1994.tb06844.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hachiya N, Mihara K, Suda K, Horst M, Schatz G, Lithgow T. Reconstitution of the initial steps of mitochondrial protein import. Nature. 1995;376:705–709. doi: 10.1038/376705a0. [DOI] [PubMed] [Google Scholar]
- Haid A, Suissa M. Immunochemical identification of membrane proteins after sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Methods Enzymol. 1983;96:192–205. doi: 10.1016/s0076-6879(83)96017-2. [DOI] [PubMed] [Google Scholar]
- Herrmann JM, Neupert W, Stuart RA. Insertion into the mitochondrial inner membrane of a polytopic protein, the nuclear-encoded Oxa1p. EMBO J. 1997;16:2217–2226. doi: 10.1093/emboj/16.9.2217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hines V, Brandt A, Griffiths G, Horstmann H, Brutsch H, Schatz G. Protein import into yeast mitochondria is accelerated by the outer membrane protein MAS70. EMBO J. 1990;9:3191–3200. doi: 10.1002/j.1460-2075.1990.tb07517.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen RE, Kinnally KW. The mitochondrial import pathway: are precursors imported through membrane channels? J Bioenerg Biomembr. 1997;29:3–10. doi: 10.1023/a:1022470303365. [DOI] [PubMed] [Google Scholar]
- Jensen RE, Yaffe MP. Import of proteins into yeast mitochondria: the nuclear MAS2 gene encodes a component of the processing protease that is homologous to the MAS1-encoded subunit. EMBO J. 1988;7:3863–3871. doi: 10.1002/j.1460-2075.1988.tb03272.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser C, Michaelis S, Mitchell A. Methods in Yeast Genetics. Plainview, NY: Cold Spring Harbor Laboratory Press; 1994. [Google Scholar]
- Káldi K, Bauer M, Sirrenberg C, Neupert W, Brunner M. Biogenesis of Tim23 and Tim17, integral components of the TIM machinery for matrix-targeted preproteins. EMBO J. 1998;17:1569–1576. doi: 10.1093/emboj/17.6.1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang PJ, Ostermann J, Shilling J, Neupert W, Craig EA, Pfanner N. Requirement for hsp70 in the mitochondrial matrix for translocation and folding of precursor proteins. Nature. 1990;348:137–143. doi: 10.1038/348137a0. [DOI] [PubMed] [Google Scholar]
- Kerscher O, Holder J, Srinivasan M, Leung RS, Jensen RE. The Tim54p-Tim22p complex mediates insertion of proteins into the mitochondrial inner membrane. J Cell Biol. 1997;139:1663–1675. doi: 10.1083/jcb.139.7.1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiebler M, Keil P, Schneider H, Vanderklei IJ, Pfanner N, Neupert W. The mitochondrial receptor complex — a central role of MOM22 in mediating preprotein transfer from receptors to the general insertion pore. Cell. 1993;74:483–492. doi: 10.1016/0092-8674(93)80050-o. [DOI] [PubMed] [Google Scholar]
- Kiebler M, Pfaller R, Söllner T, Griffiths G, Horstmann H, Pfanner N, Neupert W. Identification of a mitochondrial receptor complex required for recognition and membrane insertion of precursor proteins. Nature. 1990;348:610–616. doi: 10.1038/348610a0. [DOI] [PubMed] [Google Scholar]
- Klaus C, Guiard B, Neupert W, Brunner M. Determinants in the presequence of cytochrome b2 for import into mitochondria and for proteolytic processing. Eur J Biochem. 1996;236:856–861. doi: 10.1111/j.1432-1033.1996.00856.x. [DOI] [PubMed] [Google Scholar]
- Koehler CM, Jarosch E, Tokatlidis K, Schmid K, Schweyen RJ, Schatz G. Import of mitochondrial carriers mediated by essential proteins of the intermembrane space. Science. 1998;279:369–373. doi: 10.1126/science.279.5349.369. [DOI] [PubMed] [Google Scholar]
- Komiya T, Rospert S, Schatz G, Mihara K. Binding of mitochondrial precursor proteins to the cytoplasmic domains of the import receptors Tom70 and Tom20 is determined by cytoplasmic chaperones. EMBO J. 1997;16:4267–4275. doi: 10.1093/emboj/16.14.4267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komiya T, Sakaguchi M, Mihara K. Cytoplasmic chaperones determine the targeting pathway of precursor proteins to mitochondria. EMBO J. 1996;15:399–407. [PMC free article] [PubMed] [Google Scholar]
- Kübrich M, Rassow J, Voos W, Pfanner N, Honlinger A. The import route of ADP/ATP carrier into mitochondria separates from the general import pathway of cleavable preproteins at the trans site of the outer membrane. J Biol Chem. 1998;273:16374–16381. doi: 10.1074/jbc.273.26.16374. [DOI] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lawson JE, Douglas MG. Separate genes encode functionally equivalent ADP/ATP carrier proteins in Saccharomyces cerevisiae. Isolation and analysis of AAC2. J Biol Chem. 1988;263:14812–14818. [PubMed] [Google Scholar]
- Lawson JE, Gawaz M, Klingenberg M, Douglas MG. Structure-function studies of adenine nucleotide transport in mitochondria. I. Construction and genetic analysis of yeast mutants encoding the ADP/ATP carrier protein of mitochondria. J Biol Chem. 1990;265:14195–14201. [PubMed] [Google Scholar]
- Liu XQ, Bell AW, Freeman KB, Shore GC. Topogenesis of mitochondrial inner membrane uncoupling protein. Rerouting transmembrane segments to the soluble matrix compartment. J Cell Biol. 1988;107:503–509. doi: 10.1083/jcb.107.2.503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu XQ, Freeman KB, Shore GC. An amino-terminal signal sequence abrogates the intrinsic membrane-targeting information of mitochondrial uncoupling protein. J Biol Chem. 1990;265:9–12. [PubMed] [Google Scholar]
- Lohret TA, Jensen RE, Kinnaly KW. Tim23, a protein import component of the mitochondrial inner membrane, is required for normal activity of the multiple conductance channel, MCC. J Cell Biol. 1997;137:377–386. doi: 10.1083/jcb.137.2.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maarse AC, Blom J, Grivell LA, Meijer M. MPI1, an essential gene encoding a mitochondrial membrane protein, is possibly involved in protein import into yeast mitochondria. EMBO J. 1992;11:3619–3628. doi: 10.1002/j.1460-2075.1992.tb05446.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maarse AC, Blom J, Keil P, Pfanner N, Meijer M. Identification of the essential yeast protein MIM17, an integral mitochondrial inner membrane protein involved in protein import. FEBS Lett. 1994;349:215–221. doi: 10.1016/0014-5793(94)00669-5. [DOI] [PubMed] [Google Scholar]
- Mahlke K, Pfanner N, Martin J, Horwich AL, Hartl F-U, Neupert W. Sorting pathways of mitochondrial inner membrane proteins. Eur J Biochem. 1990;192:551–555. doi: 10.1111/j.1432-1033.1990.tb19260.x. [DOI] [PubMed] [Google Scholar]
- Mayer A, Nargang FE, Neupert W, Lill R. MOM22 is a receptor for mitochondrial targeting sequences and cooperates with MOM19. EMBO J. 1995;14:4204–4211. doi: 10.1002/j.1460-2075.1995.tb00094.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAda PC, Douglas MG. A neutral metallo endoprotease involved in the processing of an F1-ATPase subunit precursor in mitochondria. J Biol Chem. 1982;257:3177–3182. [PubMed] [Google Scholar]
- Miller BR, Cumsky MG. An unusual import pathway for the precursor to yeast cytochrome oxidase subunit Va. J Cell Biol. 1991;112:833–841. doi: 10.1083/jcb.112.5.833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller BR, Cumsky MG. Intramitochondrial sorting of the precursor to yeast cytochrome c oxidase subunit Va. J Cell Biol. 1993;121:1021–1029. doi: 10.1083/jcb.121.5.1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moczko M, Dietmeier K, Söllner T, Segui B, Steger HF, Neupert W, Pfanner N. Identification of the mitochondrial receptor complex in Saccharomyces cerevisiae. FEBS Lett. 1992;310:265–268. doi: 10.1016/0014-5793(92)81345-m. [DOI] [PubMed] [Google Scholar]
- Niman HL, Houghten RA, Walker LE, Reisfeld RA, Wilson IA, Hogle JM, Lerner RA. Generation of protein-reactive antibodies by short peptides is an event of high frequency: implications for the structural basis of immune recognition. Proc Natl Acad Sci USA. 1983;80:4949–4953. doi: 10.1073/pnas.80.16.4949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmieri F, Bisaccia F, Capobianco L, Dolce V, Fiermonte G, Iacobazzi V, Zara V. Transmembrane topology, genes, and biogenesis of the mitochondrial phosphate and oxoglutarate carriers. J Bioenerg Biomembr. 1993;25:493–501. doi: 10.1007/BF01108406. [DOI] [PubMed] [Google Scholar]
- Pfanner N, Craig EA, Meijer M. The protein import machinery of the mitochondrial inner membrane. Trends Biochem Sci. 1994;19:368–372. doi: 10.1016/0968-0004(94)90113-9. [DOI] [PubMed] [Google Scholar]
- Pfanner N, Hoeben P, Tropschug M, Neupert W. The carboxyl-terminal two-thirds of the ADP/ATP carrier polypeptide contains sufficient information to direct translocation into mitochondria. J Biol Chem. 1987;262:14851–14854. [PubMed] [Google Scholar]
- Pfanner N, Meijer M. Mitochondrial biogenesis: the Tom and Tim machine. Curr Biol. 1997;7:100–103. doi: 10.1016/s0960-9822(06)00048-0. [DOI] [PubMed] [Google Scholar]
- Pfanner N, Müller HK, Harmey MA, Neupert W. Mitochondrial protein import: involvement of the mature part of a cleavable precursor protein in the binding to receptor sites. EMBO J. 1987;6:3449–3454. doi: 10.1002/j.1460-2075.1987.tb02668.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfanner N, Neupert W. Transport of proteins into mitochondria: a potassium diffusion potential is able to drive the import of ADP/ATP carrier. EMBO J. 1985;4:2819–2825. doi: 10.1002/j.1460-2075.1985.tb04009.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfanner N, Neupert W. Distinct steps in the import of ADP/ATP carrier into mitochondria. J Biol Chem. 1987;262:7528–7536. [PubMed] [Google Scholar]
- Pfanner N, Tropschug M, Neupert W. Mitochondrial protein import: nucleoside triphosphates are involved in conferring import-competence to precursors. Cell. 1987;49:815–823. doi: 10.1016/0092-8674(87)90619-2. [DOI] [PubMed] [Google Scholar]
- Pollock RA, Hartl F-U, Cheng MY, Ostermann J, Horwich A, Neupert W. The processing protease of yeast mitochondria: the two co-operating components MPP and PEP are structurally related. EMBO J. 1988;7:3493–3500. doi: 10.1002/j.1460-2075.1988.tb03225.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roise D. Interaction of a synthetic mitochondrial presequence with isolated yeast mitochondria: mechanism of binding and kinetics of import. Proc Natl Acad Sci USA. 1992;89:608–612. doi: 10.1073/pnas.89.2.608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roise D, Horvath SJ, Tomich JM, Richards JH, Schatz G. A chemically synthesized pre-sequence of an imported mitochondrial protein can form an amphipathic helix and perturb natural and artificial phospholipid bilayers. EMBO J. 1986;5:1327–1334. doi: 10.1002/j.1460-2075.1986.tb04363.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roise D, Theiler F, Horvath SJ, Tomich JM, Richards JH, Allison DS, Schatz G. Amphiphilicity is essential for mitochondrial presequence function. EMBO J. 1988;7:649–653. doi: 10.1002/j.1460-2075.1988.tb02859.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Runswick MJ, Powell SJ, Nyren P, Walker JE. Sequence of the bovine mitochondrial phosphate carrier protein: structural relationship to ADP/ATP translocase and the brown fat mitochondria uncoupling protein. EMBO J. 1987;6:1367–1373. doi: 10.1002/j.1460-2075.1987.tb02377.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan KR, Jensen RE. Mas6p can be crosslinked to an arrested precursor and interacts with other proteins during mitochondrial protein import. J Biol Chem. 1993;268:23743–23746. [PubMed] [Google Scholar]
- Ryan KR, Leung RS, Jensen RE. Characterization of the inner membrane translocase complex: the Tim23p hydrophobic daomain interacts with Tim17p but not with other Tim23p molecules. Mol Cell Biol. 1998;18:178–187. doi: 10.1128/mcb.18.1.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan KR, Menold MM, Garrett S, Jensen RE. SMS1, a high-copy suppressor of the yeast mas6 mutant, encodes an essential inner membrane protein required for mitochondrial protein import. Mol Biol Cell. 1994;5:529–538. doi: 10.1091/mbc.5.5.529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saiki RK, Scharf S, Faloona F, Mullis KB, Horn GT, Erlich HA, Arnheim N. Enzymatic amplification of β-globin genomic sequences and restriction site analysis for diagnosis of sickle cell anemia. Science. 1985;230:1350–1354. doi: 10.1126/science.2999980. [DOI] [PubMed] [Google Scholar]
- Schatz G, Dobberstein B. Common principles of protein translocation across membranes. Science. 1996;271:1519–1526. doi: 10.1126/science.271.5255.1519. [DOI] [PubMed] [Google Scholar]
- Scherer PE, Manning-Krieg UC, Jenö P, Schatz G, Horst M. Identification of a 45-kDa protein at the protein import site of the yeast mitochondrial inner membrane. Proc Natl Acad Sci USA. 1992;89:11930–11934. doi: 10.1073/pnas.89.24.11930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiestl RH, Gietz RD. High efficiency transformation of intact yeast cells using single stranded nucleic acids as a carrier. Curr Genet. 1989;16:339–346. doi: 10.1007/BF00340712. [DOI] [PubMed] [Google Scholar]
- Schleyer M, Schmidt B, Neupert W. Requirement of a membrane potential for the posttranslational transfer of proteins into mitochondria. Eur J Biochem. 1982;125:109–116. doi: 10.1111/j.1432-1033.1982.tb06657.x. [DOI] [PubMed] [Google Scholar]
- Schlossmann J, Dietmeier K, Pfanner N, Neupert W. Specific recognition of mitochondrial preproteins by the cytosolic domain of the import receptor MOM72. J Biol Chem. 1994;269:11893–11901. [PubMed] [Google Scholar]
- Sherman F. Mutants of yeast deficient in cytochrome c. Genetics. 1964;49:39–48. doi: 10.1093/genetics/49.1.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski R, Boeke JD. In vitro mutagenesis and plasmid shuffling: from cloned gene to mutant yeast. Methods Enzymol. 1991;194:302–318. doi: 10.1016/0076-6879(91)94023-6. [DOI] [PubMed] [Google Scholar]
- Sikorski R, Hieter P. A system of shuttle vectors and host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–28. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirrenberg C, Bauer MF, Guiard B, Neupert W, Brunner M. Import of carrier proteins into the mitochondrial inner membrane mediated by Tim22. Nature. 1996;384:582–585. doi: 10.1038/384582a0. [DOI] [PubMed] [Google Scholar]
- Sirrenberg C, Endres M, Folsch H, Stuart RA, Neupert W, Brunner M. Carrier protein import into mitochondria mediated by the intermembrane proteins Tim10/Mrs11 and Tim12/Mrs5. Nature. 1998;391:912–915. doi: 10.1038/36136. [DOI] [PubMed] [Google Scholar]
- Smagula C, Douglas MG. Mitochondrial import of the ADP/ATP carrier protein in Saccharomyces cerevisiae. Sequences required for receptor binding and membrane translocation. J Biol Chem. 1988a;263:6783–6790. [PubMed] [Google Scholar]
- Smagula CS, Douglas MG. ADP-ATP carrier of Saccharomyces cerevisiae contains a mitochondrial import signal between amino acids 72 and 111. J Cell Biochem. 1988b;36:323–327. doi: 10.1002/jcb.240360402. [DOI] [PubMed] [Google Scholar]
- Söllner T, Griffiths G, Pfaller R, Pfanner N, Neupert W. MOM19, an import receptor for mitochondrial precursor proteins. Cell. 1989;59:1061–1070. doi: 10.1016/0092-8674(89)90762-9. [DOI] [PubMed] [Google Scholar]
- Söllner T, Pfaller R, Griffiths G, Pfanner N, Neupert W. A mitochondrial import receptor for the ADP/ATP carrier. Cell. 1990;62:107–115. doi: 10.1016/0092-8674(90)90244-9. [DOI] [PubMed] [Google Scholar]
- Söllner T, Rassow J, Wiedmann M, Schlossmann J, Keil P, Neupert W, Pfanner N. Mapping of the protein import machinery in the mitochondrial outer membrane by crosslinking of translocation intermediates. Nature. 1992;355:84–87. doi: 10.1038/355084a0. [DOI] [PubMed] [Google Scholar]
- Stuart RA, Gruhler A, van der Klei I, Guiard B, Koll H, Neupert W. The requirement for matrix ATP for the import of precursor proteins into the mitochondrial matrix and intermembrane space. Eur J Biochem. 1994;220:9–18. doi: 10.1111/j.1432-1033.1994.tb18593.x. [DOI] [PubMed] [Google Scholar]
- Stuart RA, Neupert W. Topogenesis of inner membrane proteins of mitochondria. Trends Biochem Sci. 1996;21:261–267. [PubMed] [Google Scholar]
- Stuart RA, Ono H, Langer T, Neupert W. Mechanisms of protein import into mitochondria. Cell Struct Funct. 1996;21:403–406. doi: 10.1247/csf.21.403. [DOI] [PubMed] [Google Scholar]
- von Ahsen O, Voos W, Henninger H, Pfanner N. The mitochondrial protein import machinery: role of ATP in dissociation of the Hsp70-Mim44 complex. J Biol Chem. 1995;270:29848–29853. doi: 10.1074/jbc.270.50.29848. [DOI] [PubMed] [Google Scholar]
- Wachter C, Schatz G, Glick BS. Protein import into mitochondria: the requirement for external ATP is precursor-specific whereas intramitochondrial ATP is universally needed for translocation into the matrix. Mol Biol Cell. 1994;5:465–474. doi: 10.1091/mbc.5.4.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witte C, Jensen RE, Yaffe MP, Schatz G. MAS1, a gene essential for yeast mitochondrial assembly, encodes a subunit of the mitochondrial processing protease. EMBO J. 1988;7:1439–1447. doi: 10.1002/j.1460-2075.1988.tb02961.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaffe MP, Ohta S, Schatz G. A yeast mutant temperature-sensitive for mitochondrial assembly is deficient in a mitochondrial protease activity that cleaves imported precursor polypeptides. EMBO J. 1985;4:2069–2074. doi: 10.1002/j.1460-2075.1985.tb03893.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang M, Jensen RE, Yaffe MP, Oppliger W, Schatz G. Import of proteins into yeast mitochondria: the purified matrix processing protease contains two subunits which are encoded by the nuclear MAS1 and MAS2 genes. EMBO J. 1988;7:3857–3862. doi: 10.1002/j.1460-2075.1988.tb03271.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zara V, Rassow J, Wachter E, Tropschug M, Palmieri F, Neupert W, Pfanner N. Biogenesis of the mitochondrial phosphate carrier. Eur J Biochem. 1991;198:405–410. doi: 10.1111/j.1432-1033.1991.tb16029.x. [DOI] [PubMed] [Google Scholar]