Abstract
G protein-coupled receptors (GPCRs) have a key role in many biological processes and are important drug targets for many human diseases. Therefore, understanding the molecular interactions between GPCRs and their ligands would improve drug design. Here, we describe an approach that allows the rapid identification of functional agonists expressed in bacteria. Transgenic Caenorhabditis elegans expressing the human chemokine receptor 5 (CCR5) in nociceptive neurons show avoidance behavior on encounter with the ligand MIP-1α and avoid feeding on Escherichia coli expressing MIP-1α compared with control bacteria. This system allows a simple activity screen, based on the distribution of transgenic worms in a binary food-choice assay, without a requirement for protein purification or tagging. By using this approach, a library of 68 MIP-1α variants was screened, and 13 critical agonist residues involved in CCR5 activation were identified, four of which (T8, A9, N22, and A25) have not been described previously, to our knowledge. Identified residues were subsequently validated in receptor binding assays and by calcium flux assays in mammalian cells. This approach serves not only for structure/function studies as demonstrated, but may be used to facilitate the discovery of agonists within bacterial libraries.
Keywords: CCR5, MIP-1α
GPCRs constitute the largest family of mammalian cell surface proteins. They have central roles in physiological processes and represent major targets for current pharmaceutical therapies. GPCRs bind a diverse set of ligands including volatile odorants, hormones, and protein ligands such as chemokines and cytokines, which represent a major medically relevant class of GPCR ligands. Caenorhabditis elegans is a bacteria-feeding soil nematode whose strategies for detecting food sources and environmental compounds include using GPCRs expressed in gustatory neurons driving “hard-wired” repulsive or attractive responses. Thus, C. elegans provides a potential means of studying GPCR activation. We have previously shown that heterologous expression of GPCRs in this system permit changes in specificity of the avoidance response. Heterologous expression of Somatostatin Receptor 2 and CCR5 in the chemosensory nociceptive neurons ASH and ADL of C. elegans resulted in avoidance behavior when the transgenic worms were exposed to somatostatin and MIP-1α, respectively (1).
The work described above used purified soluble compounds placed in the path of the worm to elicit an avoidance response. Because C. elegans feeds on bacteria including Escherichia coli, and because gustatory neurons have an important role in taste perception and food preference, we reasoned that expression of functional MIP-1α in bacteria (2) could drive an avoidance response in the transgenic animals, leading to reduced feeding on bacteria expressing the agonist. Here, we demonstrate that CCR5 transgenic C. elegans exhibit avoidance feeding behavior toward bacteria expressing functional MIP-1α compared with bacteria expressing an unrelated protein in a simple binary choice-feeding assay. We illustrate the usefulness of the approach by constructing and expressing a panel of MIP-1α variants in E. Coli and using the feeding assay to determine the contribution of individual amino acids to CCR5 activation.
These results were validated in mammalian cell-based assays and provide insight into the interaction of MIP-1α and other chemokines with CCR5. Therefore, this approach can be applied to mammalian GPCR whose functional ligand can be expressed in bacteria, serving as a powerful tool to survey GPCR-ligand interactions and provide a screen for novel agonists.
Results
CCR5 Transgenic Animals Avoid Feeding on MIP-1α Expressing Bacteria.
C. elegans has the ability to distinguish and select between different types of bacteria as food source (2), suggesting that it could detect and respond to repellents expressed in bacteria. Adult transgenic animals expressing CCR5 in nociceptive neurons were tested in a binary food-choice assay (2), by using bacterial colonies expressing either MIP-1α or a control protein in the same vector. MIP-1α is a member of the CC chemokine family and acts as an agonist for the CCR5 receptor. The potencies of different N-terminal variants have been reported. In this article, we use a variant with reduced aggregation properties due to a substitution of Asp-26 to alanine (3, 4). This version, which also lacks the first alanine residue (with Ser as the first amino acid), has been shown to have compatible potency with full-length MIP-1α (5).
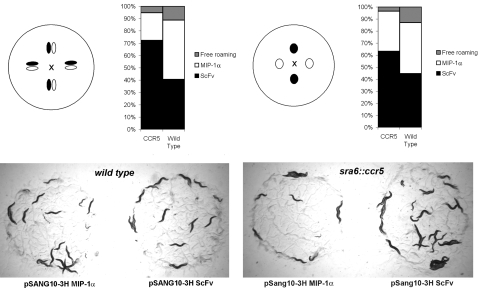
Protein expression from each construct was confirmed by Western blot analysis [supporting information (SI) Fig. S1]. The animals were tested in a binary choice-feeding configuration based on the distance between colonies and, therefore, ease of sampling of the bacterial colonies by the animals. In the “easy” arrangement, colonies expressing either single-chain Fv (scFv) or MIP-1α are directly adjacent to each other in a diamond configuration. The animals can sample the neighboring colony expressing a different protein without leaving the colony that it is currently grazing on. In the “difficult” arrangement, ScFv- and MIP-1α-expressing colonies are opposite each other in the same configuration, requiring animals to venture outside one colony to sample another.
CCR5 transgenic animals favored feeding on control bacteria compared with bacteria expressing MIP-1α (Fig. 1). This outcome is reflected as a biased distribution of the population of transgenic worms on different bacterial colonies. By using CCR5 transgenic animals (prefed with standard OP50 bacteria), we found that the number of animals feeding on each type of colony stabilized over 45 min. In the “difficult” colony arrangement, 63% of CCR5 transgenic animals fed on the bacterial colony expressing control protein (a recombinant scFv), compared with 33% on the colony expressing MIP-1α (Fig. 1 Right). In the “easy” configuration, which was used for all subsequent studies, >70% of the transgenic animals preferred to feed on the scFv expressing colonies compared with 22%, which fed on MIP-1α-expressing bacteria (Fig. 1 Left). In both cases, wild-type animals distribute evenly between the two types of expression colonies with a greater number of free roaming wild-type animals compared with transgenic animals. The same trend was observed for transgenic worms prefed with BL21 bacteria, where approximately two-thirds of the population prefer the ScFv colonies to the MIP-1α bacteria (data not shown). Thus, transgenic animals expressing mammalian GPCRs can specifically detect and avoid ligand expressed within bacterial colonies, making this assay a potentially powerful system to identify bacterial clones expressing agonist.
Fig. 1.
Agonist-directed feeding behavior in CCR5 transgenic animals. Bacterial colonies expressing MIP-1α (pSANG10-MIP-1α) or a control protein (pSANG10-scFv) were grown side-by-side in the two configurations described. Transgenic worms were placed in the center of the plate (marked with X) at equal distance from the bacterial colonies. The percentage associated with each colony was plotted. In the optimal configuration (colonies grown close together and worms prefed with standard OP50 bacteria), >70% of the transgenic animals prefer to graze on the ScFv-expressing colonies, compared with 20% that are distributed over the MIP-1α-expressing bacteria. (P ≤ 0.001; SEM for CCR5 transgenic and wild-type animals, 1.15 and 2.86, respectively.) Photograph shows the grazing animals from a pair of colonies in the closer configuration after 45 min.
Determining Function of Human MIP-1α Variants by Using The Feeding-Choice Assay.
The utility of this system was illustrated by identifying key residues involved in the interaction of MIP-1α with CCR5. MIP-1α is a member of the CC chemokine family, which share a conserved monomeric fold, consisting of the N terminus; a disorganized floppy N-loop, followed by three antiparallel β-strands arranged in a Greek key motif, separated by short loops (including the 40s loop) and C-terminal α helix (6). As with all CCs, the first two cysteines of MIP-1α are contiguous, separating the N-terminal domain from the N-loop. The N-loop is followed by the 310 turn that includes residues 20–24, which in turn precedes the first β-strand.
A panel of 68 single amino acid substituted variants of MIP-1α were generated by oligonucleotide site-directed mutagenesis and confirmed by sequencing. MIP-1α variant proteins were generated where amino acid residues were individually replaced by alanine. Alanine substitutions remove interactions beyond the β-carbon and reveal the contribution to binding made by the removed side chain (7, 8) with minimal disruption of protein structure. In positions where alanine residues are present, they were mutated to a hydrophilic-residue glutamine. Also, hydrophilic residues previously implicated in binding/activation in MIP-1α such as Arg-17, Gln-18, and Asn-22 were mutated to negatively-charged glutamic acid (hereafter represented as R17E, Q18E, and N22E). The four cysteine residues were unmodified to retain structural integrity. Expression after mutagenesis was confirmed by Western blot analysis (Fig. S1) and clones such as the P53A variant, which showed a marked reduction in expression, were removed from further analysis.
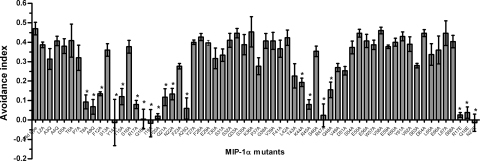
Each MIP-1α mutant was tested in the feeding assay, where transgenic animals were given a choice between colonies expressing either the MIP-1α mutant or control ScFv protein, and a “feeding avoidance index” calculated. This is calculated as the number of worms grazing on the ScFv colony minus the number of worms feeding on MIP-1α colony, divided by the total number of worms. These measurements identified 18 aa variants where avoidance was compromised to different extents (Fig. 2), corresponding to three regions of the peptide (the N terminus; the N-loop, including the 310 turn; and the 40s loop). Our results implied that mutations were tolerated at most positions with 50/68 mutations showing no significant effect. We identified four previously undescribed residues (T8, A9, N22, and A25) that contribute to the interaction between the heterologously expressed CCR5 and MIP-1α.
Fig. 2.
Sixty-one single amino acid substituted varients of MIP-1 were constructed in which the amino acid residue at each position was replaced with an alanine residue. Application to screening for MIP-1α activity in a panel of mutants. Where alanine residues were already present, a glutamine residue was introduced. Also, hydrophilic residues previously implicated in binding and activation in MIP-1α- R17, Q18, and N22 were mutated to negatively charged glutamate. Each MIP-1α variant was tested in the feeding assay and the feeding-avoidance index plotted relative to each position. The feeding-avoidance index is calculated as follows: number of worms grazing on ScFv minus number of worms feeding on MIP-1α, divided by the total number of worms. Residues that eliminate the avoidance behavior correspond to three regions within the protein (the N terminus, the charged residues within the N-loop, and the β-strand 40s loop). Each data point indicates the average of at least three independent assays for each MIP-1α variant. Error bars indicate SEM. Asterisks indicate a statistical significance between the avoidance index between the wild type vs. ScFv assay and the MIP-1α mutant vs. ScFv assay (P < 0.005).
Validation of Identified Residues by Using Calcium Signaling and Receptor Binding In Mammalian Cells.
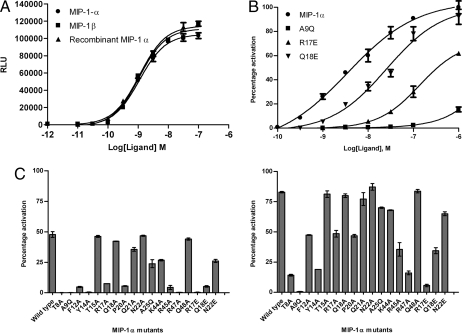
To validate the observations from the transgenic worm feeding assay, the 18 MIP-1α mutants identified were purified and tested for their ability to trigger calcium release by using CHO cells expressing human CCR5 in an aequorin assay (9, 10). A dose-response curve confirmed that the biological activity of the bacterially produced wild-type recombinant protein was equivalent to control MIP-1α. MIP-1β is an alternative agonist for CCR5 (with 70% sequence similarity to MIP-1α), which was also included as control (Fig. 3A). As a negative control, we tested a Y14A mutant, which has negligible biological activity due to its inability to fold into a proper tertiary structure (11), although its expression level is comparable with wild type. Typical dose-response curves for three variants with a range of IC50 values are shown (Fig. 3B). The activity of the rest of the MIP-1α mutants is shown in Fig. 3C measured at 1 nM (Left) and 10 nM (Right). Compromised activity was observed for 14 of the 18 amino acid variants, representing 13 out of 15 amino acid positions of MIP-1α. This result confirms the earlier findings obtained from the feeding assay for the majority of clones, although four mutants (T15A, Q18A, N22A, and Q48A) gave wild-type signaling levels.
Fig. 3.
Characterization of the functional activity of MIP-1α mutants. CCR5 expressed on CHO cells was used in aequorin calcium flux experiments. (A) Comparison of MIP-1α and MIP-1β activity with purified recombinant MIP-1α by using the aequorin assay shows that the recombinant protein has comparable activity to the commercial proteins, confirming that the purified bacterially produced proteins are biologically active. (B) Graphs of CCR5 activity for recombinant MIP-1α and several mutants, A9Q, R17E, and Q18E, that compromise activity of the peptide. Data has been normalized so that it is displayed as percentage activation compared with recombinant MIP-1α. (C) Relative ability of MIP-1α mutants to promote calcium release at 1 nM (Left) and 10 nM (Right) agonist concentration. Each bar represents the mean value of at least two experiments, with SEM shown as error bars.
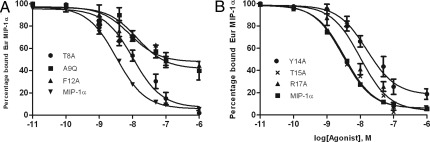
To determine whether the mutations affected ligand binding or receptor activation after binding, the binding affinity was compared by using a competition assay where unlabelled variants compete with binding of europium-labeled MIP-1α to membrane preparations from CCR5 transfected CHO cells. The assay was established by testing variable concentrations of the R17E mutant as the unlabelled competitor. Three concentrations of europium-labeled MIP-1α (5, 8, and 10 nM) were tested to ensure that an appropriate range of europium signal could be achieved within the assay and 8 nM Eur-MIP-1α was chosen for the competition binding assays (Fig. S2). The results confirmed that T15A, Q18A, N22A, and Q48A mutants bind with normal affinity, mirroring the wild-type calcium flux response in the aequorin assay (Fig. 4B and Fig. S2). The binding constant of each of the other mutants (Table 1) is consistent with their reduced ability to promote calcium release (Fig. 4 and Fig. S2), indicating that the mutations act by reducing the ability of MIP-1α to bind to the CCR5 on mammalian cells.
Fig. 4.
Binding of MIP-1α variants to CCR5. Competition binding curves were generated on membranes from CHO cells expressing CCR5 and using 8 nM Eur-MIP-1α as a trace. Example inhibition data are shown for T8A, A9Q, and F12A (A), and Y14A, T15A, and R17A, (B) and all results are summarized in Table 1. The data were normalized for nonspecific binding and maximal specific binding in the absence of competitor (100%). The results were analyzed with GraphPad PRISM software by using a single-site model. All points were run in triplicate (error bars indicate SEM), and the presented data are a representative of two independent experiments, each done in duplicate. See Fig. S2 for binding curves for other mutants.
Table 1.
Binding parameters characterizing the interaction of MIP-1α variants with CCR5
| Variant | Ki, nM |
|---|---|
| MIP-1α | 5.28 ± 0.001 |
| T8A | 18.66 ± 0.15* |
| A9Q | 20.27 ± 0.03** |
| F12A | 16.59 ± 0.07** |
| Y14A | 28.78 ± 0.11** |
| T15A | 5.47 ± 0.1 |
| R17A | 16.57 ± 0.17* |
| Q18A | 7.68 ± 0.04 |
| P20A | 25.3 ± 0.06** |
| Q21A | 8.43 ± 0.05 |
| N22A | 6.183 ± 0.13 |
| A25Q | 14.52 ± 0.17* |
| K44A | 16.81 ± 0.09* |
| R45A | 58.26 ± 0.03** |
| R47A | 52.1 ± 0.14** |
| Q48E | 6.12 ± 0.06 |
| R17E | 24.2 ± 0.08* |
| Q18E | 12.09 ± 0.1**** |
| N22E | 9.57 ± 0.09**** |
The Ki value for each mutant was obtained in a competitive binding assay by using a CCR5-expressing CHO cell line and europium-labeled MIP-1α as a tracer. The values represent the mean ± SEM resulting from at least two independent experiments. The statistical significance as compared with that of wild-type MIP-1α was calculated with Student's t test.
*, P < 0.01;
**, P < 0.001;
***, P < 0.05.
Insight into Functional Interaction of MIP-1α and The CCR5.
This study has identified 13 residues mainly within three regions of MIP-1α, which are important in the interaction of the ligand with the CCR5 (N terminus; N-loop, including 310 turn; and 40s loop). The effect on receptor binding could arise from specifically disrupting interactions between the ligand and the receptor or could arise from disrupted folding of the ligand. Fig. 3A suggests that the specific activity of wild-type MIP-1α from periplasmic expression is equivalent to that of MIP-1α and MIP-1β produced from an independent commercial source. Also, for at least 50 of 68 mutants in this study, there was little difference in activity compared with wild type (Fig. 2). However, for individual positions, there could be more generalized folding effects both in this and previous mutagenic studies.
A number of important residues identified in this study have been substantiated in other studies. For example, three N-loop residues (F12, R17, and P20) and three 40s loop residues (K44, R45, and R47) have previously been reported to be involved in binding of the structurally related chemokine, MIP-1β. These regions together form the 3D binding pocket consisting of a hydrophobic core surrounded by charged residues (Fig. 5). The hydrophobic core includes F12, whose aromatic side chain has a pivotal role in CCR5 binding (6, 12–18). Indeed, our findings show a dramatic disruption in the ability to bind CCR5, when this residue is changed to alanine, validating the importance of the aromatic side chain for its binding activity. It is possible that this residue interacts with CCR5 by interaction with the second extracellular loop (ECL2) containing seven aromatic residues (19–23). The role of P20 in maintaining local secondary structure (4) has also been highlighted in this feeding assay.
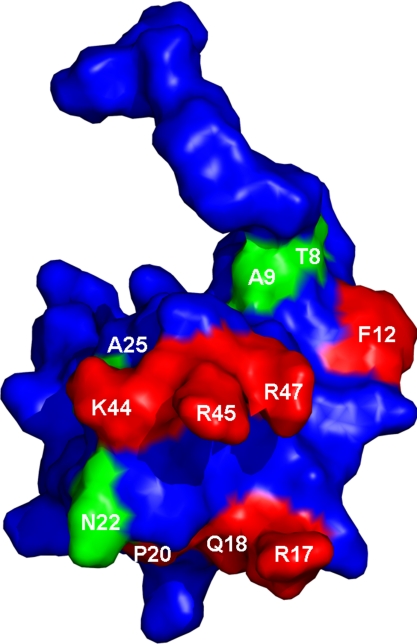
Fig. 5.
Three-dimensional structure of MIP-1α. Spatial orientation of residue side chains as derived from Czaplewski et al. (4), showing in red side chain residues that have been reported previously to be involved in CCR5 binding to structurally related MIP-1β (F12, R17, Q18, P20, K44, R45, and R47). Interactions identified from this study are highlighted in green (T8, A9, N22, and A25).
MIP-1α interacts with CCR5 by means of its charged residues in the N-loop (including the 310 turn) and 40s loop. The residues within the 310 turn contribute to binding to different extents, although R17 is the primary residue that interacts with CCR5 (11). A sharp decrease in CCR5 binding affinity was observed with R17E, whereas the effect was less severe with the R17A mutant (Table 1). Q18A and N22A mutations do not lead to considerable change in binding, although introducing a negative charge results in a significant decrease in Ki and EC50 in the binding assay and calcium mobilization assays, respectively. This result suggests that a smaller contribution could be made by Q18 and N22. In contrast, there is a less significant effect on its binding activity when Q21 is substituted with alanine. Our results also show that the highly conserved charged residues K44, R45, and R47 in the 40s loop are key mediators of CCR5 binding, of which R45 and R47 have the most important role. This finding is largely consistent with earlier reports that the corresponding basic residues in MIP-1β (K44, R45, and K47) are critical for CCR5 binding (24). These residues in the 40s loop also participate in heparin binding, although this event occurs only when the protein is in its dimeric form. Mapping the amino acid sequence of MIP-1α onto the coordinates of the closely related MIP-1β reveals that residues K44 and R45 define a turn between adjacent β-strands, and side chains of residues R17, R45, and R47 are in close proximity of each other (25). Although the sequences of MIP-1α and MIP-1β are 67% identical, the extent to which specific residues are involved in binding varies as reflected in differences between the results of our MIP-1α screen and previous studies on MIP-1β. In contrast to MIP-1β, the MIP-1α Q18A mutant is functionally active. The basic residue found in the corresponding position in MIP-1β (K18A) attenuates binding (11). In both cases, introducing a negatively charged glutamic acid results in a complete loss in binding and functional activity (Fig. 3C and Fig. S2).
In our study, we identified four residues of importance (T8, A9, N22, and A25). The replacement of N-terminal residues, T8 with alanine and A9 with glutamine, resulted in complete loss of activity, indicating the importance of a hydrophilic residue in position 8, and a hydrophobic amino acid in position 9 for receptor activation. The corresponding threonine residue in MIP-1β is important in dimer formation given its location at the hydrophobic dimer interface, where mutation to a charged residue will lead to a monomeric form (12). Interestingly, T8A mutation has no effect on the activity of RANTES with CCR1 (15). We propose that A9 could contribute to a hydrophobic core based around F12, which has been shown to have a role in CCR5 binding (Fig. 5) (12). The importance of a small hydrophobic residue (A25) at the interface between the 310 turn and first β-sheet is also highlighted, given the moderate loss of binding caused by mutation to glutamine.
Discussion
The nematode, C. elegans, has proven to be a valuable model system for increasing our understanding of many fundamental biological processes. Despite the phylogenetic distance between man and worm, it has been used to gain a deeper understanding of the mechanism of drug action and the discovery of new bioactive compounds (1, 26, 27). More recently, its tractability as a tool for drug screening and investigating receptor-ligand interactions has been realized (1, 26, 28, 29). In previous work, we have introduced four GPCRs into sensory neurons of C. elegans and have shown functional expression of all four. Expression in AWA/AWB olfactory neurons of the human hOR17-4 gene (30) and the rat I7 gene (31) result in specific behavioral response to bourgeonal and octanal, respectively (unpublished results). By switching expression to the ASH and ADL nociceptive neurons, the heterologous GPCRs were directly exposed to the aqueous environment allowing access of protein and peptide ligands to the heterologous receptors. In this way, functional expression of the somatostatin receptor and CCR5 was demonstrated, resulting in avoidance behavior of transgenic worms exposed to soluble agonists (1, 26, 28, 29). We extend this finding to show that CCR5 transgenic animals are able to sense and avoid agonists expressed in their bacterial food source, allowing a simple screen for activity in bacterial libraries.
There are a number of other methods available, based on heterologous expression of GPCRs in yeast or cultured mammalian cells, that allow screening for altered specificity or function of the GPCR itself. For example, using yeast-based systems, Armbruster et al. (32) have identified receptor variants activated by small molecules, and Li et al. (33) have identified GPCR residues causing functional perturbation. In addition, various mammalian cell-based reporter systems provide powerful drug-discovery screens. The method presented here complements these approaches, having unique strengths in the screening of protein ligands that can be expressed in bacteria. In contrast to cell-based assays, this system does not require protein-peptide ligands to be endotoxin-free. Because it is based on bacterial feeding, the system bypasses the purification of ligand candidates altogether, thereby greatly facilitating the screening process.
The value of this system was illustrated by carrying out an extensive mutagenesis screen of MIP-1α and identifying critical residues involved in CCR5 activation by analyzing individual clones in a worm feeding-choice assay. Mutants that affect receptor activation cause an elimination or reduction of the avoidance feeding bias, which resulted in the identification of 13 residues whose alteration compromised MIP-1α activity. Further studies with cell-based aequorin calcium signaling assays provided independent validation that most of these MIP-1α mutants were compromised in CCR5 activation in mammalian cells (Fig. 3 and Fig. S2) as a result of diminished receptor binding (Table 1).
Of the identified MIP-1α mutants, more than half of them correspond to residues previously highlighted to be important for CCR5 binding of the related agonist MIP-1β. Thus, our results are in good agreement with the outcome of previous site-directed mutagenesis studies validating the newly developed experimental technology. Also, we identified sites, within the N terminus T8 and A9 and the interface between the 310 turn and the beginning of the β-sheet (A25), that are important in MIP-1α function. We also highlight the relative contribution of N22 in the 310 turn for MIP-1α activity. T8 and A9 are conserved in MIP-1α, MIP-1β, and RANTES, whereas A25 is found in both MIP-1α and MIP-1β. This delineates a previously undescribed role for the N terminus residues in MIP-1α in CCR5 binding.
Four mutants identified in the original feeding-choice screen (T15A, Q18A, N22A, and Q48A) had activity levels comparable to wild-type MIP-1α in the calcium flux and receptor binding assays in mammalian cells. One possible explanation for these “false positives” is a compromised level of functional protein expressed in the bacterial colonies as a result of mutagenesis, resulting in lack of response in the feeding assay. In the mammalian cell-signaling assay, by contrast, protein was purified and input was normalized. Single amino acid mutations can have a significant effect on expression. Certainly, the yield of Q48A in shake flask culture was significantly lower than wild type (data not shown), supporting the explanation of reduced avoidance due to reduced expression. However, expression level of the other three mutants in shake flask culture was comparable with wild type. Altered expression may still be the reason, because the level of expression in an overnight shake flask culture may not truly reflect the relative expression when grown on a plate as a bacterial colony. Thus, as well as identifying variants with reduced binding or activation potential, this assay may also uncover residues important for folding and stability, which result in reduced protein expression (e.g., T15A).
We have demonstrated a screening application where distribution of a transgenic worm population is measured between test and control on a clone by clone basis. Because transgenic worms avoid feeding on bacteria that express agonist, there is the potential for using feeding selectivity to drive selection on a whole bacterial library (e.g., an expressed cDNA library), increasing the representation of bacterial clones that express agonist within the population at the expense of nonagonists clones. Although an attractive proposition, there are additional challenges to solve before this opportunity can be realized. The degree of selectivity observed is certainly sufficient for screening purposes but may be insufficient for selection, particularly if other factors are involved (e.g., variable growth within bacterial clones). Also, we have previously shown (1) that desensitization occurs on longer exposure to ligand and have also observed that the selective ratio between test and control is reduced on incubations longer than 1 h (data not shown). This erosion in performance may be exacerbated further during selection as food supplies diminish with increasing incubation time.
In summary, the approach presented allows the rapid identification of agonists expressed in bacterial colonies exemplified by identifying functionally important amino acids in MIP-1α. Besides validating previous findings in structurally related chemokines, this approach led to the identification of multiple residues in MIP-1α that were critical for function, including four previously undescribed residues. The technology described here should be applicable to other medically important GPCRs and could potentially serve to deorphanize the less well studied members of this family. Given that GPCRs represent a substantial proportion of therapeutic targets, this strategy may be of interest and relevance in drug discovery.
Methods
Transgenic Strains.
Germline transformation was carried out as described by Mello et al. (34). Transgenic strains used were previously described (1). In short, human CCR5 gene was cloned downstream of the gpa-11 promoter and upstream of the 3′ UTR of unc-54 in a pUC plasmid. This promotor drives heterologous expression in the ASH/ADL neurons. Nematodes were grown at 16°C or 20°C on E. Coli strain OP50 by using standard methods (35). Wild-type animals were C. elegans variety Bristol strain N2.
FACS Sorting Conditions.
After injection, transgenic animals were visualized by using a gut-expressed elt-2::GFP construct, allowing selection of live transgenic animals from mixed populations by using a flow sorter. Animals were washed with M9 buffer and filtered through a 100-μm mesh filter before sorting with a worm COPAS (Union Biometrica). Appropriate sorting of each developmental stage was verified by using epifluorescence microscopy. The animals were grown for a day before testing in the feeding assay.
Construction of MIP-1α Genes and Mutants.
The cDNA encoding hMIP-1α was cloned by PCR using universal human Quick-Clone cDNA (Clontech), using a nested primer strategy with the final 5′ and 3′ primers being: HMIP1αFOR_NcoI 5′ gcccagccggccatggcaTCACTTGCTGCTGACACGCCG and HMIP-1αREV_NotI 3′ atgatgtgcggatgcggccgcGGCACTCAGCTCTAGGTC.
This PCR generates a product with an NcoI and NotI site at the 5′ and 3′ end, respectively (underlined). After NcoI/NotI digestion and ligation into pSANG10 (36), the MIP-1α protein (shown in capitals) is produced in frame at 5′ and 3′ ends with the pelB leader and the hexahistidine tag of the pSANG10 vector. The N-terminal pelB leader is removed after causing secretion into the periplasmic space. This construct was further mutagenized to create pSANG10 hMIP-1α-D26A (aspartate26 substituted with alanine) to reduce aggregation during expression while still maintaining biological activity (4). As with the previously described D26A version, this one lacked the first encoded amino acid and utilized ser2 as the first amino acid. As a control, a ScFv single-chain antibody construct was also expressed from the same vector. Mutagenesis was performed according to the method of Kunkel et al. (37). Oligonucleotides used for mutagenesis were 18–30 nt in length. Each MIP-1α variant was cloned by using pSANG10 hMIP-1α D26A as a template with KOD HotStart DNA Polymerase (Novagen). Dideoxy sequencing was used to identify the correct clone for each variant.
Protein Expression and Purification.
Expression constructs were transformed into BL21 (DE3). Protein production was performed as described by Studier et al. (38) Bacterial cultures were induced overnight at 25°C. OD600 measurements were taken for each culture and normalized before spotting colonies on LB plate with 10 mM IPTG. ScFv and MIP-1α variants were purified as previously described (36).
Food Choice Assay.
Feeding behavior of mutants were analyzed by using a modified food-choice assay as described by Shtonda et al. (2). Adult animals were washed with M9 buffer; 50–100 animals were placed in the center of each assay plate in 5 μl of CTX buffer. Assays were carried out in room temperature. Animals feeding on each type of colony were counted after 45 min. Experiments were performed in triplicate.
Aequorin Assays.
Aequorin assays were carried out as previously described (9, 10). Briefly, CHO-K1 cell lines expressing CCR5, Gα16, and mitochondrial aequorin were established. A functional assay based on the luminescence of mitochondrial aequorin after intracellular Ca2+ release (9) was performed as previously described (10, 39). Results are expressed as relative light units (RLU). Our bacterially produced MIP-1α was compared with MIP-1α from an independent commercial source (R&D Systems).
Membrane Preparation.
CHO CCR5 membranes were a kind gift from Phillip Strange, University of Reading, Reading, U.K. Membranes were prepared as described in ref. 5.
MIP-1α Competition Binding Assays.
Europium-labeled MIP-1α was prepared by Perkin–Elmer. Saturation binding curves were determined with europium-labeled MIP-1α by using the Delfia filtration protocol (Perkin–Elmer). Ligand binding was conducted with 5 μg of membrane protein in a final volume of 100 μl of Delfia L*R binding buffer in Acrowell plates (Perkin–Elmer). Nonspecific binding was determined in the presence of 100-fold excess of unlabelled MIP-1α. Competition binding assays were conducted with 8 nM europium-labeled MIP-1α. All binding reactions were performed at room temperature for 90 min. Reactions were terminated by filtration of the binding reaction by centrifugation at 1800 rpm for 10 min, followed by five washes with 100 μl of Delfia wash buffer. Enhancement solution (100 μl) was added to each well and time resolved fluorescence (TRF) was quantified on the Fusion (Perkin–Elmer).
Data Analysis.
The data from saturation and dose-displacement experiments were analyzed by using curve fitting functions in GraphPad Prism software version 5.0.
Supplementary Material
Acknowledgments.
The authors thank A. Bradley, M. Dekkers, M. Dyson, and M. Bushell for helpful discussions, B. Schuster-Böckler for his assistance in protein modeling, and B. L. Ng for her expertise on flow cytometry. Rajika Perera provided technical assistance and M. Detheux from Euroscreen SA conducted the aequorin assays. We are also grateful to P. Strange (University of Reading, Reading, UK) for his generous support with providing reagents. M.S.T. was supported by the Wellcome Trust Sanger Postdoctoral Fellowship.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0803290105/DCSupplemental.
References
- 1.Teng MS, et al. Expression of mammalian GPCRs in C. elegans generates novel behavioural responses to human ligands. BMC Biol. 2006;4:22. doi: 10.1186/1741-7007-4-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shtonda BB, Avery L. Dietary choice behavior in Caenorhabditis elegans. J Exp Biol. 2006;209:89–102. doi: 10.1242/jeb.01955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hunter MG, et al. BB-10010: An active variant of human macrophage inflammatory protein-1 alpha with improved pharmaceutical properties. Blood. 1995;86:4400–4408. [PubMed] [Google Scholar]
- 4.Czaplewski LG, et al. Identification of amino acid residues critical for aggregation of human CC chemokines macrophage inflammatory protein (MIP)-1alpha, MIP-1beta, and RANTES Characterization of active disaggregated chemokine variants. J Biol Chem. 1999;274:16077–16084. doi: 10.1074/jbc.274.23.16077. [DOI] [PubMed] [Google Scholar]
- 5.Mueller A, Mahmoud NG, Strange PG. Diverse signalling by different chemokines through the chemokine receptor CCR5. Biochem Pharmacol. 2006;72:739–748. doi: 10.1016/j.bcp.2006.06.001. [DOI] [PubMed] [Google Scholar]
- 6.Clark-Lewis I, et al. Structure-activity relationships of chemokines. J Leukocyte Biol. 1995;57:703–711. doi: 10.1002/jlb.57.5.703. [DOI] [PubMed] [Google Scholar]
- 7.Cunningham BC, Wells JA. High-resolution epitope mapping of hGH-receptor interactions by alanine-scanning mutagenesis. Science. 1989;244:1081–1085. doi: 10.1126/science.2471267. [DOI] [PubMed] [Google Scholar]
- 8.Wells JA. Systematic mutational analyses of protein-protein interfaces. Methods Enzymol. 1991;202:390–411. doi: 10.1016/0076-6879(91)02020-a. [DOI] [PubMed] [Google Scholar]
- 9.Stables J, et al. A bioluminescent assay for agonist activity at potentially any G-protein-coupled receptor. Anal Biochem. 1997;252:115–126. doi: 10.1006/abio.1997.2308. [DOI] [PubMed] [Google Scholar]
- 10.Detheux M, et al. Natural proteolytic processing of hemofiltrate CC chemokine 1 generates a potent CC chemokine receptor (CCR)1 and CCR5 agonist with anti-HIV properties. J Exp Med. 2000;192:1501–1508. doi: 10.1084/jem.192.10.1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bondue A, et al. Characterization of the role of the N-loop of MIP-1 beta in CCR5 binding. Biochemistry. 2002;41:13548–13555. doi: 10.1021/bi026087d. [DOI] [PubMed] [Google Scholar]
- 12.Laurence JS, et al. CC chemokine MIP-1 beta can function as a monomer and depends on Phe13 for receptor binding. Biochemistry. 2000;39:3401–3409. doi: 10.1021/bi9923196. [DOI] [PubMed] [Google Scholar]
- 13.Gong JH, et al. RANTES and MCP-3 antagonists bind multiple chemokine receptors. J Biol Chem. 1996;271:10521–10527. doi: 10.1074/jbc.271.18.10521. [DOI] [PubMed] [Google Scholar]
- 14.Gong JH, Clark-Lewis I. Antagonists of monocyte chemoattractant protein 1 identified by modification of functionally critical NH2-terminal residues. J Exp Med. 1995;181:631–640. doi: 10.1084/jem.181.2.631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pakianathan DR, et al. Distinct but overlapping epitopes for the interaction of a CC-chemokine with CCR1, CCR3 and CCR5. Biochemistry. 1997;36:9642–9648. doi: 10.1021/bi970593z. [DOI] [PubMed] [Google Scholar]
- 16.Weber M, et al. Deletion of the NH2-terminal residue converts monocyte chemotactic protein 1 from an activator of basophil mediator release to an eosinophil chemoattractant. J Exp Med. 1996;183:681–685. doi: 10.1084/jem.183.2.681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.LiWang AC, et al. Dynamics study on the anti-human immunodeficiency virus chemokine viral macrophage-inflammatory protein-II (VMIP-II) reveals a fully monomeric protein. Biochemistry. 1999;38:442–453. doi: 10.1021/bi9812726. [DOI] [PubMed] [Google Scholar]
- 18.Kim S, Jao S, Laurence JS, LiWang PJ. Structural comparison of monomeric variants of the chemokine MIP-1beta having differing ability to bind the receptor CCR5. Biochemistry. 2001;40:10782–10791. doi: 10.1021/bi011065x. [DOI] [PubMed] [Google Scholar]
- 19.Lee B, et al. Epitope mapping of CCR5 reveals multiple conformational states and distinct but overlapping structures involved in chemokine and coreceptor function. J Biol Chem. 1999;274:9617–9626. doi: 10.1074/jbc.274.14.9617. [DOI] [PubMed] [Google Scholar]
- 20.Samson M, et al. The second extracellular loop of CCR5 is the major determinant of ligand specificity. J Biol Chem. 1997;272:24934–24941. doi: 10.1074/jbc.272.40.24934. [DOI] [PubMed] [Google Scholar]
- 21.Wu L, et al. Interaction of chemokine receptor CCR5 with its ligands: Multiple domains for HIV-1 gp120 binding and a single domain for chemokine binding. J Exp Med. 1997;186:1373–1381. doi: 10.1084/jem.186.8.1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dragic T, et al. Amino-terminal substitutions in the CCR5 coreceptor impair gp120 binding and human immunodeficiency virus type 1 entry. J Virol. 1998;72:279–285. doi: 10.1128/jvi.72.1.279-285.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Blanpain C, et al. Multiple charged and aromatic residues in CCR5 amino-terminal domain are involved in high affinity binding of both chemokines and HIV-1 Env protein. J Biol Chem. 1999;274:34719–34727. doi: 10.1074/jbc.274.49.34719. [DOI] [PubMed] [Google Scholar]
- 24.Laurence JS, et al. Importance of basic residues and quaternary structure in the function of MIP-1 beta: CCR5 binding and cell surface sugar interactions. Biochemistry. 2001;40:4990–4999. doi: 10.1021/bi002593w. [DOI] [PubMed] [Google Scholar]
- 25.Koopmann W, Ediriwickrema C, Krangel MS. Structure and function of the glycosaminoglycan binding site of chemokine macrophage-inflammatory protein-1 beta. J Immunol. 1999;163:2120–2127. [PubMed] [Google Scholar]
- 26.Kwok TC, et al. A small-molecule screen in C. elegans yields a new calcium channel antagonist. Nature. 2006;441:91–95. doi: 10.1038/nature04657. [DOI] [PubMed] [Google Scholar]
- 27.Jones AK, Buckingham SD, Sattelle DB. Chemistry-to-gene screens in Caenorhabditis elegans. Nat Rev Drug Discov. 2005;4:321–330. doi: 10.1038/nrd1692. [DOI] [PubMed] [Google Scholar]
- 28.Segalat L. Invertebrate animal models of diseases as screening tools in drug discovery. ACS Chem Biol. 2007;2:231–236. doi: 10.1021/cb700009m. [DOI] [PubMed] [Google Scholar]
- 29.Segalat L. Drug discovery: Here comes the worm. ACS Chem Biol. 2006;1:277–278. doi: 10.1021/cb600221m. [DOI] [PubMed] [Google Scholar]
- 30.Spehr M, et al. Identification of a testicular odorant receptor mediating human sperm chemotaxis. Science. 2003;299:2054–2058. doi: 10.1126/science.1080376. [DOI] [PubMed] [Google Scholar]
- 31.Milani N, et al. Functional expression of a mammalian olfactory receptor in Caenorhabditis elegans. NeuroReport. 2002;13:2515–2520. doi: 10.1097/00001756-200212200-00027. [DOI] [PubMed] [Google Scholar]
- 32.Armbruster BN, et al. Evolving the lock to fit the key to create a family of G protein-coupled receptors potently activated by an inert ligand. Proc Natl Acad Sci USA. 2007;104:5163–5168. doi: 10.1073/pnas.0700293104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li B, et al. Rapid identification of functionally critical amino acids in a G protein-coupled receptor. Nat Methods. 2007;4:169–174. doi: 10.1038/nmeth990. [DOI] [PubMed] [Google Scholar]
- 34.Mello CC, Kramer JM, Stinchcomb D, Ambros V. Efficient gene transfer in C.elegans: Extrachromosomal maintenance and integration of transforming sequences. EMBO J. 1991;10:3959–3970. doi: 10.1002/j.1460-2075.1991.tb04966.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Martin CD, et al. A simple vector system to improve performance and utilisation of recombinant antibodies. BMC Biotechnol. 2006;6:46. doi: 10.1186/1472-6750-6-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kunkel TA, Roberts JD, Zakour RA. Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol. 1987;154:367–382. doi: 10.1016/0076-6879(87)54085-x. [DOI] [PubMed] [Google Scholar]
- 38.Studier FW. Protein production by auto-induction in high density shaking cultures. Protein Expr Purif. 2005;41:207–234. doi: 10.1016/j.pep.2005.01.016. [DOI] [PubMed] [Google Scholar]
- 39.Blanpain C, et al. CCR5 binds multiple CC-chemokines: MCP-3 acts as a natural antagonist. Blood. 1999;94:1899–1905. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.