Abstract
The secondary structures of two proteins were examined by circular dichroism spectroscopy after adsorption onto a series of organically modified silica glasses. The glasses were prepared by the sol-gel technique and were varied in hydrophobicity by incorporation of 5% methyl, propyl, trifluoropropyl, or n-hexyl silane. Both cytochrome c and apomyoglobin were found to lose secondary structure after adsorption onto the modified glasses. In the case of apomyoglobin, the α-helical content of the adsorbed protein ranged from 21% to 28%, well below the 62% helix found in solution. In contrast, these same glasses led to a striking increase in apomyoglobin structure when the protein was encapsulated within the pores during sol-gel processing: the helical content of apomyoglobin increased with increasing hydrophobicity from 18% in an unmodified glass to 67% in a 5% hexyl-modified glass. We propose that proteins preferentially adsorb onto unmodified regions of the silica surface, whereas encapsulated proteins are more susceptible to changes in surface hydration due to the proximity of the alkyl chain groups.
Adsorption of proteins onto solid surfaces is an important issue for many fields of research, including the design of biomedical devices, biocatalyst and biosensor development, food manufacturing, and protein separation techniques (1). Adsorption phenomena are governed by many factors, including the charge and polarity of the interacting surfaces, the ease of desolvation, and the structural stability of the protein (2).
Porous silica materials produced by the sol-gel technique provide an interesting system for testing the effects of surface chemistry on protein adsorption. Sol-gel-derived silica glasses are obtained by hydrolysis of a tetraalkoxysilane (typically tetramethyoxysilane (TMOS) or tetraethoxysilane) followed by condensation to form a network of Si-O-Si linkages (3). The tetraalkoxysilane may be supplemented with other silane precursors, e.g., RSi(OCH3)3, where R represents any nonhydrolyzable organic group, to incorporate new chemical functionality into the silica matrix. In this manner, glasses may be engineered to alter the charge, polarity, or hydrophobicity of the surface in contact with the solvent phase. Under many conditions, these glasses result in optically transparent materials that are amenable to spectroscopic techniques and therefore the study of protein structure after adsorption.
Because of the inherently mild processing conditions involved, many investigators use the sol-gel technique to encapsulate biomolecules in porous glasses as a noncovalent means of immobilization; proteins may be added to the liquid sol before the condensation reaction that leads to formation of the matrix is initiated. This approach has led to several examples where the encapsulated protein maintains its native structure and biological activity (4,5).
In the current study, silica glasses were made with TMOS and a series of RSi(OCH3)3 reagents (where R represents a methyl, propyl, trifluoropropyl, or n-hexyl group) to study adsorption as a function of increasing hydrophobicity. The choice of these functional groups was guided by previous work indicating that a low content of hydrophobic precursor can have a striking effect on the structure of an encapsulated protein (6,7).
Because the addition of organic groups may alter the porosity of the glass in addition to the surface chemistry, it is important to check the physical properties of the bulk glass. As shown in Table 1, the 5% methyl, propyl, and hexyl glasses yield similar values for the specific surface area, pore volume, and average pore diameter. The control glass of 100% TMOS is characterized by larger pores and lower specific surface area than the alkyl-modified glasses, and the glass containing organic fluorine yields values intermediate between the two extremes. (See Figs. S1 and S2 in the Supplementary Material, Data S1, for pore size distributions and nitrogen adsorption/desorption isotherms.)
TABLE 1.
Physical properties of glasses
| Glass composition | Specific surface area (m2/g) | Pore volume (cm3/g) | Average pore diameter (nm) |
|---|---|---|---|
| 100% TMOS | 635 | 1.88 | 10.8 |
| 5% Methyl | 960 | 0.97 | 4.2 |
| 5% Propyl | 925 | 0.87 | 4.2 |
| 5% F3-propyl | 745 | 0.70 | 4.8 |
| 5% Hexyl | 919 | 0.85 | 4.0 |
Adsorption experiments were completed with two proteins: cytochrome c and apomyoglobin. The amount of adsorbed protein was inferred from the depletion of protein in the incubation buffer, as monitored by circular dichroism (CD) spectroscopy. In the far-UV region, the CD signal is a measure of secondary structure due to the differential absorbance of the peptide bonds that define the backbone. Since protein in the bulk solution is expected to maintain its native conformation, the reduced CD signal is directly proportional to the decrease in protein concentration.
Measurements taken during the first 24 h revealed no significant difference in the rate of adsorption for the five glasses. The larger pore size of the control glass did not appear to enhance adsorption kinetics, and ∼85% of the total protein in solution had adsorbed onto each glass after 24 h (see Fig. S3 of Data S1).
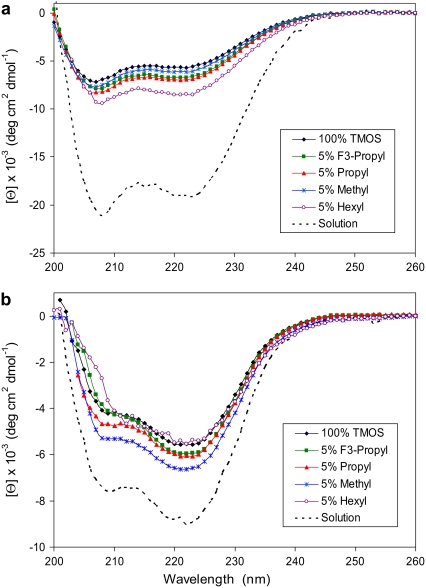
After the 24 h incubation, each glass was removed from the protein solution, analyzed by CD, and converted to units of molar ellipticity, [Θ], using a protein concentration equal to the amount of adsorbed protein divided by the total volume of the glass wafer. As shown in Fig. 1 a, adsorption onto each glass yielded a similar spectrum for apomyoglobin, indicating a similar protein structure. Furthermore, the CD spectra indicate a major loss in helical content relative to the solution structure upon binding to the glass surface.
FIGURE 1.
Molar ellipticity of apomyoglobin (a) and cytochrome c (b) after adsorption onto glasses of variable composition. Each glass wafer was analyzed by CD at 25°C in the presence of 10 mM phosphate, pH 7.0. The solution spectra were measured at t = 0 before incubation with the glass samples. (See detailed description of materials and methods in Data S1.)
The same trend was observed for cytochrome c: protein adsorption resulted in a lower molar ellipticity, indicating less structure relative to the conformation in solution (Fig. 1 b). In the case of cytochrome c, the deviation from the native state was less dramatic than apomyoglobin. This is likely due to a difference in protein stability: cytochrome c is a 104-residue protein with a heme group attached by two thioether bonds, whereas the noncovalent heme group of the 153-residue apomyoglobin has been removed. For both proteins, adsorption was essentially irreversible upon transfer to a protein-free phosphate buffer.
A general loss in native protein structure after adsorption onto solid surfaces has been observed in many systems. For example, reduced secondary structure upon binding to silica particles or silica gels has been detected by CD for serum albumin and hen eggwhite lysozyme (8), hemoglobin and myoglobin (9), T4 lysozyme (10), carbonic anhydrase II (11), subtilisin 309 (12), and cutinase and α-chymotrypsin (13). In the specific case of adsorbed apomyoglobin, a change in conformation has been detected by fluorescence techniques (14).
The results of the work presented here are remarkable because the same organically modified glasses that led to unfolding of adsorbed cytochrome c and apomyoglobin are known to enhance the secondary structure of apomyoglobin when the protein is encapsulated within the pores of the silica matrix during sol-gel formation (6,7). Table 2 compares the secondary structure of apomyoglobin under the two conditions (adsorbed versus encapsulated) using a structure analysis algorithm that estimates the helical content of the protein from the corresponding CD spectrum. For reference, the helical content of apomyoglobin in solution is 62% when calculated with the same algorithm.
TABLE 2.
Helical content of apomyoglobin
In contrast to the adsorbed protein, apomyoglobin structure varies greatly in helical content when encapsulated in the same modified glasses. For the 5% hexyl-modified glass, adsorbed apomyoglobin has less than half the helical content found in solution, whereas the encapsulated protein exceeds the helical content found in solution. It should be noted that hyperhelical conformations also have been reported for proteins adsorbed onto strongly hydrophobic surfaces, including Teflon (12,13,15) and trichloromethylsilane-coated quartz (16).
It has been hypothesized that apomyoglobin is largely unfolded in the pores of an unmodified glass due to the unfavorable influence of the silica surface on the free energy of water (17). Here we expand this hypothesis by stating that the unfolded state of encapsulated apomyoglobin is governed by an adsorption phenomenon, and this adsorption process is not mediated by electrostatic interactions or by hydrophobic-hydrophobic interactions. We suggest that protein adsorption is driven by desolvation of the silica surface and that desolvation is entropically favorable because silica can serve only as a proton acceptor, and never as a proton donor, in forming hydrogen bonds with the solvent (assuming pH > 4 when SiO− groups are present). This means that interfacial water molecules must be oriented in a similar manner with their protons pointing toward the oxygen atoms of the silica, resulting in a low-entropy state. Disturbing this layer of unfavorable water by introducing a new surface group appears to enhance the structure of neighboring protein molecules, even when the disruption is caused by a hydrophobic alkyl chain, as observed when apomyoglobin is confined to the pores of organically modified glasses by sol-gel encapsulation.
However, when free apomyoglobin adsorbs onto an organically modified glass from the bulk solution, the protein docks with the silica surface at locations with the highest free energy state of water, i.e., regions distant from any alkyl groups that disrupt the low-entropy hydration layer. After docking, the adsorbed protein will be prone to unfold on the glass surface to minimize the total number of thermodynamically unfavorable interactions between water and silica. This interpretation supports the view that water, in general, controls protein adsorption to surfaces (18).
This explanation also is consistent with the observation that many adsorbed proteins recover native-like structure as the total surface loading increases (8,9,16). At high protein loadings, regions with the most unfavorable surface energy will be occupied first, and ensuing molecules will adsorb to less-destabilizing positions. Thus, the fraction of native-like structure is expected to increase as the surface becomes more saturated with protein.
SUPPLEMENTARY MATERIAL
To view all of the supplemental files associated with this article, visit www.biophysj.org.
Supplementary Material
Acknowledgments
This work was supported by a grant to D.K.E. from the National Institutes of Health, Minority Biomedical Research Support Support of Continuous Research Excellence) program (S06 GM008192). C.T. is a National Institutes of Health Minority Access to Research Careers scholar (grant 2 T34 GM008253).
Editor: Kathleen B. Hall.
References
- 1.Nakanishi, K., T. Sakiyama, and K. Imamura. 2001. On the adsorption of proteins on solid surfaces, a common but very complicated phenomenon. J. Biosci. Bioeng. 91:233–244. [DOI] [PubMed] [Google Scholar]
- 2.Haynes, C. A., and W. Norde. 1994. Globular proteins at solid/liquid interfaces. Colloids Surf. B. Biointerfaces. 2:517–566. [Google Scholar]
- 3.Brinker, C., and G. Scherer. 1990. Sol-Gel Science: The Physics and Chemistry of Sol-Gel Processing. Academic Press, San Diego.
- 4.Pierre, A. C. 2004. The sol-gel encapsulation of enzymes. Biocatal. Biotransformation. 22:145–170. [Google Scholar]
- 5.Avnir, D., T. Coradin, O. Lev, and J. Livage. 2006. Recent bio-applications of sol-gel materials. J. Mater. Chem. 16:1013–1030. [Google Scholar]
- 6.Rocha, V. A., and D. K. Eggers. 2007. Hydrophobic, organically-modified silica gels enhance the secondary structure of encapsulated apomyoglobin. Chem. Commun.1266–1268. [DOI] [PubMed]
- 7.Menaa, B., M. Herrero, V. Rives, M. Lavrenko, and D. K. Eggers. 2008. Favourable influence of hydrophobic surfaces on protein structure in porous organically-modified silica glasses. Biomaterials. 29:2710–2718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Norde, W., and J. P. Favier. 1992. Structure of adsorbed and desorbed proteins. Coll. Surf. 64:87–93. [Google Scholar]
- 9.Kondo, A., and J. Mihara. 1996. Comparison of adsorption and conformation of hemoglobin and myoglobin on various inorganic ultrafine particles. J. Colloid Interface Sci. 177:214–221. [DOI] [PubMed] [Google Scholar]
- 10.Billsten, P., M. Wahlgren, T. Arnebrant, J. McGuire, and H. Elwing. 1995. Structural changes of T4 lysozyme upon adsorption to silica nanoparticles measured by circular dichroism. J. Colloid Interface Sci. 175:77–82. [Google Scholar]
- 11.Billsten, P., U. Carlsson, B. H. Jonsson, G. Olofsson, F. Hook, and H. Elwing. 1999. Conformation of human carbonic anhydrase II variants adsorbed to silica nanoparticles. Langmuir. 15:6395–6399. [Google Scholar]
- 12.Maste, M. C. L., W. Norde, and A. J. W. G. Visser. 1997. Adsorption-induced conformational changes in the serine proteinase savinase: a tryptophan fluorescence and circular dichroism study. J. Colloid Interface Sci. 196:224–230. [DOI] [PubMed] [Google Scholar]
- 13.Zoungrana, T., G. H. Findenegg, and W. Norde. 1997. Structure, stability, and activity of adsorbed enzymes. J. Colloid Interface Sci. 190:437–448. [DOI] [PubMed] [Google Scholar]
- 14.Lochmüller, C. H., and S. S. Saavedra. 1987. Intrinsic fluorescence characteristics of apomyoglobin adsorbed to microparticulate silica. Langmuir. 3:433–438. [Google Scholar]
- 15.Maste, M. C. L., E. H. W. Pap, A. van Hoek, W. Norde, and A. J. W. G. Visser. 1996. Spectroscopic investigation of the structure of a protein adsorbed on a hydrophobic latex. J. Colloid Interface Sci. 180:632–633. [Google Scholar]
- 16.Wu, H., Y. Fan, J. Sheng, and S.-F. Sui. 1993. Induction of changes in the secondary structure of globular proteins by a hydrophobic surface. Eur. Biophys. J. 22:201–205. [DOI] [PubMed] [Google Scholar]
- 17.Eggers, D. K., and J. S. Valentine. 2001. Crowding and hydration effects on protein conformation: a study with sol-gel encapsulated proteins. J. Mol. Biol. 314:911–922. [DOI] [PubMed] [Google Scholar]
- 18.Cha, P., A. Krishnan, V. F. Fiore, and E. A. Vogler. 2008. Interfacial energetics of protein adsorption from aqueous buffer to surfaces with varying hydrophilicity. Langmuir. 24:2553–2563. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.