Abstract
Fracture healing requires controlled expression of thousands of genes. Only a small fraction of these genes have been isolated and fewer yet have been shown to play a direct role in fracture healing. The purpose of this study was threefold: (1) to develop a reproducible open femur model of fracture healing that produces consistent fracture calluses for subsequent RNA extraction, (2) to use this model to determine temporal expression patterns of known and unknown genes using DNA microarray expression profiling, and (3) to identify and validate novel gene expression in fracture healing. In the initial arm of the study, a total of 56 wild-type C57BL/6 mice were used. An open, stabilized diaphyseal femur fracture was created. Animals were killed at 1, 5, 7, 10, 14, 21, and 35 days after surgery and the femurs were harvested for analysis. At each time point, fractures were radiographed and sectioned for histologic analyses. Tissue from fracture callus at all stages following fracture yielded reproducibly large amounts of mRNA. Expression profiling revealed that genes cluster by function in a manner similar to the histologic stages of fracture healing. Based on the expression profiling of fracture tissue, temporal expression patterns of several genes known to be involved in fracture healing were verified. Novel expression of multiple genes in fracture callous tissue was also revealed including leptin and leptin receptor. In order to test whether leptin signaling is required for fracture repair, mice deficient in leptin or its receptor were fractured using the same model. Fracture calluses of mice deficient in both leptin or leptin receptor are larger than wild-type mice fractures, likely due to a delay in mineralization, revealing a previously unrecognized role of leptin signaling in fracture healing. This novel model of murine fracture repair is useful in examining both global changes in gene expression as well as individual signaling pathways, which can be used to identify specific molecular mechanisms of fracture healing.
Keywords: animal model, cDNA microarray, fracture healing, leptin
Introduction
Nearly six million fractures occur in the United States every year. It has been estimated that nearly 10% of these fractures have delayed or disrupted healing resulting in significant morbidity and loss of productivity. Enhancing the quality and/or speed of fracture healing can translate into significant therapeutic benefit both in terms of patient outcomes and cost to society [1–3].
Many biological and biophysical interventions have been used in an attempt to accelerate fracture healing times including the use of bone graft substitutes [4, 5], application of exogenous local recombinant growth factors [6, 7], gene therapy [8], or tissue engineering approaches with biodegradable scaffolds [9, 10]. Several modalities have gone on to be clinically useful such as recombinant growth factors, but high costs continue to preclude routine clinical use. Clearly, a better understanding of the molecular events of fracture healing is necessary to develop novel therapeutics to accelerate fracture healing [11, 12].
Fracture healing occurs through a tightly regulated cascade of molecular events which are associated with histologically recognizable temporal changes at the site of the fracture. Fracture healing is typically divided into three phases: inflammatory (initial hematoma with subsequent infiltration of inflammatory cells), reparative (invasion of fibroblasts, new capillary formation, chondrogenesis, and early osteogenesis), and remodeling (continued bone formation with remodeling to osteonal bone). The relationship between these specific morphological changes and underlying gene expression at each phase is still being studied. In fact, only a fraction of the genes necessary to complete this complex process have been identified and characterized [13–15].
Multiple studies have used a “gene-by-gene” approach to examine the temporal expression of the gene or genes of interest [16–21]. This process reveals the spatial distribution of expression, but is not useful in isolating novel genes involved in fracture healing. The development of modern expression profiling using microarray technology has allowed the study of the thousands of coordinately expressed genes during the complex biological processes of fracture healing and, recently, distraction osteogenesis [22–24].
The purpose of this study was to employ a mouse open femur fracture model to collect and study the total fracture callous RNA at multiple time points. We show that an open stabilized femur fracture allows for tissue retrieval as early as 1 day following fracture. Ample tissue retrieval allows for relatively large amounts of RNA to be isolated for subsequent analyses. Pooled RNA was used for expression-profiling studies using Affymetrix microarrays (Affymetrix, Santa Clara, CA, USA) verifying the expression of several genes known to be involved with fracture healing and revealing several with novel expression including the leptin and leptin receptor. This model can be used to uncover other novel mechanisms of fracture healing.
Materials and methods
Animals
Female C57bL/6 mice (Harlan Sprague Dawley, Indianapolis, IN, USA) weighing 31.0 ± 5.5 g (mean±standard deviation) were used for this experiment. All animals were between 6 and 8 weeks old at the initiation of this study. All experimental procedures were approved by the Institutional Animal Use and Care Committee.
Surgical procedure
After appropriate anesthesia with intramuscular injection of ketamine hydrochloride (100 mg/kg) and acetylpromazine (5 mg/kg), the right leg was scrubbed with 10% povidone–iodine solution. A 1-cm right medial parapatellar incision was made. The longitudinal fibers of the quadriceps mechanism were divided anteromedially at the knee, and the patella was dislocated laterally to expose the femoral condyles. A sterile 25-gauge needle was introduced into the intramedullary canal and advanced in a retrograde fashion up the shaft of the femur to the level of the greater trochanter. Distally, the pin was cut flush with the joint so as not to interfere with the motion of the knee. Using careful blunt dissection, the diaphysis was carefully exposed taking care to maintain the periosteum and soft tissue envelop. A small right-angled wire snipper was then used to fracture the middiaphysis of the femur. The soft tissue envelop was carefully closed and the extensor mechanism was repaired with interrupted 5-0 chromic sutures in a standard fashion followed by closure of the skin with 4-0 nylon sutures (Ethicon, Somerville, NJ, USA). This closure provided a sleeve of soft tissue surrounding the fracture site.
The left femur was left unfractured. Animals were allowed free, unrestricted weight bearing after recovery from anesthesia. At the time points recorded, animals were euthanized with halothane gas (Sigma Chemical, St. Louis, MO, USA). Final radiographs of all animals were made at euthanasia.
Histologic and histomorphometric analyses
Animals used for histologic analysis underwent decalcification at 1, 5, 7, 10, 14, 21, and 35 days. The fractured and control contralateral limbs were cleanly harvested, cleaned of soft tissue, and fixed in 10% buffered formalin. The femurs were decalcified in 5% nitric acid for 24–48 h. The femurs were then dehydrated through successive grades of ethanol and were paraffin-embedded. The embedded femurs were cut sagittally into 8 μm sections and stained with hematoxylin and eosin and Goldner’s trichrome. Sections were studied under a light microscope and images were recorded for analysis of percentage area of fibrous tissue, cartilage, and bony tissue estimation using the Metamorph Image Analysis and Processing System (Version 4.0; Universal Imaging, Downington, PA, USA) computer program.
Leptin knockout and leptin receptor-deficient mice underwent analysis at 14 days. Callous height and total callus area for the wild-type, leptin-deficient, and leptin receptor-deficient groups were calculated as follows: at the histologic level of the widest diameter of fracture callus (WDFC) determined by two independent observers, a vertical measurement axis (VMA) was drawn with the Metamorph Imaging program and the height calculated in microns. The vertical measurement axis calculations were then repeated at 8-μ intervals for three contiguous sections medial and lateral to the WDFC. Similarly, at the WDFC, a freehand lasso tool in Metamorph Imaging was used to delineate the boundaries of the fracture callus as demarcated craniocaudally by four intact cortices. These data were also then repeated at 8-μ intervals for three contiguous sections both medial and lateral to the WDFC. A total of three callouses (n = 3 animals for wild-type, leptin-deficient, and leptin receptor-deficient) for each group were used and the values averaged.
Radiography
After killing the mice, the femurs were gently freed of all soft tissues. They were then placed on a 1/4-in. Plexiglas plate, and high-resolution scanning radiographs were taken using a Faxitron machine (Hewlett Packard, Palo Alto, CA, USA). Each film included the aluminum density standard. Anteroposterior and lateral images were recorded on Kodak Kodolith graphic arts film (Eastman Kodak, Rochester, NY, USA). For image acquisition, digital images of limb radiographs were obtained using a Kodak DCS 200 high-resolution digital camera. While photographing the images, the F-stop was adjusted in a way that the lower three aluminum density standards were distinctly different in their density measurements. Images were downloaded from the camera using the Adobe PhotoShop software (version 5.0.2,). Digitized images were stored in “*.tif” format for analysis on a PC. They were converted to gray scale in Adobe Photoshop.
RNA isolation and microarray analysis
Fractures were carefully opened through the same muscular interval. With careful dissection, the same interval can be identified and entered and early hematoma and soft callous can be retained. The central one third of the femur including all of the callous or hematoma was removed and flash frozen in liquid nitrogen. Total RNA was isolated from each sample using TRIzol (Invitrogen Life Technologies, Carlsbad, CA, USA) according to the manufacturer’s protocol using a Brinkman Polytron homogenizer. RNA samples were purified on Rneasy columns by Qiagen (Valencia, CA, USA). The total RNA isolated was pooled from three animals for each separate array time point.
Five micrograms of total RNA was converted to cDNA using the SuperScript Choice System (Invitrogen Life Technologies) and the T7-Oligo(dT) promoter primer kit (Affymetrix, Santa Clara, CA, USA). The cDNA was cleaned using the GeneChip® Sample Cleanup Module (Affymetrix, Santa Clara, CA, USA). Clean cDNA was used for the in vitro synthesis of biotin-labeled cRNA using the BioArray RNA transcript labeling kit (Enzo Diagnostics). cRNA was cleaned using the GeneChip® Sample Cleanup Module and fragmented into 35–200 base pair fragments using a magnesium acetate buffer. Ten micrograms of labeled cRNA were hybridized to Affymetrix GeneChip® Mouse Expression Set 430A and 430B arrays for 16 h at 45°C. The GeneChips® were washed and stained according to the manufacturer’s recommendations using the GeneChips® Fluidics Station 400 (Affymetrix, Santa Clara, CA, USA). This procedure includes washing the chips with phycoerythrin–streptavidin, signal amplification by a second staining with biotinalyted antistreptavadin, and a third staining with phycoerythrin–streptavidin Each chip was scanned using the GeneChips® Scanner 2500 (Affymetrix, Santa Clara, CA, USA).
The GeneChip Mouse Expression Set 430 contains over 45,000 probe sets for over 34,000 well-characterized mouse genes. Signal intensity and detection calls were generated using the Affymetrix GeneChip® Operating Software. The absolute intensity values of each chip were scaled to the same target intensity value of 150 in order to normalize the data for interarray comparisons.
Quantitative PCR
One microgram of the total RNA used in the microarray analysis was used for cDNA synthesis. After priming with random hexamers according to the protocol, cDNA was synthesized using the Fermentas First Strand DNA synthesis kit (#K1612). For qPCR, 25 μl reactions were set up using 2X iQ SYBR Green Supermix (Bio-Rad 170–8882), water, and 10 μM primers (Col10a1 forward tca tgc ctg atg gct tca ta 3′, 3′ cag cct act gct ggg taa gc; GAPDH 5′ ggg tgt gaa cca cga gaa at 3′, 3′ cct tcc aca atg cca aag tt 5′; Col2a1, Indian hedgehog (Ihh), leptin, and leptin receptor sequences were previously published [25]. The qPCR reaction using FAM 490 consisted of 1 cycle at 95°C for 3 min; 42 cycles at 95°C for 10 s, 60°C for 45 s; 1 cycle at 95°C for 1 min; and 50 cycles at 70°C for 10 s. The quantity of the gene of interest was determined relative to GAPDH cycle threshold (Ct).
Results
The mouse open femur fracture model produces typical transverse fracture with normal progression of fracture healing
In the initial arm of the study, 56 animals were used. Three mice at each time point (n = 21) were killed exclusively for RNA isolation and qPCR experiments. Of the remaining 35 mice, 12 died in the perioperative period. The remaining mice were killed at days 1 (n = 4), 5 (n = 4), 7 (n = 3), 10 (n = 3), 14 (n = 3), 21 (n = 3), and 35 (n = 3).
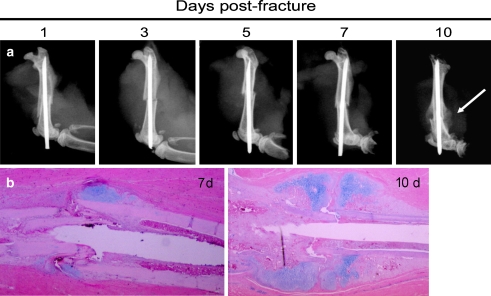
The fractures produced by the open femur fracture method were primarily transverse with occasional butterfly fragments (Fig. 1). The callouses produced were large enough to easily visualize on radiographs and easy to discern when dissecting free. Histologic examination of the healing fracture tissue revealed normal progression through the stages of fracture healing (Fig. 1).
Fig. 1.
Mouse open femur fracture model produces consistent diaphyseal fracture that follow the normal radiographic (a) and histologic (b) stages of fracture healing. Calcification of the cartilage is noted by 10 days following fracture (a) though a large percentage of the callous remains soft (noncalcified) (b)
Early fracture hematoma and fracture callous tissue were surgically dissected free and isolated from the femur without difficulty in all subsequent procedures. The callous tissue or hematoma was dissected free and total RNA isolated. RNA was not degraded and was isolated in microgram quantities even at early time points.
Expression profiling reveals clustering of genes upregulated throughout fracture healing
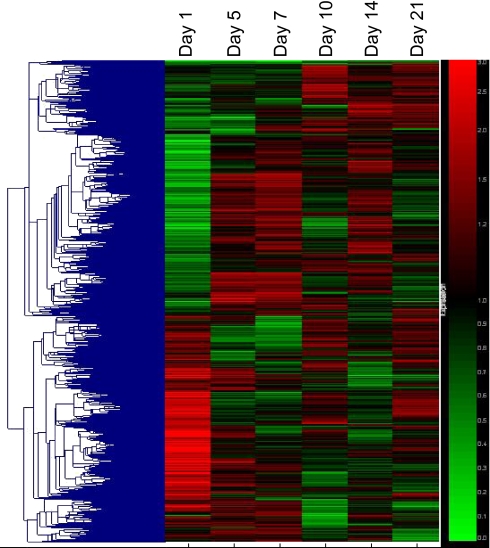
Total fracture callous RNA was evaluated using expression-profiling techniques with the Affymetrix GeneChip Mouse Expression Set 430. Five thousand five hundred forty-six genes were found to be upregulated or downregulated at least twofold during the course of fracture healing between any given two time points, not necessarily consecutive, and called “present” by the Affymetrix software in at least one of the six arrays.
Self-organizing maps of the 5,546 genes were generated using GeneSpring (Agilent Technologies, Santa Clara, CA, USA) (Fig. 2). Genes were clustered together based on temporal expression patterns alone with no bias given to a gene’s function or family. In this way, novel functions may be inferred based on the time of expression during the phases of fracture healing (inflammatory, reparative/chondrogenic, or remodeling) and possibly by understanding the functions of the genes that are temporally coexpressed.
Fig. 2.
Cluster analysis of gene expression profiles in mouse fracture callous. Five thousand five hundred forty-six genes that are “present” in at least one out of the six samples and have a greater than twofold change between one of the time points was used in this analysis. Genes differentially expressed following fracture cluster into distinct expression patterns
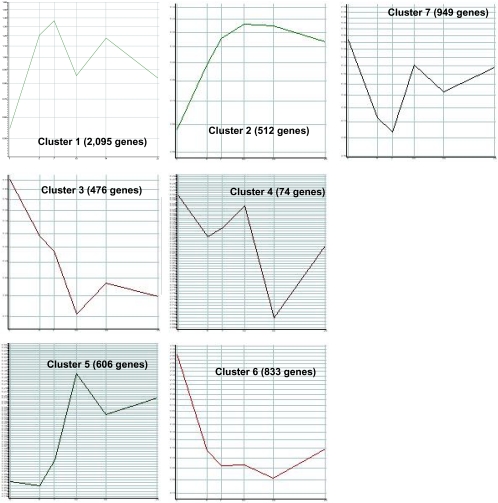
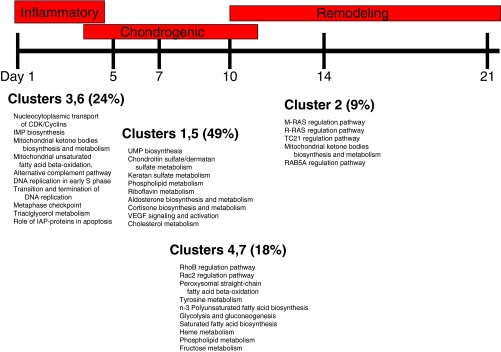
A total of seven clusters were identified (Figs. 3 and 4). Temporal clustering revealed that many genes are upregulated and downregulated as families, though they may be unrelated structurally and functionally.
Fig. 3.
Five thousand five hundred forty-six genes that are “present” in at least one out of the six samples and have a greater than twofold change between one of the time points was used in this analysis. Genes differentially expressed following fracture cluster into distinct expression patterns. Relative expression levels are shown on the y-axis with the corresponding time points on the x-axis in days. Each cluster represents the average gene expression pattern for genes within the cluster. The number of cluster members is indicated in parentheses next to the cluster number
Fig. 4.
Five thousand five hundred forty-six genes that are “present” in at least one out of the six samples and have a greater than twofold change between one of the time points was used in this analysis. Genes differentially expressed following fracture cluster into distinct expression patterns. Relative expression levels are shown on the y-axis with the corresponding time points on the x-axis in days. Each cluster represents the average gene expression pattern for genes within the cluster. The number of cluster members is indicated in parentheses next to the cluster number
Cluster 1 consisted of 2,095 genes whose expression consisted of two peaks: the first major peak at 7 days and secondary peak at 14 days. The first peak lies within the chondrogenic phase of fracture healing. Of the 2,095 genes, 196 were involved in signal transduction (∼9%), including TGF-beta, SMAD1, WNT5A, VEGF-C, FGFR2, IGF-2 receptor, IBP4 and 5, and leptin receptor. This cluster also included genes involved in cell adhesion (including osteomodulin [26]), intercellular communication (including connexin 43 [27]), and skeletal development (including BMPs 1 and 5 [28]).
Cluster 2 consisted of 512 genes whose expression increases until day 10 and then levels off, staying elevated during the later stages of fracture healing. Genes in this group have functions which include signal transduction (TGF-beta receptors type I and III [29]), transcriptional regulation (including SMAD9 and Sox4 [30, 31]), and cell transport. This cluster also contains approximately 15 genes involved in ossification and bone remodeling consistent with the histologic phase of fracture healing These genes include activin receptor type 2A precursor, which binds and activates SMAD transcriptional regulators [32]; SMADs 6 and 9, which are activated by BMPs [33, 34]; BMP6, which induces cartilage and bone formation [35]; matrix extracellular phosphoglycoprotein precursor (MEPE), which seems to play a role in mineralization [36]; IGF-1, which is a differentiation factor [37]; type I collagen and Runt-related transcription factor 2 (Runx2) [38], which interacts with STAT1 in bone remodeling and osteoblast differentiation.
Cluster 3 consists of 476 genes whose expression peaks at day 1 and decreases thereafter. The expression of these genes is within the inflammatory phase of fracture healing. Genes involved in inflammation expressed highly within this cluster include IL-2, 4, and 6 signaling including p21, NF-kappa-B inhibitor beta, and complement C3 precursor. Seven of the genes in cluster 3 are involved in bone formation including BMP-2-inducible protein kinase (BIKe) [39] that has been shown to attenuate the program of osteoblast differentiation. This group also contains genes involved in signal transduction, transcription, and cell cycle regulation (including COX-2) [40].
Cluster 4 is a small group consisting of 74 genes whose expression has a prominent double peak. Immediate upregulation promptly decreases at day 5, increases at day 10, and decreases again at day 14. Genes in this group are involved in transcription and inflammation (through the IL-2, 4, 6, and 12 signaling pathways).
Cluster 5 consists of 606 genes whose expression starts to increase at day 5 and reaches a peak at 10 days, followed by a decrease in expression. This cluster occurs within the chondrogenic phase of fracture healing. The functions of this cluster include signal transduction (including leptin receptor precursor), transport, and inflammatory response including histamine H1 receptor and IFN alpha and type I, JNK, and CREB1.
Cluster 6 shows high expression at day 1 followed by a sharp decrease. This group consists of 833 genes, 60 of which are involved in transcription and inflammation (including IL-1 and IL-6).
Cluster 7 consists of 949 genes whose expression decreases initially until day 7 and then increases. Ninety-three of the genes are involved in signal transduction and transport. There are a large number of genes are involved in protein amino acid phosphorylation (∼5%), including BMP receptor 1 and MAPK1.
Identification of novel gene expression in fracture healing
Ihh and type II collagen are known to be upregulated during fracture healing [41–43]. These genes show significant upregulation between days 7 and 14 after fracture healing.
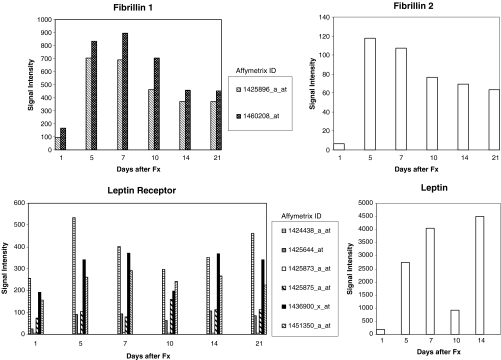
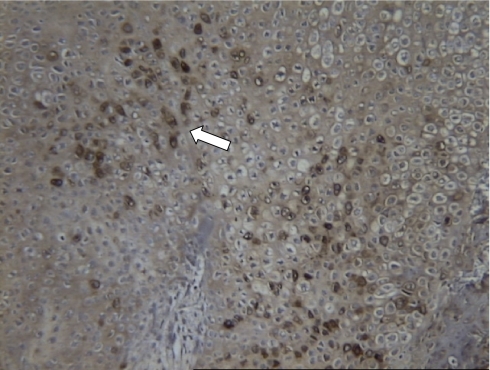
Recent studies have shown that leptin may be important in endochondral bone formation and skeletal homeostasis [44, 45]. Multiple representatives of leptin receptor showed strong upregulation throughout days 5, 7, 10, and 14 following fracture (Figs. 5 and 6). To verify the expression of these genes in healing fracture, RNA from each individual animal was used for quantitative PCR verification of the expression. Three animals from each of days 1, 5, 7, 10, 14, and 21 were used. Both leptin and leptin receptor were upregulated during the chondrogenic phase of healing fracture callus in wild-type animals. In order to verify the presence of leptin receptor protein in fracture tissue, we performed immunohistochemistry on 10-day-old fracture callus tissue. Leptin receptor was expressed in the prehypertrophic or hypertrophic chondrocytes of fracture callus (Fig. 7). This is consistent with the location of leptin and leptin receptor expression in the hypertrophic zone of the endochondral growth plate chondrogenesis as demonstrated by Kishida [44]. They found that the growth plates of ob/ob mice were fragile with reduced type X collagen expression, disturbed collagen fibril arrangement, increased chondrocyte apoptosis, and premature matrix mineralization. Their findings suggested that a physiological level of leptin plays an essential role in the regulation of chondrocyte differentiation and cartilage matrix maturation in the growth plate which may in turn alter the rate of longitudinal bone growth.
Fig. 5.
Known genes with that may be important in endochondral bone formation and bone homeostasis. Multiple probe sets are displayed for the genes leptin and leptin receptor. Data are presented as in Fig. 4
Fig. 6.
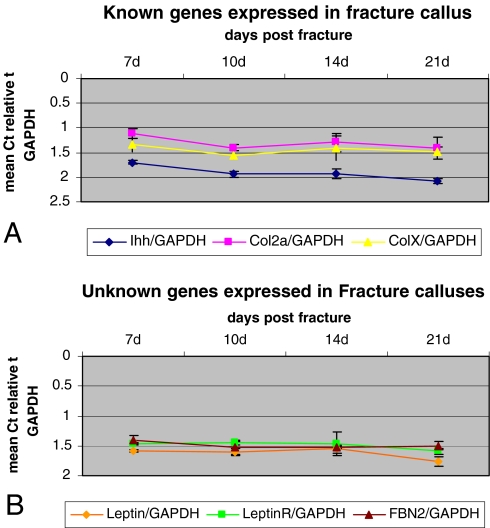
Gene expression detected by array analysis confirmed by qPCR. a Ihh, Col2a, and ColX transcripts are detected in fracture callous. b Leptin, leptin receptor, and fibrillin 2 expression, initially determined by array experiment, is verified by qPCR
Fig. 7.
Leptin receptor protein is detected in healing fracture callous. Leptin receptor protein is detected in proliferative and prehypertrophic chondrocytes (stained brown indicated by arrow) consistent with the expression in growth plate chondrogenesis
Mice deficient in leptin signaling produce larger fracture calluses indicating a regulatory role of leptin in fracture healing
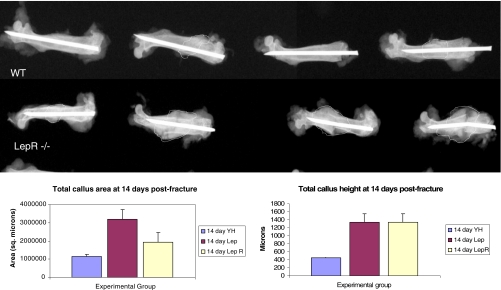
In order to test whether the leptin signaling pathway is involved in fracture healing, we produced femur fractures in animals deficient in either leptin (ob/ob) or leptin receptor (db/db) (Jax Labs, Bar Harbour, ME, USA). Fracture calluses in both leptin- and leptin receptor-deficient animals were significantly larger in size than wild-type animals at 14 days following fracture (Fig. 8). ob and db fracture callouses were predominantly composed of hypertrophic chondrocytes that persisted to the later phases of fracture healing whereas the wild-type counterparts had completed remodeling at those later times.
Fig. 8.
Radiographs of 14-day fracture calluses from wild-type (WT) and leptin receptor-deficient (LepR−/−) animals. The extent of the fracture callouses is outlined. Though there is variability, larger callouses are present in the animals deficient in leptin receptor. Animals deficient in leptin show a similar radiographic appearance (data not shown). Leptin- and leptin receptor-deficient mice produce larger fracture callouses than wild-type controls. Both fracture area (a) and total callous height (b) are statistically greater in leptin- and leptin receptor-deficient animals than in controls (*p < 0.05; **p < 0.01; n = 3 fractures per group)
Discussion
Fracture healing is a complex physiological postnatal process, which involves the cooperation of several different cell types. A large number of genes involved in this process are known, but many more remain to be discovered. Developing a reproducible model of fracture repair allows for the study of these fundamental and previously unknown genes.
In this experiment, we have shown that an open femur model of fracture healing histologically reproduces the stages of osteogenic repair while still allowing for the production of a large enough fracture calluses and, therefore, large enough amounts of soft tissue to isolate RNA for subsequent expression-profiling studies. The unique feature of this model compared to closed models or open tibia models is that a soft tissue sleeve using the thigh musculature can help hold in soft tissue and even early fracture hematoma near the site of the callus to examine global expression of genes in this localized tissue. Additionally, creation of the fractures under direct visualization improves consistency at the site of the fracture.
We have shown that, with this model, normal stages of fracture healing occur, RNA can be effectively isolated and used for expression-profiling experiments, and novel gene expression in fracture tissue can then be identified. By utilizing a murine model, we were able to use genetically identical animals with reproducible results both in RNA isolation, array analysis, and histological healing. The benefit of using this novel murine fracture model is that the function of previously unrecognized genes can be systematically studied in genetically modified mouse strains.
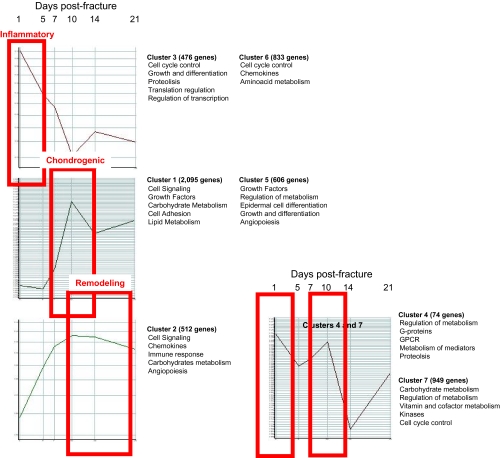
In our initial array analysis, we looked at clusters of genes and identified seven clusters of genes based upon their dynamic temporal sequence of expression, which reflects stage-specific functional needs for bone healing (Fig. 9). The primary event after fracture healing is hematoma formation and an acute inflammatory response. This response is heralded by the infiltration of inflammatory cells at the fracture site leading to full-scale preparation for bone repair. The immediate response genes were identified in clusters 1 and 6 as upregulated genes involved in signal transduction, transcription, immune response, cell transport, and cell adhesion. Furthermore, discrete genes with similar functions were identified in cluster 7; however, their expression initially decreased at the fracture site only to increase at day 7 after fracture. This indicates that the early enrichment of genes with energy derivation, transporter function, and binding activities at the fracture site prepares the wound site for the process of energy accumulation and intercellular and intracellular molecular cross-talk. It is important to note that deactivation (clusters 3 and 6) after transient activation of genes may be just as important as continued expression or timing of activation because continued expression could waste energy better utilized toward the later stages of fracture repair. After the initial inflammatory phase comes to a close, “work horse cells” such as macrophages, fibroblasts, and endothelial cells invade the fracture site to begin the process of callus formation. Expression of gene clusters that continually increased at this time point included clusters 1, 2, 4, and 5. Functional analysis of these genes suggests that the protein products derived from these groups are engaged in repair. These genes included those associated with the biological events of callus formation, both the cartilaginous phase such as collagen types VI and XI, FGFR, VEGF, PDGF as well as the bony phase such as collagen type I, V, XII, tenascin, and BMPs. Comparing gene clusters with the temporal events of fracture healing allows us to suggest that the function of the immediate response genes in our identified clusters largely centers on binding activities, energy expenditure, ion transport, and cell-to-cell communication while continually expressed genes or genes with progressive increase in expression comparable to histological time points consistent with callus formation and maturation largely function as signaling cues for reparative growth factors.
Fig. 9.
Gene clusters of expression as related to the phenotypic phases of fracture repair
It is interesting that we identified two clusters that expressed genes in a bimodal distribution (clusters 1 and 5). This may imply that the same set of genes may be required early and late along the temporal course of healing; hence, identifying which specific genes play a anabolic role depending upon the stage of fracture repair may allow us to design interventions that can be delivered early or late during the fracture healing cascade. This may be of particular relevance for treating delayed unions seen with trauma.
We verified known genes expressed during fracture repair in our array data with RT-PCR (Ihh, collagen type II) and examined new signaling pathways that have not been described during fracture repair such as the leptin signaling pathway.
Leptin is a 16-kDa hormone, primarily secreted by adipose tissue, which controls body weight through its effects on food intake and energy expenditure by negative feedback at the level of the hypothalamic nuclei. Various functions of leptin have been described including actions in the immune system, reproduction, development, hemopoiesis, angiogenesis, and most recently, in bone metabolism [46–53]. Takeda et al. [54] have demonstrated that leptin’s anorexigenic and antiosteogenic effects are performed by two distinct neuronal pathways and that the sympathetic nervous system relays leptin’s downstream signals via norepinephrine, which stimulates peripheral β2 adrenergic receptors on osteoblasts, thus reducing bone formation. Interestingly, Steppan et al. [55] and Yagasaki et al. [56] have demonstrated that peripheral leptin administration has anabolic effects on bone metabolism in leptin-deficient ob/ob mice and the demonstration that human osteoblasts in vitro express leptin receptor suggests that the bone growth-promoting effects of leptin could be direct. In addition, Cornish et al. [57] have demonstrated that, in mice, leptin tends to reduce bone fragility and that it also could contribute to the high bone mass and low fracture rates in obesity.
Fracture healing was altered in leptin and leptin receptor-deficient mice. Compared to wild-type fractures, there was significant radiographic and histologic lag in the progression of fracture repair. Leptin may regulate angiogenesis in endochondral ossification. Deficiency in leptin signaling may, therefore, lead to delayed healing and an arrest in the immature phase of endochondral phase of fracture repair. Further studies are underway to elucidate the exact nature of this defect in fracture healing.
There are several limitations with our data. The array data needs to be verified as differences in fracture gene expression between studies are common and it appears that variations in experimental approaches, in addition to biological variation in the regulation of fracture healing, can affect interpretations of gene expression changes. Indeed, the role of marrow as a contributor to the early inflammatory response could have been altered by the presence of a pin within the medullary canal of our experimental animals. We also did not have control tissue that would normalize normal baseline marrow gene expression to allow for more sensitive detection of early inflammatory gene expression.
Our goal with this new open femur fracture model was to perform experiments in genetically identical animals so that additional experiments can be performed using mice with the same genetic background. In addition, using a small animal such as a mouse allows us to utilize genetically modified animals to confirm and validate the roles of these genes in healing fracture tissue.
References
- 1.Jones A (2005) Recombinant human bone morphogenic protein-2 in fracture care. J Orthop Trauma 19(10 Supplement):S23–S25 [DOI] [PubMed]
- 2.Endo Y, Aharonoff G, Zuckerman J, Egol K, Koval K (2005) Gender differences in patients with hip fracture: a greater risk of morbidity and mortality in men. J Orthop Trauma 19(1):29–35 doi:10.1097/00005131-200501000-00006 [DOI] [PubMed]
- 3.Zlowodzki M, Obremskey W, Thomison J, Kregor P (2005) Functional outcome after treatment of lower-extremity nonunions. J Trauma Inj Infect Crit Care 58(2):312–317 doi:10.1097/01.TA.0000154302.23143.63 [DOI] [PubMed]
- 4.Chakkalakal DA, Strates BS, Garvin KL, Novak JR, Fritz ED, Mollner TJ et al (2001) Demineralized bone matrix as a biological scaffold for bone repair. Tissue Eng 7(2):161–177 Aprdoi:10.1089/107632701300062778 [DOI] [PubMed]
- 5.Hierholzer C, Sama D, Toro JB, Peterson M, Helfet DL (2006) Plate fixation of ununited humeral shaft fractures: effect of type of bone graft on healing. J Bone Jt Surg Am 88(7):1442–1447 Juldoi:10.2106/JBJS.E.00332 [DOI] [PubMed]
- 6.Jones AL, Bucholz RW, Bosse MJ, Mirza SK, Lyon TR, Webb LX et al (2006) Recombinant human BMP-2 and allograft compared with autogenous bone graft for reconstruction of diaphyseal tibial fractures with cortical defects. A randomized, controlled trial. J Bone Jt Surg Am 88(7):1431–1441 Juldoi:10.2106/JBJS.E.00381 [DOI] [PubMed]
- 7.Swiontkowski MF, Aro HT, Donell S, Esterhai JL, Goulet J, Jones A et al (2006) Recombinant human bone morphogenetic protein-2 in open tibial fractures. A subgroup analysis of data combined from two prospective randomized studies. J Bone Jt Surg Am 88(6):1258–1265 Jundoi:10.2106/JBJS.E.00499 [DOI] [PubMed]
- 8.Zhao M, Zhao Z, Koh JT, Jin T, Franceschi RT (2005) Combinatorial gene therapy for bone regeneration: cooperative interactions between adenovirus vectors expressing bone morphogenetic proteins 2, 4, and 7. J Cell Biochem 95(1):1–16 May 1doi:10.1002/jcb.20411 [DOI] [PubMed]
- 9.Lin CY, Schek RM, Mistry AS, Shi X, Mikos AG, Krebsbach PH et al (2005) Functional bone engineering using ex vivo gene therapy and topology-optimized, biodegradable polymer composite scaffolds. Tissue Eng 11(9–10):1589–1598 doi:10.1089/ten.2005.11.1589 [DOI] [PubMed]
- 10.Matziolis G, Tuischer J, Kasper G, Thompson M, Bartmeyer B, Krocker D et al (2006) Simulation of cell differentiation in fracture healing: mechanically loaded composite scaffolds in a novel bioreactor system. Tissue Eng 12(1):201–208 doi:10.1089/ten.2006.12.201 [DOI] [PubMed]
- 11.Dimitriou R, Tsiridis E, Giannoudis P (2005) Current concepts of molecular aspects of bone healing. Injury 36(12):1392–1404 doi:10.1016/j.injury.2005.07.019 [DOI] [PubMed]
- 12.Einhorn TA (2005) The science of fracture healing. J Orthop Trauma 19(10 Suppl):S4–S6 [DOI] [PubMed]
- 13.Meyer RA Jr, Desai BR, Heiner DE, Fiechtl J, Porter S, Meyer MM (2006) Young, adult, and old rats have similar changes in mRNA expression of many skeletal genes after fracture despite delayed healing with age. J Orthop Res 24(10):1933–1944 Octdoi:10.1002/jor.20124 [DOI] [PubMed]
- 14.Rundle CH, Wang H, Yu H, Chadwick RB, Davis EI, Wergedal JE et al (2006) Microarray analysis of gene expression during the inflammation and endochondral bone formation stages of rat femur fracture repair. Bone 38(4):521–529 doi:10.1016/j.bone.2005.09.015 [DOI] [PubMed]
- 15.Tsiridis E, Giannoudis PV (2006) Transcriptomics and proteomics: advancing the understanding of genetic basis of fracture healing. Injury 37(Suppl 1):S13–S19 doi:10.1016/j.injury.2006.02.036 [DOI] [PubMed]
- 16.Heiner DE, Meyer MH, Frick SL, Kellam JF, Fiechtl J, Meyer RA Jr (2006) Gene expression during fracture healing in rats comparing intramedullary fixation to plate fixation by DNA microarray. J Orthop Trauma 20(1):27–38 doi:10.1097/01.bot.0000184143.90448.aa [DOI] [PubMed]
- 17.Bostrom MP, Lane JM, Berberian WS, Missri AA, Tomin E, Weiland A et al (1995) Immunolocalization and expression of bone morphogeneic proteins 2 and 4 in fracture healing. J Orthop Res 13(3):357–367 doi:10.1002/jor.1100130309 [DOI] [PubMed]
- 18.Meyer RA Jr, Meyer MH, Tenholder M, Wondracek S, Wasserman R, Garges P (2003) Gene expression in older rats with delayed union of femoral fractures. J Bone Jt Surg Am 85-A(7):1243–1254 [DOI] [PubMed]
- 19.Kuorilehto T, Ekholm E, Nissinen M, Hietaniemi K, Hiltunen A, Paavolainen P et al (2006) NF1 gene expression in mouse fracture healing and in experimental rat pseudarthrosis. J Histochem Cytochem 54(3):363–370 Mardoi:10.1369/jhc.5A6784.2005 [DOI] [PubMed]
- 20.Gersch RP, Lombardo F, McGovern SC, Hadjiargyrou M (2005) Reactivation of Hox gene expression during bone regeneration. J Orthop Res 23(4):882–890 Juldoi:10.1016/j.orthres.2005.02.005 [DOI] [PubMed]
- 21.Imai Y, Terai H, Nomura-Furuwatari C, Matsumoto K, Nakamura T et al (2005) Hepatocyte growth factor contributes to fracture repair by upregulating the expression of BMP receptors. J Bone Miner Res 20(10):1723–1730 Oct doi:10.1359/JBMR.050607 [DOI] [PubMed]
- 22.Nakazawa T, Nakajima A, Seki N, Okawa A, Kato M, Moriya H et al (2004) Gene expression of periostin in the early stage of fracture healing detected by cDNA microarray analysis. J Orthop Res 22(3):520–525 Maydoi:10.1016/j.orthres.2003.10.007 [DOI] [PubMed]
- 23.Hatano H, Siegel HJ, Yamagiwa H, Bronk JT, Turner RT, Bolander ME et al (2004) Identification of estrogen-regulated genes during fracture healing, using DNA microarray. J Bone Miner Metab 22(3):224–235 doi:10.1007/s00774-003-0482-y [DOI] [PubMed]
- 24.Carvalho RS, Einhorn TA, Lehmann W et al (2004) The role of angiogenesis in a murine tibial model of distraction osteogenesis. Bone 34(5):849–861 Maydoi:10.1016/j.bone.2003.12.027 [DOI] [PubMed]
- 25.Abe N, Yoshioka H, Inoue H, Ninomiya Y (1994) The complete primary structure of the long form of mouse alpha 1(IX) collagen chain and its expression during limb development. Biochim Biophys Acta 1204(1):61–67 [DOI] [PubMed]
- 26.Balint E, Lapointe D, Drissi H, van der Meijden C, Young DW, van Wijnen AJ et al (2003) Phenotype discovery by gene expression profiling: mapping of biological processes linked to BMP-2-mediated osteoblast differentiation. J Cell Biochem 89(2):401–426 [DOI] [PubMed]
- 27.Kamijo M, Haraguchi T, Tonogi M, Yamane GY (2006) The function of connexin 43 on the differentiation of rat bone marrow cells in culture. Biomed Res 27(6):289–295 doi:10.2220/biomedres.27.289 [DOI] [PubMed]
- 28.Li X, Cao X (2006) BMP signaling and skeletogenesis. Ann NY Acad Sci 1068:26–40 doi:10.1196/annals.1346.006 [DOI] [PubMed]
- 29.Kanaan RA, Kanaan LA (2006) Transforming growth factor beta1, bone connection. Med Sci Monit 12(8):164–169 [PubMed]
- 30.Jadlowiec JA, Zhang X, Li J, Campbell PG, Sfeir C (2006) Extracellular matrix-mediated signaling by dentin phosphophoryn involves activation of the Smad pathway independent of bone morphogenetic protein. J Biol Chem 281(9):5341–5347 doi:10.1074/jbc.M506158200 [DOI] [PubMed]
- 31.Nonaka K, Shum L, Takahashi I, Takahashi K, Ikura T, Dashner R et al (1999) Convergence of the BMP and EGF signaling pathways on Smad1 in the regulation of chondrogenesis. Int J Dev Biol 43(8):795–807 Nov [PubMed]
- 32.Chen YG, Wang Q, Lin SL, Chang CD, Chuang J, Ying SY (2006) Activin signaling and its role in regulation of cell proliferation, apoptosis, and carcinogenesis. Exp Biol Med 231(5):534–544 [DOI] [PubMed]
- 33.Haque T, Mandu-Hrit M, Rauch F, Lauzier D, Tabrizian M, Hamdy RC (2006) Immunohistochemical localization of bone morphogenetic protein-signaling Smads during long-bone distraction osteogenesis. J Histochem Cytochem 54(4):407–415 doi:10.1369/jhc.5A6738.2005 [DOI] [PubMed]
- 34.Park SH (2005) Fine tuning and cross-talking of TGF-beta signal by inhibitory Smads. J Biochem Mol Biol 38(1):9–16 Jan 31 [DOI] [PubMed]
- 35.Indrawattana N, Chen G, Tadokoro M, Shann LH, Ohgushi H, Tateishi T et al (2004) Growth factor combination for chondrogenic induction from human mesenchymal stem cell. Biochem Biophys Res Commun 320(3):914–919 doi:10.1016/j.bbrc.2004.06.029 [DOI] [PubMed]
- 36.Lu C, Huang S, Miclau T, Helms JA, Colnot C (2004) Mepe is expressed during skeletal development and regeneration. Histochem Cell Biol 121(6):493–499 Jundoi:10.1007/s00418-004-0653-5 [DOI] [PMC free article] [PubMed]
- 37.Wildemann B, Schmidmaier G, Brenner N, Huning M, Stange R, Haas NP et al (2004) Quantification, localization, and expression of IGF-I and TGF-beta1 during growth factor-stimulated fracture healing. Calcif Tissue Int 74(4):388–397 Aprdoi:10.1007/s00223-003-0117-2 [DOI] [PubMed]
- 38.Kim S, Koga T, Isobe M, Kern BE, Yokochi T, Chin YE et al (2003) Stat1 functions as a cytoplasmic attenuator of Runx2 in the transcriptional program of osteoblast differentiation. Genes Dev 17(16):1979–1991 doi:10.1101/gad.1119303 [DOI] [PMC free article] [PubMed]
- 39.Kearns AE, Donohue MM, Sanyal B, Demay MB (2001) Cloning and characterization of a novel protein kinase that impairs osteoblast differentiation in vitro. J Biol Chem 276(45):42213–42218 doi:10.1074/jbc.M106163200 [DOI] [PubMed]
- 40.Helliwell RJ, Adams LF, Mitchell MD (2004) Prostaglandin synthases: recent developments and a novel hypothesis. Prostaglandins Leukot Essent Fat Acids 70(2):101–113 doi:10.1016/j.plefa.2003.04.002 [DOI] [PubMed]
- 41.Dillon R, Gadgil C, Othmer HG (2003) Short- and long-range effects of Sonic hedgehog in limb development. Proc Natl Acad Sci U S A 100(18):10152–10157 Sep 2doi:10.1073/pnas.1830500100 [DOI] [PMC free article] [PubMed]
- 42.Murakami S, Noda M (2000) Expression of Indian hedgehog during fracture healing in adult rat femora. Calcif Tissue Int 66(4):272–276 Aprdoi:10.1007/PL00005843 [DOI] [PubMed]
- 43.Sandberg M, Aro H, Multimaki P, Aho H, Vuorio E (1989) In situ localization of collagen production by chondrocytes and osteoblasts in fracture callus. J Bone Jt Surg Am 71(1):69–77 [PubMed]
- 44.Kishida Y, Hirao M, Tamai M, Nampei A, Fujimoto T, Nakase T et al (2005) Leptin regulates chondrocyte differentiation and matrix maturation during endochondral ossification. Bone 37(5):607–621 Novdoi:10.1016/j.bone.2005.05.009 [DOI] [PubMed]
- 45.Ben-Eliezer M, Philip M, Gat-Yablonski G (2007) Leptin regulates chondrogenic differentiation in ATDC5 cell-line through JAK/STAT and MAPK pathways. Endocrine 32(2):235–244 Octdoi:10.1007/s12020-007-9025-y [DOI] [PubMed]
- 46.Kume K, Satomura K, Nishisho S, Kitaoka E, Yamanouchi K, Tobiume S et al (2002) Potential role of leptin in endochondral ossification. J Histochem Cytochem 50(2):159–169 [DOI] [PubMed]
- 47.Zhang YR, Proenca R, Maffei M, Barone M, Leopold L, Friedman JM (1994) Positional cloning of the mouse obese gene and its human homologue. Nature 372:425–432 doi:10.1038/372425a0 [DOI] [PubMed]
- 48.Cinti S, Frederich RC, Zingaretti MC, De Matteis R, Flier JS, Lowell BB (1997) Immunohistochemical localization of leptin and uncoupling protein in white and brown adipose tissue. Endocrinology 138:797–804 doi:10.1210/en.138.2.797 [DOI] [PubMed]
- 49.Ahima RS, Flier JS (2000) Leptin. Annu Rev Physiol 62:413–437 doi:10.1146/annurev.physiol.62.1.413 [DOI] [PubMed]
- 50.Lord GM, Matarese G, Howard JK, Baker RJ, Bloom SR, Lechler RI (1998) Leptin modulates the T-cell immune response and reverses starvation-induced immunosuppression. Nature 394:897–901 doi:10.1038/29795 [DOI] [PubMed]
- 51.Considine RV, Caro JF (1997) Leptin and the regulation of body weight. Int J Biochem Cell Biol 29(11):1255–1272 doi:10.1016/S1357-2725(97)00050-2 [DOI] [PubMed]
- 52.Hoggard N, Hunter L, Duncan JS, Williams LM, Trayhurn P, Mercer JG (1997) Leptin and leptin receptor mRNA and protein expression in the murine fetus and placenta. Proc Natl Acad Sci U S A 94:11073–11078 doi:10.1073/pnas.94.20.11073 [DOI] [PMC free article] [PubMed]
- 53.Gainsford T, Willson TA, Metcalf D, Handman E, McFarlane C, Ng A et al (1997) Leptin can induce proliferation, differentiation, and functional activation of hemopoietic cells. Proc Natl Acad Sci U S A 93:14564–14568 doi:10.1073/pnas.93.25.14564 [DOI] [PMC free article] [PubMed]
- 54.Takeda S, Elefteriou F, Levasseur R, Liu X, Zhao L, Parker KL et al (2002) Leptin regulates bone formation via the sympathetic nervous system. Cell 111:305–317 doi:10.1016/S0092-8674(02)01049-8 [DOI] [PubMed]
- 55.Steppan CM, Crawford DT, Chidsey-Frink KL, Ke H, Swick AG (2000) Leptin is a potent stimulator of bone growth in ob/ob mice. Regul Pept 92:73–78 doi:10.1016/S0167-0115(00)00152-X [DOI] [PubMed]
- 56.Yagasaki Y, Yamaguchi T, Watahiki J, Konishi M, Katoh H, Maki K (2000) The role of craniofacial growth in leptin deficient (ob/ob) mice. Orthodontics & Craniofacial Research 6:233–241 doi:10.1034/j.1600-0544.2003.00260.x [DOI] [PubMed]
- 57.Cornish J, Callon KE, Bava U, Lin C, Naot D, Hill BL et al (2002) Leptin directly regulates bone cell function in vitro and reduces bone fragility in vivo. J Endocrinol 175(2):405–415 doi:10.1677/joe.0.1750405 [DOI] [PubMed]