Abstract
Alzheimer's disease and other neurodegenerative disorders of aging are characterized by clinical and pathological features that are relatively specific to humans. To obtain greater insight into how brain aging has evolved, we compared age-related gene expression changes in the cortex of humans, rhesus macaques, and mice on a genome-wide scale. A small subset of gene expression changes are conserved in all three species, including robust age-dependent upregulation of the neuroprotective gene apolipoprotein D (APOD) and downregulation of the synaptic cAMP signaling gene calcium/calmodulin-dependent protein kinase IV (CAMK4). However, analysis of gene ontology and cell type localization shows that humans and rhesus macaques have diverged from mice due to a dramatic increase in age-dependent repression of neuronal genes. Many of these age-regulated neuronal genes are associated with synaptic function. Notably, genes associated with GABA-ergic inhibitory function are robustly age-downregulated in humans but not in mice at the level of both mRNA and protein. Gene downregulation was not associated with overall neuronal or synaptic loss. Thus, repression of neuronal gene expression is a prominent and recently evolved feature of brain aging in humans and rhesus macaques that may alter neural networks and contribute to age-related cognitive changes.
Introduction
Aging is the primary risk factor for Alzheimer's disease and other prevalent neurodegenerative disorders [1], [2]. Little is known, however, about the degree to which normal brain aging is conserved among mammalian species, an issue of central importance in the biology of aging and the development of animal models of human neurological diseases [3]. Gene expression changes that appear during normal brain aging have been explored using microarrays that interrogate only part of the genome in a number of species, including mice, rats, monkeys, and humans [4], [5], [6], [7]. Comparison of the partial expression profiles of the aging mouse and human brain did not show significant overlap [8]. However, there has yet to be a systematic comparison of gene expression at a genome-wide scale in aging mice, monkeys, and humans. Recent advances in sequencing the rhesus macaque, mouse, and human genomes have enabled us to perform a genome-scale comparative analysis of gene expression in the aging mammalian brain [9], [10], [11], [12]. Although a small subset of age-related gene expression changes are conserved from mouse to man, major changes in the expression of genes involved in neurotransmission have evolved in the primate cortex that are potentially relevant to age-related changes in cognition and vulnerability to neurodegeneration.
Results
Phylogenetic Analysis of Brain Aging in Humans, Rhesus Macaques, and Mice
A central issue in a cross-species comparative analysis of aging is the identification of similar aging groups in species with very different maximal life spans. We previously defined the expression profile for age-related expression changes in the human cortex and demonstrated that these changes occur in the majority of individuals by the age of 70 years [6]. We used this expression profile as the basis for defining our aged group in humans as individuals older than 70 years of age who were not diagnosed with a neurodegenerative disorder (Table S1). To identify a similar age group in mice, we used comparative survival curves for humans and mice which suggest that a 30-month-old mouse is similar to an 81-year-old human since at these ages approximately 25% of the original populations survive. A similar survival analysis in rhesus monkeys in captivity determined that 25% survival occurred at approximately 26 years of age [13]. Hence, we chose 30 months and 28–31 years as the aged groups for mice and rhesus monkeys, respectively.
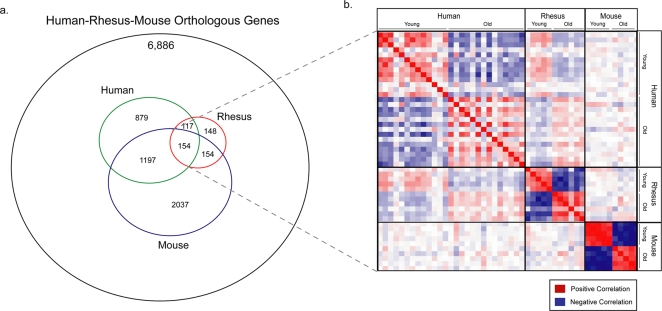
To identify age-related changes in gene expression, cortical samples from 13 young (≤40 years old) and 15 aged (≥70 years old) humans were hybridized to Affymetrix U133plus 2.0 arrays, 5-month-old (n = 5) and 30-month-old mice (n = 5) were hybridized to Affymetrix Mouse 430 2.0 arrays, and samples from 5–6-year-old (n = 5) and 28–31-year-old (n = 6) rhesus macaques were hybridized to Affymetrix whole genome rhesus arrays. Since the rhesus macaque genome has only recently been sequenced [9], the rhesus microarrays are based primarily on gene predictions. Therefore, we used an all-against-all protein BLAST to identify orthologous genes between the rhesus predictions and the other two species. For each homolog pair, we required a BLAST score of greater than 200, and at least 80% alignment of the human or mouse protein sequence with the rhesus sequence (Table S2). The final gene set was composed of genes that possess an ortholog in every species and are represented on all three array platforms. We then employed a two-sample t-test between young and aged age groups with a 1% false discovery rate (FDR) cut-off to identify 3542, 573, and 2347 age-related genes in mice, rhesus monkeys, and humans, respectively (Tables S3 –5). Among these age-related changes, only 154 were significantly associated with aging in all three species (Fig. 1a and Table S6). To assess this gene group as an indicator of brain aging, the behavior of all 154 genes was compared across age groups and species to derive Pearson correlation coefficients. The resulting correlation matrix showed that this core gene set distinguishes between young and aged samples in all three species (Fig. 1b). Furthermore, this set of age-related gene expression changes distinguished between chronological and biological age. For example, a 30-year-old rhesus monkey more closely resembled a 70-year-old human than a 30-year-old human. This set of common age related expression changes is therefore linked to the biology of the aging process in the brain.
Figure 1. Genome-wide comparison of brain aging in humans, rhesus macaques, and mice.
a. Venn diagram indicating the extent of overlap in age-related gene expression changes between the three species. The size of each circle corresponds to the number of age-related expression changes in each species. b. A group of 154 common aging genes provides an indicator of biological aging in all three species. Shown is a matrix of Pearson correlation coefficients that indicate the degree of overall similarity between any two samples (see Methods). Positively correlated sample pairs are indicated by red and negatively correlated pairs are indicated by blue. The degree of correlation correlates with color intensity. The species and age groups are indicated (Human: young ≤40 years; aged ≥70 years. Rhesus macaque: young 5–6 years; aged 28–31 years. Mouse: young 5 months; aged 30 months).
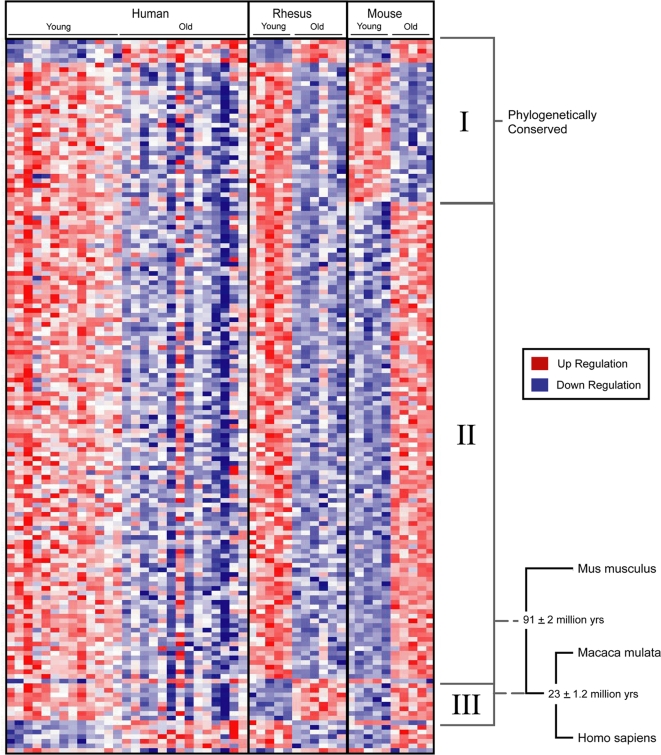
Hierarchical clustering of the common age-regulated genes demonstrated that they fall into three distinct groups: I. Age-regulated genes that are conserved among all three species. II. Genes that change with age in all 3 species but differ in directionality between mouse and rhesus (e.g., from age-downregulated to age-upregulated); and III. Age-regulated genes that change directionality between rhesus and human (Fig. 2 and Table 1). Among the category I genes conserved in all 3 species, the most robustly age-upregulated gene was the anti-oxidant lipid binding protein apolipoprotein D. The most robustly age-downregulated genes in the conserved category were CAMK4, a component of synaptic cAMP-mediated signaling, and ARPP-21, a phosphoprotein also implicated in neuronal cAMP signaling [14] (Table 1). The genes in category II were composed almost entirely of genes that are age-upregulated in mice and downregulated in both rhesus monkeys and humans, defining a set of age-related gene expression changes common to rhesus monkeys and humans. The most robustly downregulated of these primate aging genes was calbindin 1 (CALB1), a marker of cortical inhibitory interneurons (Table 1).
Figure 2. Age-regulated genes common to humans, rhesus macaques, and mice.
The transcriptional profiles of genes that are age-regulated in all three species were analyzed by hierarchical clustering. Reduced expressed with aging is indicated by a transition from red in the young to blue in the aged, and vice versa. Genes separate into three groups based on whether the direction of age-related changes (i.e., age-upregulated or age-downregulated) is conserved in all three species (category I), changes between mice and rhesus macaques (category II), or changes between rhesus macaques and humans (category III). Also indicated is the evolutionary time of divergence in years for each pair of species based on analysis of protein sequence alignments [37].
Table 1. Age-regulated genes common to humans, rhesus macaques, and mice.
| Gene Description | Gene Symbol | Q-Value (%) | Fold Change | ||||
| Human | Rhesus | Mouse | Human | Rhesus | Mouse | ||
| I. Phylogenetically Conserved Aging Genes | |||||||
| apolipoprotein D | APOD | 0.035 | 0.856 | 0.017 | 2.251 | 4.006 | 2.245 |
| G protein-coupled receptor, family C, group 5, member B | GPRC5B | 0.279 | 0.232 | 0.017 | 1.642 | 1.412 | 1.415 |
| tripeptidyl peptidase I | TPP1 | 0.390 | 0.438 | 0.017 | 1.496 | 1.247 | 1.145 |
| ribosomal protein S9 | RPS9 | 0.549 | 0.232 | 0.028 | 1.337 | 1.754 | 1.270 |
| calnexin | CANX | 0.090 | 0.232 | 0.057 | 1.200 | 1.371 | 1.677 |
| solute carrier family 35 (UDP-galactose transporter), member A2 | SLC35A2 | 0.035 | 0.438 | 0.028 | −1.136 | −1.525 | −1.447 |
| Cofactor required for Sp1 transcriptional activation, subunit 8, 34 kDa | CRSP8 | 0.195 | 0.856 | 0.120 | −1.175 | −1.323 | −1.323 |
| Hypothetical protein MGC29898 | MGC29898 | 0.279 | 0.232 | 0.776 | −1.195 | −1.967 | −1.306 |
| glutathione synthetase | GSS | 0.279 | 0.438 | 0.348 | −1.200 | −1.534 | −1.603 |
| ubiquitin-conjugating enzyme E2Q (putative) 1 | UBE2Q1 | 0.549 | 0.720 | 0.639 | −1.202 | −1.534 | −1.249 |
| tRNA methyltranferase 12 homolog (S. cerevisiae) | TRMT12 | 0.740 | 0.501 | 0.240 | −1.229 | −1.452 | −1.538 |
| eukaryotic translation termination factor 1 | ETF1 | 0.195 | 0.856 | 0.412 | −1.231 | −1.409 | −1.215 |
| hypothetical protein FLJ20232 | RP5-1104E15.5 | 0.020 | 0.943 | 0.776 | −1.244 | −1.304 | −1.283 |
| dual specificity phosphatase 14 | DUSP14 | 0.065 | 0.856 | 0.057 | −1.260 | −1.296 | −1.428 |
| member RAS oncogene family | RAB14 | 0.035 | 0.537 | 0.288 | −1.294 | −1.547 | −1.314 |
| Transmembrane protein 49 | TMEM49 | 0.020 | 0.943 | 0.017 | −1.299 | −1.535 | −1.613 |
| NEDD8-conjugating enzyme | UBE2F | 0.020 | 0.534 | 0.288 | −1.300 | −1.467 | −1.119 |
| zinc finger protein 64 homolog (mouse) | ZFP64 | 0.090 | 0.856 | 0.348 | −1.304 | −1.623 | −1.508 |
| transmembrane protein vezatin | VEZT | 0.020 | 0.943 | 0.288 | −1.318 | −1.608 | −1.293 |
| transmembrane protein 4 | TMEM4 | 0.020 | 0.537 | 0.120 | −1.351 | −2.788 | −1.307 |
| tribbles homolog 2 (Drosophila) | TRIB2 | 0.965 | 0.232 | 0.949 | −1.365 | −1.520 | −1.209 |
| Glutamine-fructose-6-phosphate transaminase 1 | GFPT1 | 0.035 | 0.639 | 0.949 | −1.385 | −1.600 | −1.923 |
| protein disulfide isomerase family A, member 6 | PDIA6 | 0.020 | 0.856 | 0.240 | −1.436 | −1.419 | −1.303 |
| Metallophosphoesterase domain containing 1 | MPPED1 | 0.020 | 0.943 | 0.120 | −1.441 | −1.457 | −1.307 |
| armadillo repeat containing 8 | ARMC8 | 0.195 | 0.438 | 0.057 | −1.454 | −1.472 | −1.331 |
| Ribonuclease H1 | RNASEH1 | 0.020 | 0.599 | 0.120 | −1.478 | −1.425 | −1.174 |
| kelch repeat and BTB (POZ) domain containing 6 | KBTBD6 | 0.020 | 0.537 | 0.057 | −1.479 | −1.548 | −1.539 |
| Membrane-associated ring finger (C3HC4) 1 | MARCH1 | 0.195 | 0.639 | 0.120 | −1.481 | −1.361 | −1.977 |
| Acetoacetyl-CoA synthetase | AACS | 0.020 | 0.775 | 0.412 | −1.517 | −1.347 | −1.456 |
| adrenergic, beta, receptor kinase 2 | ADRBK2 | 0.020 | 0.856 | 0.057 | −1.525 | −2.077 | −1.335 |
| golgi autoantigen, golgin subfamily a, 1 | GOLGA1 | 0.020 | 0.856 | 0.412 | −1.565 | −1.542 | −1.192 |
| transmembrane protein 14B | TMEM14B | 0.020 | 0.438 | 0.039 | −1.619 | −1.876 | −1.240 |
| Transforming growth factor, beta receptor associated protein 1 | TGFBRAP1 | 0.090 | 0.775 | 0.776 | −1.738 | −1.226 | −1.117 |
| Cyclic AMP-regulated phosphoprotein, 21 kD | ARPP-21 | 0.020 | 0.232 | 0.017 | −1.953 | −1.457 | −1.137 |
| Calcium/calmodulin-dependent protein kinase IV | CAMK4 | 0.020 | 0.438 | 0.193 | −2.174 | −2.111 | −1.560 |
| II. Aging Genes that Diverged Between Mice and Rhesus Monkeys | |||||||
| calbindin 1, 28 kDa | CALB1 | 0.020 | 0.880 | 0.348 | −3.722 | −1.629 | 1.281 |
| neuronal pentraxin II | NPTX2 | 0.020 | 0.880 | 0.017 | −2.340 | −1.815 | 1.323 |
| chromobox homolog 6 | CBX6 | 0.020 | 0.943 | 0.017 | −2.305 | −1.199 | 1.377 |
| adenylate cyclase 2 (brain) | ADCY2 | 0.020 | 0.856 | 0.193 | −1.973 | −1.264 | 1.109 |
| tubulin tyrosine ligase | TTL | 0.020 | 0.599 | 0.057 | −1.964 | −1.527 | 1.198 |
| 3-hydroxy-3-methylglutaryl-Coenzyme A reductase | HMGCR | 0.020 | 0.232 | 0.288 | −1.866 | −1.367 | 1.469 |
| hepatic leukemia factor | HLF | 0.020 | 0.639 | 0.017 | −1.851 | −1.250 | 1.337 |
| similar to hepatocellular carcinoma-associated antigen HCA557b | LOC151194 | 0.020 | 0.501 | 0.240 | −1.838 | −1.479 | 1.127 |
| trophoblast glycoprotein | TPBG | 0.020 | 0.438 | 0.017 | −1.809 | −1.988 | 1.563 |
| phospholipase C-like 2 | PLCL2 | 0.020 | 0.375 | 0.017 | −1.770 | −1.566 | 1.163 |
| Fusion (involved in t(12;16) in malignant liposarcoma) | FUS | 0.020 | 0.959 | 0.412 | −1.743 | −1.535 | 1.145 |
| protein phosphatase 3 (formerly 2B), catalytic subunit, beta isoform (calcineurin A beta) | PPP3CB | 0.020 | 0.720 | 0.039 | −1.725 | −1.074 | 1.202 |
| KIAA1944 protein | KIAA1944 | 0.020 | 0.232 | 0.949 | −1.687 | −1.673 | 1.169 |
| Chromosome 18 open reading frame 1 | C18orf1 | 0.020 | 0.537 | 0.949 | −1.650 | −1.299 | 1.331 |
| phosphodiesterase 4D interacting protein (myomegalin) | PDE4DIP | 0.195 | 0.534 | 0.057 | −1.640 | −1.333 | 1.292 |
| similar to aspartate beta hydroxylase (ASPH) | ASPHD2 | 0.020 | 0.232 | 0.120 | −1.628 | −1.494 | 1.154 |
| discs, large homolog 3 (neuroendocrine-dlg, Drosophila) | DLG3 | 0.020 | 0.880 | 0.085 | −1.610 | −1.228 | 1.124 |
| adrenergic, alpha-2A-, receptor | ADRA2A | 0.020 | 0.232 | 0.639 | −1.597 | −2.092 | 1.306 |
| component of oligomeric golgi complex 8 | COG8 | 0.020 | 0.775 | 0.017 | −1.557 | −1.327 | 1.274 |
| protein tyrosine phosphatase, non-receptor type 3 | PTPN3 | 0.020 | 0.856 | 0.949 | −1.538 | −1.701 | 1.201 |
| Small nuclear ribonucleoprotein polypeptide A′ | SNRPA1 | 0.020 | 0.943 | 0.146 | −1.528 | −1.217 | 1.250 |
| RAS guanyl releasing protein 1 (calcium and DAG-regulated) | RASGRP1 | 0.020 | 0.232 | 0.098 | −1.525 | −1.648 | 1.150 |
| Signal-induced proliferation-associated 1 like 2 | SIPA1L2 | 0.020 | 0.232 | 0.500 | −1.520 | −2.293 | 1.189 |
| Ubiquitin carboxyl-terminal hydrolase L5 | UCHL5 | 0.020 | 0.775 | 0.028 | −1.492 | −1.999 | 1.357 |
| neuregulin 3 | NRG3 | 0.020 | 0.959 | 0.017 | −1.491 | −1.293 | 1.216 |
| tubulin, alpha 1 (testis specific) | TUBA1 | 0.020 | 0.438 | 0.949 | −1.486 | −1.289 | 1.067 |
| solute carrier family 36 (proton/amino acid symporter), member 1 | SLC36A1 | 0.065 | 0.232 | 0.146 | −1.482 | −1.860 | 1.235 |
| opsin 3 (encephalopsin, panopsin) | OPN3 | 0.020 | 0.943 | 0.057 | −1.478 | −1.430 | 1.268 |
| bicaudal D homolog 2 (Drosophila) | BICD2 | 0.020 | 0.501 | 0.017 | −1.470 | −1.523 | 1.426 |
| p21(CDKN1A)-activated kinase 7 | PAK7 | 0.020 | 0.959 | 0.017 | −1.455 | −1.343 | 1.534 |
| chromosome 21 open reading frame 5 | DOPEY2 | 0.035 | 0.232 | 0.017 | −1.453 | −1.667 | 1.272 |
| Nuclear factor I/B | NFIB | 0.195 | 0.856 | 0.348 | −1.450 | −1.230 | 1.209 |
| membrane associated guanylate kinase, WW and PDZ domain containing 1 | MAGI1 | 0.020 | 0.639 | 0.500 | −1.450 | −1.364 | 1.167 |
| TNF receptor-associated factor 3 | TRAF3 | 0.279 | 0.232 | 0.776 | −1.447 | −1.498 | 1.112 |
| small glutamine-rich tetratricopeptide repeat (TPR)-containing, beta | SGTB | 0.090 | 0.959 | 0.017 | −1.445 | −1.806 | 1.316 |
| hypothetical protein FLJ20701 | FLJ20701 | 0.020 | 0.720 | 0.146 | −1.442 | −1.208 | 1.175 |
| LanC lantibiotic synthetase component C-like 2 (bacterial) | LANCL2 | 0.020 | 0.639 | 0.639 | −1.440 | −1.380 | 1.113 |
| Rho GTPase-activating protein | RICS | 0.065 | 0.880 | 0.240 | −1.437 | −1.507 | 1.141 |
| chromosome 10 open reading frame 9 | C10orf9 | 0.020 | 0.856 | 0.017 | −1.425 | −1.379 | 1.490 |
| dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 2 | DYRK2 | 0.195 | 0.232 | 0.039 | −1.422 | −1.821 | 1.251 |
| Zinc finger protein 148 (pHZ-52) | ZNF148 | 0.020 | 0.599 | 0.017 | −1.420 | −1.336 | 1.320 |
| similar to BcDNA∶GH11415 gene product | C3orf59 | 0.020 | 0.232 | 0.017 | −1.412 | −1.333 | 1.689 |
| importin 11 | IPO11 | 0.195 | 0.880 | 0.017 | −1.408 | −1.358 | 1.334 |
| neuronal pentraxin receptor | NPTXR | 0.090 | 0.943 | 0.288 | −1.407 | −1.199 | 1.310 |
| solute carrier family 35, member B4 | SLC35B4 | 0.020 | 0.537 | 0.017 | −1.403 | −1.780 | 1.341 |
| secretogranin III | SCG3 | 0.020 | 0.720 | 0.500 | −1.401 | −1.255 | 1.125 |
| Proprotein convertase subtilisin/kexin type 2 | PCSK2 | 0.020 | 0.232 | 0.146 | −1.399 | −1.280 | 1.243 |
| Programmed cell death 8 (apoptosis-inducing factor) | PDCD8 | 0.020 | 0.856 | 0.017 | −1.397 | −1.507 | 1.265 |
| tripartite motif-containing 44 | TRIM44 | 0.020 | 0.943 | 0.017 | −1.387 | −2.430 | 1.201 |
| v-akt murine thymoma viral oncogene homolog 3 (protein kinase B, gamma) | AKT3 | 0.020 | 0.880 | 0.017 | −1.385 | −1.155 | 1.475 |
| reticulon 4 receptor-like 1 | RTN4RL1 | 0.020 | 0.959 | 0.288 | −1.378 | −1.369 | 1.187 |
| WD repeat domain 32 | WDR32 | 0.140 | 0.959 | 0.057 | −1.378 | −1.364 | 1.324 |
| zinc finger, DHHC-type containing 4 | ZDHHC4 | 0.020 | 0.639 | 0.146 | −1.370 | −1.995 | 1.181 |
| karyopherin alpha 6 (importin alpha 7) | KPNA6 | 0.090 | 0.959 | 0.017 | −1.362 | −1.452 | 1.370 |
| tribbles homolog 1 (Drosophila) | TRIB1 | 0.279 | 0.534 | 0.017 | −1.359 | −1.758 | 1.519 |
| calmodulin regulated spectrin-associated protein 1 | CAMSAP1 | 0.020 | 0.534 | 0.017 | −1.351 | −1.267 | 1.453 |
| member RAS oncogene family | RAB22A | 0.020 | 0.524 | 0.057 | −1.345 | −1.766 | 1.303 |
| Calumenin | CALU | 0.035 | 0.856 | 0.039 | −1.344 | −1.923 | 1.292 |
| HEPIS | LOC119710 | 0.020 | 0.775 | 0.146 | −1.342 | −1.443 | 1.152 |
| Ankyrin repeat domain 6 | ANKRD6 | 0.140 | 0.943 | 0.098 | −1.341 | −1.361 | 1.220 |
| kelch repeat and BTB (POZ) domain containing 7 | KBTBD7 | 0.020 | 0.537 | 0.017 | −1.327 | −1.548 | 1.226 |
| Rho-associated, coiled-coil containing protein kinase 2 | ROCK2 | 0.020 | 0.232 | 0.120 | −1.323 | −1.773 | 1.289 |
| dynactin 4 (p62) | DCTN4 | 0.090 | 0.375 | 0.028 | −1.320 | −1.306 | 1.317 |
| UDP-glucuronate decarboxylase 1 | UXS1 | 0.279 | 0.232 | 0.017 | −1.315 | −1.943 | 1.558 |
| chromosome 1 open reading frame 21 | C1orf21 | 0.020 | 0.599 | 0.017 | −1.307 | −1.326 | 1.259 |
| proliferation-associated 2G4, 38 kDa | PA2G4 | 0.090 | 0.775 | 0.028 | −1.303 | −1.761 | 1.186 |
| isocitrate dehydrogenase 2 (NADP+), mitochondrial | IDH2 | 0.965 | 0.720 | 0.057 | −1.300 | −1.538 | 1.292 |
| ring finger protein 41 | RNF41 | 0.140 | 0.375 | 0.057 | −1.299 | −2.333 | 1.286 |
| ATPase, aminophospholipid transporter-like, Class I, type 8A, member 2 | ATP8A2 | 0.035 | 0.720 | 0.949 | −1.296 | −2.771 | 1.272 |
| zinc finger protein 697 | ZNF697 | 0.279 | 0.524 | 0.057 | −1.295 | −1.543 | 1.193 |
| makorin, ring finger protein, 1 | MKRN1 | 0.020 | 0.537 | 0.500 | −1.295 | −1.224 | 1.085 |
| eukaryotic translation initiation factor 3, subunit 12 | EIF3S12 | 0.020 | 0.501 | 0.120 | −1.294 | −2.174 | 1.187 |
| transforming, acidic coiled-coil containing protein 1 | TACC1 | 0.035 | 0.537 | 0.017 | −1.288 | −1.945 | 1.968 |
| THUMP domain containing 3 | THUMPD3 | 0.090 | 0.639 | 0.098 | −1.285 | −1.648 | 1.175 |
| mediator of RNA polymerase II transcription, subunit 8 homolog (yeast) | MED8 | 0.020 | 0.438 | 0.193 | −1.282 | −1.376 | 1.131 |
| Casein kinase 2, alpha 1 polypeptide | CSNK2A1 | 0.020 | 0.959 | 0.098 | −1.279 | −1.367 | 1.572 |
| metastasis associated 1 family, member 3 | MTA3 | 0.390 | 0.959 | 0.017 | −1.276 | −1.299 | 1.268 |
| DEAD (Asp-Glu-Ala-Asp) box polypeptide 54 | DDX54 | 0.965 | 0.959 | 0.017 | −1.272 | −1.272 | 1.322 |
| Ras-associated protein Rap1 | RBJ | 0.020 | 0.856 | 0.949 | −1.270 | −1.325 | 1.131 |
| cleavage stimulation factor, 3′ pre-RNA, subunit 3, 77 kDa | CSTF3 | 0.020 | 0.534 | 0.146 | −1.268 | −1.145 | 1.472 |
| N-myristoyltransferase 1 | NMT1 | 0.090 | 0.943 | 0.017 | −1.262 | −1.174 | 1.340 |
| Component of oligomeric golgi complex 1 | COG1 | 0.035 | 0.959 | 0.017 | −1.258 | −1.685 | 1.659 |
| SERPINE1 mRNA binding protein 1 | SERBP1 | 0.035 | 0.943 | 0.240 | −1.257 | −1.303 | 1.143 |
| kelch domain containing 3 | KLHDC3 | 0.140 | 0.720 | 0.193 | −1.252 | −1.255 | 1.194 |
| zinc finger protein 436 | ZNF436 | 0.195 | 0.232 | 0.193 | −1.249 | −1.379 | 1.122 |
| KIAA1217 | KIAA1217 | 0.965 | 0.639 | 0.240 | −1.247 | −1.953 | 1.328 |
| sideroflexin 4 | SFXN4 | 0.020 | 0.639 | 0.639 | −1.247 | −1.233 | 1.182 |
| ankyrin repeat domain 28 | ANKRD28 | 0.065 | 0.856 | 0.120 | −1.238 | −1.505 | 1.345 |
| Phosphodiesterase 8B | PDE8B | 0.140 | 0.856 | 0.017 | −1.236 | −1.964 | 1.329 |
| casein kinase 2, alpha prime polypeptide | CSNK2A2 | 0.020 | 0.943 | 0.288 | −1.231 | −1.315 | 1.137 |
| Ras association (RalGDS/AF-6) domain family 5 | RASSF5 | 0.965 | 0.524 | 0.017 | −1.222 | −1.416 | 1.304 |
| microfibrillar-associated protein 1 | MFAP1 | 0.090 | 0.501 | 0.017 | −1.221 | −1.347 | 1.280 |
| tRNA nucleotidyl transferase, CCA-adding, 1 | TRNT1 | 0.065 | 0.537 | 0.288 | −1.219 | −1.673 | 1.137 |
| golgi SNAP receptor complex member 2 | GOSR2 | 0.390 | 0.959 | 0.639 | −1.216 | −1.404 | 1.545 |
| v-ral simian leukemia viral oncogene homolog A (ras related) | RALA | 0.965 | 0.232 | 0.017 | −1.207 | −1.690 | 1.240 |
| hypothetical protein FLJ11305 | RP11-98F14.6 | 0.965 | 0.943 | 0.348 | −1.205 | −1.411 | 1.312 |
| Zinc finger protein 291 | ZNF291 | 0.279 | 0.943 | 0.017 | −1.205 | −1.342 | 1.458 |
| UDP-N-acetyl-alpha-D-galactosamine (GalNAc-T2) | GALNT2 | 0.390 | 0.537 | 0.017 | −1.202 | −1.155 | 1.388 |
| Acyl-Coenzyme A dehydrogenase family, member 9 | ACAD9 | 0.020 | 0.232 | 0.017 | −1.185 | −1.364 | 1.403 |
| deltex 4 homolog (Drosophila) | DTX4 | 0.020 | 0.599 | 0.017 | −1.180 | −1.254 | 1.314 |
| casein kinase 1, gamma 1 | CSNK1G1 | 0.279 | 0.856 | 0.017 | −1.169 | −1.430 | 1.274 |
| KIAA1698 protein | KIAA1698 | 0.279 | 0.534 | 0.017 | −1.157 | −1.405 | 1.380 |
| Yip1 domain family, member 3 | YIPF3 | 0.965 | 0.599 | 0.017 | −1.143 | −1.703 | 1.540 |
| Adducin 3 (gamma) | ADD3 | 0.065 | 0.599 | 0.085 | 1.540 | 1.329 | −1.289 |
| III. Aging Genes that Diverged Between Rhesus Monkeys and Humans | |||||||
| cell division cycle 42 (GTP binding protein, 25 kDa) | CDC42 | 0.020 | 0.524 | 0.017 | −1.688 | 1.608 | 1.186 |
| melanoma antigen family H, 1 | MAGEH1 | 0.020 | 0.232 | 0.776 | −1.504 | 1.697 | 1.203 |
| Seryl-tRNA synthetase | SARS | 0.020 | 0.501 | 0.017 | −1.453 | 1.375 | 1.650 |
| clathrin, heavy polypeptide (Hc) | CLTC | 0.020 | 0.375 | 0.057 | −1.438 | 1.056 | 1.207 |
| E-1 enzyme | MASA | 0.020 | 0.524 | 0.949 | −1.323 | 1.618 | 1.158 |
| F-box protein 28 | FBXO28 | 0.020 | 0.232 | 0.146 | −1.301 | 1.701 | 1.155 |
| abhydrolase domain containing 14A | ABHD14A | 0.390 | 0.501 | 0.017 | −1.192 | 1.318 | 1.404 |
| eukaryotic translation initiation factor 1A, X-linked | EIF1AX | 0.740 | 0.232 | 0.146 | −1.175 | 2.762 | 1.278 |
| hypothetical protein FLJ11155 | FLJ11155 | 0.140 | 0.959 | 0.085 | 1.868 | −3.539 | −1.172 |
Shown are fold changes (aged to young intensity ratio; minus sign for age-downregulated, no sign for age-upregulated) and statistical q-values (%) derived by Significance Analysis of Microarrays (SAM) as described in Methods. Category I contains genes for which age-related expression changes are conserved, both in terms of significance and direction, across all three species. Category II contains genes in which the direction of the relationship with age changes from mouse to rhesus macaque. Category III contains genes in which the direction of the relationship with age changes between rhesus macaque and human.
Cell-Type Enrichment of Age-Related Gene Expression Changes
To identify the cell types in the brain that exhibit prominent age-related changes in gene expression, we utilized the Allen Brain Atlas [15]. This database, derived by in situ hybridization and 3-dimensional imaging of the adult mouse brain (56 days old), includes genes in which expression was significantly enriched in one of five specific cell types. By combining this cell type analysis with our mouse gene expression data, a subset of age-related gene expression changes was localized to specific cell types. To determine whether the mouse brain dataset predicts the cell type distribution of these genes in the human brain, we performed microarray analysis of isolated neurons, astrocytes, and microglial cells derived from primary human cortical cultures, as previously described [16]. For each set of genes enriched in a specific cell type in mice, we determined the median fold enrichment in each of the three human cell type arrays. Genes that were predicted to be enriched in astrocytes and neurons in the mouse brain by the Allen database were also enriched in the corresponding cell types derived from the human cortex (Fig. S1 and Table S7).
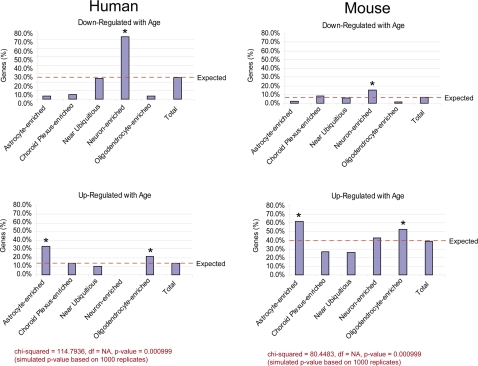
The agreement between the human and mouse cell-type enrichment datasets enabled us to use the Allen Brain Atlas to localize age-related gene expression changes in both species. The limited number of age-related changes in the rhesus dataset prevented us from conducting a comprehensive cell type analysis in rhesus macaques. To determine whether there is a relationship between age-related expression changes and cell type localization, we determined the number of age-related gene expression changes that could be localized to each cell type using the Allen Brain Atlas. A relationship between age-related expression changes and cell type localization was analyzed statistically by determining if the number of age-regulated genes enriched in specific cell types deviated significantly from the number expected if these changes were independent of cell type localization. Both the human and mouse localization analysis showed significant deviation from values expected under the independence assumption (p-value<0.05). Statistical significance was assessed using a Chi-squared test in which the null distribution was estimated based on 1,000 replications (see Methods). The primary data and hypergeometric-based estimates are provided in Tables S8 and S9, respectively. Both humans and mice exhibit a larger fraction of age-upregulated astrocyte- and oligodendrocyte-enriched genes, and age-downregulated neuron-enriched genes, than would be expected by chance alone (Fig. 3). However, relative to mice, human aging is distinguished by a dramatic increase in the proportion of neuron-enriched downregulated genes (Fig. 3). This was also observed when the data was stratified by gender (Text S2). Analysis of our data using a different cell type transcriptome database, derived by isolation of astrocytes, neurons and oligodendrocytes from transgenic mouse cortex [17], confirmed that downregulation of neuronal genes distinguishes aging humans from aging mice (data not shown).
Figure 3. Cell type localization of gene expression in the aging cortex.
Genes enriched in specific cortical cell types, based on the Allen Brain Atlas, were analyzed in the aging mouse and human gene expression profiles. The percentage of age-regulated genes enriched in each cell type is represented by the Y-axis was determined as described in Methods. The expected percentages are indicated by the dashed line. Statistically significant cell type enrichment was determined using a Chi-square test with a permutation-based p-value (1,000 replicates). Specific cell types that exhibit a statistically significant change in age-regulated genes are indicated by an asterisk.
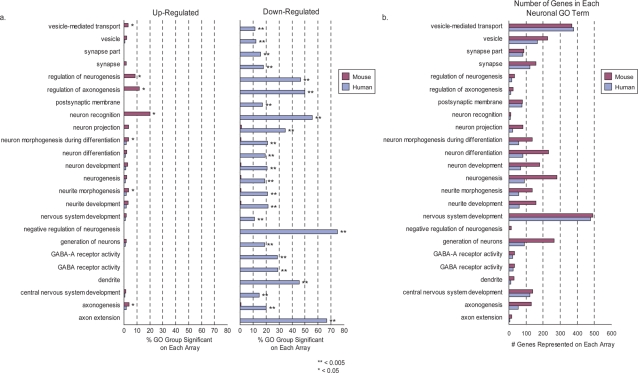
As an independent line of evidence for age-related downregulation of neuronal genes, we identified Gene Ontology (GO) groups that were significantly enriched for age-related expression changes (Table S10). In total, 24 neuronal GO groups were significantly enriched for age-related expression changes in humans (hypergeometric p-value<0.005) (Fig. 4a). In contrast, only 5 of these 24 neuronal GO terms were slightly enriched for genes significantly associated with age in mice (hypergeometric p-value<0.05), despite similar or greater gene numbers for each GO term represented on mouse versus human microarrays (Fig. 4b). Further characterization of these GO terms revealed that the vast majority of genes in the human-enriched neuronal GO terms were downregulated with age. In contrast, the significant mouse neuronal GO terms were primarily enriched for age-upregulated genes (Fig. 4a). Thus, aging reduces the expression of genes with a variety of neuronal functions to a much greater extent in humans than mice.
Figure 4. Neuronal gene ontology groups distinguish the expression profiles of the aging human and mouse cortex.
a. Neuronal gene ontology (GO) groups that are significantly enriched (p-value≤0.005; binomial approximated p-value for a hypergeometric distribution) for age-related expression changes (SAM comparison, FDR≤0.01) were identified. The X-axis represents the percentage of genes in a GO group with age-related up- or down-regulation. Multiple neuronal GO groups are enriched in the human aging profile; while only a few neuronal GO terms appear at less significant thresholds in the mouse aging profile. Age-upregulated and age-downregulated genes are shown separately. b. Number of genes in each GO group that are represented on the mouse and human microarray platforms.
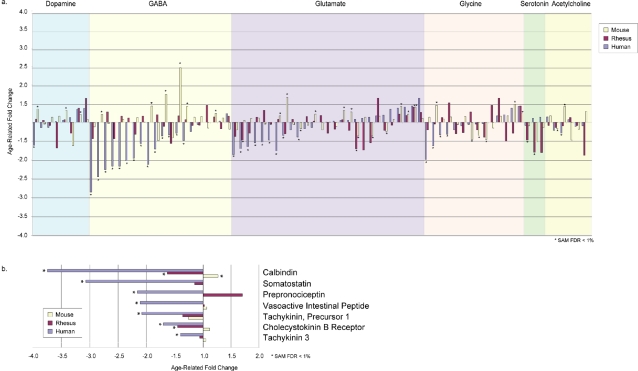
Age-Related Repression of Genes Involved in Inhibitory Neurotransmission
Aging is associated with characteristic neurophysiologic and cognitive changes attributable to specific neurotransmitter systems. An important question, therefore, is whether age-related repression of neuronal genes selectively affects specific neurotransmitter systems. We noted that the only significantly enriched GO groups relating to a specific neurotransmitter were “GABA and GABA-A receptor activity” (Fig. 4). To explore this finding further, we examined the age-regulated expression of genes related to each of the major cortical neurotransmitters, including glutamate, gamma-aminobutyric acid (GABA), dopamine, glycine, serotonin, and acetylcholine (Fig. 5a). The most robustly age-regulated group corresponded to genes involved in GABA-mediated inhibitory neurotransmission (Fig. 5a and Table S11). Multiple genes in this category were age-downregulated with large fold changes in humans, including GABA A receptor subunits alpha 1 (GABRA1), alpha 5 (GABRA5), beta 3 (GABRB3) and gamma 2 (GABRG2), the GABA vesicular transporter (SLC32A1), and the GABA biosynthetic enzymes glutamate decarboxylase 1 and 2 (GAD1 and GAD2) (Table S11). Moreover, genes for the neuropeptides calbindin 1 (CALB1), somatostatin (SST), vasoactive intestinal peptide (VIP), cholecystokinin (CCK), tachykinin (TAC1), and nociceptin (PNOC), which are markers of inhibitory neuronal subpopulations in prefrontal cortex, were significantly age-downregulated (Fig. 5b). These genes were not significantly age-downregulated in mice, although some inhibitory markers, such as calbindin 1 and GABA A receptor subunit alpha 1, were significantly age-downregulated in rhesus macaques (Fig. 5). Downregulation of several glutamate-related genes, such as the glutamate receptor subunits AMPA 1 (GRIA1) and kainate 1 (GRIK1), was also observed, but the number and magnitude of these expression changes were less than that observed for GABA-related genes (Fig. 5a and Table S11). A subset of these age-related changes, notably calbindin 1, GABA A receptor subunit β3 and AMPA 1, have been confirmed by quantitative real time RT-PCR [6]. Thus, genes associated with inhibitory neurotransmission are repressed in the aging human cortex.
Figure 5. Global repression of genes associated with GABA-mediated inhibitory neurotransmission.
Shown are age-related changes in the expression of genes that mediate major neurotransmitter systems in the cortex of humans, rhesus monkeys, and mice. a. Genes involved in specific neurotransmitters were identified based on membership in the corresponding GO groups. Age-related fold changes in genes with orthologs in all three species and represented on all three microarray platforms are shown for humans, rhesus monkeys, and mice. Gene identities are provided in Table S11. *q-value≤0.01. b. Age-related fold changes for markers of inhibitory neuronal subpopulations. Statistical significance in a specific species (q-value≤0.01) is denoted with an asterisk.
Age-Related Reduction of Neuronal Proteins Is Not Associated with Overall Neuronal or Synaptic Loss
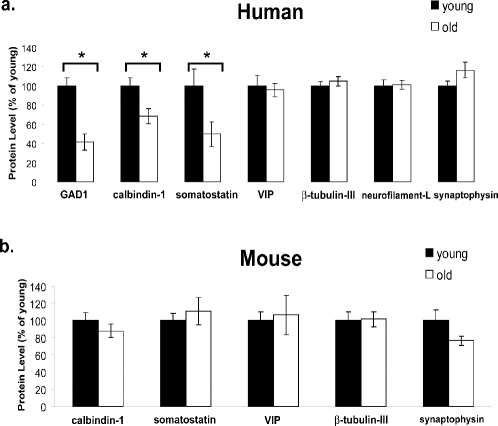
To determine whether reduced mRNA levels are associated with reduced protein levels in the aging brain, a subset of gene products expressed in GABAergic neurons was examined by quantitative Western blotting in cortical samples from young adult and aged humans and mice. The protein level of the major GABA biosynthetic enzyme in the brain, GAD1, was significantly reduced in the aging human cortex, as well as the levels of calbindin 1 and somatostatin, in agreement with the microarray data (Fig. 6a and Fig. S2a). The neuropeptide VIP did not show a significant age-related change at the protein level, in contrast to the age-related reduction in VIP mRNA. This difference may reflect limited sensitivity of the antibody used for Western blotting of VIP, or post-translational regulation of VIP levels. In contrast to aging human cortex, the aging mouse cortex did not exhibit altered levels of calbindin or somatostatin, which is also in agreement with the microarray data (Fig. 6b and Fig. S2b).
Figure 6. Reduced protein markers of inhibitory neurons in the aged human cortex.
a. GAD1, calbindin-1, and somatostatin protein levels are significantly lower in the aged (71–91 yr; white) human cortex than in the young adult (24–35 yr; black) cortex, in agreement with microarray results. VIP expression is age-stable at the protein level. The neuronal markers β-tubulin-III and neurofilament-L are age-stable at the protein level, as is the synaptic protein synaptophysin. n = 15. The primary Western blot data are shown in Figure S2a. b. Calbindin-1, somatostatin, and VIP protein levels are age-stable in the mouse cortex, in agreement with the microarray results. Likewise, β-tubulin-III and synaptophysin do not change significantly with age. Attempts to probe for mouse GAD1 and neurofilament-L were not successful. n = 6. The primary Western blot data are shown in Figure S2b. In both a and b, the level of each protein was normalized to the level of actin. Values represent the mean±S.E.M. expressed as percent of the mean young value for each protein. * P<0.05 by Student's two-tailed t-test.
Stereological cell counting studies suggest that neuronal loss is not significant in the aging human prefrontal cortex. To confirm this finding, we performed quantitative Western blotting for two established neuron-specific markers, β-tubulin III and neurofilament L chain [17]. The levels of both proteins did not change significantly in the aging human prefrontal cortex (Fig. 6a and Fig. S2a). We also examined the presynaptic marker synaptophysin, which did not show a significant age-related change in this cortical region (Fig. 6a and Fig. S2a). These results suggest that downregulation of neuronal genes in the aging human cortex cannot be attributed to overall loss of neurons or synapses.
Discussion
We have compared the protein-coding transcriptome of the aging cerebral cortex in mice, rhesus monkeys, and humans by utilizing species-specific genome-scale microarrays. As such, this study is not confounded by cross-species hybridization of RNA to microarrays, and provides a broad view of the evolution of the mammalian aging brain. Our results suggest that a relatively small subset of age-regulated gene expression changes are conserved from mouse to man. The most robustly age-upregulated of these conserved genes is apolipoprotein D, which has been shown to protect against oxidative stress and extend lifespan in Drosophila [18], [19]. Moreover, apolipoprotein D is upregulated at the protein level in the aging human brain and to a greater extent in a variety of neurological diseases, including Alzheimer's disease [20], [21]. The most robustly age-downregulated gene conserved in all three species is CAMK4, a key component of the cAMP signaling cascade that links synaptic activity to CREB-dependent transcription and modulates synaptic plasticity [14], [22]. Another key cAMP signaling gene, adenylate cyclase 2, is age-downregulated in humans and rhesus macaques. Thus, increased expression of neuroprotective genes and reduced expression of genes involved in synaptic function are conserved features of mammalian aging.
Localization of gene expression by in situ hybridization and analysis of gene ontology groups indicates that 3 cell types – astrocytes, oligodendrocytes, and neurons – exhibit significant age-dependent changes in gene expression in mice and humans. However, age-related downregulation of neuronal genes has increased dramatically from mouse to man, and is a major distinguishing feature. Several lines of evidence suggest that this is unlikely to be secondary to neuronal cell death. First, stereological analysis of neuronal cell number did not detect neuronal loss in the region of the aging human prefrontal cortex used in this study [2], [23]. Second, we have shown that expression of a number of neuron-specific genes is unaltered in the aging human prefrontal cortex at both the mRNA and protein levels. Moreover, the absence of a significant age-related change in synaptophysin levels suggests that overall synapse numbers may also be preserved. However, this does not rule out more subtle changes in synaptic or dendritic spine structure as reported in aging rhesus monkeys [24]. Finally, we showed previously that age-related gene downregulation did not correlate with postmortem interval in the range used in our study [6], consistent with the lack of an effect of postmortem interval on RNA integrity in another study [25]. In addition, we monitored brain tissue pH to exclude human cases with prolonged terminal hypoxia [26]. Taken together, these findings are consistent with a primary age-related change in the regulation of neuronal gene expression. In a previous study, we found that downregulated neuronal genes were associated with DNA damage in the aging human cortex, and that DNA damage can repress the transcription of these genes in primary neuronal cultures [6]. Another study suggested that some genes undergo age-dependent DNA methylation [27]. Thus, transcriptional repression in neurons may be a primary feature of human brain aging that has evolved in long-lived primates.
A systematic investigation of genes involved in the major cortical neurotransmitter systems suggests that the GABA system, which mediates inhibitory neurotransmission, may be particularly affected in the aging human prefrontal cortex. This is underscored by the 50–60% reduction in mRNA and protein levels of GAD1, the primary GABA biosynthetic enzyme in the brain. In addition, the marked downregulation of calbindin 1 and somatostatin suggests that specific inhibitory neuronal subpopulations may be unusually vulnerable. Reduced calbindin 1 immunocytochemical staining has also been demonstrated during normal brain aging in rhesus monkeys and humans, and becomes more pronounced in Alzheimer's disease [28]. Thus, aging of the brain may be associated with reduced inhibitory neurotransmission.
The central role of GABA in cognition and affective state raises the possibility that age-dependent downregulation of this system might contribute to neurophysiological and psychological changes in the aging population [29]. Reduced inhibitory circuit activity might increase cortical activation during the performance of routine cognitive tasks, a phenomenon that has been demonstrated in the aging human prefrontal cortex by functional imaging studies [30], [31]. This pattern of increased cortical activation may initially be compensatory, enabling aged individuals to function at a higher level [31]. However, increased excitation could predispose to excitotoxicity, a mechanism of neuronal cell death associated with a variety of age-related neurological disorders, including Alzheimer's disease [32]. Functional imaging studies have implicated cortical overactivation due to impaired inhibitory function in patients with Alzheimer's disease [33]. The relevance of overexcitation to disease pathogenesis is suggested by the clinical efficacy of the NMDA receptor antagonist memantine, currently the only treatment that delays progression of moderate to late stage Alzheimer's disease [34]. Interestingly, significant downregulation of GABA-related genes is not detected in the aging mouse cortex, which may increase resistance to excitotoxicity relative to aging humans. This may, in turn, contribute to the paucity of neuronal cell death in mouse models of neurodegenerative diseases compared with the human pathology [2]. Hence, a greater understanding of normal brain aging and its evolution may provide new insights into pathogenic mechanisms involved in age-related neurodegeneration.
Materials and Methods
Samples and Microarray Platforms
All aspects of animal housing and experimental procedures were approved by the Institutional Animal Care and Use Committees of Children's Hospital Boston and the Beth Israel-Deaconess Hospital (for rhesus macaques) and by the William S. Middleton V.A. Medical Center and the University of Wisconsin-Madison Medical School (for mice). Postmortem human tissue was procured in accordance with institutional guidelines. Detailed description of the human, rhesus macaque and murine samples and extraction protocols are supplied in Text S1 and Table S1. Postmortem human cortical samples were derived from subjects that did not carry a diagnosis of Alzheimer's disease or another neurodegenerative disease, and showed neuropathological findings within the normal range for age. In addition, human brain tissue samples with a pH>6.5 were used to exclude prolonged terminal hypoxia [26]. We generated genome-wide expression profiles of young and aged cortical samples in humans, rhesus monkeys and C57BL/6J mice using Affymetrix Human Genome U133plus 2.0 arrays, Rhesus Macaque Genome arrays and Mouse Genome 430 2.0 arrays, respectively. Affymetrix Human Genome U133plus 2.0 arrays were also used for expression profiling of neurons, microglia and astrocytes isolated from primary fetal human cortical cultures. Samples with acceptable parameters of RNA quality (Text S1) were hybridized to the corresponding Affymetrix oligonucleotide arrays, which were then scanned and expression data extracted using the standard Affymetrix Microarray Suite Software.
Gene Mapping
Predicted rhesus macaque proteins, based on the Jan. 2006 version of the rhesus genome (Baylor College of Medicine HGSC v1.0), were aligned to human Refseq protein sequences mapped to NCBI Build 36 of the human genome. The mapping was conducted using the BLAST program [35] by first creating a BLAST protein database from the predicted rhesus proteins. Using protein-BLAST, individual human Refseq proteins were then compared to the rhesus protein database. A BLAST score greater than 200, and at least 80% of the human protein aligning to the predicted rhesus protein, was required to declare an orthologous pair between the two species. A complete list of orthologous human-rhesus gene pairs is provided in Table S2.
Expression Profiling and Analytical Approaches
To compare expression across species, genes were required to have orthologs in the human, rhesus macaque and mouse genome databases, and to have probesets in the microarray platforms for each species. We also required that probes be called present using dChip software in at least 20% of the arrays for each species. Four approaches were used to analyze the genes meeting these criteria. 1. Significance Analysis of Microarrays (SAM) software was used to compare young and aged groups within each species with the following criteria for identifying age-related expression changes: 1000 permutations and median false discovery rate (FDR) ≤0.01. Significant age-related gene expression changes are listed for humans, rhesus macaques and mice in Tables S3, S4, and S5, respectively. The subset of genes that are age-regulated in all three species is provided in Table S6. These common age-related genes were also resolved by hierarchical clustering using dChip software (build date: April 11, 2007) [36]. The display range used in the hierarchical clustering was 2.0 (a value greater than 2.0 standard deviations above the mean is pure red, below is pure blue, and equal to the mean is white). 2. Correlation coefficients between samples were calculated and visualized using dChip software across the 154 genes that are significantly associated with aging in all three species. The range of observed correlation coefficients was (−0.78, 0.82), excluding a sample's correlation with itself. The display range used was 0.7 (correlation above 0.7 is pure red, below is pure blue, and 0 is white). 3. The cell type enrichment analysis of age-regulated genes was performed independently for mice and humans, and included all genes that met the above microarray criteria and were also present in the list of cell type-enriched genes in the Allen Brain Atlas (Table S8). For the analysis in humans, we required that the mouse ortholog to the human gene be present in the list. For each species, the number of significant age up- and down-regulated genes, as well as the number of non-significant genes, was determined for each of the five cell types indicated in the Allen Brain Atlas resulting in a 3-by-5 table (Table S9). Assuming independence between cell types and age-related expression changes, the expected count within each cell of the table was estimated using the row and column totals. As a result of the low count in some cells, a Monte Carlo p-value, based on 1000 replications was calculated for each species to test whether the observed count significantly deviated from what was expected by chance. 4. Gene Ontology analysis was performed independently for humans and mice using dChip software (Table S10). A Gene Ontology group is considered to be enriched in the aging database if it contains a greater number of significantly age-related genes than expected by chance. The statistical significance of GO group enrichment is determined using a binomial approximation to the hypergeometric distribution with a p-value cut-off of 0.005, as described in detail elsewhere (www.dchip.org) [36].
Analysis of Cultured Human Cortical Cell Types
Neurons, astrocytes and microglia were isolated from primary fetal human cortical cultures as described previously [16]. Expression profiling of the isolated cortical cell types was performed using Affymetrix U133 plus 2.0 arrays. To assess the concordance of gene expression profiles of corresponding human and mouse cortical cell types, we analyzed the probe sets on the human U133plus 2.0 arrays that corresponded to the Allen Brain Atlas list of mouse cell type-enriched genes (Table S7). Fold enrichment of a particular gene in a specific human cell type was calculated as follows: the intensity of the gene in one cell type was divided by the maximum of the intensities in the two remaining cell types. The median fold enrichment of a particular human cell type was then calculated over all of the genes that were called enriched in a specific mouse cell type (Allen Brain Atlas). The result was a human cell type enrichment score for every human-mouse cell type combination (Fig. S1). Median fold values greater than 1.0 indicated enrichment.
Western Blot Analysis
Human Brain
Postmortem human cortical tissue (Brodmann area 9/10) was flash frozen and stored at −150° until use. Tissue was homogenized in RIPA-DOC buffer containing protease inhibitors (Complete, Roche) with microcystin (1 µm) and Na3Vo4 (1 mM). Tissue was homogenized, sonicated, and centrifuged at 10,000 rpm at 4°C and the protein concentration in the resulting supernatant was assayed (BioRad protein assay). Samples were boiled in 1× SDS sample buffer containing DTT and resolved by 7% SDS-PAGE using the Criterion System (BioRad) and electrotranferred to PVDF membranes (Immobilon, Millipore). The primary antibodies and dilutions used to probe the PVDF membranes are described in Table S12. Secondary antibodies (Jackson ImmunoResearch) were used at 1∶2000 diluted in 5% nonfat milk. Blots were developed on film or with a gel documentation system (Syngene) and quantified with GeneTools software (Syngene).
Mouse Brain
Three young B6C3F1 mice (5 months) and three aged B6C3F1 mice (30 months) were sacrificed and the cortex was isolated and homogenized in 20 mM HEPES, 125 mM NaCl, 0.1% NP40, 0.1% Triton X-100, 1 mM EDTA, 10 mM nicotinamide, 1 µM trichostatin A, protease inhibitors (Complete, Roche), and phosphatase inhibitors (PhosSTOP, Roche). Samples were boiled in SDS sample buffer containing DTT, resolved by SDS-PAGE on 10% or 12.5% Tris-glycine gels and electrotransferred to PVDF membranes.
Supporting Information
The MIAME Checklist
(0.07 MB DOC)
Analysis of the Effects of Gender
(0.03 MB DOC)
Concordance of Gene Expression in Human and Murine Neural Cell Types
(0.63 MB TIF)
Western Blot Analysis of Age-Downregulated and Age-Stable Proteins in Human and Mouse Cortex.
(8.39 MB TIF)
Sample information
(0.03 MB XLS)
Mapping of predicted rhesus macaque and human genes
(6.28 MB XLS)
Age-Related Expresssion Changes in Humans
(1.16 MB XLS)
Age-Related Expression Changes in Rhesus Macaques
(0.42 MB XLS)
Age-Related Expresssion Changes in Mice
(1.78 MB XLS)
Genes with Significant Age-Related Expression Changes in Humans, Rhesus Macaques, and Mice
(0.08 MB XLS)
Concordant Expression Patterns in Cultured Human Cortical Cell Types and the Allen Brain Atlas
(0.07 MB XLS)
Age-Related Expression Changes of Genes Enriched in Specific Cortical Cell Types In Vivo
(0.12 MB XLS)
Calculations for Determining Significant Cell-Type Enrichment of Age-Regulated Genes
(0.03 MB XLS)
Gene Ontology Enrichment Analysis of Human and Mouse Expression Data
(0.05 MB XLS)
Age-Related Gene Expression Changes Classified by Neurotransmitter Type
(0.05 MB XLS)
Antibodies employed in the analysis of cortical protein levels
(0.02 MB XLS)
Acknowledgments
We thank Ruth Capella for her excellent assistance in the preparation of the manuscript.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work was supported by grants from the National Institute on Aging (grant numbers AG26651, AG27040, AG27916), and an Ellison Medical Foundation Senior Scholar award to B.A.Y.. P.L. was supported by NIH training grant T32GM074897.
References
- 1.Martin GM. Genetic modulation of senescent phenotypes in Homo sapiens. Cell. 2005;120:523–532. doi: 10.1016/j.cell.2005.01.031. [DOI] [PubMed] [Google Scholar]
- 2.Yankner BA, Lu T, Loerch P. The Aging Brain. Annu Rev Pathol. 2007;3:41–66. doi: 10.1146/annurev.pathmechdis.2.010506.092044. [DOI] [PubMed] [Google Scholar]
- 3.Fischer A, Sananbenesi F, Wang X, Dobbin M, Tsai LH. Recovery of learning and memory is associated with chromatin remodelling. Nature. 2007;447:178–182. doi: 10.1038/nature05772. [DOI] [PubMed] [Google Scholar]
- 4.Lee CK, Weindruch R, Prolla TA. Gene-expression profile of the ageing brain in mice. Nat Genet. 2000;25:294–297. doi: 10.1038/77046. [DOI] [PubMed] [Google Scholar]
- 5.Jiang CH, Tsien JZ, Schultz PG, Hu Y. The effects of aging on gene expression in the hypothalamus and cortex of mice. Proc Natl Acad Sci U S A. 2001;98:1930–1934. doi: 10.1073/pnas.98.4.1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lu T, Pan Y, Kao SY, Li C, Kohane I, et al. Gene regulation and DNA damage in the ageing human brain. Nature. 2004;429:883–891. doi: 10.1038/nature02661. [DOI] [PubMed] [Google Scholar]
- 7.Fraser HB, Khaitovich P, Plotkin JB, Paabo S, Eisen MB. Aging and gene expression in the primate brain. PLoS Biol. 2005;3:e274. doi: 10.1371/journal.pbio.0030274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zahn JM, Poosala S, Owen AB, Ingram DK, Lustig A, et al. AGEMAP: a gene expression database for aging in mice. PLoS Genet. 2007;3:e201. doi: 10.1371/journal.pgen.0030201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gibbs RA, Rogers J, Katze MG, Bumgarner R, Weinstock GM, et al. Evolutionary and biomedical insights from the rhesus macaque genome. Science. 2007;316:222–234. doi: 10.1126/science.1139247. [DOI] [PubMed] [Google Scholar]
- 10.Waterston RH, Lindblad-Toh K, Birney E, Rogers J, Abril JF, et al. Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420:520–562. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- 11.Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 12.Venter JC, Adams MD, Myers EW, Li PW, Mural RJ, et al. The sequence of the human genome. Science. 2001;291:1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- 13.Tigges J, Gordon TP, McClure HM, Hall EC, Peters A. Survival rate and life span of rhesus monkeys at the Yerkes regional primate research center. Am J Primatol. 1988;15:263–273. doi: 10.1002/ajp.1350150308. [DOI] [PubMed] [Google Scholar]
- 14.Greengard P. The neurobiology of slow synaptic transmission. Science. 2001;294:1024–1030. doi: 10.1126/science.294.5544.1024. [DOI] [PubMed] [Google Scholar]
- 15.Lein ES, Hawrylycz MJ, Ao N, Ayres M, Bensinger A, et al. Genome-wide atlas of gene expression in the adult mouse brain. Nature. 2007;445:168–176. doi: 10.1038/nature05453. [DOI] [PubMed] [Google Scholar]
- 16.Wang J, Gabuzda D. Reconstitution of human immunodeficiency virus-induced neurodegeneration using isolated populations of human neurons, astrocytes, and microglia and neuroprotection mediated by insulin-like growth factors. J Neurovirol. 2006;12:472–491. doi: 10.1080/13550280601039659. [DOI] [PubMed] [Google Scholar]
- 17.Cahoy JD, Emery B, Kaushal A, Foo LC, Zamanian JL, et al. A transcriptome database for astrocytes, neurons, and oligodendrocytes: a new resource for understanding brain development and function. Journal of Neuroscience. 2008;28:264–278. doi: 10.1523/JNEUROSCI.4178-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Walker DW, Muffat J, Rundel C, Benzer S. Overexpression of a Drosophila homolog of apolipoprotein D leads to increased stress resistance and extended lifespan. Curr Biol. 2006;16:674–679. doi: 10.1016/j.cub.2006.01.057. [DOI] [PubMed] [Google Scholar]
- 19.Sanchez D, Lopez-Arias B, Torroja L, Canal I, Wang X, et al. Loss of glial lazarillo, a homolog of apolipoprotein D, reduces lifespan and stress resistance in Drosophila. Curr Biol. 2006;16:680–686. doi: 10.1016/j.cub.2006.03.024. [DOI] [PubMed] [Google Scholar]
- 20.Kalman J, McConathy W, Araoz C, Kasa P, Lacko AG. Apolipoprotein D in the aging brain and in Alzheimer's dementia. Neurol Res. 2000;22:330–336. doi: 10.1080/01616412.2000.11740678. [DOI] [PubMed] [Google Scholar]
- 21.Belloir B, Kovari E, Surini-Demiri M, Savioz A. Altered apolipoprotein D expression in the brain of patients with Alzheimer disease. J Neurosci Res. 2001;64:61–69. doi: 10.1002/jnr.1054. [DOI] [PubMed] [Google Scholar]
- 22.Ho N, Liauw JA, Blaeser F, Wei F, Hanissian S, et al. Impaired synaptic plasticity and cAMP response element-binding protein activation in Ca2+/calmodulin-dependent protein kinase type IV/Gr-deficient mice. J Neurosci. 2000;20:6459–6472. doi: 10.1523/JNEUROSCI.20-17-06459.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bussiere T, Gold G, Kovari E, Giannakopoulos P, Bouras C, et al. Stereologic analysis of neurofibrillary tangle formation in prefrontal cortex area 9 in aging and Alzheimer's disease. Neuroscience. 2003;117:577–592. doi: 10.1016/s0306-4522(02)00942-9. [DOI] [PubMed] [Google Scholar]
- 24.Peters A, Sethares C, Moss MB. The effects of aging on layer 1 in area 46 of prefrontal cortex in the rhesus monkey. Cereb Cortex. 1998;8:671–684. doi: 10.1093/cercor/8.8.671. [DOI] [PubMed] [Google Scholar]
- 25.Ervin JF, Heinzen EL, Cronin KD, Goldstein D, Szymanski MH, et al. Postmortem delay has minimal effect on brain RNA integrity. J Neuropathol Exp Neurol. 2007;66:1093–1099. doi: 10.1097/nen.0b013e31815c196a. [DOI] [PubMed] [Google Scholar]
- 26.Kingsbury AE, Foster OJ, Nisbet AP, Cairns N, Bray L, et al. Tissue pH as an indicator of mRNA preservation in human post-mortem brain. Brain Res Mol Brain Res. 1995;28:311–318. doi: 10.1016/0169-328x(94)00219-5. [DOI] [PubMed] [Google Scholar]
- 27.Siegmund KD, Connor CM, Campan M, Long TI, Weisenberger DJ, et al. DNA methylation in the human cerebral cortex is dynamically regulated throughout the life span and involves differentiated neurons. PLoS ONE. 2007;2:e895. doi: 10.1371/journal.pone.0000895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Geula C, Bu J, Nagykery N, Scinto LF, Chan J, et al. Loss of calbindin-D28k from aging human cholinergic basal forebrain: relation to neuronal loss. J Comp Neurol. 2003;455:249–259. doi: 10.1002/cne.10475. [DOI] [PubMed] [Google Scholar]
- 29.Mohler H. Molecular regulation of cognitive functions and developmental plasticity: impact of GABAA receptors. J Neurochem. 2007;102:1–12. doi: 10.1111/j.1471-4159.2007.04454.x. [DOI] [PubMed] [Google Scholar]
- 30.Bäckman L, Almkvist O, Andersson J, Nordberg A, Windblad B, et al. Brain activation in young and older adults during implicit and explicit retrieval. J Cogn Neurosci. 1997;9:378–391. doi: 10.1162/jocn.1997.9.3.378. [DOI] [PubMed] [Google Scholar]
- 31.Cabeza R, Anderson ND, Locantore JK, McIntosh AR. Aging gracefully: compensatory brain activity in high-performing older adults. Neuroimage. 2002;17:1394–1402. doi: 10.1006/nimg.2002.1280. [DOI] [PubMed] [Google Scholar]
- 32.Waxman EA, Lynch DR. N-methyl-D-aspartate receptor subtypes: multiple roles in excitotoxicity and neurological disease. Neuroscientist. 2005;11:37–49. doi: 10.1177/1073858404269012. [DOI] [PubMed] [Google Scholar]
- 33.Petrella JR, Wang L, Krishnan S, Slavin MJ, Prince SE, et al. Cortical deactivation in mild cognitive impairment: high-field-strength functional MR imaging. Radiology. 2007;245:224–235. doi: 10.1148/radiol.2451061847. [DOI] [PubMed] [Google Scholar]
- 34.Cummings JL. Alzheimer's disease. N Engl J Med. 2004;351:56–67. doi: 10.1056/NEJMra040223. [DOI] [PubMed] [Google Scholar]
- 35.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 36.Li C, Wong WH. Model-based analysis of oligonucleotide arrays: expression index computation and outlier detection. Proc Natl Acad Sci U S A. 2001;98:31–36. doi: 10.1073/pnas.011404098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hedges SB. The origin and evolution of model organisms. Nat Rev Genet. 2002;3:838–849. doi: 10.1038/nrg929. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The MIAME Checklist
(0.07 MB DOC)
Analysis of the Effects of Gender
(0.03 MB DOC)
Concordance of Gene Expression in Human and Murine Neural Cell Types
(0.63 MB TIF)
Western Blot Analysis of Age-Downregulated and Age-Stable Proteins in Human and Mouse Cortex.
(8.39 MB TIF)
Sample information
(0.03 MB XLS)
Mapping of predicted rhesus macaque and human genes
(6.28 MB XLS)
Age-Related Expresssion Changes in Humans
(1.16 MB XLS)
Age-Related Expression Changes in Rhesus Macaques
(0.42 MB XLS)
Age-Related Expresssion Changes in Mice
(1.78 MB XLS)
Genes with Significant Age-Related Expression Changes in Humans, Rhesus Macaques, and Mice
(0.08 MB XLS)
Concordant Expression Patterns in Cultured Human Cortical Cell Types and the Allen Brain Atlas
(0.07 MB XLS)
Age-Related Expression Changes of Genes Enriched in Specific Cortical Cell Types In Vivo
(0.12 MB XLS)
Calculations for Determining Significant Cell-Type Enrichment of Age-Regulated Genes
(0.03 MB XLS)
Gene Ontology Enrichment Analysis of Human and Mouse Expression Data
(0.05 MB XLS)
Age-Related Gene Expression Changes Classified by Neurotransmitter Type
(0.05 MB XLS)
Antibodies employed in the analysis of cortical protein levels
(0.02 MB XLS)