Abstract
Dynamins are 100-kDa GTPases that are essential for clathrin-coated vesicle formation during receptor-mediated endocytosis. To date, three different dynamin genes have been identified, with each gene expressing at least four different alternatively spliced forms. Currently, it is unclear whether these different dynamin gene products perform distinct or redundant cellular functions. Therefore, the focus of this study was to identify additional spliced variants of dynamin from rat tissues and to define the distribution of the dynamin family members in a cultured rat epithelial cell model (Clone 9 cells). After long-distance reverse transcription (RT)-PCR of mRNA from different rat tissues, the full-length cDNAs encoding the different dynamin isoforms were sequenced and revealed four additional spliced variants for dynamin I and nine for dynamin III. Thus, in rat tissues there are a total of at least 25 different mRNAs produced from the three dynamin genes. Subsequently, we generated stably transfected Clone 9 cells expressing full-length cDNAs of six different spliced forms tagged with green fluorescent protein. Confocal or fluorescence microscopy of these transfected cells revealed that many of the dynamin proteins associate with distinct membrane compartments, which include clathrin-coated pits at the plasma membrane and the Golgi apparatus, and several undefined vesicle populations. These results indicate that the dynamin family is more extensive than was originally predicted and suggest that the different dynamin proteins are localized to distinct cytoplasmic or membrane compartments.
INTRODUCTION
Dynamin is a large GTPase that was first identified from mammalian brain based on its ability to bind microtubules in a nucleotide-dependent manner (Shpetner and Vallee, 1989). Dynamin’s GTPase can be stimulated in vitro through interaction with effector molecules at its proline-rich C-terminal domain. Insights into dynamin function emerged by demonstrating substantial identity with the Drosophila shibire (shits) gene product (Chen et al., 1991; van der Bliek and Meyerowitz, 1991), which expresses a temperature-sensitive mutation in the GTP-binding domain of the fly dynamin. At the restrictive temperature, cells exhibit a defect in endocytosis (Grigliatti et al., 1973; Kosaka and Ikeda, 1983a,b). Further work revealed that shits mutants are generally deficient at an early step in endocytosis, mainly, the ability to form coated vesicles at the plasma membrane (Kosaka and Ikeda, 1983b; Kessell et al., 1989; Masur et al., 1990). In support of these studies, transient overexpression of dominant-negative GTP-binding mutants of dynamin blocked clathrin-mediated endocytosis (Herskovits et al., 1993; van der Bliek et al., 1993; Damke et al., 1994, 1995). More recently, dynamin has been localized to clathrin-coated pits at the plasma membrane in cultured cells (Damke et al., 1994) and to the necks of membrane invaginations and clathrin-coated pits in an isolated synaptosomal preparation (Takei et al., 1995). For recent reviews on dynamin, see Robinson et al. (1994), De Camilli et al. (1995), Warnock and Schmid (1996), and Urrutia et al. (1997).
Numerous biochemical studies have suggested that the dynamins bind to several different cellular proteins (Vallee et al., 1995). Some of these binding proteins include microtubules (Maeda et al., 1992; Shpetner and Vallee, 1992), phospholipids (Tuma et al., 1993), and a subset of Src homology 3 domain-containing proteins (Gout et al., 1993; Herskovits et al., 1993; Scaife et al., 1994). From these numerous interactions it is possible that dynamin may perform a variety of different processes at distinct cellular locations. Indeed, dynamin has recently been implicated in several unique functions distinct from clathrin-mediated endocytosis at the plasma membrane. These functions include internalization of caveolae (Schnitzer et al., 1996; Henley et al., 1998; Oh et al., 1998), sorting of toxins from internal membrane compartments (Llorente et al., 1998), and formation of nascent secretory vesicles from the trans-Golgi network (Henley and McNiven, 1996; Maier et al., 1996; Jones et al., 1998). Therefore, it is tempting to predict that different members of the dynamin family are targeted to various cellular compartments to perform a similar function, mainly, the liberation of newly formed coated vesicles.
It has become increasingly clear that the dynamin family of proteins is larger than once predicted. Subsequent to its initial identification from mammalian brain, dynamin was thought to be expressed only in neuronal tissues (Scaife and Margolis, 1990; Nakata et al., 1991). However, recent identification and cloning of two additional dynamin genes from epithelial tissue (Nakata et al., 1993; Cook et al., 1994; Sontag et al., 1994) have demonstrated that dynamin isoforms are expressed in all cells. Conventional brain dynamin, dynamin I (Dyn 1),1 is indeed neuronal specific (Nakata et al., 1991, 1993; Cook et al., 1994; Sontag et al., 1994), dynamin II (Dyn 2) is ubiquitously expressed (Cook et al., 1994; Sontag et al., 1994), and dynamin III (Dyn 3) is restricted to testis, brain, and lung (Nakata et al., 1993; Cook et al., 1996; reviewed by Robinson et al., 1994; Urrutia et al., 1997). From these studies it is known that each dynamin has at least four alternatively spliced variants, which may result in the expression of 12 different dynamin proteins in brain alone. Using long-distance RT-PCR from mRNA of various rat tissues, we have conducted an extensive characterization of the expression of the dynamin isoforms in different tissue types and have identified >25 different spliced variants of the three genes. In an attempt to define the subcellular localization of the different dynamin family members, we have coupled green fluorescent protein (GFP) to six distinct dynamin cDNAs (two splice variants for each of the three genes) for expression in a cultured, nonpolarized hepatocyte cell line (Clone 9). To ensure that the GFP tag did not alter the targeting or distribution of a specific dynamin, each dynamin protein examined was expressed with GFP at either the N- or C-terminal end, or expressed without a GFP tag. Subsequently, these transfected cells were stained with isoform-specific dynamin antibodies or membrane compartment-specific antibodies and analyzed by confocal microscopy. Interestingly, we found that the six different spliced forms of dynamin-GFP fusion proteins were localized to several distinct membrane or cytoplasmic compartments. These various locations include clathrin-coated buds at the plasma membrane and/or the Golgi apparatus, the cytosol, and various populations of presently unidentified membrane vesicles. These findings provide the first evidence that the dynamin gene products are differentially distributed in mammalian cells where they may participate in various membrane trafficking events.
MATERIALS AND METHODS
Tissues used for this study were harvested from 90- to 100-g male Sprague Dawley rats (Harlan, Madison, WI). Miniprep Express and Luria-Bertani medium were from BIO 101 (Vista, CA). Restriction enzymes were from Boehringer Mannheim (Indianapolis, IN) and Life Technologies (Gaithersburg, MD). 1-kb and 1-kb plus DNA ladder was from Life Technologies (Grand Island, NY). A polyclonal antibody to GFP was purchased from Clontech (Palo Alto, CA), and a monoclonal antibody to GFP was a gift from Boehringer Mannheim. An anti-clathrin monoclonal antibody (X22) was collected from the supernatant of the X22 hybridoma cell line (American Type Culture Collection [ATCC], Rockville, MD). Secondary antibodies used were Texas Red (TR)- or FITC-conjugated goat anti-rabbit or goat anti-mouse and TR-dextran for immunocytochemical staining (Molecular Probes, Eugene, OR). HRP-conjugated goat anti-rabbit and goat anti-mouse (Bio-source International, Camarillo, CA) were used for Western blot analysis. The Golgi-specific monoclonal antibody TGN38 was a gift from Dr. K.E. Howell (University of Colorado School of Medicine, Denver, CO). The anti-Dyn 2 antibody was raised against a peptide sequence representing amino acids 761–785 of Dyn 2, which is unique to the four Dyn 2 isoforms, sharing only 20–26% identity with the other dynamin gene products (Henley et al., 1998). The anti-Pan MC65 antibody was raised against a conserved region of dynamin (Dyn 1 and Dyn 2). It was generated in rabbits and affinity purified as described previously (Henley et al., 1998). The anti-Dyn 3 antibody was raised against a C-terminal peptide, RLTLSAPLPRPASSRGPAPAIPSPGPHS, and recognizes Dyn 3 specifically by immunoblot and immunofluorescence analysis. All other chemicals and reagents unless otherwise stated were from Sigma (St. Louis, MO).
Isolation of Total RNA and Synthesis of First-Strand cDNA
Total RNAs were extracted from brain, heart, kidney, liver, lung, pancreas, spleen, and testis. Rat tissues were removed, quickly frozen in liquid nitrogen, and ground in 7 ml of 4 M guanidinium thiocyanate (5 Prime-3 Prime, Boulder, CO) in a tissue homogenizer. Homogenates were passed five or six times through a 26-gauge 1/2 A needle and then centrifuged for 15 min at 12,580 × g, 20°C, in a Beckman centrifuge (Beckman Instruments, Palo Alto, CA). The supernatants were layered over 4 ml of a 5.7 M CsCl cushion in polyallomer tubes. The samples were centrifuged for 22 h at 35,000 × g, 20°C, in a Beckman SW41 rotor. The RNA pellets were resuspended in 0.3 M sodium acetate (pH 6.0). Total RNAs were also extracted from Clone 9, PC-12, and rat dorsal root ganglion (DRG) cells (pure and mixed) by the mRNA single-step method (Chomczynski and Sacchi, 1987). First-strand cDNA synthesis by RT was performed using a SuperScript II RNase H− reverse transcriptase kit (Life Technologies). RT was used to transcribe 1–5 μg of total RNA with 1 μl of 500 μg/ml oligo(dT)12–18 primer. The reaction conditions were according to the manufacturer.
PCR
Specific oligonucleotide primers (synthesized on an Applied Biosystems, Foster City, CA, 394 DNA/RNA synthesizer) for the three dynamin isoforms were designed (MacVector, New Haven, CT) using the different dynamin isoform cDNA sequences from GenBank (Accession numbers X54531, L25605, and D14076). Full-length cDNAs encoding the different dynamin isoforms were amplified by long-distance RT-PCR. The primers’ design is shown in Figure 1A. The 5′ PCR primers were complementary to the initiation sequence. The 3′ PCR primers included the corresponding stop codon for each dynamin-coding region, thus discriminating the two alternative C termini of Dyn 1 and Dyn 3. The PCRs were performed using the XL PCR kit (Perkin Elmer-Cetus, Branchburg, NJ). The PCR cycle conditions were as follows: 28 cycles of 94°C for 1 min, 68°C (for Dyn 1), 65°C (for Dyn 2), 60°C (for Dyn 3) for 5 min, and finally 72°C for 7 min. The reaction products were analyzed by agarose gel electrophoresis (Figure 1B). Other specific oligonucleotide primers were designed to amplify partial fragments of dynamin sequences from DRG cells (Dyn 1 5′ primer [5′-ACACGCTGCCGGGACTTC-3′], 3′ primer [5′-CAGCTGCCGGTAATCCTT-3′]; Dyn 2 5′ primer [5′-GAAGAGGGCCATACC-3′], 3′ primer [5′-AGTTGCGGATGGTCTC-3′]; Dyn 3 5′ primer [5′-ACTTCCCCAGACTTTGTG-3′], 3′ primer [5′-ACGTCCCGGACTTTCAGG-3′]). The PCR reactions were performed in 20 mM Tris-HCl (pH 8.4), 50 mM KCl, 5 mM MgCl2, and 2.5 U of Taq DNA polymerase using 30 PCR cycles (94°C for 1 min, 60°C for 2 min, and 72°C for 1 min). After PCR amplification, the reaction products were analyzed by agarose gel electrophoresis (see Figure 3B). The PCR fragments were ligated into the eukaryotic expression vector pCR3.1 (Invitrogen, Carlsbad, CA).
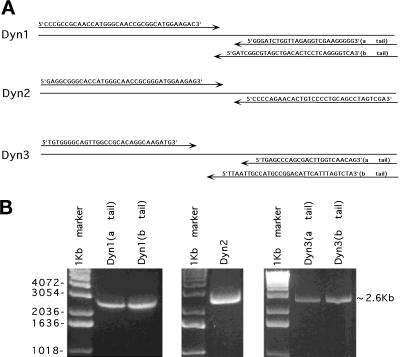
Figure 1.
Amplification of three different dynamin isoforms (Dyn 1–3) from rat brain by long-distance RT-PCR. cDNA was synthesized from 10 μg of total RNA from rat brain using an oligo(dT)n primer. Amplification of the genes encoding full-length dynamin isoforms was performed using long-distance RT-PCR. Specific primers (A) were designed by MacVector and using the dynamin cDNA sequences from GenBank. The 5′ PCR primers were complementary to the initiation sequence. The 3′ PCR primers included the corresponding stop codon for each dynamin-coding region, thus discriminating the two alternative C termini of Dyn 1 and Dyn 3. After long-distance PCR, the reaction products were analyzed by agarose gel electrophoresis (B). Approximately 2.6-kb bands represent each dynamin isoform.
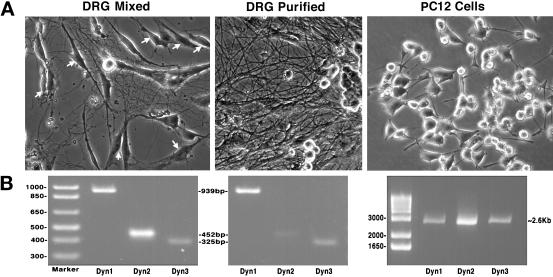
Figure 3.
RT-PCR detects only Dyn 1 and Dyn 3 transcripts in purified populations of rat sensory neurons (DRGs). (A) Phase microscopic images of isolated neuronal cell populations used to test for the expression of all three dynamin genes. Cell types examined: a mixed culture of rat sensory neurons (DRGs) that includes glial cells and fibroblasts (arrows), a highly enriched culture of rat DRGs in which the supportive cells have been selectively eliminated, and a widely used, rat neoplastic adrenal cell line (PC-12). (B) RT-PCR of total mRNA isolated from each population of cells depicted in A. All three dynamin gene products were detected in the mixed population of DRGs. In contrast, removal of the supportive, nonneuronal cells reduced the amount of Dyn 2 to nearly undetectable levels, but the levels of Dyn 1 and Dyn 3 remained unchanged. PC-12 cells expressed nearly equal amounts of all three dynamin transcripts.
cDNA Cloning and Dynamin-GFP Expression Vector
For subcloning of the full-length cDNA corresponding to Dyn 1, Dyn 2, and Dyn 3, reamplification of the different isoforms was performed using PCR. The 5′ and 3′ PCR primers for Dyn 1 and Dyn 2 were designed to contain restriction sites HindIII and EcoRI, respectively. For Dyn 3 constructs, EcoRI sites were designed at the 5′ end, and BamHI sites were designed at the 3′ end. The dynamin inserts from the pCR3.1 constructs were excised by digestion with the corresponding enzymes and subcloned into the expression vectors pEGFP-N1 and pEGFP-C1 (Clontech, Palo Alto, CA). The constructs inserted into pEGFP-N1 had no intervening stop codons. All the DNA constructs were verified by restriction enzyme analysis and sequencing (The Mayo Molecular Biology Core [ABI PRISM 377 DNA sequencer, Perkin Elmer-Cetus]). Sequences were analyzed using DNA* analysis software (DNA star, Madison, WI).
Cell Culture and Transfection
Rat DRG cells were isolated and cultured as published elsewhere (Conti et al., 1997). These preparations provided mixed cultures of DRGs and surrounding supportive cells. To obtain highly enriched (95%) DRGs without glial cells or fibroblasts, cultures were treated with antimetabolites (5 ng/ml NGF, 10% calf bovine serum, 10−5 M fluorodeoxyuridine, and 10−5 M uridine) for 5 d, and then the medium was replaced by medium without antimetabolites. This yielded a stable population of neurons without supporting Schwann cells or fibroblasts (Wood et al., 1980). PC-12 cells, a pheochromocytoma cell line from rat adrenal glands (ATCC CRL-1721), were maintained in 82.5% F-12K medium with 2 mM l-glutamine adjusted to contain 1.5 g/l sodium bicarbonate, 15% horse serum, 2.5% fetal bovine serum, 100 U/ml penicillin, and 100 μg/ml streptomycin (Life Technologies) in 5% CO2 and 95% air at 37°C. Cells were cultured in T-75 flasks (Fisher Scientific, Pittsburgh, PA). Clone 9 cells, an epithelial cell line isolated from normal rat liver (ATCC CRL-1439), were maintained in Ham’s F-12K medium supplemented with 10% fetal bovine serum (Life Technologies), 100 U/ml penicillin, and 100 μg/ml streptomycin in 5% CO2 and 95% air at 37°C. Cells were cultured in T-75 flasks (Fisher Scientific) and on 22-mm coverslips for transfections and immunocytochemistry, respectively. Plasmid DNA containing dynamin construct was purified by equilibrium centrifugation in CsCl-ethidium bromide gradients (Sambrook et al., 1989). Transfection of cells was performed by electroporation in a Gene Pulser II system (Bio-Rad, Hercules, CA). Confluent cells were trypsinized (1× trypsin-EDTA, 0.25% Trypsin, 1 mM 4Na-EDTA; Life Technologies), resuspended in 400 μl of PBS (134 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 1.8 mM KH2PO4, pH 7.4) at ∼3 × 106 cells/ml, and transferred to a sterile electroporation cuvette (0.4 cm) with 20–50 μg of plasmid DNA. After a 20 min incubation on ice, cells were electroporated at 0.3 kV, 250 μF, and subsequently transferred to 100-mm tissue culture dishes. After 24 h the cells were placed under selective pressure with a medium containing Geneticin (G418, 400 μg/ml; Life Technologies) for 10–15 d. Resistant clones were isolated using cloning cylinders (Bellco Glass, Vineland, NJ) and transferred for expansion and analysis.
Protein Extraction
Stably transfected Clone 9 cells were harvested for biochemical analysis using standard procedures. Cells from one confluent 100-mm Petri dish were rinsed two or three times with cold PBS. Cells were scraped from the dishes with a cell lifter (Costar, Cambridge, MA) and homogenized over 10 min in radioimmunoprecipitation assay buffer (50 mM Tris, pH 8.1, 150 mM NaCl, 0.5% deoxycholate, 1% NP-40, 0.1% SDS) followed by a protease inhibitor mixture consisting of 1 mM phenylmethylsulfonyl fluoride, 20 μg/ml soybean trypsin inhibitor, 20 μg/ml leupeptin, and a 1000-fold dilution of a nuclease solution (20 mg/ml RNase A and 10 mg/ml DNase I). Lysates were then clarified by centrifugation at 16,000 × g for 10 min. All procedures were carried out at 4°C. Proteins were solubilized by boiling in 4× Laemmli sample buffer for 3 min.
SDS-PAGE and Immunoblotting
Protein concentrations were determined with bicinchonic acid according to the manufacturer (Pierce, Rockford, IL), using BSA as a standard. Cellular proteins were separated by continuous SDS-PAGE under reducing conditions (Laemmli, 1970) on 7.5% polyacrylamide gels using a bisacrylamide/acrylamide ratio of 1:37.5. Protein bands were electrophoretically transferred to polyvinyldifluoride membranes (Millipore, Bedford, MA) for Western blotting (Towbin et al., 1979). Membranes containing transferred proteins were blocked in PBS-Towbin (150 mM NaCl, 10 mM NaH2PO4, pH 7.2) containing 5% milk and washed in PBS-Towbin before incubation with primary antibodies. Immunoreactive bands were detected with an appropriate secondary antibody conjugated to HRP. After each step, the membranes were washed with PBS-Towbin containing 0.3% Tween 20. Membranes were developed with enhanced chemiluminescence (Amersham, Arlington Heights, IL) and were exposed to autoradiographic film (Eastman Kodak, Rochester, NY) to detect HRP. To estimate molecular weights, bands were compared with the following prestained standards: myosin, β-galactosidase, BSA, and ovalbumin (Bio-Rad).
Immunofluorescence and Confocal Microscopy
For immunofluorescence and confocal microscopy, cells were grown on cover glasses for 1–2 d, rinsed with Dulbecco’s PBS (D-PBS; 8.1 mM Na2HPO4, 1.2 mM KH2PO4, pH 7.2, 138 mM NaCl, 2.7 mM KCl, 0.9 mM CaCl2, 0.5 mM MgCl2) at room temperature and fixed for 20 min with 2.5% formaldehyde in piperazine-N,N′-bis(2-ethanesulfonic acid) buffer [0.1 M piperazine-N,N′-bis(2-ethanesulfonic acid), pH 6.95, 3 mM MgSO4, 1 mM EGTA]. After rinsing with D-PBS, cells were permeabilized with 0.1% Triton X-100 in D-PBS for 2 min, rinsed with D-PBS, and incubated in blocking buffer (5% normal goat serum and 5% glycerol in D-PBS) for 1 h at 37°C. Cells were incubated in primary antibodies (1–40 μg/ml) for 2 h at 37°C and rinsed repeatedly with D-PBS before incubating in the appropriate fluorescein-labeled secondary antibody (1–5 μg/ml) for 1 h at 37°C. Cells were then washed extensively with D-PBS, rinsed briefly with distilled water, and mounted on a glass slide in mounting reagent (ProLong; Molecular Probes). Stably transfected dynamin-GFP cells were incubated in medium containing 200 mM TR-conjugated dextran (molecular weight, 3000; Molecular Probes) to label late endosomes and lysosomes for 1, 2, 4, 8, or 12 h at 37°C. The cells were rinsed with medium and with two changes of D-PBS containing 0.5 M NaCl. After rinsing with four changes of HBSS, the cells were fixed by the same procedure. Cells were viewed with an Axiovert 35 epifluorescence microscope (Carl Zeiss, Thornwood, NY) equipped with a 100-W mercury lamp using a 100× objective lens (Zeiss Plan-Neofluar, 1.30) and a confocal microscope (Zeiss LSM-310 equipped with an argon–krypton laser) using a 100× objective lens; excitations were at 488/568 nm. Images were acquired on the microscope at 1024 × 1024 resolution over a 32-s exposure period. Contrast and intensity for each image were manipulated uniformly using Adobe (Mountain View, CA) Photoshop software.
RESULTS
Expression of Multiple Dynamins in Different Tissues
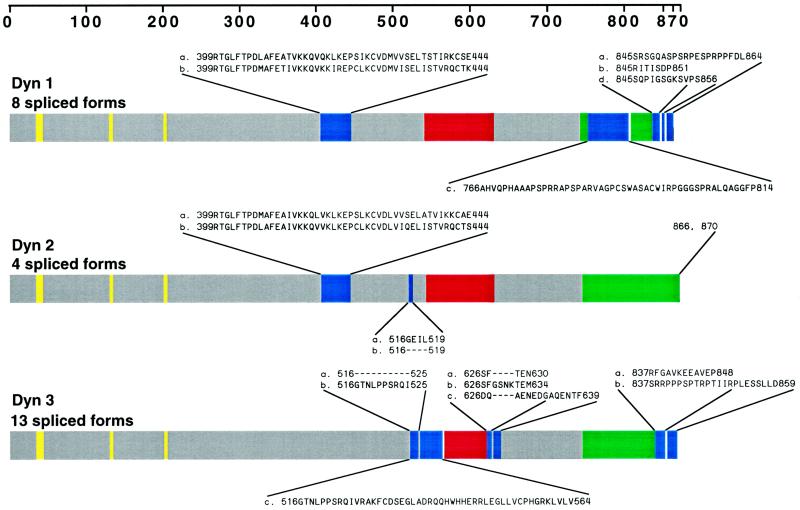
To define the cellular localization of dynamin family members, we first attempted to establish the number of distinct spliced variants produced from the three known dynamin genes. Full-length cDNAs for the different dynamin isoforms were amplified by long-distance RT-PCR with oligonucleotides complementary to dynamin sequences (Nakata et al., 1991, 1993; Robinson et al., 1993; Cook et al., 1994, 1996; Sontag et al., 1994). Figure 1A shows the specific oligodeoxynucleotide primers used in this study. The 5′ PCR primers were complementary to the initiation sequence, and the 3′ PCR primers included the two alternative C termini of Dyn 1 and Dyn 3. Figure 1B shows these RT-PCR products by agarose gel electrophoresis. The sequencing results of several cDNA clones encoding different dynamin isoforms revealed four new spliced forms for Dyn 1 and nine new spliced forms for Dyn 3 (Figure 2). We have denoted clusters of splicing regions for each dynamin isoform with two splicing regions for Dyn 1 and Dyn 2 and three splicing regions for Dyn 3. As shown previously (Robinson et al., 1993), the first splicing region of Dyn 1 contains either one of two sequences (a and b forms) of identical size but distinct nucleotide sequences. In addition, we have identified two new spliced variants in the second splicing region (tail region) making at least four distinct tails for Dyn 1 (Figure 2). These consist of an a tail of 20 amino acids, a b tail of 7 amino acids as reported by Robinson et al., 1993, a c tail of 49 amino acids, and a d tail of 12 amino acids (Figure 2). It is of interest that the c tail of Dyn 1 has a premature stop codon attributable to a 26-nucleotide deletion, and the d tail has a 4-nucleotide insertion. Therefore, Dyn 1 has at least eight spliced variants which we refer to as aa, ba, ab, bb, ac, bc, ad, and bd. For Dyn 3 we have identified an additional region of alternative splicing, making three distinct splicing regions. We now know that within the first splicing region there is a 10-amino-acid insertion (b form) and an additional variant encoding 48 amino acids (c form) with a premature stop codon attributable to a 46-nucleotide insertion (Figure 2). In addition, the novel second splicing region that we report here has three splice variants, a–c. The b form has an extra 4-amino-acid insertion, and c has 14 amino acids. The b form at this second splicing region has a 12-nucleotide insertion, whereas c and a are identical in size but distinct in nucleotide sequence. Interestingly, the c spliced form at this region is lung specific. Therefore, Dyn 3 has at least 13 spliced variants (aaa, aba, bba, baa, bbb, bab, aab, abb, aca, acb, bca, bcb, and c). Despite the identification of numerous additional spliced variants for Dyn 1 and Dyn 3, we have not identified any variants for Dyn 2 in addition to those originally reported by us and others (Cook et al., 1994; Robinson et al., 1994; Sontag et al., 1994).
Figure 2.
More than 25 distinct dynamin mRNAs are expressed in rat tissues: schematic illustration of the dynamin family of proteins. The three known dynamin gene products are diagrammed, and the corresponding alternatively spliced sites are indicated. Amino acid sequences of 12 of the spliced variants were obtained from previously published studies (Nakata et al., 1991, 1993; Robinson et al., 1993; Cook et al., 1994, 1996; Sontag et al., 1994) and were published previously in two separate reviews (Robinson et al., 1994; Urrutia et al., 1997). Sequence information for the additional 13 inserts or substitutions was obtained in this study by long-distance RT-PCR of total mRNA from several rat tissues. We have denoted the clustered sites of alternative splicing within each transcript as “splicing regions,” which are blue. Dyn 1 and Dyn 2 each have two splicing regions, whereas Dyn 3 has three splicing regions. Note that Dyn 1 and Dyn 3 each have a spliced insert encoding premature terminations in splicing regions 2 or 1 and 2, respectively. Yellow represents the GTP-binding sites; red and green represent the pleckstrin homology (PH) and proline-rich (PR) domains, respectively.
Based on the PCR studies described above, at least 25 different spliced variants of the three currently identified dynamin genes are expressed in mammalian tissues. Table 1 summarizes these findings by listing the different dynamin forms found in seven different rat tissues, one type of epithelial cell (Clone 9), and three different populations of neuronal cells. There are 21 of these variants found in rat brain alone, which appears to be the only tissue expressing all three of the dynamin genes and the only tissue that expresses Dyn 1. Furthermore, the expression of Dyn 3 is highly variable and is dependent on the specific spliced variant and the tissue examined. For example, only select forms of Dyn 3 are expressed in brain, heart, lung, and testis. It is also interesting to note that lung expresses several unique forms of Dyn 3 not found in any other tissue examined and that Dyn 3 is expressed in heart (our unpublished results). Most surprising is the observation that PCR methods do not detect Dyn 2 in total mRNA prepared from highly enriched cultures of rat peripheral sensory neurons (DRGs) (Figure 3). We were able to detect all three dynamin gene products from total mRNA from rat brain (Figure 1), cultured PC-12 cells, and mixed cultures of DRGs (Figure 3), which contain glial cells and fibroblasts. However, in a purified population of DRGs, Dyn 2 PCR products were greatly reduced to near undetectable levels, whereas products for Dyn 1 and Dyn 3 remained unchanged. This suggests, but does not prove, that the Dyn 2 is expressed by the supporting cells and not the neurons. Future detailed studies applying multiple techniques to distinct neuronal and glial cell populations will be needed to clearly define the expression of the different dynamin family members in mammalian brain.
Table 1.
Detection of dynamin isoforms in different rat tissues by PCR
| Tissue/Cell | Br | H | K | Li | Lg | P | T | Clone 9 | DRG (mixed) | DRG (pure) | PC-12 |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Dyn1 | |||||||||||
| aa | + | − | − | − | − | − | − | − | + | + | + |
| ab | + | − | − | − | − | − | − | − | + | + | + |
| ac | + | − | − | − | − | − | − | − | + | + | + |
| ad | + | − | − | − | − | − | − | − | + | + | + |
| ba | + | − | − | − | − | − | − | − | + | + | + |
| bb | + | − | − | − | − | − | − | − | + | + | + |
| bc | + | − | − | − | − | − | − | − | + | + | + |
| bd | + | − | − | − | − | − | − | − | + | + | + |
| Dyn2 | |||||||||||
| aa | + | + | + | + | + | + | + | + | + | − | + |
| ab | + | + | + | + | + | + | + | + | + | − | + |
| ba | + | + | + | + | + | + | + | + | + | − | + |
| bb | + | + | + | + | + | + | + | + | + | − | + |
| Dyn3 | |||||||||||
| aaa | + | + | − | − | + | − | + | − | + | + | + |
| aab | + | + | − | − | + | − | + | − | + | + | + |
| aba | + | + | − | − | + | − | − | − | + | + | + |
| abb | + | + | − | − | + | − | − | − | + | + | + |
| baa | + | + | − | − | + | − | + | − | + | + | + |
| bab | + | + | − | − | + | − | + | − | + | + | + |
| bbb | + | + | − | − | + | − | − | − | + | + | + |
| bba | + | + | − | − | + | − | − | − | + | + | + |
| c | + | + | − | − | + | − | + | − | + | + | + |
| aca | − | − | − | − | + | − | − | − | − | − | − |
| acb | − | − | − | − | + | − | − | − | − | − | − |
| bca | − | − | − | − | + | − | − | − | − | − | − |
| bcb | − | − | − | − | + | − | − | − | − | − | − |
Establishment of Stable Epithelial Cell Lines Expressing Six Different Dynamin-GFP Fusion Proteins
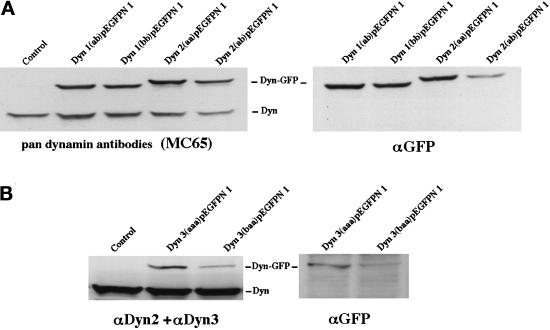
Upon detection of the additional alternatively spliced dynamin transcripts described above, our goal was to test whether the corresponding dynamin proteins occupy distinct cytoplasmic locations. In other words, might the spliced inserts of the different dynamin variants provide targeting, positional, or functional information, which could direct these proteins to distinct cytoplasmic locations? To this end we have stably transfected a nonpolarized rat hepatocyte cell line (Clone 9), which normally expresses only Dyn 2, with full-length cDNAs encoding six different dynamin spliced forms [Dyn 1(ab) and Dyn 1(bb), Dyn 2(aa) and Dyn 2(ab), and Dyn 3(aaa) and Dyn 3(baa)]. These cDNAs were tagged with GFP at either the N- or C-terminal end. As an additional control we also generated stable Clone 9 cells transfected with the same cDNAs but subcloned into a mammalian expression vector (pCR3.1) without the GFP tag (see MATERIALS AND METHODS). To confirm that transfected Clone 9 cells expressed Dyn-GFP, total proteins from homogenates of the six different dynamin-expressing cell lines were separated by SDS-PAGE and subjected to immunoblot analysis with a Pan-dynamin antibody (MC65), isoform-specific antibodies for Dyn 2 and Dyn 3, or a GFP antibody. As shown in Figure 4, immunoblotting of control cells with the Pan-dynamin antibody revealed a single band of the appropriate molecular mass (100 kDa). In contrast, immunoblots of cells expressing the four different Dyn 1- and Dyn 2-GFP fusion proteins displayed a conventional dy-namin band and one of greater molecular mass. Subsequent blotting of these cell homogenates with a GFP-specific antibody confirmed that the higher-molecular-mass bands represented the dynamin-GFP fusion proteins. Surprisingly, the Pan-dynamin antibody used to detect the Dyn 1 and Dyn 2 fusion proteins did not recognize either of the expressed Dyn 3 variants. It was unexpected that a Pan-antibody made to a peptide conserved in all three of the dynamin isoforms did not detect an unfolded protein immobilized on polyvinyldifluoride membranes. However, upon closer inspection, we noted that of the 23 amino acids that constituted the MC65 Pan-antibody peptide antigen, there were no mismatched residues with Dyn 2 but two and five amino acids mismatched for Dyn 1 and Dyn 3, respectively. As a result, homogenates of Dyn 3-GFP-expressing cells were immunoblotted with a mixture of isoform-specific antibodies to Dyn 2 and Dyn 3 (Figure 4B). These antibodies detected the conventional dynamin band and the higher-molecular-mass dynamin-GFP bands as confirmed with the GFP antibody. These manipulations confirmed the expression of the different dynamin proteins in the Clone 9 cells and, importantly, demonstrated that the levels of Dyn 1-GFP and Dyn 2-GFP expression did not significantly exceed that of the endogenous Dyn 2 normally expressed by these cells.
Figure 4.
Expression of six distinct dynamin-GFP constructs in stably transfected epithelial cells. To test whether transfected Clone 9 cells were expressing dynamin-GFP, total protein from homogenates of the six different dynamin-expressing cell lines was separated by SDS-PAGE and subjected to immunoblot analysis with the Pan-dynamin antibody (MC65), isoform-specific antibodies for Dyn 2 and Dyn 3, or a GFP antibody. (A) Immunoblots of homogenates from cells expressing two spliced variants of either Dyn 1-GFP or Dyn 2-GFP. The Pan-dynamin antibody (MC65) recognized both the endogenous dynamin protein band at 100 kDa and the dynamin fusion protein band at 120 kDa. As expected, the higher-molecular-mass band was only seen when blotted with the anti-GFP antibody. For immunoblot analysis of Dyn 3-GFP-expressing cells (B), a combination of isoform-specific antibodies to Dyn 2 and Dyn 3 was used, because the Dyn 3-GFP fusion protein was not recognized by the Pan-MC65 antibody. With these antibodies both the endogenous dynamin and expressed Dyn 3-GFP were seen.
Distinct Cellular Distributions of the Dynamin Isoforms
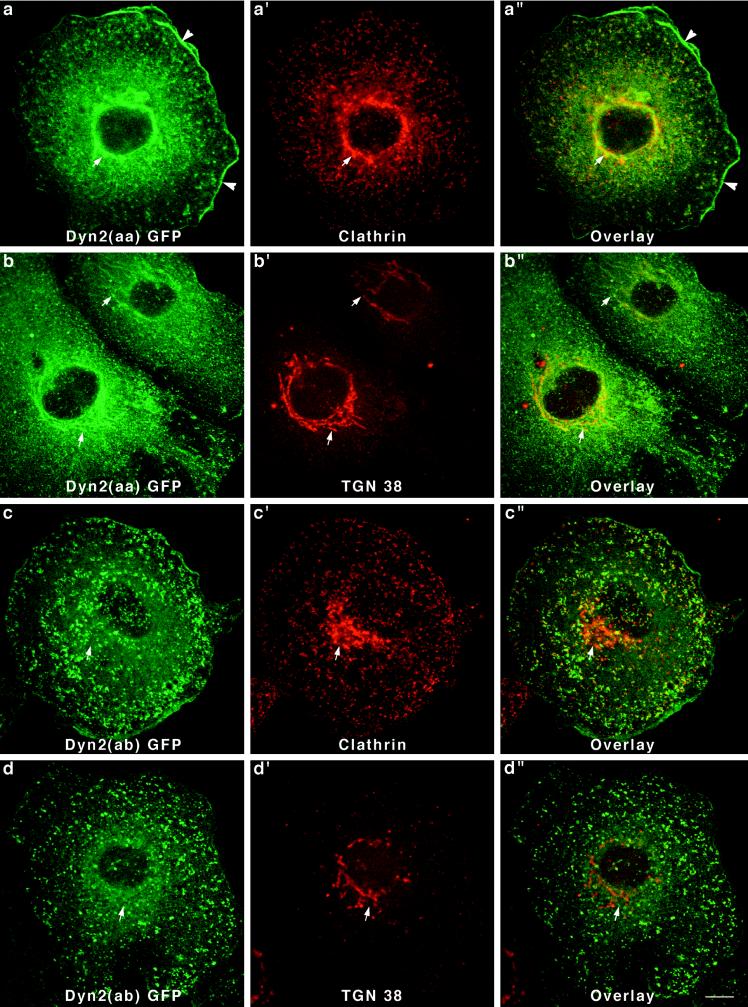
After establishing six Clone 9 cell lines, each expressing a different dynamin-GFP fusion protein, we next tested whether the cytoplasmic distribution of these dynamins was similar or distinct. After fixation and permeabilization, cells were mounted on slides and viewed by confocal microscopy. Cells transfected with a pEGFP-N1 or -C1 vector alone displayed a diffuse fluorescence throughout the cytoplasm and nucleus (our unpublished results). In contrast, cells expressing five of the six different dynamin-GFP proteins revealed a striking localization to vesicular membrane compartments. Perhaps the most dramatic differences in the cellular distribution of the dynamins were displayed by cells expressing the two forms of Dyn 2 (Figure 5). Because Dyn 2 is the only dynamin identified in Clone 9 cells to date, the localization of these endogenous proteins may also be the most relevant. As shown in Figure 5, a and b, cells expressing Dyn 2(aa)-GFP exhibited a prominent labeling of numerous punctate spots in close association with either the plasma membrane or reticular tubules about the nucleus. When these cells were double stained with antibodies to the clathrin heavy chain, most of the Dyn 2-GFP structures were labeled. From this we concluded that Dyn 2(aa) was sequestered to clathrin-coated pits and vesicles at the plasma membrane and the Golgi apparatus. This same clonal cell line was also used in an earlier report demonstrating that dynamin participates in Golgi function (Jones et al., 1998). To confirm that this perinuclear structure was indeed the Golgi, cells were double labeled with an antibody specific for a trans-Golgi–specific protein of unknown function called TGN38 (Crosby et al., 1992; Ponnambalam et al., 1996). As predicted, the TGN38 antibody staining of the perinuclear Dyn 2-GFP-positive tubules was nearly complete (Jones et al., 1998). Finally, in addition to the association of Dyn 2(aa)-GFP with clathrin-associated vesicles at the Golgi and the plasma membrane, we also observed a prominent accumulation of the fusion protein in cortical membrane ruffles and lamellapodia. This localization was not seen in any of the cells expressing the other dynamin-GFP proteins.
Figure 5.
Two variants of Dyn 2-GFP show dramatic differences in affinity for the Golgi apparatus. Double-immunofluorescence staining of cells expressing Dyn 2(aa)-GFP with antibodies to clathrin (a–a") or TGN38 (b–b") revealed that Dyn 2(aa)-GFP has a strong affinity for membrane tubules of the Golgi apparatus (arrows). Interestingly, Dyn 2(aa) was also localized in the cortical ruffles of transfected cells (a and a", arrowheads). In sharp contrast to the Dyn 2(aa) spliced variant that associated with clathrin at both the plasma membrane and the Golgi apparatus, Dyn 2(ab)-GFP, a form different by only a four-amino-acid deletion in the second splicing region, localized to clathrin-coated pits at the plasma membrane only. No affinity for the Golgi apparatus (arrows) in cells stained with a clathrin antibody (c–c") or a TGN38 antibody (d–d") was seen. Bar, 10 μm.
To test whether alternatively spliced variants expressed from the same dynamin gene might exhibit a distinct cytoplasmic localization in cells, we next viewed Clone 9 cells expressing Dyn 2(ab)-GFP. This spliced form of Dyn 2 makes a particularly interesting comparison with the Dyn 2(aa)-GFP shown in Figure 5. Dyn 2(ab) form is missing four amino acids (GEIL) at the second splicing region b. Therefore, we were surprised to find that cells expressing the modestly shorter Dyn 2 spliced form, Dyn 2(ab), revealed a distinctly different staining pattern. As shown in Figure 5, c and d, Dyn 2(ab)-GFP is localized to numerous punctate spots on the plasma membrane. Many, but not all, of these foci stain positive for clathrin (Figure 5, c′ and c"), although there are numerous clathrin pits that do not colocalize with the Dyn 2(ab)-GFP. Most striking was that none of the dynamin-GFP in these transfected cells appeared to associate with the Golgi apparatus, as confirmed by staining with antibodies to clathrin (Figure 5c′) and TGN38 (Figure 5d′). Thus, the insertion of just four amino acids appears to have profound effects on whether Dyn 2 is, or not, associated with the Golgi apparatus.
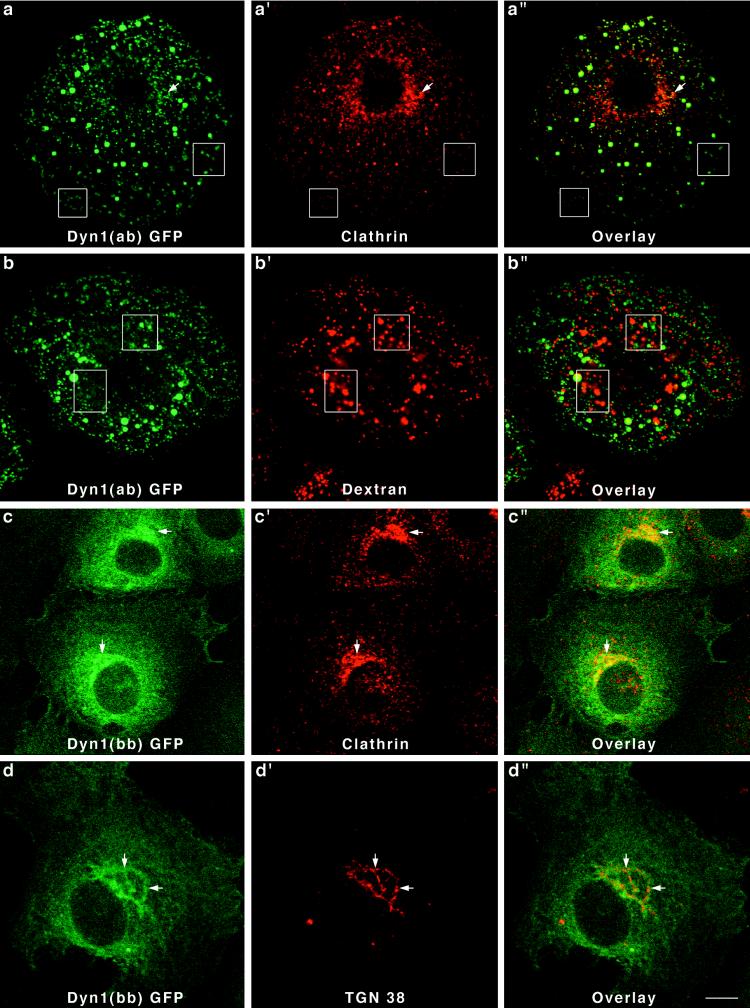
Like the Dyn 2 spliced variants, two different forms of Dyn 1 expressed in Clone 9 cells also localized to strikingly different cytoplasmic structures. In this study we chose to express Dyn 1(ab)-GFP and Dyn 1(bb)-GFP. These spliced variants are identical except for a 46-amino-acid substitution between amino acids 399 and 444 (Figure 2). Only 14 of the 46 amino acids constituting this insert are different between the two Dyn 1 forms. Expression of Dyn 1(ab)-GFP in Clone 9 cells resulted in a prominent labeling of numerous punctate foci of various sizes distributed along the plasma membrane and throughout the cytoplasm (Figure 6, a and b). Despite the vast difference in size between the different dynamin structures, most appeared to costain with clathrin antibodies. Few if any GFP-positive structures could be seen in the perinuclear Golgi region. It is of interest that these larger punctate foci were clathrin positive. Such large clathrin structures are normally not observed in these cells except for ones expressing this particular dynamin form. Thus, the expression of a foreign dynamin form in these cells, Dyn 1(ab), appears to have marked effects on clathrin distribution. To test whether the larger GFP-positive structures might be lysosomal organelles, transfected cells were incubated with TR-dextran for 1–12 h before fixation and fluorescence microscopy. As shown in Figure 6, b′ and b" (4 h), none of the large Dyn 1(ab)-GFP structures appeared to contain endocytosed dextran. In contrast to the Dyn 1(ab)-GFP form, Dyn 1(bb)-GFP exhibited a diffuse cytoplasmic distribution in transfected cells with some localization to the Golgi apparatus (Figure 6, arrows) as confirmed by double-immunofluorescence staining for clathrin (Figure 6c–c") and the trans-Golgi protein TGN38 (Figure 6d–d"). Thus, like the Dyn 2 spliced variants, both of the Dyn 1 proteins expressed, which differ by only 14 of 851 amino acids, reside on drastically different cytoplasmic compartments.
Figure 6.
Different distributions for two forms of Dyn 1-GFP in Clone 9 cells. Stably transfected Clone 9 cells expressing Dyn 1(ab)-GFP were immunostained with antibodies to clathrin (a–a") or labeled with internalized fluorescent dextran (b–b") to identify the endosomal and lysosomal compartments. Whereas the Dyn 1(ab)-GFP appeared to colocalize precisely with clathrin-coated pits at the plasma membrane (boxed regions), there was no colocalization with clathrin at the Golgi apparatus. Large vesiclular structures associated with Dyn 1-GFP patches were seen in these transfected cells and had no colocalization with endocytosed dextran (b–b") suggesting that these structures do not represent a lysosomal compartment. In contrast to Dyn 1(ab)-GFP, Dyn 1(bb)-GFP exhibited a diffuse cytoplasmic distribution in transfected cells with some localization to the Golgi apparatus (arrows), as confirmed by double-immunofluorescence staining for clathrin (c–c") and the trans-Golgi protein TGN38 (d–d"). Bar, 10 μm.
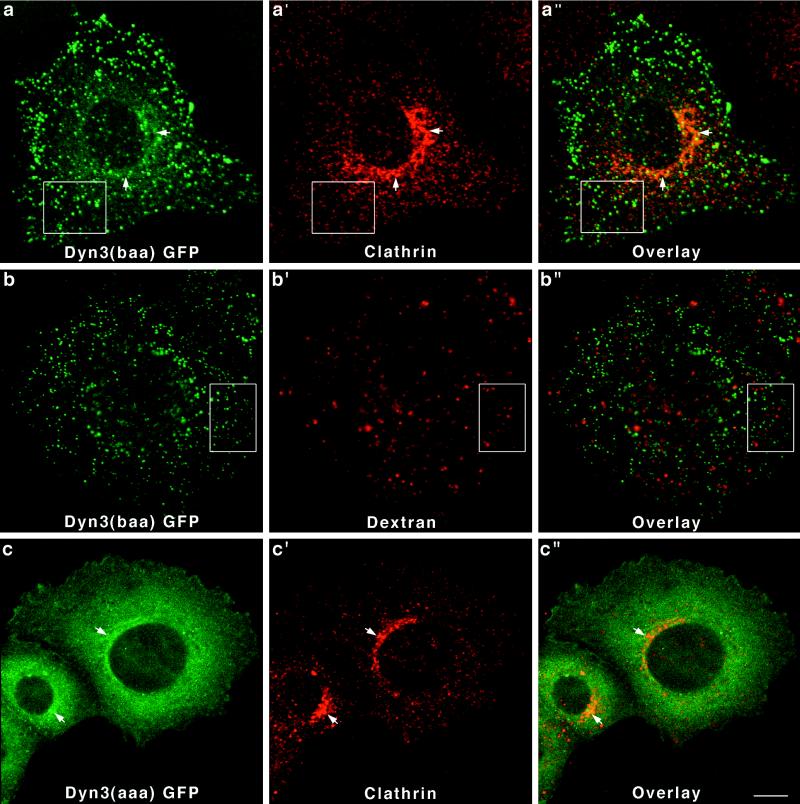
Finally, to complete this initial study on the distribution of the different dynamin isoforms in epithelial cells, we have expressed two distinct spliced forms of Dyn 3-GFP [Dyn 3(aaa) and Dyn 3(baa)] in Clone 9 cells. As shown in Figure 2, the Dyn 3 transcript undergoes the most extensive alternative splicing of the three dynamin genes with at least 13 different forms produced from three distinct sites, including a premature stop codon insertion. The two Dyn 3 forms analyzed in this study differ by only a 10-amino-acid insertion at the first splicing region located at residue 516. As for the Dyn 1 and Dyn 2 gene products, the two Dyn 3-GFP fusion proteins also exhibited different distributions in Clone 9 cells. Cells transfected with Dyn 3(baa)-GFP revealed a bright, punctate, vesicular-like staining, which did not colocalize with clathrin either at the plasma membrane or the Golgi area (Figure 7a–a"). Furthermore, these brightly stained vesicular structures did not appear to be lysosomal organelles, because they did not label with endocytosed TR-dextran (Figure 7b–b"). In contrast, Dyn 3(aaa)-GFP in Clone 9 cells was distributed diffusely throughout the cytoplasm and had some modest association with the Golgi apparatus when viewed in cells double stained for clathrin (Figure 7c–c").
Figure 7.
Dyn 3-GFP forms expressed in Clone 9 cells distribute diffusely to the cytoplasm or to unidentified vesicular structures. Fluorescence images of Dyn 3(baa)-GFP-transfected cells reveal a prominent population of brightly labeled vesicles, which did not colocalize with clathrin (a–a") at either the plasma membrane (boxes) or the Golgi apparatus (arrows). These dynamin-associated vesicles did not colocalize with endocytosed dextran (boxed regions), suggesting they are not late endocytic compartments (b–b"). In contrast, cells expressing Dyn 3(aaa)-GFP [a form differing from Dyn 3(baa) by only a 10-amino-acid insertion at the first splicing region] showed only a diffuse cytoplasmic fluorescence with a slight association with the Golgi apparatus, as demonstrated by costaining with an antibody to clathrin (c–c"). Bar, 10 μm.
DISCUSSION
Tissue-specific Expression of Multiple Dynamin Variants
The central focus of this study was twofold. First, our aim was to identify and define the number of spliced variants produced from the three dynamin genes previously cloned and sequenced from mammalian cells (Nakata et al., 1991, 1993; Robinson et al., 1993; Cook et al., 1994, 1996; Sontag et al., 1994). Second, we tested whether the different dynamin gene products are directed to various cytoplasmic membranous organelles where they might perform similar but distinct functions. Previous to this study, it had been reported that there were four different spliced variants produced from each dynamin gene, resulting in a total of 12 forms (for reviews, see Robinson et al., 1994; Urrutia et al., 1997). Using a PCR-based approach (Figure 1), we have identified an additional 13 spliced variants, making a total of 25 spliced forms, 8 forms for Dyn 1 and 13 forms for Dyn 3. We have not identified any additional forms of Dyn 2, which remains at four variants (Figure 2). The tissue distribution of these dynamin forms is summarized in Table 1. Brain and lung express the most isoforms, with 21 and 17 forms, respectively, whereas only the four forms of Dyn 2 are detected in epithelial-based tissues such as kidney, liver, and pancreas. Why an epithelial tissue such as testis expresses nine forms of Dyn 3 and Dyn 2 is unclear.
We were surprised to find that little, if any, Dyn 2 is expressed in highly enriched cultures of rat peripheral sensory neurons (DRGs) when compared with the same cultures containing substantial numbers of nonneuronal cells such as glial cells and fibroblasts (Figure 3). Whereas all three dynamin transcripts were detected in the mixed cultures, the Dyn 2 transcript was almost totally ablated upon removal of the supportive cells. To our knowledge, this is the first study aimed at defining the expression of the different dynamin gene products in an isolated neuronal population compared with the intact brain. It suggests that the Dyn 2 detected in brain by us and others may come from other cell types found in this complex tissue. Alternatively, Dyn 2 may be selectively expressed only in specific subpopulations of neurons such as motor versus sensory neurons versus neuroendocrine cells. In contrast, we did identify all three dynamin gene products (21 transcripts) in PC-12 cells (Figure 3). Whether the expression of the dynamins in these neuronally derived cells originating from a pheochromocytoma is truly representative of differentiated neuroepithelial cells or has been altered by neoplasia is unclear. Examination of distinct neuronal cell populations at different developmental stages using isoform-specific probes combined with in situ hybridization techniques will prove informative.
Altered Cytoplasmic Distributions for the Dynamin Proteins: Does Location Correlate with Function?
In an attempt to test whether the different dynamin proteins associate with multiple cellular organelles, we have expressed dynamin-GFP fusion proteins in a normal rat hepatocyte cell line, Clone 9 cells. These cells express all four forms of Dyn 2 (Table 1). Although all of the images displayed in this study (Figures 5–7) represent cell lines expressing GFP-coupled to the C termini of the different dynamin proteins, we have also closely examined and compared cells expressing the same dynamin cDNAs with GFP coupled to the N-termini and without GFP (our unpublished results). The cells expressing untagged dynamin forms were stained with isoform-specific antibodies to Dyn 1, Dyn 2, and Dyn 3 as well as with the Pan-antibodies MC63 and MC65. Despite the GFP modifications we did not observe any changes in the cellular distribution of a specific dynamin protein based on the attachment site of the GFP tag, suggesting that GFP does not hinder appropriate targeting of a dynamin to a membranous organelle. Finally, the expression levels of dynamin-GFP in these cells were not high. As shown in Figure 4, levels of the expressed fusion proteins were equal or less than levels of the endogenous dynamin. This modest level of expression makes it unlikely that the dynamins would be forced to interact with membranous organelles in a random, nonspecific manner. Because our Pan-dynamin antibodies did not recognize both the endogenous Dyn 2 and the expressed Dyn 3-GFP in the same cell, we cannot make any direct conclusions about expression levels for Dyn 3. Although the Dyn 3 bands are significantly weaker than those of the Dyn 2 in the same cells (Figure 4B), this could be due to differences in antibody sensitivity. Based on the expression levels for the other four forms of Dyn 1 and Dyn 2, which used the same expression vectors, we indirectly conclude that the levels of Dyn 3 in these cells do not exceed endogenous levels of Dyn 2.
Because the substantial size of the dynamin family prohibits detailed study of all forms, we have selected two variants expressed from each of the three genes. The dynamin proteins (Dyn 1–3) are strongly related, 75–80% similar depending on the variants compared, and spliced variants generated from each dynamin gene may differ by only several amino acids. Our goal in this study was to test not only whether the related dynamin gene products might target to distinct cellular locations but also whether modest insertional or substitutional changes within a spliced variant could alter distribution of the protein. The spliced forms manipulated in this study are highly similar to each other. Dyn 1(ab) and Dyn 1(bb) differ from each other by only 14 amino acids within a 46-residue insertional substitution. Dyn 2(aa) and Dyn 2(ab) are identical except for the four-amino-acid insertion GEIL at residue 516. Finally Dyn 3(baa) and Dyn 3(aaa) differ by only 10 amino acids, also at residue 516 (Figure 2). Despite these seemingly small differences in primary sequence, the contrasts in cellular localizations between these expressed fusion proteins could not be more striking. Indeed, at least four distinct morphological phenotypes of a possible six were generated from the different fusion protein–expressing cell lines (Table 2). These include a strong association with clathrin-coated vesicles at the plasma membrane (Figures 5a–d and 6, a and b) or at the plasma membrane and the Golgi apparatus (Figure 5, a and b), a diffuse cytosolic distribution (Figures 6, c and d, and 7c), an association with medium-sized vesicular structures, which are not clathrin positive (Figure 7, a and b), and a localization of dynamin into membrane ruffles or lamellapodia (Figure 5a). Whether the diffuse distribution of some of the dynamin proteins represents a truly cytosolic localization or an association to vesicles too small to be resolved is undefined. Perhaps the most graphic example of positional information provided by a small-sized insertional variant is demonstrated by the Dyn 2-GFP-expressing cells. Although both Dyn 2 proteins show a strong colocalization with clathrin-coated pits, only Dyn 2(aa) is associated with the Golgi apparatus, whereas Dyn 2(ab) shows little, if any, Golgi affinity. As mentioned above, these two Dyn 2 variants differ by only four amino acids (GEIL). The fact that only four amino acids, which are not situated in either the pleckstrin homology domain or the proline-rich domains of the dynamin protein, have such an effect makes this observation surprising. Currently, we are conducting point mutational analysis of this small insert to test whether we can enhance or abolish the Dyn 2(aa)–Golgi interaction. Finally, it will be interesting to test whether two specific insertional variations, Dyn 1(ac) and Dyn 1(bc), are expressed in neurons and whether they associate with clathrin-coated vesicles. Both of these dynamin proteins would have substantial alterations in a nine-amino-acid sequence near the C terminus (aa 786–794), which has been demonstrated by Shpetner and colleagues (1996) to be essential for clathrin association.
Table 2.
The distribution of GFP-dynamin proteins in Clone 9 cells
| PM-Clathrin | Golgi | Cytosol | Non-clathrin | |
|---|---|---|---|---|
| Dyn 1(ab) | ++ | − | − | − |
| Dyn 1(bb) | − | + | + | − |
| Dyn 2(aa) | ++ | ++ | − | − |
| Dyn 2(ab) | ++ | − | − | − |
| Dyn 3(aaa) | − | ± | + | − |
| Dyn 3(baa) | − | − | − | + |
Although surprising, there is precedence for such small insertional variations changing the cytoplasmic distribution of a protein. For example, the mammalian family of plasma membrane calcium pumps comprises four distinct genes with four spliced variants for each (Penniston et al., 1997). It has been shown that most, if not all, of these mRNAs are expressed in different cell types and are positioned at distinct membrane domains where they may respond differently to a given cell stimulus. A very relevant example of different cellular locations for proteins arising from the same gene has been reported by Montmayeur and Borrelli (1994), who demonstrated that an alternatively spliced C terminus of the heterotrimeric GTP-binding protein α-subunit is responsible for changing the distribution of this protein from plasma membrane to the Golgi apparatus. Thus, for the dynamin family, we predict that the different gene products are targeted to discrete membranous organelles where they may participate in the formation of nascent vesicles. In the simplest scenario one could postulate that each dynamin protein functions at a distinct cellular site not unlike that observed for the kinesin (Hirokawa, 1998) and myosin (Mermall et al., 1998) families of molecular motors. Support for this basic premise comes not only from the data presented in this study but also from other reports that dynamin proteins appear to play a role in caveolar scission (Schnitzer et al., 1996; Henley et al., 1998; Oh et al., 1998), liberation of clathrin- and nonclathrin-coated vesicles from the trans-Golgi network (Jones et al., 1998), transport of toxins from the endosome to the Golgi (Llorente et al., 1998), and vesicle traffic from the endoplasmic reticulum (Yoon et al., 1998). Most recently, a study using morphological and biochemical techniques has demonstrated the localization of Dyn 2 to membrane ruffles and lamellapodia (Figure 5a) in cultured cells, where it interacts specifically with the actin-binding protein cortactin (McNiven, Kim, Krueger, Cao, and Wong, unpublished results). Finally, recent observations made in Madin–Darby canine kidney cells expressing either Dyn 1 or Dyn 2 (S. Schmid, personal communication) suggest that mutants in Dyn 2 alter endocytosis from both the apical and basal plasma membranes, whereas a Dyn 1 mutant affects endocytosis only at the apical plasma membrane. Although Madin–Darby canine kidney cells do not express Dyn 1, these findings support the model that different dynamins are targeted to and function at different cellular sites. Although a one-dynamin protein-per-organelle model is attractive for cells and tissues that express a dozen different dynamins (neurons, lung, and testis), such a scenario is likely to be oversimplified. Although neurons perform a variety of specialized functions, it is difficult to predict why a neuronal cell would require up to four times more dynamin forms than would other polarized cells such as the hepatocyte or nephrocyte, which also perform numerous sophisticated trafficking functions. Certainly, future studies combining GFP expression systems with site-directed mutations of specific dynamin variants will prove informative.
ACKNOWLEDGMENTS
We thank Dr. A.M. Conti and J. Podratz of Dr. Windebank’s lab for isolating and culturing DRG cells and Dr. K.E. Howell for providing the monoclonal antibody TGN38. We are especially grateful to B. Oswald for antibody purification and other technical supports. We are also grateful to E. Krueger for helping with imaging techniques and R.R. Torgerson and K.R. Pitts for comments and critical reading of the manuscript. This work was supported by grant (DK44650) from National Institutes of Health to M.A.M.
Abbreviations used:
- ATCC
American Type Culture Collection
- DRG
dorsal root ganglion
- D-PBS
Dulbecco’s PBS
- Dyn 1
dynamin I
- Dyn 2
dynamin II
- Dyn 3
dynamin III
- GFP
green fluorescent protein
- RT
reverse transcription
- TR
Texas Red
REFERENCES
- Chen MS, Burgess CC, Vallee RB, Wadsworth SC. Developmental stage- and tissue-specific expression of shibire, a Drosophila gene involved in endocytosis. J Cell Sci. 1992;103:619–628. doi: 10.1242/jcs.103.3.619. [DOI] [PubMed] [Google Scholar]
- Chen MS, Obar RA, Schroeder CC, Austin TW, Poodry CA, Wadsworth SC, Vallee RB. Multiple forms of dynamin are encoded by shibire, a Drosophila gene involved in endocytosis. Nature. 1991;351:583–586. doi: 10.1038/351583a0. [DOI] [PubMed] [Google Scholar]
- Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem, 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- Conti AM, Fischer SJ, Windebank AJ. Inhibition of axonal growth from sensory neurons by excess nerve growth factor. Ann Neurol. 1997;42:838–846. doi: 10.1002/ana.410420604. [DOI] [PubMed] [Google Scholar]
- Cook T, Mesa K, Urrutia R. Three dynamin-encoding genes are differently expressed in developing rat brain. J Neurochem. 1996;67:927–931. doi: 10.1046/j.1471-4159.1996.67030927.x. [DOI] [PubMed] [Google Scholar]
- Cook TA, Urrutia R, McNiven MA. Identification of dynamin 2, an isoform ubiquitously expressed in rat tissues. Proc Natl Acad Sci USA. 1994;91:644–648. doi: 10.1073/pnas.91.2.644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crosby JR, Jones SM, Howell KE. TGN38 forms a macromolecular complex with small GTP-binding proteins: functional studies using a cell-free assay. Mol Biol Cell. 1992;3:A308. (Abstract). [Google Scholar]
- Damke H, Baba T, van der Bliek AM, Schmid SL. Clathrin-independent pinocytosis is induced in cells overexpressing a temperature-sensitive mutant of dynamin. J Cell Biol. 1995;131:69–80. doi: 10.1083/jcb.131.1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damke H, Baba T, Warnock DE, Schmid SL. Induction of mutant dynamin specifically blocks endocytic coated vesicle formation. J Cell Biol. 1994;127:915–934. doi: 10.1083/jcb.127.4.915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Camilli P, Takei K, McPherson PS. The function of dynamin in endocytosis. Curr Opin Neurobiol. 1995;5:559–565. doi: 10.1016/0959-4388(95)80059-x. [DOI] [PubMed] [Google Scholar]
- Gout I, Dhand R, Hiles ID, Fry MJ, Panayotou G, Das P, Truong O, Totty NF, Hsuan J, Booker GW. The GTPase dynamin binds to and is activated by a subset of SH3 domains. Cell. 1993;75:25–36. [PubMed] [Google Scholar]
- Grigliatti TA, Hall L, Rosenbluth R, Suzuki DT. Temperature-sensitive mutations in Drosophila melanogaster XV: a selection of immobile adults. Mol Gen Genet. 1973;120:107–114. doi: 10.1007/BF00267238. [DOI] [PubMed] [Google Scholar]
- Henley JR, Krueger EWA, Oswald BJ, McNiven MA. Dynamin-mediated internalization of caveolae. J Cell Biol. 1998;141:85–99. doi: 10.1083/jcb.141.1.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henley JR, McNiven MA. Association of a dynamin-like protein with the Golgi apparatus in mammalian cells. J Cell Biol. 1996;133:761–775. doi: 10.1083/jcb.133.4.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herskovits JS, Burgess CC, Obar RA, Vallee RB. Effects of mutant rat dynamin on endocytosis. J Cell Biol. 1993;122:565–578. doi: 10.1083/jcb.122.3.565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirokawa N. Kinesin and dynein superfamily proteins and the mechanism of organelle transport. Science, 1998;279:519–526. doi: 10.1126/science.279.5350.519. [DOI] [PubMed] [Google Scholar]
- Jones SM, Howell KE, Henley JR, Cao H, McNiven MA. Role of dynamin in the formation of transport vesicles from the trans-Golgi network. Science. 1998;279:573–577. doi: 10.1126/science.279.5350.573. [DOI] [PubMed] [Google Scholar]
- Kessell I, Holst BD, Roth TF. Membranous intermediates in endocytosis are labile, as shown in a temperature-sensitive mutant. Proc Natl Acad Sci USA. 1989;86:4968–4972. doi: 10.1073/pnas.86.13.4968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosaka T, Ikeda K. Possible temperature-dependent blockage of synaptic vesicle recycling induced by a single gene mutation in Drosophila. J Neurobiol. 1983a;14:207–225. doi: 10.1002/neu.480140305. [DOI] [PubMed] [Google Scholar]
- Kosaka T, Ikeda K. Reversible blockage of membrane retrieval and endocytosis in the garland cell of the temperature-sensitive mutant of Drosophila melanogaster, shibirets1. J Cell Biol, 1983b;97:499–507. doi: 10.1083/jcb.97.2.499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Llorente A, Rapak A, Schmid SL, van Deurs B, Sandvig K. Expression of mutant dynamin inhibits toxicity and transport of endocytosed ricin to the Golgi apparatus. J Cell Biol, 1998;140:553–563. doi: 10.1083/jcb.140.3.553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda K, Nakata T, Noda Y, Sato-Yoshitake R, Hirokawa N. Insertion of dynamin with microtubules: its structure and GTPase activity investigated by using highly purified dynamin. Mol Biol Cell. 1992;3:1181–1194. doi: 10.1091/mbc.3.10.1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier O, Lobbert J, Kaufmehl K, Streckert HJ, Werchau H. Dynamin II binds to the trans-Golgi network. Biochem Biophys Res Commun. 1996;223:229–233. doi: 10.1006/bbrc.1996.0876. [DOI] [PubMed] [Google Scholar]
- Masur SK, Kim YT, Wu CF. Reversible inhibition of endocytosis in cultured neurons from the Drosophila temperature-sensitive mutant shibirets1. J Neurogenet. 1990;6:191–206. doi: 10.3109/01677069009107110. [DOI] [PubMed] [Google Scholar]
- Mermall V, Post PL, Mooseker MS. Unconventional myosins in cell movement, membrane traffic, and signal transduction. Science. 1998;279:527–533. doi: 10.1126/science.279.5350.527. [DOI] [PubMed] [Google Scholar]
- Montmayeur JP, Borrelli E. Targeting of G alpha i2 to the Golgi by alternative spliced carboxyl-terminal region. Science. 1994;263:95–98. doi: 10.1126/science.8272874. [DOI] [PubMed] [Google Scholar]
- Nakata T, Iwamoto A, Noda Y, Takemura R, Yoshikura H, Hirokawa N. Predominant and developmentally regulated expression of dynamin in neurons. Neuron. 1991;7:461–469. doi: 10.1016/0896-6273(91)90298-e. [DOI] [PubMed] [Google Scholar]
- Nakata T, Takemura R, Hirokawa N. A novel member of the dynamin family of GTP-binding proteins is expressed specifically in the testis. J Cell Sci. 1993;105:1–5. doi: 10.1242/jcs.105.1.1. [DOI] [PubMed] [Google Scholar]
- Oh P, McIntosh DP, Schnitzer JE. Dynamin at the neck of caveolae mediates their budding to form transport vesicles by GTP-driven fission from the plasma membrane of endothelium. J Cell Biol. 1998;141:101–114. doi: 10.1083/jcb.141.1.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penniston JT, Enyedi A, Verma AK, Adamo HP, Filoteo AG. Plasma membrane Ca2+ pumps. Ann NY Acad Sci. 1997;834:56–64. doi: 10.1111/j.1749-6632.1997.tb52225.x. [DOI] [PubMed] [Google Scholar]
- Ponnambalam S, Girotti M, Yaspo ML, Owen CW, Perry ACF, Suganuma T. Primate homologues of rat TGN38: primary structure, expression and functional implications. J Cell Sci. 1996;109:675–685. doi: 10.1242/jcs.109.3.675. [DOI] [PubMed] [Google Scholar]
- Robinson PJ, Liu J-P, Powell KA, Fykse EM, Sudhof TC. Phosphorylation of dynamin I and synaptic-vesicle recycling. Trends Neurosci. 1994;17:348–353. doi: 10.1016/0166-2236(94)90179-1. [DOI] [PubMed] [Google Scholar]
- Robinson PJ, Sontag JM, Liu JP, Fykse EM, Slaughter C, McMahon H, Sudhof TC. Dynamin GTPase regulated by protein kinase C phosphorylation in nerve terminals. Nature. 1993;365:163–166. doi: 10.1038/365163a0. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd ed. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- Scaife R, Gout I, Waterfield MD, Margolis RL. Growth factor-induced binding of dynamin to signal transduction proteins involves sorting to distinct and separate proline-rich dynamin sequences. EMBO J. 1994;13:2574–2582. doi: 10.1002/j.1460-2075.1994.tb06547.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scaife R, Margolis RL. Biochemical and immunochemical analysis of rat brain dynamin interaction with microtubules and organelles in vivo and in vitro. J Cell Biol. 1990;111:3023–3033. doi: 10.1083/jcb.111.6.3023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnitzer JE, Oh P, McIntosh DP. Role of GTP hydrolysis in fission of caveolae directly from plasma membranes. Science. 1996;274:239–242. doi: 10.1126/science.274.5285.239. [DOI] [PubMed] [Google Scholar]
- Shpetner HS, Herskovits JS, Vallee RB. A binding site for SH3 domains targets dynamin to coated pits. J Biol Chem. 1996;271:13–16. doi: 10.1074/jbc.271.1.13. [DOI] [PubMed] [Google Scholar]
- Shpetner HS, Vallee RB. Identification of dynamin, a novel mechanochemical enzyme that mediates interactions between microtubules. Cell. 1989;59:421–432. doi: 10.1016/0092-8674(89)90027-5. [DOI] [PubMed] [Google Scholar]
- Shpetner HS, Vallee RB. Dynamin is a GTPase stimu-lated to high levels of activity by microtubules. Nature. 1992;355:733–735. doi: 10.1038/355733a0. [DOI] [PubMed] [Google Scholar]
- Sontag J-M, Fykse EM, Ushkaryov Y, Liu J-P, Robinson PJ, Sudhof TC. Differential expression and regulation of multiple dynamins. J Biol Chem. 1994;269:4547–4554. [PubMed] [Google Scholar]
- Takei K, McPherson PS, Schmid SL, De Camilli P. Tubular membrane invaginations coated by dynamin rings are induced by GTP. Nature. 1995;374:186–190. doi: 10.1038/374186a0. [DOI] [PubMed] [Google Scholar]
- Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuma PL, Stachniak MC, Collins CA. Activation of dynamin GTPase by acidic phospholipids and endogenous rat brain vesicles. J Biol Chem. 1993;268:17240–17246. [PubMed] [Google Scholar]
- Urrutia R, Henley JR, Cook T, McNiven MA. The dynamins: redundant or distinct functions for an expanding family of related GTPases? Proc Natl Acad Sci USA. 1997;94:377–384. doi: 10.1073/pnas.94.2.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallee RB, Vaughan KT, Echeverri CJ. Targeting of cytoplasmic dynein to membranous organelles and kinetochores via dynactin. Cold Spring Harbor Symp Quant Biol. 1995;60:803–811. doi: 10.1101/sqb.1995.060.01.086. [DOI] [PubMed] [Google Scholar]
- van der Bliek AM, Meyerowitz EM. Dynamin-like protein encoded by the Drosophila shibire gene associated with vesicular traffic. Nature. 1991;351:411–414. doi: 10.1038/351411a0. [DOI] [PubMed] [Google Scholar]
- van der Bliek AM, Redelmeier TE, Damke H, Tisdale EJ, Meyerowitz EM, Schmid SL. Mutations in human dynamin block an intermediate stage in coated vesicle formation. J Cell Biol. 1993;122:553–563. doi: 10.1083/jcb.122.3.553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warnock DE, Schmid SL. Dynamin GTPase, a force-generating molecular switch. Bioessays. 1996;18:885–893. doi: 10.1002/bies.950181107. [DOI] [PubMed] [Google Scholar]
- Wood P, Okada E, Bunge R. The use of networks of dissociated rat dorsal root ganglin neurons to induce myelination by oligodendrocytes in culture. Brain Res. 1980;196:247–252. doi: 10.1016/0006-8993(80)90732-5. [DOI] [PubMed] [Google Scholar]
- Yoon Y, Pitts KR, Dahan S, McNiven MA. A novel dynamin-like protein associates with cytoplasmic vesicles and tubules of the endoplasmic reticulum in mammalian cells. J Cell Biol. 1998;140:1–14. doi: 10.1083/jcb.140.4.779. [DOI] [PMC free article] [PubMed] [Google Scholar]