Abstract
Multiple human cancer Ags have been identified, although little is known concerning which would be most effectively used in cancer immunotherapy. To gain insight into the selection of appropriate Ags, the immunologic reactivity of a patient who had a durable complete regression of melanoma metastases was measured. PBMCs were directly cloned using the monoclonal anti-CD3 Ab OKT3 and IL-2 without any bias introduced by previous culture. A lymphocyte clone recognized a previously unknown shared melanoma Ag that was identified as the BING-4 protein encoded in a gene-rich region of the extended class II MHC. The HLA-A2-restricted BING-4 immunodominant peptide was translated from a 10-aa-long alternative open reading frame. In vitro sensitization against this peptide generated lymphocytes reactive against HLA-A2+ melanomas. Real-time semiquantitative RT-PCR analysis revealed that 8 of 15 melanoma cell lines overexpressed BING-4, and this correlated with recognition by lymphocytes. Overexpression was not found in normal tissues or other tumor types. Thus, BING-4 represents another candidate Ag for possible use in the immunotherapy of patients with melanoma.
In recent years, attempts to understand the nature of the tumor-host interaction in humans has focused on the identification of the genes encoding cancer Ags. Multiple cancer Ags, either uniquely expressed or over-expressed on cancers have been identified, and in many cases the immunodominant peptides presented on both class I and class II MHC molecules have been determined (reviewed in Ref. 1). The identification of this large number of tumor Ags has opened new opportunities for the development of cancer immunotherapies, but has also led to important questions concerning the choice of Ags to be used for immunotherapy. Are all Ags equally important as targets of immunotherapeutic attack, or do some Ags have unique qualities of immune stimulation that would facilitate their successful application for active immunization? How can one choose which Ags to use for active immunization from the large number of available cancer Ags?
Most tumor Ags have been identified using lymphocytes obtained from tumor-infiltrating lymphocyte cultures or from mixed lymphocyte tumor interactions. The use of tumor-infiltrating lymphocytes associated with tumor regression when adoptively transferred into cancer patients has improved the likelihood that the Ags identified may be suitable targets for immunotherapy (2). In the current study, we have taken a different approach to identify Ags that might be particularly important for use in active immunization. We have identified patients with metastatic melanoma that have experienced complete and durable regressions following immunotherapy. Extensive cloning of fresh, uncultured circulating PBLs from these patients has been performed using OKT3 plus IL-2 to attempt to identify circulating lymphocytes reactive with tumor without any bias introduced by artificial tumor stimulation or selective growth conditions by prior culture in vitro.
In the present paper, we describe studies of the circulating lymphocytes of a patient who received active immunization with a recombinant adenovirus and underwent complete regression of biopsy-proven s.c. and mediastinal metastases from melanoma, and remains continuously disease-free >5 years later (3). By directly cloning circulating lymphocytes from this patient, we have identified reactivity against a new shared melanoma Ag translated from a very short alternative open reading frame of a gene in the extended region of the class II MHC.
Materials and Methods
Source of lymphocytes for cloning
In June 1989, patient TC underwent excision of a 0.9-mm-thick primary melanoma from the left thigh. A palpable lymph node containing melanoma was excised in November 1991, and she was disease-free until July 1995 when recurrent melanomas developed in the left breast and left arm. Both masses were excised, and in November 1995 new left breast and mediastinal masses appeared, and she was referred to the Surgery Branch, National Cancer Institute. Fine needle aspiration confirmed the diagnosis of metastatic melanoma. In December 1995, the patient was entered into a clinical protocol approved by the National Cancer Institute Institutional Review Board, and on December 22, 1995, and January 1, 1996, she received the s.c. injection of 108 PFU of a recombinant adenovirus encoding the melanoma Ag recognized by T cells 1 (MART-1)2 (3). On follow-up 1 mo after the second injection, there was partial regression of both the mediastinal and breast melanoma metastases, and she received two additional injections of the same recombinant adenovirus on February 15, 1996, and on March 13, 1996. Shortly after the last injection, all of her disease regressed and she has remained continuously disease-free as of October 2001. The PBLs used for the present studies were obtained on January 17, 1996, 26 days after her first injection of recombinant adenovirus. This patient was the only one of 16 patients similarly treated with recombinant adenovirus who experienced cancer regression.
Cloning lymphocytes with antitumor activity
Lymphocytes were thawed into medium and cloned in round-bottom 96-well plates using techniques similar to those previously described (4). Twenty-five plates each contained 30 cells/well or 100 cells/well (total of 312,000 cells cultured). Each well also contained 5 × 104 irradiated (5000 cGy) PBMCs combined from three non-HLA-A2 donors, 30 ng/ml OKT3 (Orthoclone, Ortho-biotech, Raritan, NJ) and 300 IU IL-2 (Chiron, Emeryville, CA) per milliliter in a total volume of 0.2 ml/well. The medium was changed at 7 days and approximately every 3 days thereafter. By 3 wk after initiating the culture, 120 wells (5.0%) plated at 30 cells/well and 250 wells (10.4%) plated at 100 cells/well exhibited growth. Two-thirds of the cells in each of these 370 wells were transferred to a separate plate and tested for IFN-γ release when incubated overnight with 5 × 104, 624.38 mel, or 624.28 mel. These two culture lines were clones derived in our laboratory from the parental 624 line. However, the 624.28 mel line had selectively lost HLA-A2 Ag expression (data not shown). Six of these 370 clones exhibited reactivity against 624.38 mel, and not 624.28 mel cells. These six individual clones were expanded in a rapid expansion protocol using OKT3 and 300 IU IL-2/ml, as previously described (5). Following the expansion, retesting of the six clones revealed that three recognized the HLA-A*0201-restricted MART-1:27–35 peptide from the known MART-1 melanoma Ag (6, 7), one recognized the HLA-A*0201-restricted NY-ESO-1:157–165 peptide from the NY-ESO-1 melanoma Ag (8, 9), and one had no recognition of melanoma cell lines. The sixth clone reacted with 624.38 mel cells but had no reactivity against known Ags. It was recloned at one and five cells/well (three plates each) and 16 (0.2%) of the wells exhibited growth. A total of 7 of the 12 wells growing at five cells/well exhibited selective reactivity with 624.38 mel, and not 624.28 mel, and after expansion exhibited no reactivity against the COS-A2 cell line transfected with the MART-1, gp100, tyrosinase, tyrosinase-related protein (TRP)1, TRP2, MAGE-1, or NY-ESO-1 genes. One of these subclones, termed 10-B5, recognized several HLA-A2+, but not HLA-A2− melanoma cell lines, and was used to screen a cDNA library from the 624 mel cell line.
Screening and testing of cDNA library
A cDNA expression library from the 624 mel cell line was prepared by techniques previously described (9). In brief, total RNA was extracted, poly(A) RNA was purified and converted to cDNA using an oligo(dT) primer, ligated into the pEAK 8 vector (Edge Biosystems, Gaithersburg, MD), and electroporated into DH10B cells. Pools containing ~100 cDNA clones were prepared from bacteria and the plasmid DNA transfected into 293-A2 cells. Following transfection, the plates were incubated for 24 h and 5 × 104 10-B5 lymphocytes were added to each well. The supernatant from each well was harvested after 24 h and IFN-γ was measured using an ELISA.
Peptide synthesis and testing
Peptides were synthesized using a solid-phase method based on standard F-moc chemistry on a multiple peptide synthesizer (Gilson, Worthington, OH). The identities of the peptides were verified by mass spectrometry (Biosynthesis, Lewisville, TX). Lyophilized peptides were solubilized in DMSO and 1 μM peptide was added to T2 cells and used to stimulate the 10-B5 T cell clone in an overnight coculture assay containing 5 × 104 cells of each type. IFN-γ secretion was measured by ELISA.
Results
Ag-specific reactivity of lymphocyte clone 10-B5
The specificity of clone 10-B5 was tested against a variety of normal and malignant HLA-A2+ and HLA-A2− cell lines shown in Table I. Reactivity was seen against the three HLA-A2+ melanomas tested, but not the HLA-A2− melanomas, nor against multiple HLA-A2+ EBV-B lines, fibroblasts, human umbilical vein endothelial cell lines, or cultures of breast, colon, or prostate cancer. Thus, clone 10-B5 appeared to recognize an HLA-A2-restricted Ag expressed by melanoma cells, but not normal cells, nor cells from other nonmelanoma tumors tested.
Table I.
Recognition of melanoma and normal cells by lymphocyte clone 10-B5
| Stimulator
|
||||
|---|---|---|---|---|
| Tissue type | Tissue culture line | HLA-A2 expression | IFN-γ (pg/ml)
|
|
| Expt. 1 | Expt. 2 | |||
| None | − | − | 10 | 10 |
| Melanoma | 624 | + | *a | 1747 |
| 624.38 | + | >2000 | >2000 | |
| 526 | + | * | 1759 | |
| 624.28 | − | 16 | 13 | |
| 888 | − | * | 14 | |
| 938 | − | * | 15 | |
| EBV-B | 697 | + | 24 | 20 |
| Ba | + | 31 | 17 | |
| 1760 | + | 21 | 19 | |
| 888 | − | 18 | 17 | |
| Me | − | 28 | 23 | |
| Sl | − | 22 | 17 | |
| Fibroblast | 1102 | + | 20 | * |
| 1760 | + | 28 | * | |
| 888 | − | 29 | * | |
| HUVEC | 751675 | + | 20 | * |
| 16309 | + | 11 | * | |
| 15595 | − | 11 | * | |
| Breast | MCF-7 | + | 27 | * |
| BT20 | − | 19 | * | |
| Colon | Li | + | 15 | * |
| Sp | + | 20 | * | |
| Me | − | 15 | * | |
| Prostate | LnCap | + | 15 | * |
| Pr 22-B7 | + | * | 14 | |
| 1550 | + | * | 14 | |
| 1669 | + | * | 13 | |
*, Not done.
Cloning the gene encoding the Ag recognized by lymphocyte clone 10-B5
Transfectants of the 293-A2 cell line with 384 cDNA pools, including ~38,400 cDNA clones from the 624 library, were screened using the lymphocyte clone 10-B5, and four positive pools were identified. A total of 2 of 192 subclones from the most positive well stimulated cytokine release from the 10-B5 lymphocyte following transfection into 293-A2 cells, and one of these clones was then sequenced.
The cDNA clone contained 1162 bp that represented a partial cDNA encoding the last 330 aa of the BING-4 gene (GenBank accession number BC000388; Refs. 10–13), although presumably only the last 288 aa were translated from an initiating methionine encoded within this cDNA clone. The full-length product was then cloned using primers based on the known sequence of BING-4. Although several bands were found using 624 mel RNA, probably representing alternatively spliced variants, the full-length BING-4 transcript (~2 kb) was identical to the BING-4 sequence in Gen-Bank. The first nucleotide in the clone we isolated represented nt 839 with respect to the start site of the full-length BING-4 protein. BING-4 is a 610 aa protein of unknown function present in a gene-rich segment of chromosome six immediately centromeric to the classical class II MHC that contains many genes involved in Ag processing (Fig. 1; Refs. 10–12). A database search revealed the presence of expressed sequence tags encoding BING-4 in infant brain, retinoblastoma, melanoma, choriocarcinoma, prostate cancer, liver cancer, bone marrow, and tonsillar B cell cDNA libraries.
FIGURE 1.
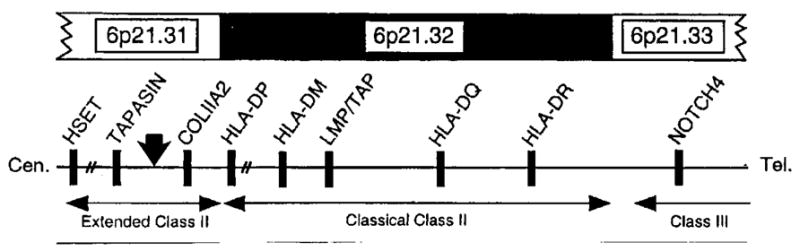
Structure of the MHC-class II region on human chromosome six (modified from Ref. 11). The solid vertical arrow indicates the approximate position of the BING-4 gene. This region just centromeric to the classical MHC class II has been considered an extended region of the class II MHC.
Identification of the BING-4 peptide recognized by the 10-B5 lymphocyte clone
Using algorithms that predict the binding of peptides to HLA-A2 within the last 288 aa of the BING-4 protein (the segment in our original cDNA clone), a total of 88 peptides (35 9-mers and 43 10-mers) were synthesized. These peptides were individually pulsed onto the HLA-A2+ T2 cell line and none were recognized by lymphocyte clone 10-B5 (data not shown). Because of the inability to detect a reactive peptide in the normal open reading frame of the BING-4 protein, alternative open reading frames of the cDNA clone that started with a methionine codon and that encoded a minimum of 9 aa were then evaluated. This cDNA encoded nine potential open reading frames (Table II) that varied between 9 aa and the 288 aa, which represented the normal BING-4 open reading frame. Twenty-five peptides conforming to an extended HLA-A2 Ag-binding motif from all of the remaining open reading frames were tested for recognition by the 10-B5 lymphocyte clone. A single 9-aa peptide, CQWGRLWQL, from a 10 aa open reading frame conferred a high degree of reactivity against lymphocyte 10-B5 (Table III). No other peptide tested within any of the open reading frames showed reactivity against the lymphocyte clone with the exception of the overlapping 10-mer, MC-QWGRLWQL, which was similar to the 9-mer peptide in its ability to stimulate clone 10-B5 (Fig. 2).
Table II.
Open reading frames in the BING-4 clone recognized by the lymphocyte clone TC-10B
| Open Reading Frame | Base Paira | No. of Peptides Tested | Amino Acid Sequence of the Open Reading Frame |
|---|---|---|---|
| 1 | 70–99 | 1 | MCQWGRLWQL |
| 2 | 103–129 | 0 | MLELGGSML |
| 3 | 129–992 | 88 | MSQNPYNAVIHLGHSNGTVSLWSPAMKEPL |
| AKILCHRGGVRAVAVDSTGTYMATSGLDHQ | |||
| LKIFDLRGTYQPLSTRTLPHGAGHLAFSQR | |||
| GLLVAGMGDVVNIWAGQGKASPPSLEQPYL | |||
| THRLSGPVHGLQFCPFEDVLGVGHTGGITS | |||
| MLVPGAGEPNFDGLESNPYRSRKQRQEWEV | |||
| KALLEKVPAELICLDPRALAEVDVISLEQG | |||
| KKEQIERLGYDPQAKAPFQPKPKQKGRSST | |||
| ASLVKRKRKVMDEEHRDKVRQSLQQQHHKE | |||
| AKAKPTGARPSALDRFVR | |||
| 4 | 148–204 | 5 | MPSSISDTAMVLCLYGVQL |
| 5 | 191–319 | 0 | MESSYEGATGKDSLSSWWGPGCGSRFYRHV |
| HGHLWPRPPAEDL | |||
| 6 | 367–666 | 7 | MEQGTWPSPRGDCWWREWVTLSTSGQGRAR |
| PAHPPLNSPTSPTGSQALCMAFSSAPLKMC | |||
| WGWGTLGASPACWSLGPVSPTSMAWRVIHT | |||
| EAGSSARSGR | |||
| 7 | 736–771 | 0 | MSSPWSRERRSR |
| 8 | 787–858 | 0 | MTRRLRLPSSQSQSRRAAAPRQAW |
| 9 | 883–1161 | 10 | MRNTGTRSGRAFSSSIIRRRRPSPRGPGHL |
| PWTDLCAEPDSRVAWEQSLPKITCREMSVP | |||
| WNKEVGAVWPLPQLGVDSCLLGWVGIKEES | |||
| DFL |
The first base of the recognized BING-4 clone corresponds to base 892 of the GenBank BING-4 nucleotide sequence (accession no. BC 000388).
Table III.
Recognition of BING-4 peptides from alternative open reading frames
| Open Reading Frame | Amino Acid No. | Peptides Tested | IFN-γ (pg/ml) |
|---|---|---|---|
| 1 | 2–10 | CQWGRLWQL | >2000 |
| 2 | 9–17 | AMVLCLYGV | 28 |
| 2 | 11–19 | VLCLYGVQL | 27 |
| 2 | 4–12 | SISDTAMVL | 29 |
| 2 | 10–19 | MVLCLYGVQL | 30 |
| 2 | 8–17 | TAMVLCLYGV | 27 |
| 4 | 49–57 | CMAFSSAPL | 29 |
| 4 | 82–90 | SMAWRVIHT | 29 |
| 4 | 58–66 | KMCWGWGTL | 27 |
| 4 | 47–55 | ALCMAFSSA | 27 |
| 4 | 67–75 | GASPACWSL | 28 |
| 4 | 74–85 | SLGPVSPTSM | 31 |
| 4 | 66–75 | LGASPACWSL | 28 |
| 7 | 51–59 | KITCREMSV | 27 |
| 7 | 73–81 | QLGVDSCLL | 27 |
| 7 | 35–43 | LCAEPDSRV | 29 |
| 7 | 85–93 | GIKEESDFL | 27 |
| 7 | 67–76 | AVWPLPQLGV | 29 |
| 7 | 43–52 | VAWEQSLPKI | 28 |
| 7 | 34–43 | DLCAEPDSRV | 27 |
| 7 | 75–84 | GVDSCLLGWV | 28 |
| 7 | 56–65 | EMSVPWNKEV | 27 |
| 7 | 59–68 | VPWNKEVGAV | 27 |
| None | 27 | ||
| 888 mel (A2−) | 25 | ||
| 824.28 mel (A2−) | 26 | ||
| 526 mel (A2+) | 835 | ||
| 624 mel (A2+) | 507 | ||
| 624.38 mel (A2+) | 1852 |
FIGURE 2.
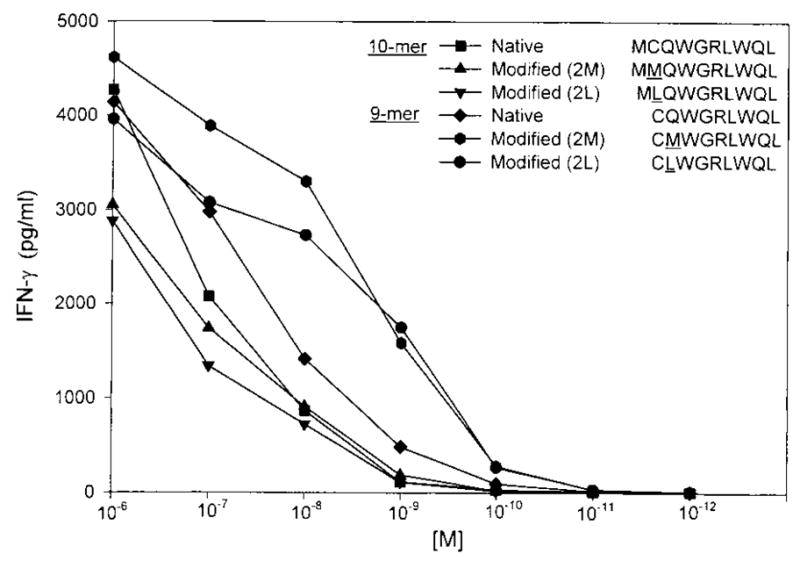
Titration of peptides from a 10-aa alternative open reading frame of the BING-4 gene. Both the native 9- and 10-mer peptides were recognized by lymphocyte clone 10-B5. Modifications of the 9-mer with either a methionine or leucine in place of glutamine at anchor position 2 provided increased recognition by clone 10-B5.
Because the glutamine (Q) in anchor position 2 of the 9-mer and cysteine (C) in position 2 of the 10-mer were not ideal amino acids for HLA-A2 binding, peptides were synthesized with substitutions of methionine (M) and leucine (L) in the second positions and tested using the 10-B5 clone to determine whether a more highly reactive peptide could be identified (Fig. 2). Modifications of the 10-mer did not improve recognition, although 9-mer peptides with either a methionine or leucine substitution at the second position were significantly better recognized than the native peptide. Significant recognition above background was seen when these modified peptides were pulsed onto T2 cells at a concentration of 0.1 nM.
Expression of BING-4 mRNA on multiple cell lines and tissues
Although the 10-B5 lymphocyte clone that selectively recognized the BING-4 peptide recognized HLA-A2+ melanomas and not other HLA-A2+ normal cells or other tumor cell lines and because Abs against BING-4 are not available, the presence of BING-4 transcripts in expressed sequence tag libraries led us to more extensively explore the expression of BING-4 mRNA. Only low levels of BING-4 expression were detected when RNA derived from multiple normal human tissues were tested using a sensitive RT-PCR assay (data not shown). A real-time semiquantitative RT-PCR assay was then performed on multiple melanoma cell lines as well as lines from other tumors and normal tissues to determine the number of BING-4 mRNA copies relative to mRNA encoding the GAPDH housekeeping gene (Fig. 3). Many melanoma cell lines expressed relatively high levels of BING-4 mRNA, whereas all nonmelanoma tumors as well as EBV-B cells, fibroblast, and melanocyte-cultured cells had relatively low levels of BING-4 expression.
FIGURE 3.
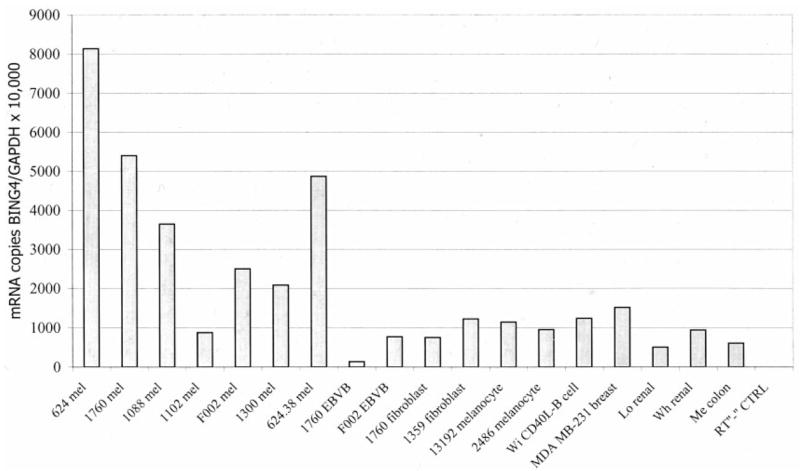
Real-time RT-PCR analysis of the number of mRNA copies encoding the BING-4 gene compared with mRNA encoding the GAPDH housekeeping gene. Many melanomas (mel) had increased expression of BING-4 mRNA compared with EBV-B cells, fibroblasts, melanocytes, CD40 ligand-stimulated B cells, and a variety of nonmelanoma tumors (breast, renal, and colon).
We next tested the correlation of BING-4 mRNA levels with recognition of melanoma cell lines by lymphocyte clone 10-B5 (Fig. 4). A significant correlation existed between BING-4 mRNA expression and immune recognition of these 15 cell lines (p < 0.05). Thus, it appears that threshold levels of BING-4 mRNA expression were required for lymphocyte recognition, and that these levels were not reached by normal cell lines or nonmelanoma tumors, but were achieved in 8 of 15 melanoma cell lines. All of these melanoma cell lines were recognized when pulsed with 1 μM native BING-4 peptide, indicating an adequate expression of HLA-A2 by the melanomas.
FIGURE 4.
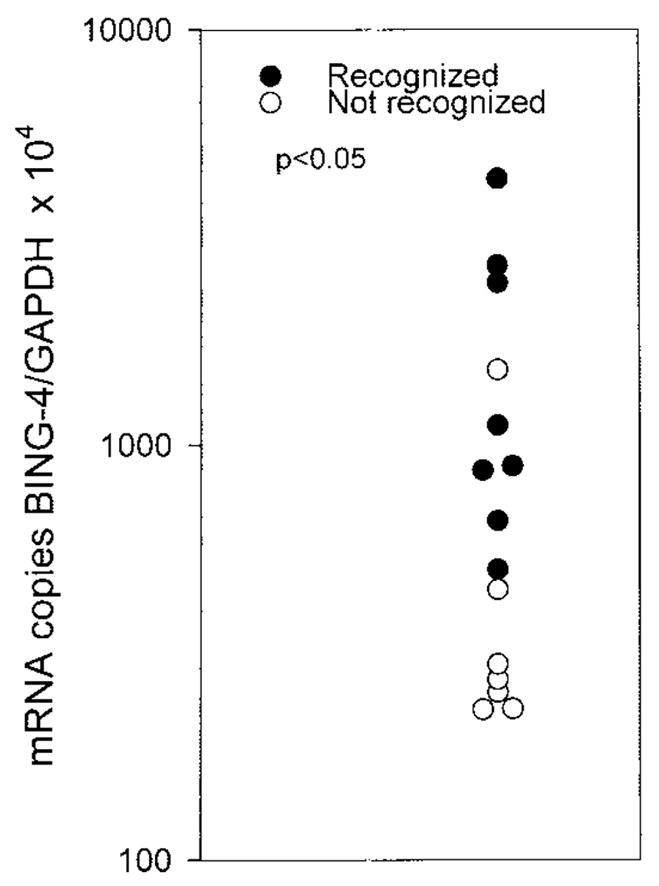
Fifteen melanoma lines were tested for recognition by lymphocyte clone 10-B5 as well as for the number of mRNA copies of the BING-4 gene and the GAPDH gene. A correlation existed between increased expression of the BING-4 gene and recognition by 10-B5 reactive lymphocytes (p < 0.05). Thus, it appears that a threshold level of BING-4 expression is an important factor enabling lymphocyte recognition.
In vitro sensitization of lymphocytes from nonimmunized melanoma patients using the BING-4 peptide
To estimate the relative immunogenicity of the BING-4 peptide, lymphocytes from 10 patients were in vitro sensitized against both the native BING-4 9-mer peptide, CQWGRLWQL, or the BING-4 peptide with methionine substituted for glutamine at the second position using techniques similar to those previously described (14). Peptide (10 μM) was added to 3 × 106 preimmune PBMC in 2 ml containing 500 mg/ml CD40 ligand. Four weekly restimulations were performed using PBMC pulsed with BING-4 peptide. On the day after each restimulation, 300 IU IL-2/ml was added to each culture. After four restimulations, PMBC derived from 7 of 10 patients that were sensitized in vitro to the native peptide developed antipeptide reactivity and in three of these patients specific reactivity was seen against HLA-A2+ BING-4 expressing tumor cells as well as 293-A2 cells transfected with the full-length BING-4 gene (Table IV). Following in vitro sensitization against the modified peptide, the PBMC from six patients also developed antipeptide reactivity, although none developed reactivity against melanoma lines (data not shown).
Table IV.
Reactivity of lymphocytes obtained by in vitro sensitization against the BING-4 peptide
| IFN-γ (pg/ml)
|
||||
|---|---|---|---|---|
| Stimulator | HLA-A2 | Patient 1 | Patient 2 | Patient 3 |
| None | 70 | 25 | 53 | |
| T2 pulsed with | ||||
| HBV peptide (10−6 M) | + | 71 | 25 | 47 |
| BING-4 peptide | ||||
| 10−6 M | + | 2986 | 1595 | 1884 |
| 10−7 M | + | 2557 | 1586 | 1684 |
| 10−8 M | + | 939 | 551 | 1677 |
| 10−9 M | + | 181 | 135 | 702 |
| 10−10 M | + | 102 | 43 | 203 |
| 10−11 M | + | 85 | 25 | 92 |
| 888 mel | − | 74 | 26 | 50 |
| 624.28 mel | − | 66 | 26 | 47 |
| 624.38 mel | + | 1365 | 600 | 146 |
| 1300 mel | + | 1372 | 390 | 198 |
| 293-A2 transfected with | ||||
| GFP | + | 115 | 51 | 48 |
| BING-4 (full length) | + | 999 | 186 | 81 |
Discussion
To attempt to identify Ags that may be of particular benefit for use in immunotherapy, we have identified a group of patients with metastatic cancer that have undergone dramatic and durable cancer regressions following active immunization. Identification of the Ags to which these patients develop strong responses might provide important clues to the choice of Ags for use in future studies. In the present paper, we have studied the circulating lymphocytes of a 35-year-old female who underwent a complete regression of metastatic melanoma to the breast and mediastinum ongoing for >5 years. This patient received immunization with a recombinant adenovirus encoding the MART-1 melanoma Ag, and she was the only patient among 16 patients similarly treated who achieved an objective cancer regression (3). Thus, it was hypothesized that the immunization protocol used in this patient triggered immune responses against an Ag(s) capable of mediating the complete destruction of her cancer. We set out to identify the reactivities in her circulating lymphocytes reactive with melanoma.
To avoid bias resulting from prolonged in vitro culture of lymphocytes, we used an approach to identify melanoma-reactive T cells generated by limiting dilution analysis of circulating lymphocytes that were stimulated with OKT3 and IL-2. Thus, we were able to grow lymphocyte clones without a specific antigenic stimulus. Because the patient’s autologous tumor was not available, screening of the clones was performed using the 624.38 and 624.28 mel cell lines both derived from the parental 624 mel cell line. These lines differed only in the loss of expression of HLA-A2 from the 624.28 cell line. A total of 312,000 cells were cultured, and thus any reactivities that were detected would likely be present in high frequency. The starting lymphocyte population used in this study was derived 26 days after the first injection of recombinant adenovirus at a time just before any obvious tumor regression was noted.
Reactivity against three Ags was detected in the circulating lymphocytes from this patient. Reactivity against the MART-1 melanoma Ag may have been the result of immunization with the recombinant adenovirus encoding MART-1. As we have reported previously, many melanoma patients have preexisting reactivity against the MART-1 immunodominant peptide and reactivity does not appear to correlate with cancer regression (15). Reactivity was also found against the NY-ESO-1:157–165 peptide. This patient also had circulating Ab reactive with the NY-ESO-1 protein, and we were able to generate HLA-DP-restricted T cells from the lymphocytes of this patient by in vitro sensitization against the HLA class II-restricted NY-ESO-1:161–180 peptide (16). Patient TC had not been immunized against the NY-ESO-1 Ag, and thus, this reactivity was either preexisting or was somehow stimulated by the adenoviral immunization. The secretion of IFN-γ by immune lymphocytes reactive to MART-1 and/or NY-ESO-1 Ags may have led to up-regulation of BING-4 Ag expression and the development of reactivity to this previously unrecognized tumor Ag.
There are several unique aspects to the detection of BING-4 as a tumor Ag in this patient. BING-4 is a protein of unknown function containing 610 aa encoded by 15 exons within a gene-rich region of chromosome six considered to represent an extended part of the class II MHC (Fig. 1; Refs. 10–12). This region contains a variety of genes including tapasin, a gene required for presentation of Ag by MHC class I molecules, DAXX, which encodes an effector of fas that stimulates apoptosis through the Jun kinase pathway as well as other novel genes. Particularly unusual is the presence of the BING-5 gene within intron 11 of the BING-4 gene. BING-5 is transcribed from the opposite strand to that encoding BING-4 (11). The function of BING-4 is unknown, although there are numerous nuclear localization signals situated near both the amino and carboxyl termini of the protein suggesting its localization to the nucleus. Proteins sharing conserved regions with BING-4 have been found in a variety of other species including the mouse, rat, Saccharomyces, Caenorhabditis, and others (11, 13). The location of BING-4 in this region and its close proximity to genes involved in Ag presentation suggest that it may be involved in immunologic phenomena.
Other tumor Ags such as TRP1 and NY-ESO-1 contain epitopes translated from alternative open reading frames (9, 17). However, the immunoreactive peptide presented on the surface of melanoma cells from the BING-4 protein is translated from an alternative open reading frame only 10 aa long. This finding emphasizes the need to explore all open reading frames when searching for presented peptides within putative tumor Ags (Table II).
Expression of BING-4 mRNA was found in virtually all normal tissues. Real-time semiquantitative RT-PCR analysis revealed that the levels of expression of the BING-4 transcript in normal tissues, including melanocytes, as well as in nonmelanoma tumors, were low relative to the levels found in melanomas. The correlation between high levels of expression of the BING-4 mRNA and recognition of tumors by BING-4-reactive T cells strongly suggests that a threshold level of BING-4 expression is necessary to mediate tumor recognition (Figs. 3 and 4). The high level of BING-4 expression on melanomas compared with other cancer types is unexplained.
It is not known whether the BING-4 Ag played a role in the tumor destruction in this patient. To test the immunogenicity of BING-4, we performed in vitro sensitizations against the BING-4 peptide using PBLs from 10 nonimmunized HLA-A2+ melanoma patients. A total of 7 of 10 patients were successfully sensitized in vitro to the native peptide, and in three of these patients a high enough level of reactivity was developed to recognize HLA-A2+ BING-4-positive tumor cells. This suggests that immunization with the BING-4 immunodominant peptide may be an effective means of immunizing patients with melanoma.
The present study was undertaken to obtain clues to explain the factors important in patients undergoing regression of metastatic cancer following immunotherapy. The reactivity of patient TC to the NY-ESO-1 Ag as well as to the new BING-4 tumor Ag in the absence of any deliberate immunization to these Ags suggests that these reactivities may have played a role in the tumor destruction. The mechanism of in vivo tumor destruction by immune lymphocytes is unclear, however, and other lymphocyte functions such as lysis and secretion of additional cytokines by BING-4-reactive cells in patients need to be evaluated. Similar analyses of other patients who have undergone significant durable cancer regressions are underway to determine whether common factors will emerge in these successfully treated patients.
Footnotes
Abbreviations used in this paper: MART-1, melanoma Ag recognized by T cells 1; TRP, tyrosinase-related protein.
References
- 1.Rosenberg SA. Progress in human tumour immunology and immunotherapy. Nature. 2001;411:380. doi: 10.1038/35077246. [DOI] [PubMed] [Google Scholar]
- 2.Rosenberg SA, Yannelli JR, Yang JC, Topalian SL, Schwartzentruber DJ, Weber JS, Parkinson DR, Seipp CA, White DE. Treatment of patients with metastatic melanoma using autologous tumor-infiltrating lymphocytes and interleukin-2. J Natl Cancer Inst. 1994;86:1159. doi: 10.1093/jnci/86.15.1159. [DOI] [PubMed] [Google Scholar]
- 3.Rosenberg SA, Zhai Y, Yang JC, Schwartzentruber DJ, Hwu P, Marincola FM, Topalian SL, Restifo NP, Seipp CA, Einhorn JH, et al. Immunizing patients with metastatic melanoma using recombinant adenoviruses encoding MART-1 or gp100 melanoma antigens. J Natl Cancer Inst. 1998;90:1894. doi: 10.1093/jnci/90.24.1894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dudley ME, Nishimura MI, Holt AKC, Rosenberg SA. Antitumor immunization with a minimal peptide epitope (G9-209-2M) leads to a functionally heterogeneous CTL response. J Immunother. 1999;22:288. doi: 10.1097/00002371-199907000-00002. [DOI] [PubMed] [Google Scholar]
- 5.Riddell SR, Watanabe KS, Goodrich JM, Li CR, Agha ME, Greenberg PD. Restoration of viral immunity in immunodeficient humans by the adoptive transfer of T cell clones. Science. 1992;257:238. doi: 10.1126/science.1352912. [DOI] [PubMed] [Google Scholar]
- 6.Kawakami Y, Eliyahu S, Delgado CH, Robbins PF, Rivoltini L, Topalian SL, Miki T, Rosenberg SA. Cloning of the gene coding for a shared human melanoma antigen recognized by autologous T cells infiltrating into tumor. Proc Natl Acad Sci USA. 1994;91:3515. doi: 10.1073/pnas.91.9.3515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kawakami Y, Eliyahu S, Sakaguchi K, Robbins PF, Rivoltini L, Yannelli JR, Appella E, Rosenberg SA. Identification of the immunodominant peptides of the MART-1 human melanoma antigen recognized by the majority of HLA-A2 restricted tumor infiltrating lymphocytes. J Exp Med. 1994;180:347. doi: 10.1084/jem.180.1.347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jager E, Chen YT, Drijfhout JW. Simultaneous humoral and cellular immune response against cancer-testis antigen NY-ESO-1:definition of human histocompatibility leukocyte antigen (HLA)-A2-binding peptide epitopes. J Exp Med. 1998;187:265. doi: 10.1084/jem.187.2.265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang R-F, Johnston SL, Zeng G, Topalian SL, Schwartzentruber DJ, Rosenberg SA. A breast and melanoma-shared tumor antigen: T cell responses to antigenic peptides translated from different open reading frames. J Immunol. 1998;161:3596. [PubMed] [Google Scholar]
- 10.Herberg JA, Beck S, Trowsdale J. TAPASIN, DAXX, RGL2, HKE2 and four new genes (BING 1, 3 to 5) form a dense cluster at the centromeric end of the MHC. J Mol Biol. 1998;277:839. doi: 10.1006/jmbi.1998.1637. [DOI] [PubMed] [Google Scholar]
- 11.Herberg JA, Sgouros J, Jones T, Copeman J, Humphray SJ, Sheer DCP, Beck S, Trowsdale J. Genomic analysis of the TAPASIN gene, located close to the TAP loci in the MHC. Eur J Immunol. 1998;28:459. doi: 10.1002/(SICI)1521-4141(199802)28:02<459::AID-IMMU459>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 12.Stephens R, Horton R, Humphray S, Rowen L, Trowsdale J, Beck S. Gene organisation, sequence variation and isochore structure at the centromeric boundary of the human MHC. J Mol Biol. 1999;291:789. doi: 10.1006/jmbi.1999.3004. [DOI] [PubMed] [Google Scholar]
- 13.Walter LGE. Physical mapping and evolution of the centromeric class I gene-containing region of the rat MHC. Immunogenetics. 2000;51:829. doi: 10.1007/s002510000219. [DOI] [PubMed] [Google Scholar]
- 14.Parkhurst MR, Salgaller ML, Southwood S, Robbins PF, Sette A, Rosenberg SA, Kawakami Y. Improved induction of melanoma reactive CTL with peptides from the melanoma antigen gp100 modified at HLA-A* 0210 binding residues. J Immunol. 1996;157:2539. [PubMed] [Google Scholar]
- 15.Marincola FM, Rivoltini L, Salgaller ML, Player M, Rosenberg SA. Differential anti-MART-1/MelanA CTL activity in peripheral blood of HLA-A2 melanoma patients in comparison to healthy donors: evidence for in vivo priming by tumor cells. J Immunother. 1996;19:266. doi: 10.1097/00002371-199607000-00003. [DOI] [PubMed] [Google Scholar]
- 16.Zeng G, Wang X, Robbins PF, Rosenberg SA, Wang R-F. CD4+ T cell recognition of MHC class II-restricted epitopes from NY-ESO-1 presented by a prevalent HLA DP4 allele: association with NY-ESO-1 antibody production. Proc Natl Acad Sci USA. 2001;98:3963. doi: 10.1073/pnas.061507398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang R-F, Parkhurst MR, Kawakami Y, Robbins PF, Rosenberg SA. Utilization of an alternative open reading frame of a normal gene in generating a novel human cancer antigen. J Exp Med. 1996;183:1131. doi: 10.1084/jem.183.3.1131. [DOI] [PMC free article] [PubMed] [Google Scholar]


