Abstract
Reactive oxygen species (ROS) are generated by mitochondrial respiration and can react with and damage cellular components. According to the free radical theory of aging, oxidative damage from mitochondrial ROS is a major cause of cellular decline during aging. Mitochondrial uncoupling proteins (UCPs) uncouple ATP production from electron transport and can be stimulated by free radicals, suggesting UCPs may perform a cytoprotective function. The nematode, Caenorhabditis elegans, contains one UCP-like protein, encoded by the ucp-4 gene. We have investigated the genetic requirement for ucp-4 in normal aging and stress resistance. Consistent with the hypothesis that ucp-4 encodes a putative uncoupling protein, animals lacking ucp-4 function contained elevated ATP levels. However, the absence of ucp-4 function did not affect adult lifespan or survival in the presence of thermal or oxidative stress. Together, these results demonstrate that ucp-4 is a negative regulator of ATP production in C. elegans, but is not required for normal lifespan.
Keywords: Caenorhabditis elegans, Mitochondria, Oxidative stress, Lifespan
1. Introduction
Mitochondrial electron transfer produces a proton gradient across the mitochondrial inner membrane that can be dissipated by the F0/F1 ATP synthase during the conversion of ADP into ATP. Alternatively, the proton gradient can be dissipated by uncoupling proteins (UCPs), a family of mitochondrial transporter proteins that allow proton movement across the mitochondrial inner membrane (Erlanson-Altbertsson, 2003). UCPs appear to be evolutionarily conserved in many species, including plants, invertebrates and mammals. However, relatively little is known about the functions of UCPs through evolution. In mammals, UCP1 expression in brown fat is induced by cold temperature and is necessary for thermogenesis (Enerback et al., 1997). Mammals also express several other UCPs that are dispensable for thermogenesis, and, thus, their functions remain mysterious (Arsenijevic et al., 2000; Vidal-Puig et al., 2000). One hypothesis is that these non-thermogenic UCPs may help to protect cells from oxidative stress (Brand et al., 2004). Studies of UCP knockout mice have generally supported a role in limiting ROS production. Macrophagic ROS production is enhanced in ucp2−/− knockout mice, consistent with a role for UCP2 in limiting ROS (Arsenijevic et al., 2000). ucp3−/− knockout mice also display enhanced ROS production in skeletal muscle, the predominant site for UCP3 expression (Vidal-Puig et al., 2000). Furthermore, exogenous superoxide has been reported to activate UCP-mediated uncoupling in vitro (Echtay et al., 2002; Krauss et al., 2003).
Although UCPs from mice and humans have been the focus of the most intensive study, UCP-like proteins have been identified in the genomes of Caenorhabditis elegans, Drosophila melanogaster, and Dictyostelium discoideum, as well as in plants (Hanak and Jezek, 2001). A phenotypic analysis of these invertebrate genes may shed light on ancestral functions of these putative UCPs. For example, invertebrates are poikliotherms and are unable to regulate internal body temperature to the same extent as mammals, which are homiotherms. An analysis of the importance of the C. elegans putative UCP-like protein for survival at cold temperature may be informative as to whether the ancestral UCPs were primarily required for adaptation to cold temperatures. Furthermore, it is important to investigate the role of invertebrate UCPs in coping with oxidative stress. Since ROS production has been implicated as a contributor to cellular decline associated with aging, it is also important to examine whether the putative UCPs in invertebrates affect longevity.
For these reasons, we have undertaken a phenotypic characterization of the only UCP-like protein encoded in the C. elegans nematode genome (Hanak and Jezek, 2001). The C. elegans UCP-like protein is most closely related to mammalian UCP4, and the two proteins share 46% sequence identity (Hanak and Jezek, 2001). Therefore, the C. elegans gene is referred to as Cel-ucp-4, or simply ucp-4. In this report, we describe the genetic analysis of ucp-4 function in C. elegans. The objectives of this study were to identify cell types where ucp-4 might function and to determine whether ucp-4 has any obvious function in stress tolerance and longevity in C. elegans. Expression analysis indicated that ucp-4 functions in muscles. Mutants lacking ucp-4 function contained elevated ATP levels, consistent with an uncoupling function of Cel-UCP-4. Interestingly, ucp-4 may be necessary for survival at cold temperature, although ucp-4 function was dispensable for resistance to oxidative and thermal stress and ucp-4 mutants had normal lifespan. These findings suggest that ancestral UCP-like proteins may have evolved to balance ATP synthesis in muscles, and possibly, other tissues, but may not have been essential for resistance to oxidative stress. These studies should complement further analysis of mitochondrial function in ucp-4 mutants.
2. Materials and methods
2.1. Nematode growth and mutant strains
Nematodes were grown according to standard protocols (Sulston and Hodgkin, 1988). Animals were cultured on NGM agar plates seeded with Escherichia coli strain OP50. The following strains were used in this study: N2 (Bristol), wildtype; TK22, mev-1(kn1); and CY121, ucp-4(ok195). The ucp-4(ok195) deletion was generated by the C. elegans Gene Knockout Consortium. Once isolated, the ok195 deletion was backcrossed five times into the wildtype N2 background, to create CY121. PCR with a set of nested primer pairs was used for genotyping the deletion. The first PCR product was amplified using primers EL1 (agtcctgaacggagctttga) and ER1 (tacaatggcagcagcaagtc) and the products were re-amplified with a nested primer set, IL1 (tcgcacattggtttgttgtt) and IR1 (aacggcatgagttagccaat). The deleted region in the ok195 mutation was determined by sequencing the ucp-4 locus in ok195 animals.
2.2. Expression analysis
A 2-kilobase (kb) fragment upstream of the ucp-4 presumptive ATG start was PCR-amplified with primers containing unique HindIII and BamHI restriction sites. The PCR product was digested with HindIII and BamHI to produce a 1.8-kb fragment that was ligated to the HindIII and BamHI sites of the pPD95.81 vector, which contains a C. elegans-optimized GFP, to create pCAW129. Animals were transformed by standard microinjection-mediated transformation at an injection concentration of 50 ng/μL for the ucp-4: GFP plasmid with the pRF4 rol-6 coinjection marker at 80 ng/μL (Mello et al., 1991). GFP fluorescence was examined using a Nikon E800 microscope with appropriate filter sets and images were collected with a Hamamatsu ORCA-ER digital camera. For Fig. 2(c) and (d), serial Z-sections were collected and out-of-focus light removed by deconvolution using OpenLab software (Improvision Inc.).
Fig. 2.
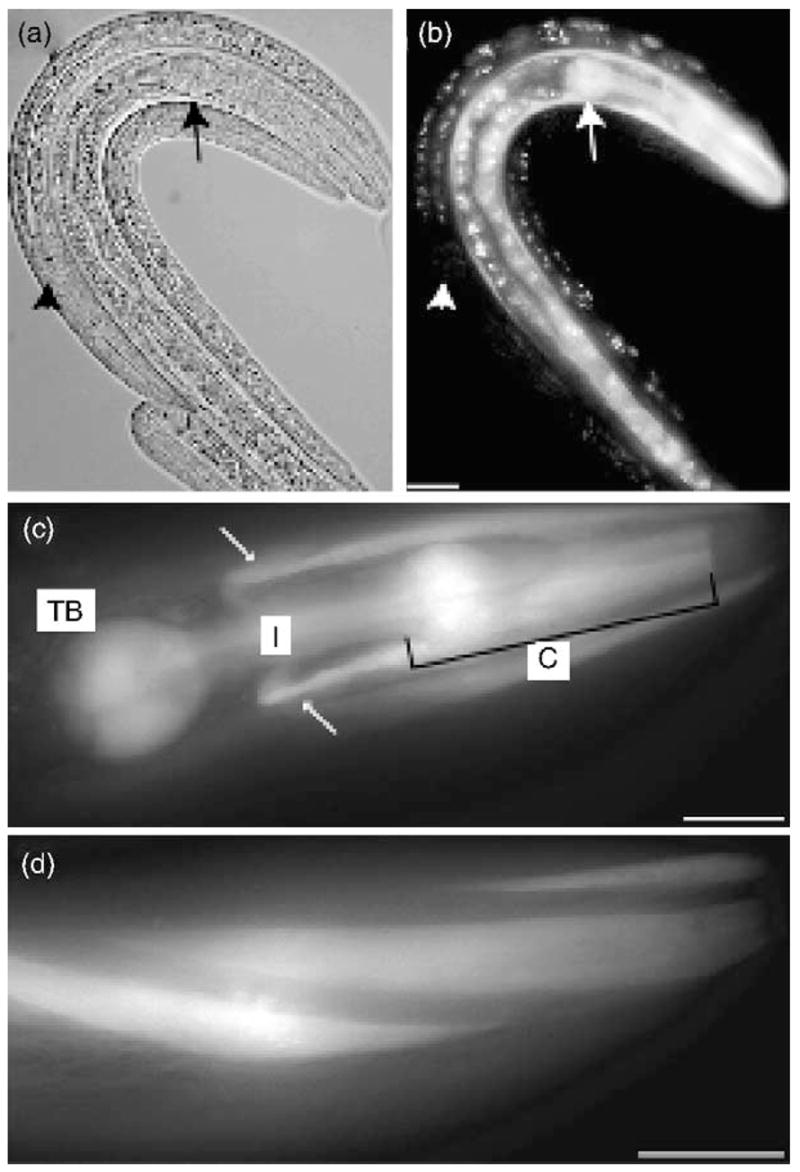
Expression analysis of ucp-4. (a) Nomarski image of three larvae at approximately the third larval stage, (b) Same animals as (a) showing fluorescence from ucp-4p::GFP expression in one transgenic animal (center), but not in two non-transgenic control animals (left and right), ucp-4p:GFP fluorescence was detected in the pharynx (large arrow) and head muscles in transgenic animals carrying the ucp-4p:GFP construct, but not in non-transgenic controls (large white arrowhead), (c) Higher magnification view of ucp-4p::GFP expression in head showing fluorescence in the pharyngeal corpus (C), isthmus (I) and terminal bulb (TB). Arrows point to UCP-4p:GFP expression in head muscles, (d) UCP-4p:GFP fluorescence in head muscles. All scale bars represent 20 μm.
2.3. ATP determinations
ATP levels in purified eggs and L4 larvae were measured as described (Ronner et al., 1999). Eggs were harvested from adult animals grown on NGM plates using bleach/hypochlorite treatment (Sulston and Hodgkin, 1988). For larval populations, eggs were plated on NGM plates with bacterial food and hatched larvae were allowed to develop to the L4 stage (approximately 40 h at 25 °C). For ATP determination, eggs or larvae were suspended in extraction medium (0.1 M NaOH, 0.5 mM EDTA) and incubated at 60 °C for 20min, then frozen at −80 °C. Lysates were diluted and added to the assay solution (glycylglycine (250 mM, pH 7.4), EGTA (2 mM), MgCl2 (2 mM), BSA (0.4 g/L), DTT (7.5 mM)) with luciferin (0.015 mM) and luficerase (10 μg/mL). Light output was measured for 10 s on a Victor luminescence counter and ATP concentrations were calculated relative to known standards.
2.4. JC-1 staining
Mitochondria membrane potential was examined using the stain JC-1 (5,5′,6,6′-tetrachloro-1,1′,3,3′-tetraethylben-zimidazolylcarbocyanide iodide, Molecular Probes). JC-1 was dissolved in DMF at a concentration of 1 mg/mL and was added to young adult animals in M9 buffer (34 mM KH2PO4, 42 mM Na2HPO4, 86 mM NaCl, 1 mM MgSO4) at concentrations of 0.066, 0.2, 0.66 and 2 μg/mL. Animals were incubated in stain for 3 h at room temperature, mounted on 2% agar pads, and then paralyzed with levamisole. Duplicate images of intestinal cell mitochondria were taken at 40 × magnification, first with an Endow GFP filter then a TRITC filter set (Chroma Filters). OpenLab v. 3.5 (Improvision Inc., Lexington, MA) was used to measure the intensity of red- or green-channel fluorescence for individual intestinal mitochondria and the red:green ratio was calculated. For each concentration of JC-1, at least five worms were photographed, and at least 10 mitochondria measurements were taken for each worm.
2.5. Phenotypic characterization
For all assays, we examined the ucp-4(ok195) allele only after at least four backcrosses against wildtype (N2) animals. For lifespan assays, overnight egg lays were performed to obtain synchronized populations. After development into young adults, animals were transferred to NGM plates spread with OP50 E. coli and supplemented with 5-fluorodeoxyuracil (FUDR; 50–100 mg/L) to inhibit progeny growth and maintained at 20 °C. The number of animals alive was scored every 1–3 days. Animals that did not respond to a gentle touch with a platinum wire were scored as dead.
To test survival under cold stress, synchronous populations were grown at 20 °C. Young adult animals on the first day of egg-laying were transferred to 2.5 °C on standard NGM plates. After 24 h, animals were removed and placed at room temperature (25 °C) for 1 h to recover. Worms were scored as alive if they were able to respond to a gentle prod with a platinum wire.
For heat stress assays, survival of 4-day adults at 35 °C was determined by the ability to respond to a gentle stroke with a platinum wire. Animals that did not respond to this mechanical stimulus were scored as dead.
Two assays were used to test for sensitivity to paraquat-induced oxidative stress. To assay development in the presence of paraquat, adult hermaphrodites were transferred to paraquat-containing agar medium and allowed to lay eggs for 6 h. The developmental stage of the hatched larvae was then scored after 48 h at 25 °C. Animals that crawled off the plates during the assay were censored. To assay survival of adult animals in the presence of paraquat, young adult animals were transferred to NGM plates containing 10 mM paraquat, which were supplemented with 50 μg/mL FUDR to inhibit progeny production. Survival was scored at 24-h intervals as ability to respond to gentle prodding with a platinum wire.
3. Results
3.1. Expression of a C. elegans UCP-like protein
In order to determine potential functions for uncoupling proteins in C. elegans, we characterized the sole C. elegans putative UCP homolog, which has been designated Cel-ucp-4, or more simply, ucp-4 (Hanak and Jezek, 2001). To determine the cell types where ucp-4 might function, we constructed a transcriptional reporter by expressing green fluorescent protein (GFP) from a 1.8-kb fragment containing the presumptive ucp-4 promoter (Fig. 1(a)). GFP expression from this construct was primarily observed in head muscles and the pharynx, where bacterial food is ingested and crushed (Fig. 2). Non-transgenic animals lacking the ucp-4:GFP construct did not display fluorescence in any of these cells (Fig. 2(b)). In some animals, GFP expression was also observed in the body wall muscles, which are used for locomotion. We found no evidence that GFP expression from the ucp-4 promoter was affected by temperature (not shown).
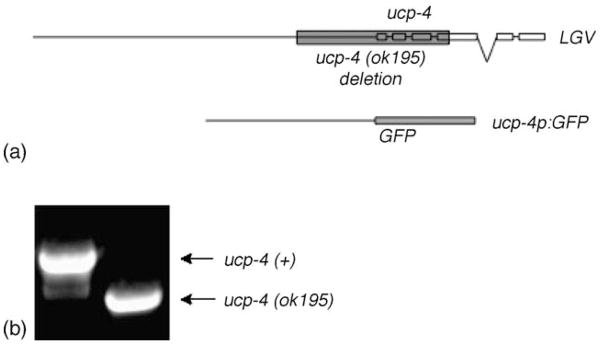
Fig. 1.
(a) Genomic organization of the ucp-4 locus, within a 5-kb fragment of chromosome V. The position of the 1.5-kb deleted region in ok195 is shown as a gray bar. The 1.8-kb promoter fragment used for the transcriptional fusion to GFP is shown below, (b) Genotypic analysis by PCR using primers flanking the ok195 deletion produced a 1.5 kb smaller product from the ok195 deletion allele than from wildtype genomic DNA.
3.2. ATP levels and development in ucp-4(ok195) animals
To determine whether ucp-4 activity could affect metabolism in vivo, we examined animals lacking ucp-4 function due to a 1.5-kb deletion in the ucp-4 gene. Sequencing revealed that the ucp-4(ok195) deletion removed 777 base pairs of the presumptive ucp-4 promoter and 641 base pairs from the coding region, including exons 1–3 and part of exon 4 (Fig. 1). To determine if mitochondrial function was altered in ucp-4(ok195) mutants, ATP levels were measured in eggs and in fourth stage (L4) larvae, prior to the adult molt. Consistent with the expectation that ucp-4 can dissipate the mitochondrial proton gradient to antagonize F0/F1 ATP synthase, ATP levels were significantly higher in ucp-4(ok195) eggs and larvae (Fig. 3(a and b)).
Fig. 3.
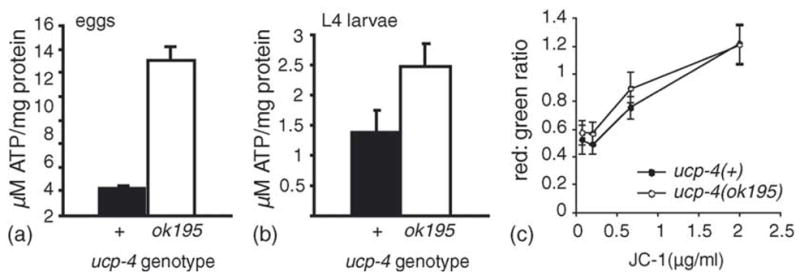
Elevated ATP levels in ucp-4(ok195) animals. ATP levels in wildtype (filled) and ucp-4(ok195) (open) eggs (a) and L4 larvae (b). The error bars represent S.E.M. from two experiments, and four replicates of each genotype were performed, (c) JC-1 staining was performed to compare mitochondrial membrane potential of wildtype and ucp-4(ok195) animals. JC-1 mitochondrial accumulation is mitochondrial membrane potential dependent, and greater JC-1 concentrations promote dye aggregation, shifting fluorescence from green to red. The red:green ratio of mitochondrial JC-1 fluorescence reflects mitochondrial membrane potential and tended to be slightly greater in ucp-4(ok195) animals than in wildtype animals, although this difference was only statistically significant at 0.66 μg/mL (p = 0.01, t-test).
We used the potential-sensitive dye, JC-1, to determine if ucp-4(ok195) animals had higher mitochondrial potential than wildtype animals. At low concentrations, JC-1 exists in the green-fluorescent monomeric form. At higher concentrations, the dye forms red-fluorescent aggregates. Thus, the ratio of red:green JC-1 fluorescence in mitochondria can be a reflection of the relative mitochondrial potential (Senoo-Matsuda et al., 2003). To compare the relative membrane potential of wildtype and ucp-4(ok195) mitochondria, intact animals at the young adult stage were incubated for 3 h with JC-1. Over a range of JC-1 concentrations, there was a trend for a slightly greater ratio of red:green fluorescence in intestinal mitochondria from ucp-4(ok195) animals, consistent with slightly higher mitochondrial membrane potential in the absence of ucp-4 activity (Fig. 3(c)). However, this difference was statistically significant at only one concentration (0.66 μg/mL, p = 0.01, t-test). These findings suggest that mitochondrial membrane potential may be slightly elevated in ucp-4(ok195) animals, as would be expected for greater mitochondrial coupling in this strain.
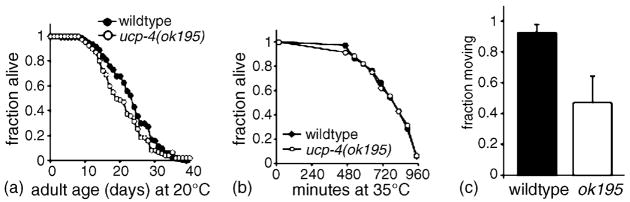
Development of ucp-4(ok195) animals was slightly retarded compared with wildtype animals, as ucp-4(ok195) larvae developed into young adults a few hours after wildtype animals when grown at 20 °C (not shown). However, aside from this modest difference in developmental rate, we observed no overt differences between wildtype and ucp-4(ok195) animals. Adult lifespan was identical between wildtype and ucp-4(ok195) animals (Fig. 4(a)). Adult fertility was examined under both normal growth conditions and at the slightly stressful temperature of 27 °C. Although egg production tended to be slightly reduced in ucp-4(ok195) animals at 27 °C, this difference was not statistically significant (p > 0.05, t-test) (not shown). ucp-4(ok195) animals exhibited normal development into and recovery from dauer larval arrest, a developmental diapause optimized for long-term survival under harsh environmental conditions (data not shown).
Fig. 4.
Lifespan and development in ucp-4(ok195) animals, (a) Adult lifespan of wildtype (filled symbols, n = 167) and ucp-4(ok195) (open symbols, n = 116) animals at 20 °C. Curve shows the average of two independent trials; consistent results were obtained in a third trial, (b) Survival at high temperature (35 °C) of wildtype (filled symbols) and ucp-4(ok195) (open symbols). Shown is average of two trials with 51–56 animals/trial, (c) Survival of wildtype and ucp-4(ok195) young adults after 24 h incubation at 2.5 °C; average survival for six trials ± S.E.M., n = 40–100 animals/trial; p = 0.046, t-test.
3.3. Requirement of ucp-4 for resistance to cold and heat stress
We next investigated whether ucp-4(ok195) animals had defects in survival at high or low temperatures, ucp-4 was not required for survival at high temperature. Wildtype and ucp-4(ok195) animals displayed similar survival at a stressful temperature, 35 °C, with 50% survival at approximately 720 min for both strains (Fig. 4(b)). Survival during cold stress was examined by placing wildtype or ucp-4(ok195) young adults at 2.5 °C, a temperature which arrests growth but was not lethal for wildtype animals. While nearly 100% of wildtype animals survived a 24-h incubation at 2.5 °C, survival of ucp-4(ok195) animals was significantly reduced (Fig. 4(c)). This observation suggests that ucp-4 may be required for survival in low-temperature environments.
3.4. Requirement of ucp-4 for resistance to oxidative stress
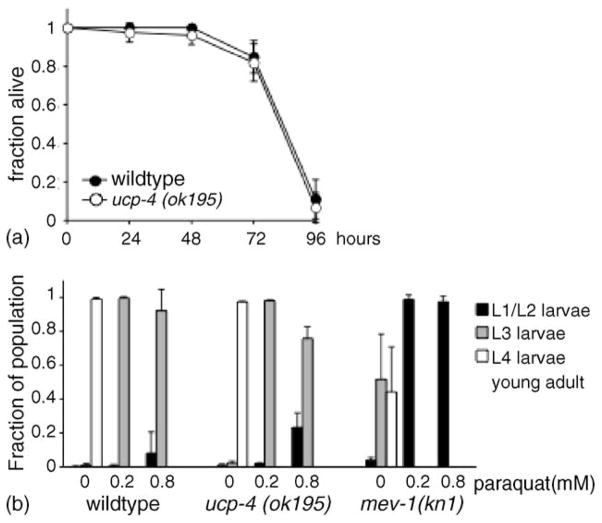
To examine whether ucp-4 was required for resistance to oxidative stress, survival of wildtype and ucp-4(ok195) animals was examined in the presence of 10 mM paraquat, an intracellular free radical generating compound. Adult wildtype and ucp-4(ok195) animals treated with 10 mM paraquat exhibited similar rates of survival (100% death after 96 h) (Fig. 5(a)). Resistance to oxidative stress was also assayed as the effect of paraquat on developmental rates of wildtype and ucp-4(ok195) larvae. This assay was previously used to examine paraquat sensitivity in strains defective for components of the mitochondrial electron transport chain (Feng et al., 2001). The presence of paraquat in growth medium was associated with a significant slowing of wildtype development (Fig. 5(b)). After 48 h, untreated wildtype animals had progressed through the fourth larval stage, while larvae grown in the presence 0.2 mM paraquat had only progressed to the third larval stage. Paraquat did not fully arrest development, as most wildtype animals developed into fertile adults after 72 h. In the presence of paraquat, development of ucp-4(ok195) animals was not significantly different from wildtype animals, indicating that ucp-4(ok195) animals were not more sensitive to paraquat. As a positive control for paraquat hypersensitivity, we examined mev-1(kn1) animals, which contain elevated superoxide levels as the result of a defect affecting mitochondrial complex II (Ishii et al., 1998; Senoo-Matsuda et al., 2001). Under normal conditions, mev-1(kn1) larvae developed slightly slower than wildtype animals. However, paraquat treatment severely affected development of mev-1(kn1) animals. After 48 h in the presence of paraquat, nearly all mev-1(kn1) animals were arrested as L1 larvae (Fig. 5(b)). At the concentrations tested, paraquat caused developmental arrest of mev-1(kn1) animals, in contrast to wildtype and ucp-4(ok195) animals, whose development was slowed, but not arrested.
Fig. 5.
Oxidative stress tolerance of ucp-4(ok195) animals, (a) Survival of adult wildtype (filled) or ucp-4(ok195) (open) animals in the presence of the free-radical generating compound, paraquat (10 mM) at 25 °C. Results are average of four trials; (n) wildtype, 73 animals total in all trials; ok195, 72 animals in all trials, (b) Larval development in the presence of paraquat. Eggs were laid onto NGM agar medium containing 0, 0.2 or 0.8mM paraquat and allowed to develop for 48 h at 25 °C. Development of wildtype (left, average n = 140 ± 61 animals/dose/trial), ucp-4(ok195) (center, average n = 102 ± 45 animals/dose/trial), and mev-1(kn1) (right, average n = 46 ± 24 animals/dose/trial) animals is shown. Graphs show the means and standard deviations of results from four trials conducted on two different days. Paraquat concentrations greater than 0.8 mM severely impaired larval development in all strains.
4. Discussion
We have described the genetic characterization of a putative C. elegans UCP homolog, ucp-4. Our analysis of the ucp-4-transcriptional GFP reporter indicated that major sites for UCP-4 expression include the muscles of the head, pharynx and body. ucp-4(ok195) embryos and larvae contained elevated ATP levels, consistent with a role in mitochondrial function. Together, these findings suggest that ucp-4 may function in muscles, and possibly other cells, to antagonize ATP production. Our analysis also indicated that deletion of ucp-4 function was not detrimental for stress resistance or lifespan, although ucp-4 mutant animals exhibited reduced survival at low temperature.
While the homology between the C. elegans UCP-4 and mammalian UCP4 proteins is suggestive of shared function, a direct examination of mitochondrial coupling in ok195 animals would be required to establish this fact. While it is feasible to isolate mitochondria from C. elegans nematodes, the purpose of this study was to characterize whole animal phenotypes associated with ucp-4. Using the potential-sensitive dye, JC-1, we did not observe a dramatic increase in JC-1 aggregate formation in ucp-4(ok195) animals, which would have been expected if the absence of uncoupling protein activity resulted in greater mitochondrial membrane potential. It is possible that any increase of the mitochondrial proton gradient in ucp-4(ok195) animals was dissipated through the f0/f1 ATP synthase. Consistent with this hypothesis, ATP levels were elevated in ucp-4(ok195) animals. Future studies of mitochondrial function in ucp-4(ok195) animals will be needed to fully ascertain this gene’s role in mitochondrial uncoupling and in ROS production.
Five different UCPs, with different tissue specificities, have been identified in mammals, UCP1–4 and BMCP. The C. elegans UCP-4 protein is most similar in sequence to mammalian UCP4, which is predominately expressed in the brain (Hanak and Jezek, 2001). Analysis of the tissue specificity of ucp-4 expression indicates that this protein may function in the body wall and pharyngeal muscles. Although transgene expression patterns do not necessarily reflect the full tissue specificity of the endogenous gene, they are useful for identifying at least a portion of the gene’s expression pattern. The pharynx is the feeding organ of the animal and pumps in rhythmic contractions approximately 250 times/min in larvae and young adults. The body wall muscles drive locomotion of the nematode. These muscles likely have high metabolic demands, which may benefit from UCP-4 function under certain circumstances. Our analyses did not reveal any overt deficiencies in pharyngeal function or locomotion in ucp-4(ok195) animals (CAW, unpublished observations).
Interestingly, ucp-4(ok195) animals exhibited a defect in survival in cold environments. This phenotype suggests that UCP-4 is necessary for maintaining homeostasis at low temperature in C. elegans. This finding is confounding, as C. elegans are poikliothermic and, as such, are not able to regulate internal body temperature in response to external temperature conditions. C. elegans nematodes are found in temperate climates and may experience periods of cold temperature during the winter. The pathways necessary for overwintering in C. elegans have not been intensively studied. One possibility that emerges from our findings is that UCP-4 may perform a thermogenic function that augments other organismal or developmental strategies for cold-temperature survival.
Genetic ablation of UCP function in mammalian cells has been correlated with increased cellular ROS levels and oxidative stress (Kim-Han et al., 2001; Krauss et al., 2003; Vidal-Puig et al., 2000). In ucp-4(ok195) animals, we observed increased ATP levels, but no deleterious effects on lifespan or stress tolerance. In other C. elegans mutants, longevity has also been found to be independent of ATP levels (Braeckman et al., 1999). Although levels of ROS or oxidative byproducts were not directly measured for this study, previous studies of mev-1(kn1) animals showed that elevated superoxide levels can correlate with increased sensitivity to paraquat-induced oxidative stress (Hartman et al., 2001; Senoo-Matsuda et al., 2001). The finding that ucp-4(ok195) animals were not more sensitive to paraquat than wildtype animals suggests that ok195 animals may not be subject to significantly increased ROS levels. One possible explanation is that any elevation of ROS in ucp-4(ok195) animals was minor as compared with the levels needed to raise sensitivity to paraquat and to shorten lifespan, as has been hypothesized for mammalian cells lacking UCP2 (Krauss et al., 2003). The genetic characterization of ucp-4 presented here provides a starting point for detailed investigation of mitochondrial function in these animals.
Acknowledgments
We thank the C. elegans Gene Knockout Consortium for isolating the ucp-4(ok195) deletion allele and the Caenor-habditis Genetics Center for providing strains. The GFP reporter plasmid, pPD95.81, was provided by Dr. A. Fire (Stanford University).
References
- Arsenijevic D, Onuma H, Pecqueur C, Raimbault S, Manning BS, Miroux B, Couplan E, Alves-Guerra MC, Goubern M, Surwit R, Bouillaud F, Richard D, Collins S, Ricquier D. Disruption of the uncoupling protein-2 gene in mice reveals a role in immunity and reactive oxygen species production. Nat Genet. 2000;26:435–439. doi: 10.1038/82565. [DOI] [PubMed] [Google Scholar]
- Braeckman B, Houthoofd K, De Vreese A, Vanfleteren J. Apparent uncoupling of energy production and consumption in long-lived Clk mutants of Caenorhabditis elegans. Curr Biol. 1999;9:493–496. doi: 10.1016/s0960-9822(99)80216-4. [DOI] [PubMed] [Google Scholar]
- Brand MD, Affourtit C, Esteves TC, Green K, Lambert AJ, Miwa S, Pakay JL, Parker N. Mitochondrial superoxide: Production, biological effects, and activation of uncoupling proteins. Free Radical Biol Med. 2004;37:755–767. doi: 10.1016/j.freeradbiomed.2004.05.034. [DOI] [PubMed] [Google Scholar]
- Echtay KS, Roussel D, St-Pierre J, Jekabsons MB, Cadenas S, Stuart JA, Harper JA, Roebuck SJ, Morrison A, Pickering S, Clapham JC, Brand MD. Superoxide activates mitochondrial uncoupling proteins. Nature. 2002;415:96–99. doi: 10.1038/415096a. [DOI] [PubMed] [Google Scholar]
- Enerback S, Jacobsson A, Simpson EM, Guerra C, Yamashita H, Harper ME, Kozak LP. Mice lacking mitochondrial uncoupling protein are cold-sensitive but not obese. Nature. 1997;387:90–94. doi: 10.1038/387090a0. [DOI] [PubMed] [Google Scholar]
- Erlanson-Altbertsson C. The role of uncoupling proteins in the regulation of metabolism. Acta Physiol Scand. 2003;178:405–412. doi: 10.1046/j.1365-201X.2003.01159.x. [DOI] [PubMed] [Google Scholar]
- Feng J, Bussiere F, Hekimi S. Mitochondrial electron transport is a key determinant of life span in Caenorhabditis elegans. Devel Cell. 2001;1:633–644. doi: 10.1016/s1534-5807(01)00071-5. [DOI] [PubMed] [Google Scholar]
- Hanak P, Jezek P. Mitochondrial uncoupling proteins and phylogenesis—UCP4 as the ancestral uncoupling protein. FEES Lett. 2001;495:137–141. doi: 10.1016/s0014-5793(01)02338-9. [DOI] [PubMed] [Google Scholar]
- Hartman PS, Ishii N, Kayser EB, Morgan PG, Sedensky MM. Mitochondrial mutations differentially affect aging, mutability and anestetic sensitivity in Caenorhabditis elegans. Mech Ageing Dev. 2001;122:1187–1201. doi: 10.1016/s0047-6374(01)00259-7. [DOI] [PubMed] [Google Scholar]
- Ishii N, Fujii M, Hartman PS, Tsuda M, Yasuda K, Senoo-Matsuda N, Yanase S, Ayusawa D, Suzuki K. A mutation in succinate dehydrogenase cytochrome b causes oxidative stress and ageing in nematodes. Nature. 1998;394:694–697. doi: 10.1038/29331. [DOI] [PubMed] [Google Scholar]
- Kim-Han JS, Reichert SA, Quick KL, Dugan LL. BMCP1: a mitochondrial uncoupling protein in neurons which regulates mito-chondrial function and oxidant productions. J Neurochem. 2001;79:658–668. doi: 10.1046/j.1471-4159.2001.00604.x. [DOI] [PubMed] [Google Scholar]
- Krauss S, Zhang C-Y, Scorrano L, Dalgaard LT, St-Pierre J, Grey ST, Lowell BB. Superoxide-mediated activation of uncoupling protein 2 causes pancreatic ß cell dysfunction. J Clin Inves. 2003;112:1831–1842. doi: 10.1172/JCI19774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mello CC, Kramer JM, Stinchcomb D, Ambros V. Efficient gene transfer in C. elegans: Extrachromosomal maintenance and integration of transforming sequences. EMBO J. 1991;10:3959–3970. doi: 10.1002/j.1460-2075.1991.tb04966.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronner P, Friel E, Czerniawski K, Frankle S. Luminometric assays of ATP, phosphocreatine, and creatine for estimation of free ADP and free AMP. Anal Biochem. 1999;275:208–216. doi: 10.1006/abio.1999.4317. [DOI] [PubMed] [Google Scholar]
- Senoo-Matsuda N, Hartman PS, Akatsuka A, Yoshimura S, Ishii N. A complex II defect affects mitochondrial structure, leading to ced-3- and cec-4-dependent apoptosis and aging. J Biol Chem. 2003;278:22031–22036. doi: 10.1074/jbc.M211377200. [DOI] [PubMed] [Google Scholar]
- Senoo-Matsuda N, Yasuda K, Tsuda M, Ohkubo T, Yoshimura S, Nakazawa H, Hartman PS, Ishii N. A defect in the cytochrome b large subunit in complex II causes both superoxide anion overproduction and abnormal energy metabolism in Caenorhabditis elegans. J Biol Chem. 2001;276:41553–41558. doi: 10.1074/jbc.M104718200. [DOI] [PubMed] [Google Scholar]
- Sulston J, Hodgkin J. Methods. In: Wood WB, editor. The nematode Caenorhabditis elegans. Cold Spring Harbor Press; Plain-view, NY: 1988. pp. 587–606. [Google Scholar]
- Vidal-Puig AJ, Grujic D, Zhang CY, Hagen T, Boss O, Ido Y, Szczepanik A, Wade J, Mootha V, Cortright R, Muoio DM, Lowell BB. Energy metabolism in uncoupling protein 3 gene knockout mice. J Biol Chem. 2000;275:16258–16266. doi: 10.1074/jbc.M910179199. [DOI] [PubMed] [Google Scholar]