Abstract
The photolysis of adenosylcobalamin (coenzyme B12) results in homolytic cleavage of the Co-C5′ bond, forming cob(II)alamin and the 5′-deoxyadenosyl radical. In the presence of molecular oxygen, it has been proposed that the primary reaction is interception of the 5′-deoxyadenosyl radical by O2 to form adenosine-5′-aldehyde as the product (Hogenkamp, H. P. C., Ladd, J. N., and Barker, H. A. (1962) J. Biol. Chem. 237, 1950—1952). 5′-Peroxyadenosine is here found to be the initial nucleoside product of this reaction and that it decomposes to adenosine-5′-aldehyde. Evidence indicates that 5′-peroxyadenosine arises from the hydrolysis of 5′-peroxyadenosylcobalamin, with the formation of cob(III)alamin. 5′-Peroxyadenosine undergoes further decomposition to adenosine-5′-aldehyde as the major final product of aerobic photolysis, as well as to adenosine and adenine as minor products. In a cobalamin-dependent process, 5′-peroxyadenosine becomes re-ligated to cob(III)alamin to form 5′-peroxyadenosylcobalamin, which quickly decomposes to adenosine-5′-aldehyde and cob(III)alamin. This is supported by spectrophotometric observations of both rapidly photolyzed adenosylcobalamin and of the reaction of cob(III)alamin with excess 5′-peroxyadenosine. 5′-Peroxyadenosine also slowly undergoes cobalamin-independent decomposition to adenosine-5′-aldehyde and the minor products adenosine and adenine. The present study provides a detailed description of the products initially formed when aqueous, homolytically cleaved adenosylcobalamin reacts with molecular oxygen and of the behavior of those products subsequent to photolysis.
Enzymes dependent upon the vitamin B12-coenzyme adenosylcobalamin1 catalyze intriguingly diverse chemical reactions, including carbon-skeleton rearrangements, reductive heteroatom eliminations, and intramolecular 1,2-migrations of heteroatomic substituents; and they proceed by mechanisms involving organic free radicals as intermediates (1,2). Adenosylcobalamin incorporates a cobalt ion in an octahedral arrangement with the pyrroline and pyrrolidine groups of a corrin ring system making up the equatorial ligands, the nitrogen from a dimethylbenzimidazole moiety as the lower axial ligand, and a 5′-deoxadenosyl moiety forming a cobalt-carbon (Co-C5′) bond in the sixth, upper axial ligand position (3).
The key to the function of adenosylcobalamin in enzymatic catalysis is the reversible homolytic scission of the Co-C5′ bond, generating cob(II)alamin and a transiently formed 5′-deoxyadenosyl radical (4–9). The bond dissociation energy of the Co-C5′ bond is 30–33 kcal mol−1, and in aqueous solution at 25 °C, thermolysis proceeds quite slowly with a rate constant on the order of 10−9 s−1, corresponding to a half-life of 22 years (10–15). The rate of Co-C5′ homolysis is drastically increased at enzymatic sites, proceeding with a rate enhancement of approximately 1012 (12,16). The mechanism employed to enhance the rate of thermal dissociation enzymatically is poorly understood.
The non-enzymatic, photolytic decomposition of adenosylcobalamin, also resulting in the formation of cob(II)alamin and the 5′-deoxyadenosyl radical (17–19), has been regarded as a model system for the enzymatic, thermal decomposition of the coenzyme. Anaerobically, cob(II)alamin and 5′-deoxy-5′,8-cycloadenosine are formed, while aerobically cob(III)alamin and adenosine-5′-aldehyde are reported as the major products of photolysis (20,21).
The present study investigates the oxidized nucleoside products derived from the aerobic photodissociation of the Co-C5′ bond in adenosylcobalamin. We study this process in an atmosphere of 100% O2 to suppress the anaerobic process. In addition to adenosine-5′-aldehyde and cob(III)alamin, we find that interception of cob(II)alamin and the 5′-deoxyadenosine radical by O2 leads initially to 5′-peroxyadenosine and cob(III)alamin, as well as minor amounts of adenine and adenosine. Furthermore, it was determined that 5′-peroxyadenosine decomposes to adenosine-5′-aldehyde and that this process is catalyzed by cob(III)alamin. The results shed further light on the aerobic photolysis of adenosylcobalamin and also create another foundation to explore the behavior of the coenzyme in its enzymatic process.
MATERIALS AND METHODS
Materials
Adenine, adenosine, 5′-deoxyadenosine, adenosylcobalamin, hydroxocobalamin hydrochloride (cob(III)alamin), EPPS, and MES were purchased from Sigma. Monobasic phosphate buffer was purchased from Fisher. Oxygen gas (99.5%) was purchased from Aldrich. Columns used in HPLC analysis were filled with Altima-HP C18 reverse phase resin, purchased pre-packed from Alltech. The column used in LC/MS analysis was filled with Inertsil C18 reverse phase resin (GL Sciences), packed in-house. The FOX assay was performed using reagents from the Peroxoquant kit (Pierce).
Photolysis of Adenosylcobalamin in Oxygen Equilibrated Water
A steady stream of oxygen gas was bubbled through 4 mL of water for 30 minutes. Oxygenated water (990 µL) was placed in a glass test tube 1 cm in diameter and made to be 100 µM adenosylcobalamin with the addition of 10 µL of a 10 mM solution. Oxygen was again bubbled through the solution for 5 minutes in the dark. The sample was irradiated for 4 minutes with a 100 W flood lamp placed approximately 1 cm from the test tube, with continued oxygen sparging for the duration of the photolysis.
After photolysis, 100 µL aliquots were placed in small-volume auto-sampler target vials and sealed with airtight caps. A standard HPLC method was employed to separate and quantify the nucleoside products. Sample vials were loaded into the HPLC auto-sampler, and injections were scheduled at indicated times. All procedures were performed under dim lighting.
Analytical Methods
A standard method was developed to separate and quantify the nucleoside products of adenosylcobalamin photolysis using a Beckman Coulter System Gold HPLC apparatus equipped with a model 128 diode array detector. Reaction products were separated on a 4.6 × 250 mm Altima-HP analytical C18 reverse phase column. Sample vials were loaded into the auto-sampler and 50 µL injections were scheduled. Separation proceeded by use of the following elution program: flow 1 mL min−1, isocratic 100% H2O for 5 min; ramp to 95% H2O/5% CH3CN over 1 min, isocratic 95% H2O/5% CH3CN for 25 min; ramp to 70% H2O/30% CH3CN over 10 min, isocratic 70% H2O/30% CH3CN for 10 min. If the sample contained cobamides, the column was reconditioned with multiple washes, cycling from 5% to 95% CH3CN at 25 minutes per cycle. Nucleoside product concentrations were determined by integration of total peak area using the Beckman Coulter 32 Karat software (v 7.0.1048) from the absorbance output measured at 260 nm and compared to a standard curve.
A standard curve was created by using known concentrations of adenine and adenosine and running them by the standard HPLC method. Integration of total peak area from the absorbance output of the HPLC diode array detector at 260 nm was used to relate total absorbance to moles of standard in a 50 µL injection. The standard curve used to determine the concentration of any experimental peak was chosen by proximity of elution time to the standard. The adenine curve was used for early peaks, and the adenosine curve was used for later peaks.
Mass spectral analysis was performed using an Agilent LC/MSD ESI-TOF mass spectrometer in the positive ion mode. A reaction sample was prepared as described (see photolysis of adenosylcobalamin in oxygen equilibrated water) and separated on a 2.1 ×200 mm Inertsil C18 reverse phase column in-line with the ESI-TOF detector. Separation was performed isocratically at 95% H2O/5% CH3CN. Peaks were analyzed for m/z by the detector as they emerged. Mass spectrometry was done at the Mass Spectrometry Facility (Biotechnology Center, University of Wisconsin-Madison).
1H NMR analysis was performed on an Avance DMX-600 NMR spectrometer (Bruker Biospin Corp) using a CryoProbe operated at 600.13 MHz or an Avance DMX-400 NMR spectrometer (Bruker Biospin Corp) operated at 400.13 MHz. This work was performed at the National Magnetic Resonance Facility at Madison.2
Peroxides were detected by ferrous oxidation in xylenol orange (FOX assay) (22) in samples from reverse phase HPLC purification (see preparation of 5′-peroxyadenosine for kinetic analysis).
Absorption spectra were recorded on a Cary 50 UV/Vis spectrophotometer using the Cary WinUV software in the scanning kinetics mode.
Characterization of Nucleosides
Photolysis of adenosylcobalamin in oxygen equilibrated water produced four major non-cobamide products. Of these four compounds, two constituted the bulk of the products observed and were named nucleoside A and nucleoside B for their order of elution upon C18 reverse phase HPLC. Both products contained an adenine moiety, as determined spectrophotometrically from the absorbance output of the HPLC detector (λmax 206 nm and 259 nm, λmin 227 nm).
Samples of photolyzed adenosylcobalamin containing enough material for NMR analysis of nucleoside B were prepared and separated on a 10 ×250 mm Altima-HP analytical C18 reverse phase column. In a small test tube, 9 mg of adenosylcobalamin was dissolved in 2 mL of oxygen equilibrated water. Oxygen gas was bubbled through the solution for 5 minutes in the dark. The sample was irradiated for two and a half minutes with a 100 W flood lamp placed approximately 1 cm from the test tube, with continued oxygen sparging for the duration of the photolysis. The sample was quickly cooled and immediately injected into a 2 mL loop on the HPLC apparatus. Separation proceeded with a flow rate of 5 mL min−1, isocratically at 95% H2O/5% CH3OH. After elution of the nucleoside, successive washes with 95% CH3OH removed remaining cob(III)alamin. Fractions containing nucleoside B from 6 repetitions were pooled and dried by rotary evaporation at 30 °C. The sample was further dried on a lyophilizer and resuspended in DMSO-d6: 1H NMR (600 Mhz, DMSO-d6) δ-values 4.06 (dd, 1H), 4.11 (d, 1H), 4.15 (d, 1H), 4.16 (dd, 1H), 4.63 (d, 1H), 5.32 (d, 1H), 5.52 (d, 1H), 5.89 (d, 1H), 7.28 (brs, 2H), 8.15 (s, 1H), 8.32 (s, 1H), 11.92 (s, 1H). The mass of nucleoside B was 283.099 amu. The spectroscopic analysis indicated that nucleoside B is 5′-peroxyadenosine.
Samples of photolyzed adenosylcobalamin with enough material for NMR analysis of nucleoside A were prepared as described above. The photolysis mixture was stored overnight in the dark to allow for conversion of nucleoside B into nucleoside A and separated on a 10 × 250 mm Altima-HP analytical C18 reverse phase column. Samples were injected into a 2 mL loop on the HPLC apparatus. Separation proceeded with a flow rate of 5 mL min−1, isocratically at 95% H2O/5% CH3CN. After elution of the nucleoside, successive washes with 95% CH3CN removed remaining cob(III)alamin. Fractions containing nucleoside A from 3 repetitions were pooled and dried by rotary evaporation at 30 °C. The sample was further dried on a lyophilizer and resuspended in DMSO-d6. Hydrated species: 1H NMR (400 Mhz, DMSO-d6) δ-values 3.82 (d, 1H), 4.14 (d, 1H), 4.64 (d, 1H), 4.70 (d, 1H), 5.18 (d, 1H), 5.75 (d, 1H), 5.88 (d, 1H), 6.31 (d, 2H), 7.33 (brs, 2H), 8.13 (s, 1H), 8.31 (s, 1H). Dehydrated species: 1H NMR (400 Mhz, DMSO-d6) δ-values 4.12 (d, 1H), 4.44 (d, 1H), 4.92 (d, 1H), 5.36 (d, 1H), 5.80 (d, 1H), 6.04 (d, 1H), 7.35 (brs, 2H), 8.14 (s, 1H), 8.40 (s, 1H), 9.75 (s, 1H). The mass of nucleoside A was identified as 283.099 amu. The spectroscopic analysis indicated that nucleoside A is adenosine-5′-aldehyde.
Samples of photolyzed adenosylcobalamin were co-injected with commercial standards. Only two standards, adenine and adenosine, co-migrated with minor peaks found in the photolysis reaction mixture. The masses of these two peaks were determined to be 135.062 amu and 267.104 amu, confirming their identities as adenine and adenosine, respectively.
Anaerobic Decomposition of Nucleoside Products Derived from the Aerobic Photolysis of Adenosylcobalamin
An experiment was performed to monitor the decomposition of nucleoside B (5′-peroxyadenosine) in an anaerobic solution. A sample was prepared as described (see photolysis of adenosylcobalamin in oxygen saturated water). Immediately after irradiation, the sample was placed in a glass cuvette sealed with a stopcock, where it was subjected to partial evacuation for 20 seconds and flushed with argon. This procedure was repeated 6 times. The sample was sealed in a cuvette under a blanket of argon and passed into an anaerobic glove box. Several 100 µL aliquots were placed in small-volume auto-sampler target vials and sealed with airtight caps. These samples were brought out of the glove box and subjected to the standard HPLC method at indicated times. A similar experiment was repeated wherein the sample remained aerobic for the duration of the incubation.
Purification of 5′-Peroxyadenosine for Kinetic Analysis
Nucleoside B, provisionally identified as 5′-peroxyadenosine, was separated from cobamides and other oxidized nucleoside products on an analytical C18 reverse phase column. Water was equilibrated with oxygen gas. Into 1 mL of oxygen equilibrated water, 4.4 mg of adenosylcobalamin was dissolved, and oxygen was bubbled through the solution for 5 minutes in the dark. The sample was irradiated for 4 minutes with a 100 W flood lamp placed approximately 1 cm from the test tube, and sparging with oxygen was continued. The reaction mixture (50 µL) was loaded onto a 4.6 × 250 mm Altima-HP C18 reverse phase column and subjected to the standard HPLC method. The column was visibly overloaded but still provided sufficient separation from all other products of the photolysis.
Determination of Rates of 5′-Peroxyadenosine Decomposition Under Various Conditions
Experiments were performed to observe the rates of decomposition of 5′-peroxyadenosine at 20 °C, 60 °C, and in the presence of 200 µM cob(III)alamin. Purified 5′-peroxyadenosine, from the analytical C18 reverse phase column (see preparation of 5′-peroxyadenosine for kinetic analysis), was adjusted to either pH 5.4 or 8.4 by the addition 1 M phosphate at an appropriate pH to a final concentration of 20 mM. At selected intervals over 24 h at 20 °C, samples were withdrawn, sealed in small-volume target vials, and subjected to the standard HPLC method at periodic times over 24 h. Rates in the presence of 200 µM cob(III)alamin were measured in the same way except for the addition of cob(III)alamin.
For rate measurements at 60 °C, solutions of 5′-peroxyadenosine were prepared identically to those at room temperature but placed in a 60 °C water bath. Approximately every 45 minutes, a 100 µL aliquot was removed and placed in the 250 µL small-volume insert for the auto-sampler target vial. The insert was then transferred into an auto-sampler target vial that contained 1.5 mL of water at room temperature. After 4 minutes, the sample was run using the standard HPLC method at the indicated times.
An experiment was performed in which 200 µM adenosylcobalamin was photolyzed, and the rate of decomposition of unpurified 5′-peroxyadenosine was observed over 20 hours. The reaction mixture was prepared as described above (see photolysis of adenosylcobalamin in oxygen equilibrated water), but included the addition of 20 µL of 10 mM adenosylcobalamin instead of 10 µL.
Absorption Spectroscopy in the Photolysis of Adenosylcobalamin
A stream of oxygen gas was bubbled through water for 30 minutes, and 1 mL was placed in a glass test tube 1 cm in diameter and brought to 80 µM adenosylcobalamin. Oxygen bubbling continued through the solution for 5 minutes in the dark. The sample was irradiated for 45 seconds with a 100 W flood lamp placed approximately 1 cm from the test tube, with continued oxygen sparging for the duration of the photolysis. Immediately after photolysis, the sample was adjusted to pH 8.5 in 10 mM EPPS and placed in the spectrophotometer, where absorption spectra were periodically acquired over 20 hours.
A complementary sample was prepared containing 80 µM cob(III)alamin, 10 mM EPPS at pH 8.5, and 400 µM 5′-peroxyadenosine. Immediately after the addition of 5′-peroxyadenosine, the sample was placed in the spectrophotometer where absorption spectra were acquired periodically over 1 hour. In this and the preceding experiment, the absorption spectrum of 80 µM cob(III)alamin in 10 mM EPPS at pH 8.5 was obtained for comparison with spectra recorded of test solutions.
RESULTS
Photolysis of Adenosylcobalamin in O2-Saturated Water
Photolysis products in O2-saturated water are cob(III)alamin, identified spectrophotometrically, and products derived from the 5′-deoxyadenosyl moiety. Figure 1 shows the HPLC elution profile of the adenine nucleosides. The two major products of aerobic photolysis are designated nucleosides A and B, named for the order of elution upon C18 reverse phase chromatography. Other products are detectable amounts of adenine (135 amu) and adenosine (267 amu), identified by spectrophotometry (λmax 206 nm and 259 nm, λmin 227 nm) by chromatographic co-migration with standards, and by mass analysis through LC/MS (Figure 1).
Figure 1. HPLC elution profile of aerobically photolyzed adenosylcobalamin.
Adenosylcobalamin (100 µM) was irradiated with a 100 W flood lamp for 4 minutes and subjected to reverse phase chromatography. (A) Chromatography immediately after photolysis. Above each peak is the mass as acquired by ESI-TOF mass spectrometry. (B) Sample made anaerobic immediately after photolysis and aged 14 hours before chromatography. The compounds identified are: RT 12.6 min, adenosine-5′-aldehyde hydrate; RT 12.9 min, adenine; RT 16.9 min, 5′-peroxyadenosine; RT 17,4 min, adenosine. Inset: Decomposition of 5′-peroxyadenosine under aerobic ( , 2 µM/h) and anaerobic (
, 2 µM/h) and anaerobic ( , 4 µM/h) conditions in the presence of cob(III)alamin from photolysis.
, 4 µM/h) conditions in the presence of cob(III)alamin from photolysis.
Figure 1A is the reverse phase HPLC elution profile of adenine-products immediately after photolysis, showing a significant amount of nucleoside A, traces of adenine and adenosine, and the major product nucleoside B. With the passage of time after photolysis, HPLC analysis shows increasing amounts of nucleoside A, adenine, and adenosine and decreasing amounts of nucleoside B, which is clearly decomposing. Figure 1B is the reverse phase HPLC profile 14 h after photolysis, when most of the nucleoside B is decomposed and the amount of nucleoside A is dramatically increased, with lesser increases in adenine and adenosine. Clearly, nucleoside B gradually decomposes, mainly to nucleoside A but also to smaller amounts of adenine and adenosine.
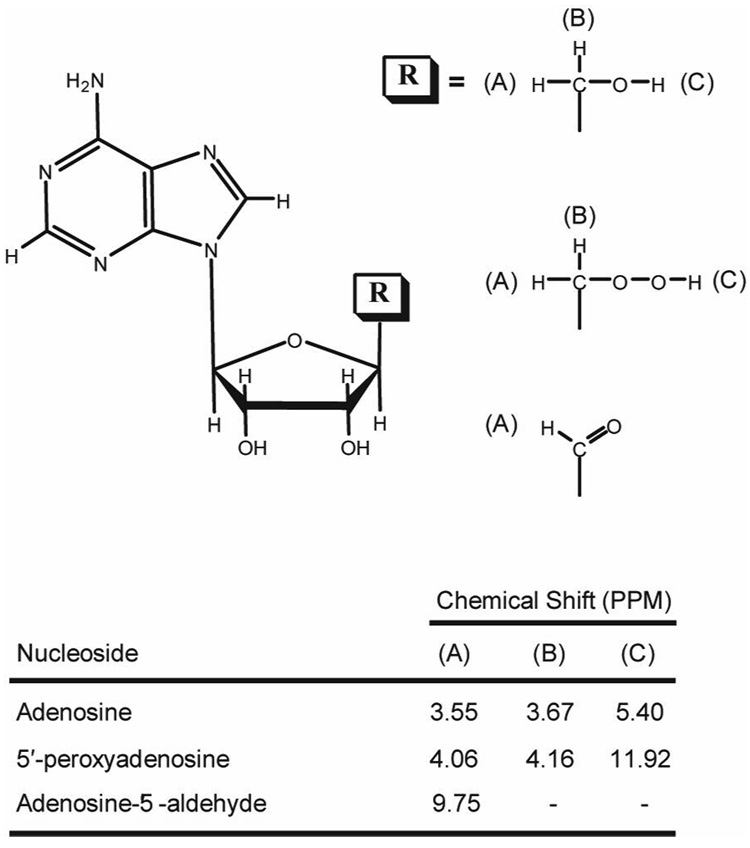
Mass spectral analysis of nucleoside A indicates a mass of 283.099 amu (Figure 1), which is in agreement with the identification of this peak as the hydrate of adenosine-5′-aldehyde (20). In addition, sodium borohydride reduction of adenosine-5′-aldehyde would be expected to yield adenosine, and adenosine is produced when 0.5 mL of 80 µM nucleoside A is reduced with 2 mg of NaBH4 for 30 min. Co-injection of the resultant reaction mixture with standards in reverse phase chromatography shows that the reduced nucleoside is identical with adenosine, as reported previously (20). 1H NMR spectroscopy performed on adenosine-5′-aldehyde (nucleoside A) in DMSO-d6 shows a clear signal for a single proton at 9.75, diagnostic of an aldehydic proton. A comparison of NMR chemical shifts for the C5′ (H), C5′ (OH), and C5′ (OOH) protons in nucleoside products derived from the aerobic photolysis of adenosylcobalamin is presented in Figure 2.
Figure 2. Structures of nucleoside products from the aerobic photolysis of adenosylcobalamin. I.
In the table are the 1H NMR chemical shifts for each R group labeled in the structure.
Identification of Nucleoside B
Nucleoside B is unstable and decomposes over time under a variety of conditions. The major decomposition product of nucleoside B is nucleoside A, adenosine-5′-aldehyde, together with minor but detectable amounts of adenosine and adenine (Figure 1B). This decomposition is inhibited by the presence of oxygen and progresses at a faster velocity when the solution is made anaerobic (Figure 1, insert).
Nucleoside B has a mass identical with that of the hydrate of adenosine-5′-aldehyde (Figure 1). The most obvious isomer of the hydrate is 5′-peroxyadenosine; a reasonable structure, taking into consideration its observed decomposition to adenosine-5′-aldehyde. 1H NMR analysis of nucleoside B in DMSO-d6 is consistent with 5′-peroxyadenosine and shows that the 5′-peroxyadenosine hydroperoxy proton resonates far downfield at 11.92 ppm relative to the 5′-peroxyadenosine hydroxy proton of adenosine at 5.40 ppm (21) (Figure 2). This is somewhat further downfield than expected for a hydroperoxide proton, but can be explained by an unusually low pKa for this substituent and/or participation in internal hydrogen bonding. In addition, the two C5′-protons in nucleoside B appear downfield compared to those in adenosine (Figure 2). This behavior would be expected for a transformation of the C5′-substituent from an alcohol to a hydroperoxide.
An assay was performed in which purified nucleoside B gave a rapid and strong positive response to ferrous oxidation in xylenol orange (FOX assay) (22). In this assay, 25 mM Fe2+ in 2.5 M sulfuric acid is oxidized to Fe3+ by the hydroperoxide. Xylenol orange then forms a complex with Fe3+ giving a purple product with a maximum absorbance at 560 nm. Sorbitol is included in this reaction mixture to increase assay sensitivity. No other compounds tested, adenosine or a variety of carboxylic acids, gave a positive result in this assay.
Initial work with 5′-peroxyadenosine was performed in a buffer system comprised of either EPPS or MES, both of which are tertiary amine containing sulfonic acids. The properties of 5′-peroxyadenosine were obscured when work in these buffers at 60 ° C yielded unexpectedly high yields of adenosine as a cobalamin-independent decomposition product, compared to those of unbuffered solutions or in phosphate buffer. It is known that amines and hydroperoxides can react to form N-oxides and the corresponding alcohol. This result further supports the identity of nucleoside B as 5′-peroxyadenosine.
A very minor product of aerobic photolysis is a peak that elutes with the highest retention time in our chromatographic system. The mass of this peak is 249.094 amu, in agreement with previous work identifying 5′-deoxy-8, 5′-cycloadenosine as the product of anaerobic photolysis of adenosylcobalamin (21). The present photolysis experiments in pure O2 are intended to minimize the yield of this product.
One of the two major nucleoside products of photolysis of adenosylcobalamin has been reported to be adenosine-5′-carboxylic acid (24). We did not find this compound as a photolytic product of adenosylcobalamin.
Transformation of 5′-Peroxyadenosine into Adenosine-5′-aldehyde
5′-Peroxyadenosine decomposes to adenosine-5′-aldehyde under a variety of conditions. In order to study this reaction, larger amounts of 5′-peroxyadenosine were purified on a C18 reverse phase HPLC column, eluted isocratically at 5% CH3CN in water. The purified compound was divided and incubated at two temperatures and at pH 5.8 and pH 8.4. The initial rates of production of adenosine-5′-aldehyde and adenosine were monitored by reverse phase HPLC (Table I). Adenine production was also observed, but the rate was too slow for accurate measurement by the HPLC assay.
Table 1.
Initial rate of formation of adenosine and adenosine-5′-aldehyde from the decomposition of 5′-peroxyadenosinea
| pH 5.8 |
pH 8.4 |
|||
|---|---|---|---|---|
| Conditionb | Ado-5′-CHO (µM min−1) | Ado(µM min−1) | Ado-5′-CHO (µM min−1) | Ado(µM min−1) |
| 20 °C | 7 ×10−4 | 1 × 10−4 | 3 × 10−4 | 4 × 10−4 |
| 60 °C | 1.3 × 10−2 | 1.2 × 10−3 | 2.1 × 10−3 | 7.5 × 10−3 |
| Cbl(III), 20 °C | 2.9 × 10−2 | 5 × 10−4 | 3.2 × 10−2 | 1.9 × 10−3 |
All reactions run with chromatographically purified 5′-peroxyadenosine at approximately 45 µM.
5′-Peroxyadenosine was incubated at 20 °C (room temperature), at 60 °C, and in the presence of 200 µM cob(III)alamin at 20 °C (room temperature).
In the cobalamin-independent reactions, the rate of appearance of decomposition products increases approximately 10 to 20-fold at both pHs with a 40 °C increase in temperature (Table 1). Adenosine-5′-aldehyde and adenosine production are slightly sensitive to pH, both exhibiting small differences in rates of formation at the two pHs. The trend for the change in the initial rate of formation due to pH is observed at 20 °C and 60 °C for both products. Interestingly, adenosine-5′-aldehyde production is 6-fold slower at the higher pH at 60 °C, while adenosine production is accelerated by the same magnitude at the higher pH (Table 1). The most striking result of the pH-study is the remarkable insensitivity to pH: the rates decrease or increase 2-to 6-fold with the 400-fold increase in hydroxide concentration between pHs 5.8 and 8.4.
When 5′-peroxyadenosine decomposition is monitored within the photolysis reaction mixture, which includes cob(III)alamin, adenosine-5′-aldehyde formation is much faster than in the transformation of the purified nucleoside. Addition of authentic cob(III)alamin to the purified 5′-peroxyadenosine reproduces these faster rates, as documented in Table 1.
The rate of formation of adenosine-5′-aldehyde and adenosine from 5′-peroxyadenosine incubated with cob(III)alamin showed even less dependence on pH than in the absence of cob(III)alamin. At the two pHs studied, the appearance of adenosine-5′-aldehyde in the cobalamin-dependent process was similar at pH 5.8 and pH 8.4. In the formation of adenosine, the magnitude of rate increase between pH 5.8 and pH 8.4 was the same for the cobalamin-dependent and independent processes. The production of adenosine-5′-aldehyde was never seen to be reversible under any conditions studied. Increasing the ionic strength to 0.5 elicited a very small decrease in rate (< 10%) at 20 °C, an effect inconsistent with a polar transition state.
Absorption Spectrum of an Intermediate in the Photolysis of Adenosylcobalamin
Figure 3 shows the changes in the absorption spectra associated with the photolysis of adenosylcobalamin and the subsequent cobalamin-dependent decomposition of free 5′-peroxyadenosine. Figure 3A shows the initial absorption spectrum and its decay of 80 µM adenosylcobalamin at pH 8.5 after exposure to a short period of photolysis. Over time, the initial cob(III)alamin-related spectrum decays, nearly quantitatively, to that of cob(III)alamin (- - -).3 The decay occurs in a biphasic manner, with a rapid change over ca. 30 minutes and a subsequent slower decay over a period of nearly 20 h (Figure 3A, inset). The spectral change, and the biphasic time course, indicate the formation and decay of an intermediate.
Figure 3. Changes to the absorption spectra of cob(III)alamin after the photolysis of adenosylcobalamin and from the addition of 5′-peroxyadenosine.
(A) Photolyzed adenosylcobalamin (80 µM) held for 20 h at pH 8.5. The initial spectrum decays to that of cob(III)alamin (- - -). Inset: Biphasic decay of reaction intermediate monitored spectrophotometrically at 312 nm. (B) Cob(III)alamin in the presence (80 µM) of 400 µM 5′-peroxyadenosine and held 25 min at pH 8.5. The initial absorption spectrum of 80 µM cob(III)alamin at pH 8.5 is ( - - -).
In a second experiment, 80 µM cob(III)alamin at pH 8.5 was incubated with 400 µM 5′-peroxyadenosine. Figure 3B shows the evolution of the cob(III)alamin absorption spectrum (- - -) to one similar to that of the apparent intermediate initially observed following the photolysis of adenosylcobalamin. The maximal change in the absorption spectrum occurs at approximately 25 minutes. At lower pHs, the spectral change occurs to a much smaller extent (data not shown). The observed spectral changes implicate the formation of a complex between cob(III)alamin and 5′-peroxyadenosine that is similar or identical to that initially generated by aerobic photolysis of adenosylcobalamin.
DISCUSSION
Photolysis of Adenosylcobalamin
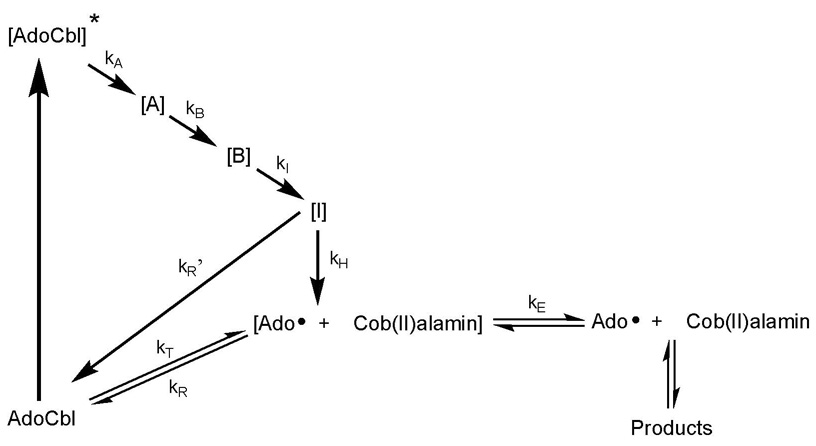
Exposure to visible light decomposes adenosylcobalamin (25), producing a radical pair (17–19). Photodissociation serves as a model for the thermal homolysis of the Co-C5′ bond in adenosylcobalamin, and Scheme 1 illustrates the proposed relationship between the photolytic and thermolytic pathways (17,25–30). Photolysis begins with activation of the Co-C5′ bond and proceeds through excited states [A] and [B] displaying absorption spectra evolving from cob(III)alamin-like to cob(II)alamin-like species and leading to an intermediate [I], which either re-forms adenosylcobalamin or relaxes to a geminate radical pair [Ado• + Cob(II)alamin], similar to that formed upon thermolysis of the Co-C5′ bond. The radical pair is the intersection of the two pathways. In aqueous solution, the radical pair reacts either by recombination or escape from the solvent cage and quenching. Anaerobically, cob(II)alamin and 5′-deoxy-8, 5′-cycloadenosine are formed (21). Aerobically, the major reported products are adenosine-5′-aldehyde and cob(III)alamin (20).
Scheme 1.
Products from the Photolysis of Adenosylcobalamin
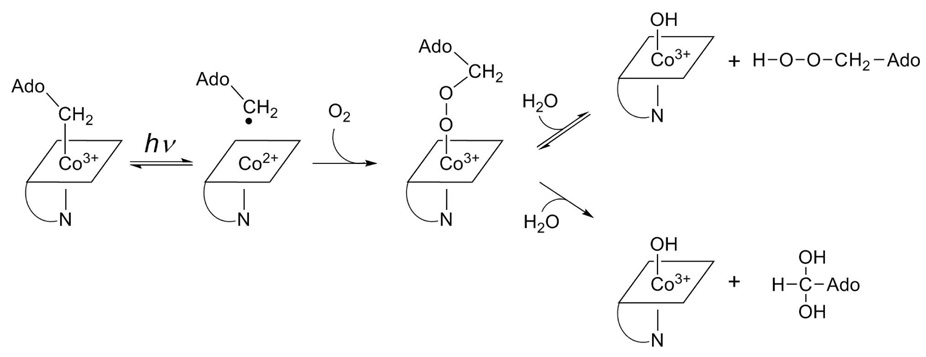
Scheme 2 outlines a minimal mechanism for the major reactions of the geminate radical pair resulting from homolytic cleavage of the Co-C5′ bond in air. Molecular oxygen intercepts the alkyl radical within the radical pair to form an alkylperoxy radical. This reaction occurs at the diffusional limit for the methyl radical (4.7 × 109 M−1 s−1) (31), much faster than the reaction of O2 with cob(II)alamin (t1/2 = 21 min) (32). Formation of the methylperoxy radical is proposed to be the major secondary reaction in the photolysis of alkylcobalamins (33). Reaction of cob(II)alamin with the methylperoxy radical leads to concomitant electron transfer from Co2+ and ligation to the upper-axial position of cob(III)alamin, leading directly to 5-peroxyadenosylcobalamin.
Scheme 2.
Four lines of evidence implicate 5′-peroxyadenosylcobalamin as an intermediate in the formation of adenosine-5′-aldehyde: a) In photolysis, a small amount of adenosine-5′-aldehyde appears in a burst (Figure 1A), followed by much slower accumulation over hours, implicating an intermediate in the mechanism. b) Hydroxocobalamin accelerates the decomposition of purified 5′-peroxyadenosine (Table 1). It is difficult to imagine how this could occur except through complexation, and the obvious complex is one with 5′-peroxyadenosine ligated to cob(III)alamin in the upper axial position. c) The cob(III)alamin-dependent decomposition is slower in the presence of O2 than in its absence (Figure 1B, inset), which we take to indicate competition of O2 with 5′-peroxyadenosine for binding to cob(III)alamin. d) The transient absorption spectra in Figure 3 clearly show the formation and decomposition of an intermediate. The electronic spectral evidence is very powerful because of the well-known effects of axial ligands on the electronic spectra (38). Taken together, the evidence implicates an intermediate, and the simplest and most reasonable structure is 5′-peroxyadenosylcobalamin.
Precedents for the mechanism in Scheme 2 have been published. Based on the production of cob(III)alamin, formaldehyde, and hydroxide ion in the photolysis of methylcobalamin, and the behavior of alkyl radicals in the presence of O2 (34,35), the methyl peroxy radical was proposed as an intermediate (33). Moreover, the methyl peroxy radical reacts rapidly with the model compound, Co-([14]tetraeneN4)2+ (36). Kinetic analysis monitoring absorbance changes at 470 nm were attributed to the loss of cob(II)alamin due to formation of this alkylperoxocobalamin. Rate constants were 2.4 × 109 M−1 s−1 for methylcobalamin (37) and 3 × 109 M−1 s−1 for adenosylcobalamin (18). Spectrophotometric evidence for the initial formation of 5′-peroxyadenosylcobalamin is provided in Figure 3. Spectral differences for cob(III)alamin derivatives in the main absorption bands (i.e. α, 535 nm; β 510 nm; and γ, 360 nm) of the spectrum arising from differences in the upper axial ligand of cob(III)alamin have been primarily attributed to differences in the sum of the σ-donor strength of the axial ligands (38).
Two major products from the aerobic photolysis of adenosylcobalamin likely arise from decomposition of 5′-peroxyadenosylcobalamin (Scheme 2). First, free 5′-peroxyadenosine can appear by substitution of water or hydroxide ion into the upper axial position of 5′-peroxyadenosylcobalamin. This allows for the reversible association of 5′-peroxyadenosine with cob(III)alamin. 5′-Peroxyadenosine accounts for 89% of the nucleoside products immediately following photolysis. Second, 5′-Peroxyadenosylcobalamin decomposes to adenosine-5′-aldehyde and cob(III)alamin in an apparently irreversible reaction.
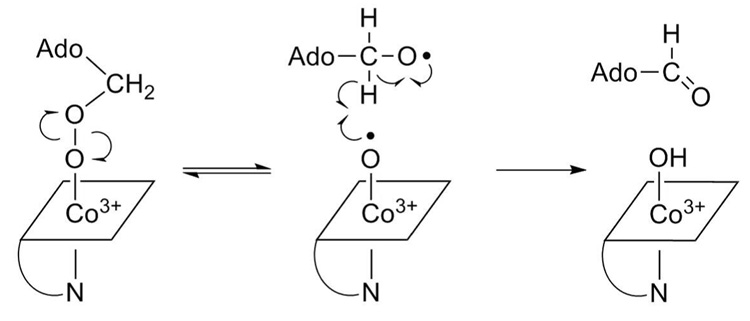
Adenosine-5′-aldehyde likely results from homolytic scission of the O-O bond in the hydroperoxide. In the 5′-peroxyadenosylcobalamin intermediate, cleavage of this bond generates an alkoxyl radical and an oxocobalamin radical species. Abstraction of a C5′-hydrogen atom by the oxocobalamin radical in a cage reaction would yield both the aldehyde and cob(III)alamin (Scheme 3). Alternatively, a concerted reaction involving 5′-peroxyadenosylcobalamin could lead directly to adenosine-5-aldehyde through a 4-membered cyclic transition state (Scheme 4). The aldehyde initially accounts for 9% of the nucleoside products in photolysis, one-tenth of the initial yield of 5′-peroxyadenosine. Small amounts of adenosine and adenine are also observed, each accounting for <2% of the nucleoside products and arise by unknown mechanisms.
Scheme 3.
Scheme 4.
Based on available information, a polar reaction mechanism for the production of adenosine-5′-aldehyde from 5′-peroxyadenosylcobalamin appears unlikely. Such a mechanism would involve base catalysis in the abstraction of a proton from the 5′-methylene group, either by a base in solution or intramolecularly by the peroxyanion ligated to Co(III). A base-catalyzed mechanism appears unlikely because of the absence of a substantial rate enhancement at pH 8.4 relative to pH 5.8. Furthermore, high ionic strength would stabilize a polar transition state, but high ionic strength does not enhance the rate.
Cob(III)alamin-Catalyzed Decomposition of 5 ′-Peroxyadenosine
Catalysis of the isomerization of free 5′-peroxyadenosine to adenosine-5′-aldehyde by cob(III)alamin depends upon the reversible ligation of 5′-peroxyadenosine to cob(III)alamin to generate 5′-peroxyadenosylcobalamin (Cob(III)-OH + Ado-5′-OOH ⇌ Cob(III)-O–O-5′-Ado + H2O). The catalytic effect of cob(III)alamin on the process of decomposition is likely brought about by the weakness of the O—O bond in the latter complex. Conceivably, participation of occupied non-bonding p-orbitals on the Co-coordinated oxygen atom interacting with the lowest unoccupied d orbitals in cob(III)alamin would weaken the peroxide bond, promoting homolysis and increasing the rate of aldehyde formation. Decomposition to adenosine-5′-aldehyde proceeds either by a caged radical mechanism as in Scheme 3 or by the concerted, internal rearrangement in Scheme 4.
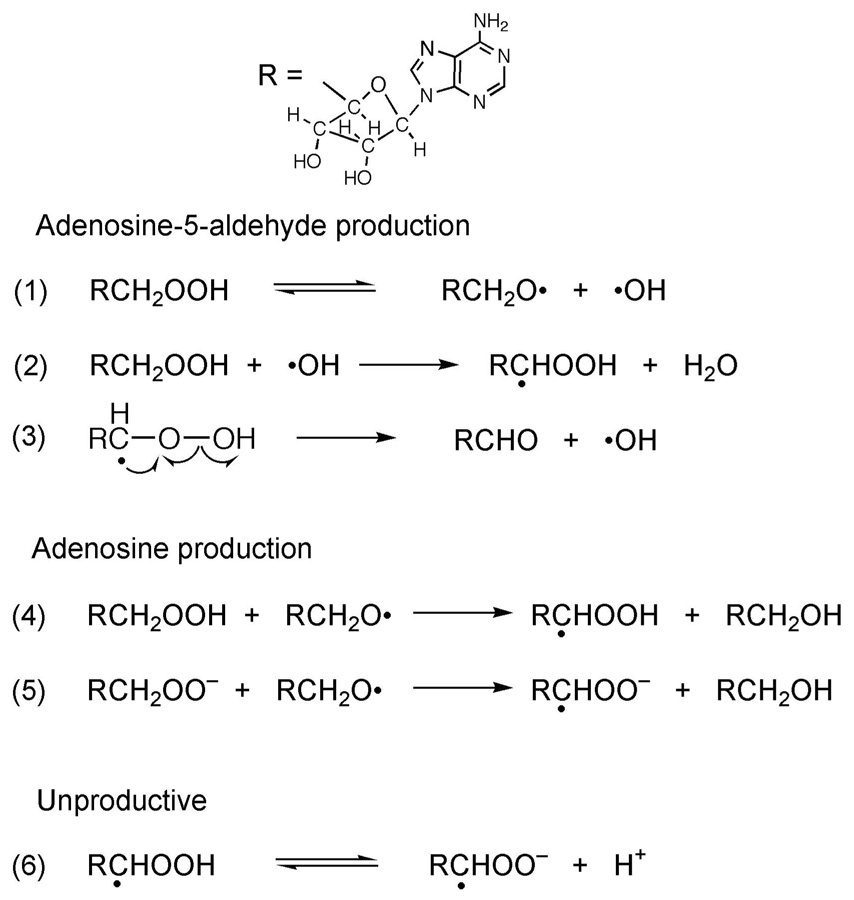
Cobalamin-Independent Decomposition of 5 ′-Peroxyadenosine
In the much slower, cobalamin-independent decomposition, 5′-peroxyadenosine also undergoes transformation into adenosine-5′-aldehyde as the major product or into adenosine as a minor product. Reactions 1–6 in Figure 4 provide a reasonable mechanistic rationale for the formation of these two products. The process likely occurs through a radical chain mechanism because of the apparent slightly autocatalytic production of both adenosine-5′-aldehyde (Figure S1, Supplemental Information) and adenosine (data not shown). The reactions in Figure 4 leading to observed products are selected based on the following criteria: radical coupling reactions are expected to be rare, as the short lifetimes of the radical intermediates would not allow them to accumulate. Abstraction of hydrogen atoms from the O—H bond of 5′-peroxyadenosine is not expected to contribute significantly to product formation, because the O—H bond is stronger than C—H bonds. Hydrogen abstractions from the C5′ of 5′-peroxyadenosine are selected because the C5′—H bonds are weaker, less sterically hindered, and provide a more stable radical product than cleavage of other C—H bonds. Products arising from radical-abstraction of other hydrogens within the nucleoside are not observed. The rare chain terminating radical couplings do not contribute significantly to the product profile.
Figure 4. Radical chain reactions leading to the cob(III)alamin-independent decomposition of 5-peroxyadenosine.
Reactions 1 –3 lead to the transformation of 5′-peroxyadenosine into adenosine-5′-aldehyde as the major product, and reactions 4 and 5 account for the production of smaller amounts of adenosine.
Reactions 1 – 3 in Figure 4 account for adenosine-5′-aldehyde formation, and reactions 4 and 5 account for adenosine formation. Reaction 5 can explain the pH related increase in the rate of adenosine production under basic conditions. Fractional deprotonation of the alkyl hydroperoxide at the higher pH leads to the hydroperoxy anion, reaction of which with the alkoxyl radical leads to adenosine and a C5′-centered radical in the hydroperoxy anion, which is stabilized through a dative effect from the electron-releasing oxyanion substituent. This product-stabilization would lead to an increase in the rate of reaction 5 relative to 4, and could account for the increase in the observed rate of appearance of adenosine (Table 1).
Reaction 6 in Figure 4 is unproductive and provides a rationale for the pH related decrease in the formation of aldehyde under basic conditions. Should the alkyl hydroperoxide species with the C5′-centered radical undergo fractional deprotonation to form the hydroperoxy anion, which is stabilized by the dative effect described above, it would lead to a “dead-end” regarding product accumulation.
The process leading to formation of adenine is not known at this time. Adenine production was too slow for accurate rate measurements by the HPLC assay, either in the presence or absence of cob(III)alamin.
The identification of 5′-peroxyadenosine as the primary product of photo-induced homolysis of the Co-C5′ bond in the presence of O2 sheds new light on the reactivity of adenosylcobalamin. The formation of the hydroperoxide and the cob(III)alamin-catalyzed decomposition of this compound provides chemical support for existence of the 5′-peroxyadenosylcobalamin intermediate in the aerobic photolysis of adenosylcobalamin.
Supplementary Material
Supporting information includes Figure S1 showing the progress curves for the transformation of 5′-peroxyadenosine into adenosine-5′-aldehyde under various conditions. This material is available free of charge via the Internet at http://pubs.acs.org.
ACKNOWLEDGEMENTS
We thank Dr. Mark Anderson at the NMRFAM for assistance in acquiring 1H NMR spectra. We also thank Jim Brown at the Mass Spectrometry Facility (Biotechnology Center, University of Wisconsin-Madison) for his assistance in LC/MS analysis.
Footnotes
This research was supported by NIH grant DK28607 from the National Institute of Diabetes and Digestive and Kidney Diseases.
Abbreviations: Adenosylcobalamin, 5′-deoxyadenosylcobalamin; Ado, adenosine; cob(III)alamin, hydroxocobalamin and/or aquocobalamin; 5′-deoxyadenosyl radical, 5′-deoxyadenosyl-5′-yl; DMSO, dimethyl sulfoxide; EPPS, N-[2-Hydroxyethyl]piperazine-N'-3-propanesulfonic acid; ESI, electrospray ionization; FOX, ferrous oxidation in xylenol orange assay; HPLC, high performance liquid chromatography; LC/MS, liquid chromatography-mass spectrometry; MES, 2-[N-morpholino]ethanesulfonic acid; NMR, nuclear magnetic resonance; RT, retention time; TOF, time-of-flight.
This study made use of the National Magnetic Resonance Facility at Madison, which is supported by National Institutes of Health grants P41RR02301 (Biomedical Research Technology Program, National Center for Research Resources) and P41GM66326 (National Institute of General Medical Sciences). Equipment in the facility was purchased with funds from the University of Wisconsin, the National Institutes of Health (P41GM66326, P41RR02301, RR02781, RR08438), the National Science Foundation (DMB-8415048, OIA-9977486, BIR-9214394), and the U.S. Department of Agriculture.
The pKa for cob(III)alamin is 7.8. Hydroxocobalamin and aquocobalamin in aqueous solution have different absorption spectra.
REFERENCES
- 1.Dolphin D, editor. B12 Vols. 1 and 2. Wiley-Interscience: 1982. p. New York. [Google Scholar]
- 2.Banerjee R, editor. Chemistry and Biochemistry of B12. New York: Wiley-Interscience; 1999. [Google Scholar]
- 3.Ouyang L, Rulis P, Ching WY, Nardin G, Randaccio L. Accurate redetermination of the X-ray structure and electronic bonding in adenosylcobalamin. Inorg. Chem. 2004;43:1235–1241. doi: 10.1021/ic0348446. [DOI] [PubMed] [Google Scholar]
- 4.Babior BM. The Mechanism of action of ethanolamine deaminase. VI. Ethylene glycol, a quasi-substrate for ethanolamine deaminase. J. Biol. Chem. 1970;245:1755–1766. [PubMed] [Google Scholar]
- 5.Finlay TH, Valinsky JE, Mildvan AS, Abeles RH. Electron spin resonance studies with dioldehydrase; Evidence for radical intermediates in reactions catalyzed by coenzyme B12. J. Biol. Chem. 1973;248:1285–1290. [PubMed] [Google Scholar]
- 6.Orme-Johnson WH, Beinert H, Blakley RL. Cobamides and ribonucleotide reduction. XII. The electron paramagnetic resonance spectrum of “active coenzyme B12". J. Biol. Chem. 1974;24:2338–2343. [PubMed] [Google Scholar]
- 7.Babior BM. Mechanism of cobalamin-dependent rearrangements. Acc. Chem. Res. 1975;8:376–384. [Google Scholar]
- 8.Abeles RH, Dolphin DH. The vitamin B12 coenzyme. Acc. Chem. Res. 1976;9:114–120. [Google Scholar]
- 9.Banerjee R. Radical carbon skeleton rearrangements: catalysis by coenzyme B12-dependent mutases. Chem. Rev. 2003;103:2083–2094. doi: 10.1021/cr0204395. [DOI] [PubMed] [Google Scholar]
- 10.Halpern J, Kim SH, Leung TW. Cobalt-carbon bond dissociation energy of coenzyme B12. J. Am. Chem. Soc. 1984;106:8317–8319. [Google Scholar]
- 11.Finke RG, Hay BP. Thermolysis of adenosylcobalamin: a product, kinetic, and cobalt-carbon (C5′) bond dissociation energy study. Inorg. Chem. 1984;23:3041–3043. [Google Scholar]
- 12.Hay BP, Finke RG. Thermolysis of the cobalt-carbon bond of adenosylcobalamin. 2. Products, kinetics, and cobalt-carbon bond dissociation energy in aqueous solution. J. Am. Chem. Soc. 1986;108:4820–4829. [Google Scholar]
- 13.Hay BP, Finke RG. Thermolysis of the cobalt-carbon bond in adenosylcorrins. 3. Quantification of the axial base effect in adenosylcobalamin by the synthesis and thermolysis of axial base-free adenosylcobinamide. Insights into the energetics of enzyme-assisted cobalt-carbon bond homolysis. J. Am. Chem. Soc. 1987;109:8012–8018. [Google Scholar]
- 14.Hay BP, Finke RG. Thermolysis of the Co---C bond in adenosylcobalamin (coenzyme B12)-IV. Products, kinetics and Co---C bond dissociation energy studies in ethylene glycol. Polyhedron. 1988;7:1469–1481. [Google Scholar]
- 15.Garr CD, Finke RG. Adocobalamin (AdoCbl or coenzyme B12) cobalt-carbon bond homolysis radical-cage effects: product, kinetic, mechanistic, and cage efficiency factor (Fc) studies, plus the possibility that coenzyme B12-dependent enzymes function as "ultimate radical cages" and "ultimate radical traps". Inorg. Chem. 1993;32:4414–4421. [Google Scholar]
- 16.Marsh ENG, Ballou DP. Coupling of cobalt-carbon bond homolysis and hydrogen atom abstraction in adenosylcobalamin-dependent glutamate mutase. Biochemistry. 1998;37:11864–11872. doi: 10.1021/bi980512e. [DOI] [PubMed] [Google Scholar]
- 17.Walker LA, II, Shiang JJ, Anderson NA, Pullen SH, Sension RJ. Time-resolved spectroscopic studies of B12 coenzymes: the photolysis and geminate recombination of adenosylcobalamin. J. Am. Chem. Soc. 1998;120:7286–7292. [Google Scholar]
- 18.Endicott JF, Netzel TL. Early events and transient chemistry in the photohomolysis of alkylcobalamins. J. Am. Chem. Soc. 1979;101:4000–4002. [Google Scholar]
- 19.Hogenkamp HPC. In: B12. Dolphin D, editor. Vol. 1. New York: Wiley-Interscience; 1982. pp. 295–323. [Google Scholar]
- 20.Hogenkamp HPC, Ladd JN, Barker HA. The identification of a nucleoside derived from coenzyme B12. J. Biol. Chem. 1962;237:1950–1952. [PubMed] [Google Scholar]
- 21.Hogenkamp HPC. A cyclic nucleoside derived from coenzyme B12. J. Biol. Chem. 1963;238:477–480. [PubMed] [Google Scholar]
- 22.Jiang Z-Y, Woollard ACS, Wolff SP. Hydrogen peroxide production during experimental protein glycation. FEBS Lett. 1990;268:69–71. doi: 10.1016/0014-5793(90)80974-n. [DOI] [PubMed] [Google Scholar]
- 23.Pouchert CJ, Behnke J. The Aldrich Library of 13C and 1H FT-NMR Spectra. Vol. 3. Milwaukee: Sigma-Aldrich; 1993. p. 222. [Google Scholar]
- 24.Johnson AW, Shaw N. Some reactions of the vitamin B12coenzyme. J. Chem. Soc. 1962:4608–4614. [Google Scholar]
- 25.Chen E, Chance MR. Nanosecond transient absorption spectroscopy of coenzyme B12. Quantum yields and spectral dynamics. J. Biol. Chem. 1990;265:12987–12994. [PubMed] [Google Scholar]
- 26.Chagovitz AM, Grissom CB. Magnetic field effects in adenosylcob(III)alamin photolysis: Relevance to B12 enzymes. J. Am. Chem. Soc. 1993;115:12152–12157. [Google Scholar]
- 27.Brown KL. Chemistry and enzymology of vitamin B12. Chem. Rev. 2005;105:2075–2150. doi: 10.1021/cr030720z. [DOI] [PubMed] [Google Scholar]
- 28.Chen E, Chance MR. Continuous-wave quantum yields of various cobalamins are influenced by competition between geminate recombination and cage escape. Biochemistry. 1993;32:1480–1487. doi: 10.1021/bi00057a011. [DOI] [PubMed] [Google Scholar]
- 29.Cole AG, Yoder LM, Shiang JJ, Anderson NA, Walker LA, II, Banaszak Holl MM, Sension RJ. Time-resolved spectroscopic studies of B12 coenzymes: a comparison of the primary photolysis mechanism in methyl-, ethyl-, n-propyl-, and 5′-deoxyadenosylcobalamin. J. Am. Chem. Soc. 2002;124:434–441. doi: 10.1021/ja011628s. [DOI] [PubMed] [Google Scholar]
- 30.Yoder LM, Cole AG, Walker LA, II, Sension RJ. Time-resolved spectroscopic studies of B12 coenzymes: influence of solvent on the photolysis of adenosylcobalamin. J. Phys. Chem. B. 2001;105:12180–12188. [Google Scholar]
- 31.Thomas JK. Pulse radiolysis of aqueous solutions of methyl iodide and methyl bromide. The reactions of iodine atoms and methyl radicals in water. J. Phys. Chem. 1967;71:1919–1925. [Google Scholar]
- 32.Pratt JM. The chemistry of vitamin B12. Part II. Photochemical reactions. J. Chem. Soc. 1964:5154–5160. [Google Scholar]
- 33.Hogenkamp HPC. The Photolysis of methylcobalamin. Biochemistry. 1966;5:417–422. doi: 10.1021/bi00866a005. [DOI] [PubMed] [Google Scholar]
- 34.Heicklen J, Johnston HS. Photochemical oxidations. II. Methyl iodide. J. Am. Chem. Soc. 1962;84:4030–4039. [Google Scholar]
- 35.Heicklen J, Johnston HS. Photochemical oxidations. I. Ethyl iodide. J. Am. Chem. Soc. 1962;84:4394–4403. [Google Scholar]
- 36.Mok CY, Endicott JF. The photochemistry of organocobalt complexes containing tetraaza macrocyclic ligands. Cobalt-methyl homolysis and the nature of the cobalt-carbon bond. J. Am. Chem. Soc. 1978;100:123–129. [Google Scholar]
- 37.Endicott JF, Ferraudi GJ. A flash photolytic investigation of low energy homolytic processes in methylcobalamin. J. Am. Chem. Soc. 1977;99:243–245. doi: 10.1021/ja00443a043. [DOI] [PubMed] [Google Scholar]
- 38.Pratt JM. The roles of Co, corrin, and protein. I. Co-ligand bonding and the trans effect. In: Banerjee R, editor. Chemistry and Biochemistry of B12. New York: Wiley-Interscience; 1999. pp. 73–112. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting information includes Figure S1 showing the progress curves for the transformation of 5′-peroxyadenosine into adenosine-5′-aldehyde under various conditions. This material is available free of charge via the Internet at http://pubs.acs.org.