Abstract
We present a proof-of-principle for a fully automated bottom-up approach to protein characterization. Proteins are first separated by capillary electrophoresis. A pepsin microreactor is incorporated into the distal end of this capillary. Peptides formed in the reactor are transferred to a second capillary, where they are separated by capillary electrophoresis and characterized by mass spectrometry. While peptides generated from one digestion are being separated in the second capillary, the next protein fraction undergoes digestion in the microreactor. The migration time in the first dimension capillary is characteristic of the protein while migration time in the second dimension is characteristic of the peptide. Spot capacity for the two-dimensional separation is 590. A MS/MS analysis of a mixture of cytochrome C and myoglobin generated Mascot MOWSE scores of 107 for cytochrome C and 58 for myoglobin. The sequence coverages were 48% and 22%, respectively.
Introduction
Mass spectrometric analyses of proteins are usually classified as either top-down or bottom-up1. In top-down methods, intact proteins are separated and then introduced into a mass spectrometer for fragmentation and identification2–4. Top-down approaches are ideal for the characterization of post-translational modifications, since the entire protein is introduced into the mass spectrometer for analysis. However, the high cost of instrumentation and challenges in protein fragmentation have discouraged widespread application of this technology.
In bottom-up methods, proteins are digested into peptides, and those peptides are analyzed by mass spectrometry1,5–7. Peptides are much easier to handle than proteins with most instruments, and bottom-up approaches are much more common than top-down analyses.
Classically, intact proteins are separated by two-dimensional electrophoresis and spots are isolated and digested before bottom-up analysis5. The protein separation allows recognition of post-translational modifications through shifts in spot position during electrophoresis. However, two-dimensional electrophoresis is painfully tedious and labor-intensive and suffers from poor reproducibility. More recently, LC-MS/MS-based methods, such as MudPIT6 and ICAT7, have been developed where the complex protein mixture is digested into a much more complex peptide mixture, which is then separated by multidimensional liquid chromatography before mass spectrometric analysis. This multidimensional separation and mass spectrometric analysis is automated, which is of tremendous advantage in large-scale proteomics projects. However, these methods suffer from two challenges. They require high separation resolution because of the complex sample produced by digestion. More importantly, not all peptides produced from a protein are detected, and some post-translational modifications or alternative splice forms of genes may go undetected.
This paper describes a new example of bottom-up protein analysis, wherein a mixture of intact proteins is first separated, fractions are then digested with an on-line microreactor, and the resulting peptides are then separated in a second dimension before being introduced into a mass spectrometer for bottom-up analysis. This approach shares advantages of both classic two-dimensional gels and of multidimensional separations of peptides. Like gel electrophoresis of proteins, this method employs a separation dimension of intact proteins, which places fewer constraints on the separation efficiency than do peptide separations. Also, by separation of intact proteins, the presence of post-translational modifications may be detected as a mobility shift of the protein. Like chromatographic-based bottom-up methods, this method is fully automated, which greatly simplifies sample handling.
The key to our approach is the development of an integrated on-column microreactor for rapid digestion of protein fractions into peptides. There have been a number of examples of microreactors coupled with capillary electrophoresis; however, it has proven difficult to design a microreactor-electrophoresis system that is fast and that produces efficient separations. Kuhr immobilized trypsin on the inner surface of a capillary to form a microreactor that was coupled with a separation capillary via a fluid joint8–9. This approach was used successfully for on-line digestion and mapping of proteins with CE. However, an overall digestion time of several hours was required in this system due to the low surface-to-volume ratio of the microreactor. Waldron developed an on-line system allowing digestion of proteins, followed by preconcentration, separation, and detection of the tryptic fragments in 4 h10. Their enzymatic reactor was prepared by packing trypsin immobilized glass beads into a 530 μm ID capillary. However, separation efficiency was poor due to a multiple valve design and dispersion of the 60 nL desorption plug. Thibault developed a microfluidic system on a chip in which an electrospray interface to a mass spectrometer was integrated with a CE channel, an injector, and a protein digestion bed on a substrate11. The protein digestion bed was formed by packing a cavity in the chip with trypsin immobilized agarose beads. They found that cytochrome C or bovine serum albumin could be completely consumed within 3–6 min. Unfortunately, theoretical plate counts were less than 1000 for the peptide separation. Lee reported a reactor based on adsorption of trypsin on a PCDF membrane12. Proteins were forced through pores in this membrane, and the digested proteins were focused by transient isotachophoresis, separated by CE, and detected by electrospray mass spectrometry. The rather large dead volume of this device resulted in poor separation efficiency for the digested peptides; theoretical plate counts were about 1500 for the peptides resulting from digestion of cytochrome C. Toyo’oka reported a trypsin encapsulation technique using the sol-gel method for the preparation of an on-line enzyme reactor integrated with CE13–14. In their initial publication, microreactor digestions were restricted to proteins that could fit through the pores that contained the encapsulated trypsin. This problem was later solved by first preparing a methacryloxypropyltrimethoxysilane (MPTMS) monolith with relatively large through-pores, and then coating the monolith surface with a sol-gel that contained the enzyme pepsin. Only the inlet end of a capillary was modified with the monolith, and the rest of the capillary was used for separation of the digests of lysozyme and insulin. This system was on-line connected to ESI-MS.
Continuous rods of macroporous poly(glycidyl methacrylate-co-ethylene dimethacrylate) have been used as affinity chromatographic media as well as the support for enzyme reactors because of their high hydrophilicity and easy modification15. We have described the production of a macroporous monolith with immobilized trypsin that was coupled to capillary electrophoresis for peptide mapping, which produced over 300,000 theoretical plates for peptide separation16. That system employed post-column labeling with fluorescence detection but did not separate proteins before digestion. In this paper, we incorporate two-dimensional separations with mass spectrometric detection. We also employ pepsin as the digestion enzyme, which allows use of a volatile acetic acid-ammonium acetate buffer.
MATERIALS AND METHODS
Materials
Bovine heart cytochrome C, horse heart myoglobin, pepsin from porcine stomach mucosa, 2,2′-azobisisobutyronitrile (AIBN) (98%), cyclohexanol (99%), dodecanol (98%), ethylene glycol dimethacrylate (EDMA) (98%), glacial acetic acid, 50% glutaraldehyde in water, γ-methacryloxypropyltrimethoxysilane (γ-MAPS) (98%), glycidyl methacrylate (GMA) (97%), glycine ethyl ester hydrochloride (99%), hydroquinone (99+%), poly(vinyl alcohol) (PVA) (avg. MW 89,000–98,000 Da), sodium hydroxide (98%), and sodium periodate (99%) were purchased from Sigma. Acetonitrile (HPLC grade) and formic acid (88%) were from Fisher Scientific. Ammonium acetate (puriss. p.a. for mass spectroscopy) and sodium cyanoborohydride (≥95%) were from Fluka. Methanol (HPLC grade) was from EMD Chemicals. Hydrochloric acid (38%) was from J.T. Baker. Water was distilled and deionized by a Nanopure system. Helium gas (99.99%) was from Airgas.
Buffers and sheath liquids were passed through 0.22 μm filters. Amber flasks were used for buffers containing hydroquinone to minimize photodegradation. Fresh hydroquinone buffers were prepared every 1–2 weeks. Buffers and sheath liquids were He sparged for 5–10 min shortly before use. 1 mL of the 30 mM acetic acid-ammonium acetate (pH 2.7) buffer for the first dimension was centrifuged for 5 min at 15,000 rpm in a 1.6 mL Eppendorf tube. 50 μL of the solution was transferred to a cut-off PCR tube inside an injection/buffer reservoir device. The second dimension buffer was 50 mM acetic acid-ammonium acetate + 20 mM hydroquinone (pH 3.9). Hydroquinone was added to alleviate the reduction or oxidation of water, which had caused CE current instabilities17. Buffers in the reservoirs were exchanged for each experiment.
Protein sample preparation
A stock solution of each protein was prepared in ddH2O and frozen at −80°C. Thawed solutions were used to prepare 200 or 300 μM mixtures in 10 mM acetic acid-ammonium acetate (pH 2.7). 50 μL aliquots of the mixtures were frozen in 0.6 mL Eppendorf tubes at −80°C until use. An aliquot was then thawed in the refrigerator, briefly mixed by pipetting, and spun down at 8,000 rpm for 5 min. Next, 40 μL of the supernatant was transferred to a cut-off PCR tube inside the sample holder well of the injection/buffer reservoir device. The sample in the injection block was covered with a piece of clean Parafilm and stored in the refrigerator between injections. A fresh aliquot was thawed each day.
Microreactor preparation
Integrated monolithic microreactors were prepared in the ends of 48 μm ID, 142 μm OD capillaries. The initial length of the capillaries was ~ 45 cm. A ~ 5 cm long monolith was prepared at one end of the capillary using the same reagents employed in our earlier procedure16. Initially, the whole capillary was coated with γ-methacryloxypropyltrimethoxysilane (γ-MAPS), which decreased analyte-capillary wall interactions in the part of the capillary that was not subsequently filled with the monolith. One end of the capillary was next filled with the monomer reaction solution via capillary action. Both ends of the capillary were stoppered with silicon rubber. Polymerization proceeded for 12 hours at 50°C. The capillary was unstoppered and flushed from the monolith-filled end with MeOH for 20 min, followed by ddH2O for another ~ 20 min.
Pepsin immobilization proceeded by first reducing the epoxide groups on the monolith by flushing a pH 0.6 HCl solution through the capillary for 20 min. Next, the ends were stoppered and the capillary was stored at room temperature for 48 hours. The capillary was then flushed with ddH2O, followed by a freshly prepared 2% (w/v) NaIO4 solution for 60 min to oxidize the monolith. The monolith was then rinsed with ddH2O and 0.1 M acetic acid-ammonium acetate (pH 4.0) buffer. A freshly prepared 3 mg/mL pepsin in 0.1 M acetic acid-ammonium acetate (pH 4.0) solution was pumped through the microreactor for 10 hours at 4°C, followed by a 0.1 M acetic acid-ammonium acetate (pH 4.0) rinse. A solution of freshly prepared 25 mM sodium cyanoborohydride + 10 mM glycine ethyl ester hydrochloride in 0.1 M acetic acid-ammonium acetate (pH 4.0) was then pumped through the capillary for 4 hours, followed again by a 0.1 M acetic acid-ammonium acetate (pH 4.0) rinse. The microreactor capillary (48 μm ID, 142 μm OD) was cut to a final total length of 31.2 cm, with the microreactor occupying 1.1 cm at one end.
Poly(vinyl alcohol) coating procedure
The peptide separation, or second dimension, capillary was coated with poly(vinyl alcohol) (PVA) to reduce analyte-capillary wall interactions. The coating procedure was adapted from Belder18 and Fogarty19. PVA was ground with a mortar and pestle for ~ 5 min. An initial 2.5% (w/w) PVA suspension was prepared by adding 37.5 mg of the ground PVA, 1.312 mL of ddH2O, and 150 μL of 50% (v/v) ddH2O-concentrated HCl to a 1.6 mL Eppendorf tube. The tube was shaken and vortexed for 3 min, then sonicated for 15 min. The PVA did not completely dissolve, so the suspension was allowed to stand for 1 min, and then 300 μL of the supernatant was transferred into a 0.6 mL Eppendorf tube. The concentration of the final PVA suspension is unknown.
Several capillaries could be PVA-coated at one time. The capillaries were cleaned by pumping 1 M NaOH, followed by ddH2O, through the capillaries for 30 min at 4 psi. The capillaries were dried by flowing He gas for 30 min at 4 psi.
Next, a glutaraldehyde-HCl solution was prepared by adding 100 μL of a 50% glutaraldehyde solution to 150 μL of 10% (v/v) ddH2O-concentrated HCl. The PVA suspension was briefly vortexed 10 min before starting the glutaraldehyde-HCl flush. The glutaraldehyde solution was flushed through the dried capillaries at 20 psi until the solution exited from the distal end of the capillary. Next, the PVA suspension was flushed through the capillaries at 60 psi for 1 min, which was followed by He gas at 60 psi for 10 min. After the 10 min, the gas pressure was reduced to 5 psi and the flushing continued for 5 hours.
Last, ddH2O was flushed through the capillaries. Capillaries were stored with their ends in ddH2O. Shortly before use, the capillaries were cut to the desired length.
2D CE-microreactor-CE instrumentation
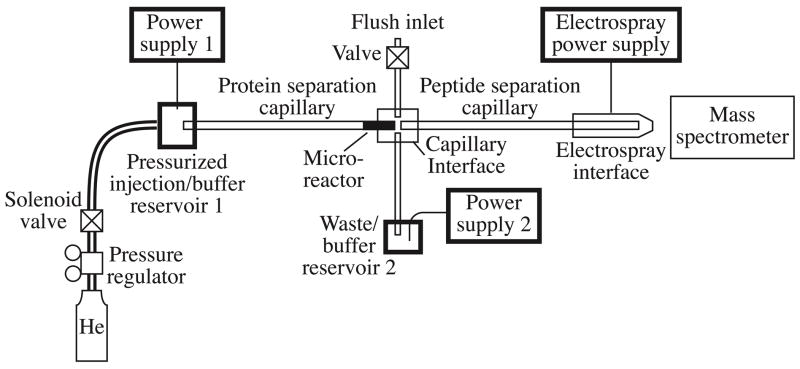
A diagram of the system is shown in Figure 1. Two 0–30 kV positive dc power supplies (UltraVolt, Ronkonkoma, NY) provided the potential for CE separations; the mass spectrometer provided potential for the electrospray interface. The ground returns of the power supplies were connected to the power outlet on the back of the mass spectrometer. As a safety feature, the CE system was enclosed in a Plexiglas box equipped with a safety-interlocked door. The liquid levels of the samples and buffers were adjusted to the height of the inlet orifice of the MS to avoid siphoning.
Figure 1.
Block diagram. The first dimension capillary for protein separation contains a short reactor containing immobilized pepsin. Two power supplies are under computer control and are used to drive sample from the protein separation capillary to the peptide separation capillary and from the peptide separation capillary to the mass spectrometer. Helium is used to pump buffer through the capillaries through an injection block; a solenoid-valve is used to provide timed pulses of gas to assist sample injection in some experiments. A capillary interface (Figure 2) connects the two separation capillaries with a buffer reservoir connected to power supply 2. An inlet is provided to the interface to flush contaminants between runs; this line is closed with a valve during analysis. An electrospray interface delivers sample from the peptide separation capillary to the mass spectrometer.
Sample injector
Sample, or the separation buffer for the protein separation dimension, were contained an injection block, described previously20. A pulse of helium pressure was used in some experiments to increase the amount of sample injected. A solenoid valve was placed in the gas line and was controlled by a timer.
Capillary interface
The protein and peptide separation capillaries were joined by an interface, Figure 2. Perpendicular channels were macromachined into a ¼″ thick Plexiglas piece, and glass microcaps (0.037 inch OD, VWR) and glass pipets (0.046 inch OD, VWR) were fixed into the channels with UV glue. The channels’ depths corresponded to the glass microcaps’ and pipets’ outer diameters. 150 μm ID, 360 μm OD fused silica capillary sleeves were glued into the pair of microcaps. The protein and peptide separation capillaries were glued into these sleeves later. A piece of glass microscope slide was then glued on top of the Plexiglas piece to seal the interface.
Figure 2.

Interface between capillaries. The dark microreactor can be seen in the left capillary, and the glass pipets can be seen above and below the interface.
Next, high purity Teflon tubing was carefully twisted over, and glued to, the glass pipets for connections to the flush inlet and the buffer reservoir/waste. 70% EtOH was flushed through the interface for a few min to dissolve and flush out any uncured UV glue, followed by ddH2O and air. The interface was allowed to dry overnight.
The microreactor-containing end of the protein separation capillary was cut to give a desired microreactor length, and the other end was cut to obtain the desired capillary length (e.g. 31.2 cm). Next, ~ 1–2 mm of polyimide was shaved from the microreactor-containing end of the protein separation capillary and from one end of a 48 μm ID, 142 μm OD, 31 cm long PVA-coated peptide separation capillary; the shaved ends were inserted into the capillary interface.
The gap between the two capillaries in the interface was adjusted to ~ 30–50 μm, Figure 2, under a microscope. The capillaries were secured into the interface by placing drops of UV glue at the ends of the sleeve capillaries. The protein separation capillary was replaced once a week due to loss of activity of pepsin.
A valve was placed in-line with the flush inlet Teflon tubing to allow flushing of the system between runs. The flush valve was closed during the analysis.
CE-MS interface
A triple quadrupole mass spectrometer (2000 Q TRAP, ABI/MDS Sciex) was used as the detector. The Q TRAP was operated using the Q TRAP’s Analyst software version 1.4. The electrospray ion source used was a NanoSpray ion source with an x-y-z translational stage for electrospray tip positioning.
The peptide separation capillary was inserted into a 5.4 cm stainless steel electrode, or needle, with a 150 μm ID tapered tip, which was seated inside a stainless steel MicroIonSpray head sheath flow CE-MS interface. The capillary’s end was positioned very close to the needle outlet, yet did not protrude. The MicroIonSpray head had an electrospray voltage applied to it that closed the circuit for CE operation.
A nanofluidic module pump (Upchurch) was used to deliver the sheath flow of He sparged 90% MeOH at 1300 nL/min. A 20 μm ID, 360 μm OD fused silica capillary was used between the pump and the MicroIonSpray head. A stainless steel tee with 750 μm through holes was used in the MicroIonSpray head.
Instrument operation and voltage program
Figure 3 presents a typical voltage program applied to each electrode during the experiment; see figure captions for specific voltages used in each experiment. Sample was electrokinetically injected onto the protein separation capillary by applying 10 kV and 5 kV to power supplies 1 and 2, respectively, for 10 s. After injection, a preliminary separation of proteins was performed in the first capillary, so that the fastest moving components have traveled roughly 75% of the protein separation capillary’s length. Following the preliminary separation, cycles of digestion of proteins in the microreactor and separation of peptides in the second capillary began. A fraction from the protein separation capillary was moved into the microreactor and, simultaneously, the contents of the microreactor were transferred into the peptide separation capillary. Once a fraction was transferred, its components were separated in that second dimension capillary. Simultaneously, by applying about the same potential to the inlet and outlet of the protein separation capillary, sample did not flow in this capillary and a protein fraction was parked in the microreactor for digestion. The voltage applied at the injection end of the protein separation capillary was slightly lower than that at the capillary interface because there was a voltage drop across the Teflon tubing between the capillary interface and buffer reservoir 2. The cycles of fraction transfer, online digestion, and peptide separation were repeated for 25–40 times under computer control.
Figure 3.
Example voltage timing diagram. See figure captions for specific voltages used in each experiment. The potentials are chosen to create zero voltage drop across the protein separation capillary during the peptide separations.
The Q TRAP was run in linear ion trap scan mode with a scan rate of 1000 amu/sec. The electrospray voltage was set to +2250 V, the declustering potential to 30 V, the entrance potential to 10 V, the ion detector to 2300 V, the curtain gas to 15.0, and the nebulizer gas to 0.0. The nebulizer gas flow was set to 10.0 during a 0.2 min prerun of an acquisition batch in the Q TRAP’s Analyst software to blow off excess liquid. The m/z range scanned was from 300–1100 amu.
Data analysis
Data were analyzed with the Q TRAP’s Analyst software and Matlab. There were periodic dropouts of the electrospray during which no ions were detected with MS. These data dropouts were replaced in Matlab with average MS intensities of the whole run.
Information dependent acquisition to obtain tandem mass spectra and Mascot analysis
CE-ESI-MS/MS analyses were performed using the Q TRAP’s information dependent acquisition (IDA) capability. The MS parameters were set as follows: for survey scans: electrospray voltage: +2250 V; curtain gas flow: 15.0; nebulizer gas flow: 0.0; m/z range: 400–1200 amu; scan rate: 4000 amu/sec. For enhanced resolution scans, conditions were the same as for survey scans, except the m/z range was set based on the m/z peak selected, and scan rate: 250 amu/sec. For enhanced product ion scans, conditions were the same as for survey scans, except m/z range: 100–1300 amu; scan rate: 1000 amu/sec. For all scans, two m/z range scans were collected and summed before going on to the next scan. IDA criteria were set so that the 2 most intense mass peaks (with intensities > 10,000 counts per second) were selected in the 400–1200 amu range of the survey scans; ions with 2+ to 5+, or unknown, charge states were submitted for fragmentation; ions that had been fragmented twice were excluded from fragmentation for 60 sec. One mass spectrometry data acquisition cycle lasted 6.5182 sec.
The Mascot search parameters were as follows: PepsinA chosen as enzyme. The database searched was SwissProt. Up to 3 missed cleavages were allowed. Peptide mass tolerance was ± 0.8 Da. The MS/MS mass tolerance was ± 0.6 Da.
RESULTS AND DISCUSSION
High electrophoresis resolution CE-microreactor-CE-MS analysis of myoglobin
Figure 4 presents the raw CE-microreactor-CE-MS data of the analysis of a myoglobin sample as a false color image. The vertical axis corresponds to mass spectral scan number or overall electrophoresis time. The horizontal axis is the m/z axis. Intensity is color coded with the relative abundance scale shown on the right. The instrument generated brief signal drop-outs where the signal dropped to near zero. These drop-outs were replaced with the average signal at each mass and are seen at scans 1800, 3000, and 4100. No spray was seen during the signal dropout times.
Figure 4.
CE-microreactor-CE-MS analysis of a 300 μM solution of myoglobin – raw data. Inserts show close-ups of regions generated by peptides of different m/z ratios. Samples were injected for 10 s; power supply 1 was at a potential of +16 kV and power supply 2 was at 1 kV. The preliminary protein separation was performed for 150 s; power supply 1 was at 18 kV and power supply 2 was at 3 kV. Fractions were transferred for 10 s; power supply 1 was at 25 kV and power supply 2 was at 10 kV. Peptide separation was performed for 180 s; power supply 1 was at 12.95 kV and power supply 2 was at 13 kV. The electrospray voltage was 2.25 kV for the entire procedure.
The inserts show close-ups of data generated at m/z ~ 401, 620, 763, and 985 AMU. The data consist of a set of spots that reappear every 140 cycles, corresponding to the detection of the same peptide during successive transfers. The width of the spot in the x-dimension is related to the mass spectrometer resolution, the spot width in the y-dimension is related to the second dimension (peptide separation) electrophoresis efficiency, and the number of spots observed is related to the efficiency of the first dimension (protein) separation and dispersion induced by the microreactor.
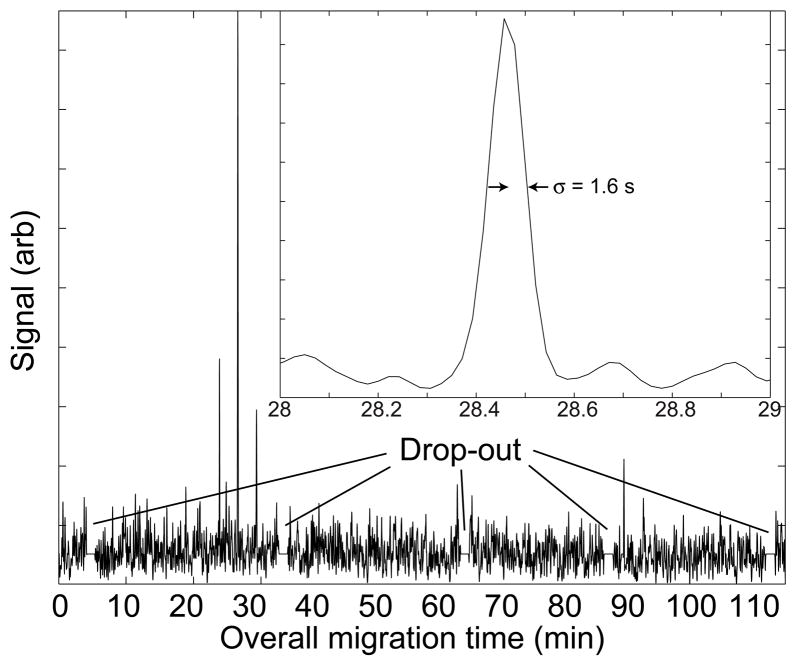
Figure 5 presents the intensity trace generated at m/z ~ 985. A set of three peaks is observed; one main peak and two satellite peaks that arrive 140 transfers before and after the main peak. The number of satellite peaks reflects the efficiency of both the protein separation and the microreactor. The center of this set of peaks is determined by the migration time of the intact protein in the first capillary. The spacing of the peaks is determined by the duration of the second dimension (peptide) separation. The width of those peaks is determined by the second dimension electrophoresis efficiency.
Figure 5.
Raw data generated at m/z = ~ 985. A close-up of the most intense peak is shown in the insert. The peak width, expressed as the standard deviation of a Gaussian function, is 1.6 s. Periods when the signal dropped out are noted; the data during the drop-outs have been replaced with the mean intensity.
Each mass spectral channel can be converted to an image. Successive second-dimension peptide separations are converted to intensity traces, and the set of intensity traces rastered to create an image. Figure 6 presents the 985 AMU data as a gel image, where the vertical distance corresponds to the fractions transferred from the protein separation capillary, the horizontal distance corresponds to the migration time of the peptide in the second capillary, and optical density is related to intensity. This peptide migrates in the 9th fraction transferred from the protein separation capillary, with a nominal migration time of 90 s in the peptide separation dimension.
Figure 6.
Gel image of data generated at m/z ~ 985.
Figure 7 presents six electropherograms generated at m/z ~ 401 (GKVEADIA), 510 (GADAQGAMTKA), 620 (TKHKIPIKYL), 763 (GLSDGEW), 930 (GGILKKKGHHEAELKPL), and 985 (RLFTGHPETLEKFDKF) AMU. These data are shown as landscapes, which tend to better present the dynamic range of the measurements than the gel-image. All spots are centered at the 9th fraction transfer from the protein separation capillary. The spots have different migration times in the peptide electrophoresis dimension, which minimizes ion-suppression effects that would be observed if peptides were simply ionized directly from the microreactor. We characterize these spots by fitting them with a Gaussian surface. The spots have an average width (expressed as standard deviation of the Gaussian) of 0.5 ± 0.1 transfers in the protein separation dimension. Spots are also remarkably sharp in the peptide separation dimension, with a width of 1.4 ± 0.1 s. The microreactor does not introduce excessive band broadening.
Figure 7.
Landscape images generated at six different m/z ratios for the data of Figure 4.
The peak capacity of a one-dimensional separation and the spot capacity of a two-dimensional separation measure the number of components that can be resolved without overlap. Peak capacity is given by the separation window (the time from the first to last migrating components) divided by four times the standard deviation of the Gaussian peak. Peak capacity in the protein separation dimension is 18.5 and in the peptide dimension is 32. The spot capacity is given by the product of the peak capacities and equals 590.
Low electrophoresis resolution CE-microreactor-CE-MS/MS of a mixture
We attempted to use information dependent data acquisition to obtain MS/MS data from the system that generated Figures 4–7. Unfortunately, the electrophoresis peaks were too fast for successful MS/MS generation.
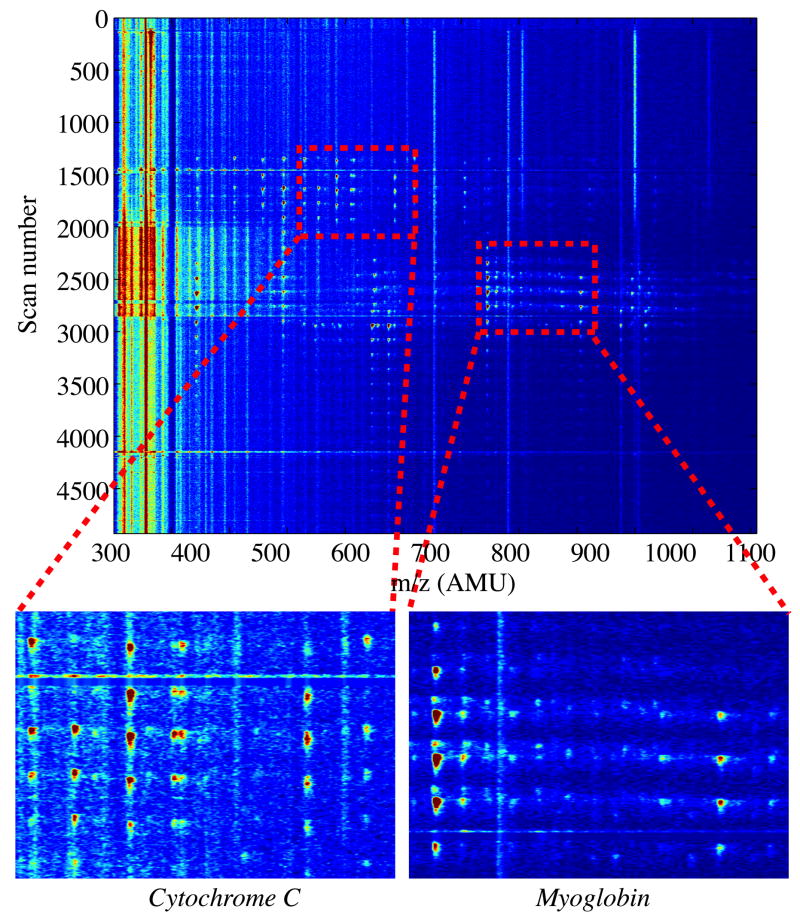
One of our analyses employed a PVA-coated peptide separation capillary that was several weeks old. This system generated relatively broad peaks that were suitable for MS/MS analysis. Figure 8 presents data for this analysis of a mixture of cytochrome C and myoglobin. One group of spots is generated by digestion of cytochrome C and is observed in the region from 1400 to 2000 scans. The second set of spots is generated by myoglobin and is found in the 2400 to 3200 region.
Figure 8.
CE-microreactor-CE-MS data for a mixture of 200 μM cytochrome C and myoglobin. Samples were injected for 10 s; during injection, power supply 1 was at a potential of +9.7 kV, power supply 2 was at 1 kV, and a pressure of 22 PSI was applied to the injector. The preliminary protein separation was performed for 150 s; power supply 1 was at 14.1 kV and power supply 2 was at 3 kV. Fractions were transferred for 10 s; power supply 1 was at 25 kV and power supply 2 was at 10 kV. Peptide separation was performed for 180 s; power supply 1 was at 12.95 kV and power supply 2 was at 13 kV. The electrospray voltage was 2.25 kV for the entire procedure.
Information dependent data acquisition for CE-microreactor-CE-MS/MS
The peaks in Figure 8 are broad, ~ 30 seconds in the peptide separation dimension, which provides ample time to perform information dependent MS/MS analysis. Data were submitted to Mascot, which returned correct identifications of both cytochrome C and myoglobin with Mowse scores of 107 for cytochrome C and 58 for myoglobin. Bovine cytochrome C was identified based on 4 different peptides, and horse heart myoglobin based on 2. The sequence coverages were 48% and 22%, respectively. The identified sequences of the four cytochrome C peptides were GRKTGQAPGF, IAYLKKATNE, AGIKKKGEREDL, and MEYLENPKKYIPGTKMIF, and the sequences of the two myoglobin peptides were TALGGILKKKGHHEAEL and FRNDIAAKYKELGFQG.
Conclusion
This proof-of-principle system offers several potential advantages. First, the system relies on electrophoretic separation of intact proteins in the first dimension. In principle, changes in post-translational modifications should be detected through a mobility shift of the intact proteins. Second, the system is fully automated, which allows unattended operation once the sample is loaded. Third, since the system is based on capillary electrophoresis, separations are fast and can demonstrate high efficiency.
However, narrow peak widths also place constraints on the mass spectrometer performance for MS/MS analysis. Peak widths can be as narrow as 1.4 seconds, which is challenging for information dependent data acquisition, particularly when using electrospray ionization. The technology would be better suited for MALDI-based analysis, where mass spectrometer analysis time is decoupled from electrophoresis time.
We did not investigate detection limits for the system. These experiments injected a few picomoles of protein and generated good signal-to-noise ratios. Undoubtedly, improved mass spectrometry and sample handling will allow analysis of much smaller amounts of protein.
As demonstrated elsewhere, two-dimensional capillary electrophoresis with laser-induced fluorescence can produce highly reproducible and relatively rapid analyses of complex biological samples21–22. Ultimately, it will be of interest to analyze complex protein samples using the mass spectrometer. However, capillary electrophoresis separation of proteins often suffers from component adsorption to the capillary walls. Addition of surfactant can enhance the protein solubility and decrease adsorption. The use of acid-labile surfactants will be attractive in this system to provide efficient electrophoretic separation in a mass spectrometer compatible buffer23.
Acknowledgments
This work was funded by a contract from ABI/MDS Sciex and a grant from the National Institutes of Health (R01GM071666). Darren Lewis of Upchurch generously donated the microfluidic pump employed in this experiment. Charles Liu of MDS Sciex and Applied Biosystems provided the MicroIonSpray head and suggested the use of poly(vinyl alcohol). We also gratefully acknowledge the support and encouragement of William Davidson on this and many other projects.
Literature cited
- 1.Bogdanov B, Smith RD. Mass Spectrom Rev. 2005;24:168–200. doi: 10.1002/mas.20015. [DOI] [PubMed] [Google Scholar]
- 2.Sze SK, Ge Y, Oh H, McLafferty FW. Proc Natl Acad Sci (USA) 2002;99:1774–1779. doi: 10.1073/pnas.251691898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Reid GE, McLuckey SA. J Mass Spectrom. 2002;37:663–667. doi: 10.1002/jms.346. [DOI] [PubMed] [Google Scholar]
- 4.Kelleher NL. Anal Chem. 2004;76:197A–203A. [PubMed] [Google Scholar]
- 5.Clauser KR, Hall SC, Smith DM, Webb JW, Andrews LE, Tran HM, Epstein LB, Burlingame AL. Proc Natl Acad Sci (USA) 1995;92:5072–5076. doi: 10.1073/pnas.92.11.5072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Link AJ, Eng J, Schieltz DM, Carmack E, Mize GJ, Morris DR, Garvik BM, Yates JR. Nat Biotechnol. 1999;17:676–678. doi: 10.1038/10890. [DOI] [PubMed] [Google Scholar]
- 7.Gygi SP, Rist B, Gerber SA, Turecek F, Gelb MH, Aebersold R. Nat Biotechnol. 1999;17:994–999. doi: 10.1038/13690. [DOI] [PubMed] [Google Scholar]
- 8.Amankwa LN, Kuhr WG. Anal Chem. 1993;65:2693–2697. [Google Scholar]
- 9.Licklider L, Kuhr WG, Lacey MP, Keough T, Purdon MP, Takigiku R. Anal Chem. 1995;67:4170–4177. [Google Scholar]
- 10.Bonneil E, Waldron KC. Talanta. 2000;53:687–699. doi: 10.1016/s0039-9140(00)00554-3. [DOI] [PubMed] [Google Scholar]
- 11.Wang C, Oleschuk R, Ouchen F, Li J, Thibault P, Harrison DJ. Rapid Commun Mass Spectrom. 2000;14:1377–1383. doi: 10.1002/1097-0231(20000815)14:15<1377::AID-RCM31>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 12.Gao J, Xu J, Locascio LE, Lee CS. Anal Chem. 2001;73:2648–2655. doi: 10.1021/ac001126h. [DOI] [PubMed] [Google Scholar]
- 13.Sakai-Kato K, Kato M, Toyo’oka T. Anal Chem. 2002;74:2943–2949. doi: 10.1021/ac0200421. [DOI] [PubMed] [Google Scholar]
- 14.Kato M, Sakai-Kato K, Jin H, Kubota K, Miyano H, Toyo’oka T, Dulay Maria T, Zare RN. Anal Chem. 2004;76:1896–1902. doi: 10.1021/ac035107u. [DOI] [PubMed] [Google Scholar]
- 15.Svec F, Huber CG. Anal Chem. 2006;78:2100–2107. doi: 10.1021/ac069383v. [DOI] [PubMed] [Google Scholar]
- 16.Ye M, Hu S, Schoenherr RM, Dovichi NJ. Electrophoresis. 2004;25:1319–1326. doi: 10.1002/elps.200305841. [DOI] [PubMed] [Google Scholar]
- 17.Moini M, Cao P, Bard AJ. Anal Chem. 1999;71:1658–1661. doi: 10.1021/ac9811266. [DOI] [PubMed] [Google Scholar]
- 18.Belder D, Deege A, Husmann H, Koehler F, Ludwig M. Electrophoresis. 2001;22:3813–3818. doi: 10.1002/1522-2683(200109)22:17<3813::AID-ELPS3813>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- 19.Fogarty K, Van Orden A. Anal Chem. 2003;75:6634–6641. doi: 10.1021/ac035022t. [DOI] [PubMed] [Google Scholar]
- 20.Krylov SN, Starke DA, Arriaga EA, Zhang Z, Chan NW, Palcic MM, Dovichi N. J Anal Chem. 2000;72:872–877. doi: 10.1021/ac991096m. [DOI] [PubMed] [Google Scholar]
- 21.Michels DA, Hu S, Schoenherr RM, Eggertson MJ, Dovichi NJ. Mol Cell Proteomics. 2002;1:69–74. doi: 10.1074/mcp.t100009-mcp200. [DOI] [PubMed] [Google Scholar]
- 22.Kraly JR, Jones MR, Gomez DG, Dickerson JA, Harwood MM, Eggertson M, Paulson TG, Sanchez CA, Odze R, Feng Z, Reid BJ, Dovichi NJ. Anal Chem. 2006;78:5977–5986. doi: 10.1021/ac061029+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Meng F, Du Y, Miller LM, Patrie SM, Robinson DE, Kelleher NL. Anal Chem. 2004;76:2852–2858. doi: 10.1021/ac0354903. [DOI] [PubMed] [Google Scholar]