Abstract
2′-Deoxy-2′-fluoro-arabinonucleic acid (FANA) and ANA paired to RNA are substrates of RNase H. The conformation of the natural DNA:RNA hybrid substrates appears to be neither A-form nor B-form. Consistent with this the conformations of FANA and ANA were found to be intermediate between the A- and B-forms. However, FANA opposite RNA is preferred by RNase H over ANA, and the RNA affinity of FANA considerably exceeds that of ANA. By investigating the conformational boundaries of FANA and ANA residues in crystal structures of A- and B-form DNA duplexes at atomic resolution, we demonstrate that FANA and ANA display subtle conformational differences. The structural data provide insight into the structural requirements at the catalytic site of RNase H. They also allow conclusions with regard to the relative importance of stereoelectronic effects and hydration as modulators of RNA affinity.
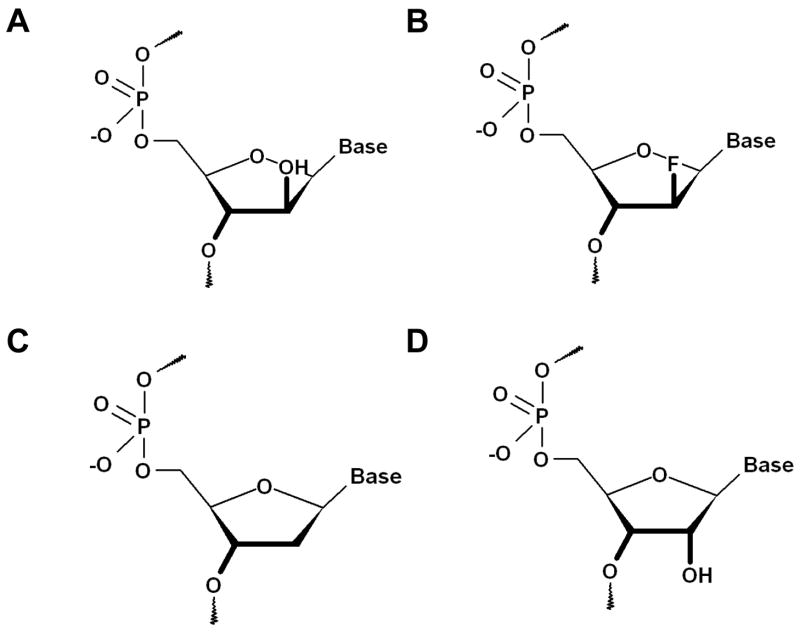
Gene silencing via the antisense strategy has proven a formidable challenge in practice, requiring nucleic acid modifications with high RNA affinity and nuclease resistance that are capable of permeating the cell wall and being recognized as substrates by RNase H (1–3). The last criterion in particular has only been satisfied by very few analogues, among them phosphorothioate DNA (PS-DNA) (4). Therefore clinical trials have for the most part been conducted with antisense PS-DNAs or so-called gap-mers, oligos featuring various modifications in the flanks and central PS-DNA windows (5–8). However, despite favorable properties including nuclease resistance and mediation of RNase H action, PS-DNAs also exhibit limitations with regard to RNA-binding, pharmacokinetics, pharmacodynamics and toxicology (reviewed in refs. 9 and 10). Among the second-and third-generation candidate modifications for antisense (11), arabinonucleic acids (ANAs; Figure 1) constitute an exception as all-modified oligonucleotides hybridized to RNA are substrates of RNase H (12).
FIGURE 1.
Structures of nucleosides. (A) Arabinonucleic acid (ANA); (B) 2′-Fluoro-arabinonucleic acid (FANA); (C) DNA; (D) RNA.
The 2′-fluoro-ANA (FANA) analogue displays increased RNA affinity compared to both DNA and PS-DNA (13). By comparison, the corresponding hybrid duplexes formed by ANA oligonucleotides are of lower stability relative to DNA and PS-DNA (13, 14). Thus, the observed trend for the stability of heteroduplexes between RNA and antisense oligonucleotides (AONs) is as follows: FANA > RNA > DNA > PS-DNA ≫ ANA (56% pyrimidine content) (13). Moreover, arabinonucleic acids have nuclease resistance to serum and cellular nucleases that exceed those by DNA but do not match the stability seen with PS-DNA (12). Although PS-FANA AONs showed only relatively weak inhibition of cellular target expression, mixed-backbone oligomers with PS-DNA cores flanked by PS-FANA stretches were found to have potent antisense activity (15). So-called FANA/DNA altimers, consisting of alternating FANA and DNA residues (16), and FANA constructs containing acyclic nucleotide inserts (17) were also capable of promoting efficient cleavage of RNA targets by RNase H.
CD-spectra of DNA:RNA, FANA:RNA and ANA:RNA duplexes in solution, although displaying close similarity, show distinct positive bands near 270 nm (13). Both X-ray crystallography and NMR solution studies indicated the O4′-endo (East (18)) conformation as preferred by FANA sugars (19, 20). This conformation is also adopted by ANA residues in the structure of an ANA:RNA duplex determined by NMR (21). The latter finding contradicts the results of earlier computational simulations that suggested a preference for the C2′-endo pucker by arabinose (22). Although the above experimental data are consistent with similar conformational tendencies by FANA and ANA, the conformation of ANA was demonstrated to vary significantly depending on the geometry of the parent duplex (23–29).
RNase H1 binds RNA duplexes and DNA:RNA hybrids with similar affinities but recognizes only the latter as substrates (30). The precise interactions between RNase H and the heteroduplex and the conformation of the DNA:RNA duplex at the active site of the enzyme have been the subject of numerous investigations over the last 15 years (see for example refs. 31–34 and cited literature). The enzyme was expected to use the availability of 2′-hydroxyl groups in the RNA strand and the absence thereof in the DNA strand to discriminate between DNA:RNA and RNA:RNA duplexes (31). Based on NMR investigations in solution, it was concluded that sugars of the DNA strand in the hybrid duplex adopted Eastern pucker. It was also suggested that the enzyme would recognize the narrowing of the minor groove in a heteroduplex with such a conformation relative to an RNA:RNA duplex (32). A modeled duplex between an RNA strand with C3′-endo riboses and a FANA strand with O4′-endo arabinoses exhibits intact base-pairing and stacking interactions and a slightly contracted minor groove compared with an RNA duplex (35) (Figure 2). However, in the structure of HIV-1 reverse transcriptase in complex with a polypurine tract DNA:RNA, the minor groove is indeed contracted but deoxyriboses of the duplex portion bound to the RNase H domain exhibit Southern puckers (33). And in the recent crystal structure of the complex between a bacterial RNase H and a DNA:RNA hybrid, riboses adopt C3′-endo pucker and deoxyriboses adopt C2′-endo and C1′-exo puckers (36). The minor groove of the hybrid duplex is also narrower compared to a canonical A-form duplex and five 2′-hydroxyl groups of the RNA strand are contacted directly by the enzyme.
FIGURE 2.
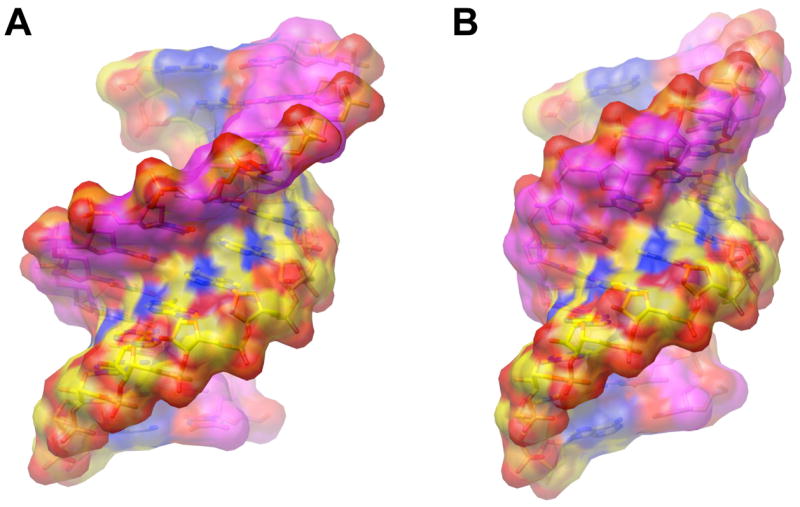
Dimensions and groove shapes of DNA:RNA heteroduplexes as a function of the sugar pucker in the two strands. (A) The DNA and RNA adopting O4′-endo and C3′-endo conformation, respectively. Carbon atoms in DNA are colored in magenta and carbon atoms in RNA are colored in yellow. (B) Both, the DNA and RNA adopting C3′-endo conformation. Color scheme identical to panel (A). The van der Waals surfaces are shown in a semi-transparent mode and illustrate the narrower minor groove of the duplex with O4′-endo puckers of 2′-deoxyriboses. The models were generated with the program TURBO-FRODO (47).
No structural data are available at this time for an inhibitor complex (RNase H-RNA:RNA). In addition, there is an unmet need to understand a wealth of available functional data with regard to the dependence of RNase H cleavage activity on location and nature of chemical modifications in the DNA strand (for examples, see ref. 37). Currently, it is unclear if the enzyme tolerates a limited range of conformations of the DNA or AON strand paired to RNA and whether the minor groove width is really the central recognition feature exploited by RNase H (see the discussion in ref. 33). Interestingly, all-FANA strands and isolated FANA residues in B-form DNA appear to be restricted to the O4′-endo conformation and a typical Southern pucker may be unfavorable. To explore the capacity of the FANA analogue for conformational variability and to work out potential differences in the conformational behavior of FANA and ANA that relate to their recognition by RNase H, we have determined high-resolution crystal structures of DNA duplexes containing arabino nucleosides. The structural data demonstrate that FANA and ANA differ conformationally, depending on the duplex environment, and that the FANA analogue disfavors the Southern and South-Eastern pucker ranges.
Experimental Section
Reagents
The synthesis of the 1-[2-Deoxy-2-fluoro-5-O-(4,4′-dimethoxytrityl)-3-O-(β-cyanoethyl-N,N-diisopropylphosphoramidite)-β-D-arabinofuranosyl] thymine monomer was previously described (13). 1-(2-O-benzoyl-5-O-(4,4′-dimethoxytrityl)-3-O-(β-cyanoethyl-N,N-diisopropylphosphoramidite)-β-D-arabino-furanosyl] uracil was synthesized from 2,2′-anhydro-1-(β-D-arabinofuranosyl) uracil (RI Chemicals, Orange, CA) according to published procedures (14). 1-(2-O-Methyl-5-O-(4,4′-dimethoxytrityl)-3-O-(β-cyanoethyl-N,N-diisopropylphosphor-amidite)-β-D-ribofuranosyl] adenine, 2′-deoxynucleotide phosphoramidites and 3′-terminal nucleoside controlled pore glass (CPG) support were purchased from Glen Research (Sterling, VA). All other chemicals for solid phase oligonucleotide synthesis were purchased from Glen Research (Sterling, VA) as well.
Oligonucleotide Synthesis and Purification
The ANA-/2′-O-methyl-modified DNA decamer with sequence 5′-GCGT-2′OMeA-aU-ACGC-3′ (aU=arabino-U), the ANA-modified DNA dodecamers 5′-CGCGAA-aU-TCGCG-3′ and 5′-CGCGAAT-aU-CGCG-3′, and the FANA-/2′-O-methyl-modified DNA decamer 5′-GCGT-2′OMeA-faT-ACGC-3′ (faT=2′-fluoroarabino-T) were all synthesized on an Applied Biosystems 381A DNA synthesizer following slight modifications to published procedures (14). Monomer coupling times were 90 seconds for 2′-deoxynucleotide phosphoramidites and 10 minutes for the 2′-OMe-ribo, ANA and FANA phosphoramidites. Deprotection and cleavage of the oligonucleotides from the solid support were achieved using 28% NH4OH, 55°C for 8 hours. The sequences were analyzed and purified by strong anion exchange (SAX) HPLC using a DIONEX DNAPAC PA-100 analytical column (4 × 25 mm) purchased from Dionex Corp (Sunnyvale, CA). For preparatory runs, 10 O.D. units were purified at a time on an analytical column using a gradient of 25 mM TrisHCl (pH = 7.8) to 1 M NaCl over 30 minutes with a flow rate of 1.0 mL/min. Oligomers purified by HPLC were desalted on SEP PAK cartridges (Waters Inc.). The cartridge was pre-equilibrated with acetonitrile followed by water. The oligonucleotide solution was applied to the cartridge, washed with water and a solution of 75% methanol in water was then used to elute the desalted oligomer. Molecular weights of the sequences were determined by MALDI-TOF mass spectrometry.
Crystallization and X-ray Diffraction Data Collection
The stock solutions of all chemically modified oligos were adjusted to concentrations of between 1 and 2 mM. Crystallizations were performed by the vapor diffusion technique using hanging drops. The commercially available Nucleic Acid Mini Screen™ (Hampton Research, Aliso Viejo, CA) (38) was used to establish crystallization conditions. Droplets containing oligonucleotide and buffer in 1:1 and 1:2 ratios were equilibrated against 0.5 ml 35% (v/v) 2′-methyl-2,4-pentanediol (MPD) at 18°C. Crystals of the ANA-/2′-O-methyl-modified DNA decamer were grown using condition 4 (final droplet composition: 1 mM oligonucleotide, 10% MPD, 20 mM sodium cacodylate, pH 5.5, 10 mM cobalt hexamine, 20 mM LiCl and 10 mM MgCl2). Crystals of the FANA-/2′-O-methyl-modified DNA decamer were grown using condition 3 (final droplet composition: 1 mM oligonucleotide, 10% MPD, 20 mM sodium cacodylate, pH 5.5, 10 mM cobalt hexamine, 6 mM NaCl, and 80 mM KCl). Crystals for both ANA-modified DNA dodecamers were obtained from droplets containing 0.6 mM oligonucleotide, 20 mM sodium cacodylate, pH 7, 6.3 mM Mg(OAc)2, and 3 mM spermine tetrahydrochloride.
We also undertook crystallization experiments with the following DNA oligonucleotides that contained FANA modifications: 5′-G-faC-GTA-2′OMeT-ACGC-3′, 5′-GCGTA-2′OMeT-A-faC-GC-3′, 5′-CG-faC-GCG-3′ and 5′-faC-G-faC-G-faC-G-3′. However, either no crystals were obtained or the crystals that were produced diffracted only to relatively low resolution and structure determination was then not further pursued.
Crystals were mounted in nylon loops and frozen in liquid nitrogen. Diffraction data were collected either on the insertion device beamlines of the DuPont-Northwestern-Dow Collaborative Access Team (DND-CAT, 5-ID) or the South-East Regional Collaborative Access Team (SER-CAT, 22-ID) at the Advanced Photon Source, Argonne National Laboratory (Argonne, IL), using MarCCD-300 detectors. The wavelengths for the data collections varied between ca. 0.9 Å and 1.0 Å. In all cases high- and low-resolution data sets were collected separately. To avoid overloads in the low-resolution frames the beam was attenuated. Raw data were integrated and scaled with either AUTOMAR (MarResearch Inc.) or XDS (39). Selected crystal data and data collection and processing parameters for all four structures are summarized in Table 1.
Table 1.
Selected crystal data, data collection and refinement parameters
| Structure | A-ANA | A-FANA | B-ANA1 | B-ANA2 |
|---|---|---|---|---|
| DNA Sequence | GCGT-2′OMeA-aU-ACGC | GCGT-2′OMeA-faT-ACGC | CGCGAA-aU-TCGCG | CGCGAAT-aU-CGCG |
| Space Group | P212121 | P21 | P212121 | P212121 |
| Crystal System | orthorhombic | monoclinic | Orthorhombic | orthorhombic |
| Cell Constants (Å/°) |
a = 32.28
b = 32.47 c = 66.75 |
a = 32.73
b = 69.18 c = 32.83 β= 92.2 |
a = 25.47
b = 39.76 c = 65.35 |
a = 25.70
b = 39.92 c = 65.56 |
| Temperature (°C) | −170 | |||
| Strands per Asymm. Unit | 2 | 4 | 2 | 2 |
| No. of Ions, Solvent Molecules | 3 Co(NH3)63+, 1 Cl−, 131 H2O | 7 Co(NH3)63+, 177 H2O | 1 Mg2+, 179 H2O | 1 Mg2+, 131 H2O |
| Unique data [I/σ(I) ≥ 0) | 21,545 | 15,464 | 23,845 | 19,780 |
| Completeness, Overall (%) | 93.1 (33-1.19 Å) | 94.0 (35-1.69 Å) | 96.3 (50-1.13 Å) | 99.8 (30-1.24 Å) |
| Completen., Last Shell (%) | 88.0 (1.27-1.19 Å) | 89.6 (1.80-1.69 Å) | 59.5 (1.17-1.13 Å) | 99.5 (1.34-1.24 Å) |
| R-merge | 0.070 | 0.094 | 0.033 | 0.077 |
| Resolution (Å) | 1.19 | 1.69 | 1.13 | 1.24 |
| R-work | 0.157 | 0.194 | 0.188 | 0.155 |
| R-free | 0.169 | 0.211 | 0.240 | 0.212 |
| R.m.s. Bond Lengths (Å) | 0.007 | 0.015 | 0.011 | 0.012 |
| R.m.s. Bond Angles (°/Å)a | 1.2 | 2.7 | 0.019 | 0.019 |
| PDB ID code | 2FIJ | 2FIL | 2FIH | 2FII |
Structure Determination and Refinement
All structures were determined by the Molecular Replacement technique. For the ANA-modified B-form dodecamers, native B-DNA dodecamer search models in combination with the program CNS (40) were used. For the ANA-/2′-O-methyl-modified and FANA-/2′-O-methyl-modified DNA decamers, a high resolution native A-form structure was used as the MR search model (41) and the solutions were found by using EPMR (42). The structures of the modified B-form duplexes were initially refined with CNS and at a later stage with the program SHELX-97 (43). In the cases of the A-ANA and A-FANA decamers, refinements were performed initially with simulated annealing by using CNS, followed by geometric constraint/maximum-likelihood refinement and isotropic temperature factors refinement for individual atoms by using the program REFMAC 5.0 (44–46). In later refinement stages, solvent water molecules were added and anisotropic temperature factors refinement was carried out. All four structures were refined as all-DNA molecules at the beginning until R-factor values decreased to around 30%. At that stage the arabinonucleic acid residues were introduced into the duplexes based on Fourier (2Fo-Fc) sum and (Fo-Fc) difference electron density maps and corresponding geometry/topology files were adapted. The program TURBO-FRODO (47) was used to display electron density maps and to manually rebuild portions of the duplex models and add/delete water molecules. To calculate the R-free (48), 5% randomly chosen reflections were set aside in all four cases. Selected final refinement parameters for the four structures are listed in Table 1.
RESULTS AND DISCUSSION
Structure Determinations
To study the conformational properties of arabino nucleosides we used decamer and dodecamer DNA template sequences. The decamer sequence 5′-GCGTATACGC was locked in an A-form by replacing the second adenosine with a 2′-O-methylated A (2′OMeA). A single ribonucleoside or 2′-O-methylated ribonucleoside is sufficient to drive the conformational equilibrium for the decamer toward the A-form (49, 50) (see ref. 51 for another case of a B- to A-form DNA duplex conversion driven by a single 2′-OH). The 3′-adjacent T was then replaced by either ANA-U or FANA-T and the resulting sequences GCGT-2′OMeA-aU-ACGC (aU=arabino-U) and GCGT-2′OMeA-faT-ACGC (faT=2′-fluoroarabino-T) used for crystal structure determination. We will refer to these duplexes and structures as A-ANA and A-FANA, respectively. The crystallographic asymmetric unit of the A-FANA structure contains two independent duplexes (Table 1), termed A-FANA1 and A-FANA2. Residues in the decamer duplexes are numbered 1 to 10 in strand 1 and 11 to 20 in strand 2. Residues of A-FANA-2 strands are numbered 21 to 30 and 31 to 40. The dodecamer sequence 5′-CGCGAATTCGCG served as the B-form environment to examine the conformational preferences of ANA residues. Both Ts were separately replaced with ANA-U, an approach that had previously provided insight into the conformation of FANA-T (19). The resulting dodecamers CGCGAAUU-aU-TCGCG and CGCGAAT-aU-CGCG were crystallized using standard methods and their structures determined at near atomic resolution (Table 1). The two structures and duplexes will be referred to as B-ANA1 and B-ANA2, respectively, and residues are numbered 1 to 12 in strand 1 and 13 to 24 in strand 2.
X-ray diffraction data collections for all four crystals were carried out on the 5-ID beamline at the Advanced Photon Source (APS, Argonne, IL) and the structures were determined with the Molecular Replacement technique. Selected crystal data and data collection and processing parameters are summarized in Table 1. The ANA-modified B-DNA dodecamers crystallize in the standard orthorhombic lattice favored by the Mg2+-form of the Dickerson-Drew dodecamer (52). However, the ANA- and FANA-modified decamers crystallize in new P212121 and P2 1 forms, respectively, unlike other decamers of the same sequence containing 2′-O-modified residues (see for example refs. 49, 53–58), that crystallize in an orthorhombic form with approximate unit cell constants of 25, 44 and 45 Å. It is noteworthy that the decamer duplex with 2′-O-methyl-A at positions 5 (strand 1) and 15 (strand 2) crystallizes in the ‘regular’ orthorhombic form (53). Therefore the presence of the ANA and FANA modifications triggers a change in the lattice for these two decamers. The a and b cell constants of A-ANA crystals are rather similar (Table 1) and the diffraction data can be indexed and merged with reasonably low figures of merit in tetragonal space groups (P4 and P422). However, all attempts to phase the tetragonal data using Molecular Replacement and A-form models failed and it was concluded that the correct space group is orthorhombic P212121 with a single duplex constituting the asymmetric unit.
Refinements of the structures were performed either with the programs CNS (40) and SHELX-97 (43) (B-ANA1 and B-ANA2) or REFMAC (44, 45) (A-ANA and A-FANA), using anisotropic temperature factors for all DNA atoms, ions and the majority of solvent molecules. Final refinement parameters for all structures are listed in Table 1 and examples of the quality of the final electron density are depicted in Figures 3A, B.
FIGURE 3.

Quality of the electron density and overall conformations of ANA- and FANA-modified duplexes. (A) Sum (2Fo-Fc) Fourier electron density (1.5 σ level) around residues 2′OMeA5 and aU6 in the A-ANA structure. Note the unstacking of bases as a result of a strong roll. (B) Sum (2Fo-Fc) Fourier electron density (1.5 σ level) around residues T7 and aU8 in the B-ANA2 structure. (C) The A-FANA1 duplex viewed into the major groove. Residues faT6 and faT16 are highlighted in magenta and 2′-fluorine atoms are green. (D) The B-ANA2 duplex viewed into the major groove. Residues aU8 and aU20 are highlighted in blue and 2′-oxygen atoms are red. (E) Different degrees of bending into the major groove exhibited by ANA- and FANA-modified DNA decamer duplexes: A-FANA1 (green) > A-FANA2 (red) > A-ANA (blue). The duplexes were superimposed by aligning the helical axes in their bottom halves (base pairs G1:C20 – T4:A17). The view is across the major (left) and minor (right) grooves and the molecular dyad is horizontal. 2′-Fluorine (A-FANA1 and A-FANA2) and 2′-oxygen atoms (A-ANA) are highlighted as spheres with matching colors.
Overall Helix Geometries
As expected the global geometries of the A-ANA and A-FANA duplexes are of the A-form and those of the B-ANA1 and B-ANA2 duplexes are B-form (Figures 3C, D). Therefore, incorporation of isolated ANA or FANA residues does not bring about a change in the duplex type intrinsically preferred by a particular DNA sequence, and stabilized by 2′-O-methylated nucleosides in the case of the DNA decamer investigated here. In the A-ANA and A-FANA duplexes the arabino-U and -T residues, respectively, occupy positions on opposite strands at the central base-pair step (Figure 3C), and in the B-form duplexes ANA-Us are separated by the two central A:T base pairs (Figure 3D). It appears that the effects of ANA residues on the overall geometry of the helix in the case of B-ANA1 and B-ANA2 are only minor. Compared to a native DNA duplex of the same sequence, isolated ANA residues do not significantly affect backbone geometry, helix diameter, and/or groove widths and no bend at the local level is induced (data not shown). Conversely, analysis of the helical parameters for all three A-form duplexes, A-ANA, A-FNA1 and A-FANA2, with the program CURVES (59) reveals that they exhibit significant distortions globally and locally compared to a reference duplex. As a reference duplex we chose the decamer of identical sequence GCGTAT*ACGC and with a single 2′-O-methyl-3′-methylene-T (T*) modification per strand (PDB ID code 1dpl; http://www.rcsb.org). Its structure was determined at ultra-high resolution (0.83 Å), yielding precise helical parameters and detailed insight into the water structure (41). In the reference duplex all sugars adopt C3′-endo (Northern) pucker (18, 60). A localized kink of about 13° occurs at the T4pA5 (T*16pA17) step and divides the duplex into canonical A-form tetramers and hexamers. The kink is into the major groove, narrows that groove and chiefly affects the roll between the fourth and fifth base pairs. Thus, the duplex is shortened by about 5% relative to a hypothetical straight form. A similar distortion albeit of varying magnitude is observed in all other A-form decamers with single 2′-O-modified thymidines in the aforementioned P212121 lattice (i.e. refs. 55, 57, 58). However, the A-ANA, A-FANA1 and A-FANA2 helices display markedly different distortions from canonical A-form geometry.
In the A-ANA duplex, like in the reference one, the bend is localized, but it occurs between base pairs in the center of the duplex. The kink amounts to about 26° and is the result of a large roll between the 2′OMeA5:aU16 and aU6:2′OMeA15 pairs combined with a 1.5 Å slide (y-displacement). This leads to shortening of the duplex by about 4% and, since the kink is into the major groove, a narrower major groove (by ca. 1 Å) and a slightly wider minor groove near and at the center of the duplex. Nevertheless, the A-ANA duplex exhibits typical features of an A-form duplex such as strongly inclined base pairs relative to the helix axis (avg. 20°). A wire-diagram in stereo of the A-ANA helix along with the global axis to illustrate the kink is depicted in Figure 3E, and the two A-FANA duplexes are superimposed for comparison. The two latter duplexes exhibit bending that involves strong rolls between four consecutive base pairs (T4:A17, 2′OMeA5:faT16, faT6:2′OMeA15, and A7:T14). In A-FANA1, the rolls between these pairs are 25° (18°), 14° (11°) and 17° (13°), respectively (global axis curvature angles at the respective sites are given in parentheses). In A-FANA2, the rolls are smaller by comparison and amount to 13° (11°), 10° (7°), and 19° (15°), respectively (axis bend angles in parentheses). The bends result in considerably shorter duplexes compared to an ideal straight helix (10% and 5% for A-FANA1 and A-FANA2, resp.). In both cases, x- and y-displacements between adjacent pairs make only minor contributions to the distortion of the helix compared with A-ANA. Similar to the A-ANA duplex, the A-FANA helices are bent into the major groove (Figure 3E) and views into the minor groove along the molecular dyad reveal more or less straight helix axes (not shown). The different helix geometries of A-FANA1 and A-FANA2 evident from the different degrees of bending are also manifested in deviating inclinations (5° and 11°, resp.).
In summary, incorporation of ANA residues into B-form DNA does not lead to obvious changes in the local or global helical geometry. On the other hand, incorporation of both ANA and FANA residues into A-form duplexes triggers notable conformational changes at the local (vide infra) and global levels. ANA and FANA appear to be accommodated differently and the two crystallographically independent A-FANA duplexes also adopt different geometries (Figure 3E). Although A-ANA and A-FANA crystals grow in different crystal systems (Table 1), unit cell constants and packing interactions are very similar (the latter will be discussed in more detail later and also resemble those in crystals of the reference duplex). We may conclude that the differences in the geometries of the A-form helices are not simply due to crystal packing effects but to a significant extent the direct result of the individual and apparently deviating conformational preferences of ANA and FANA nucleosides.
Conformations of ANA and FANA Nucleosides in A- and B-DNA Helices
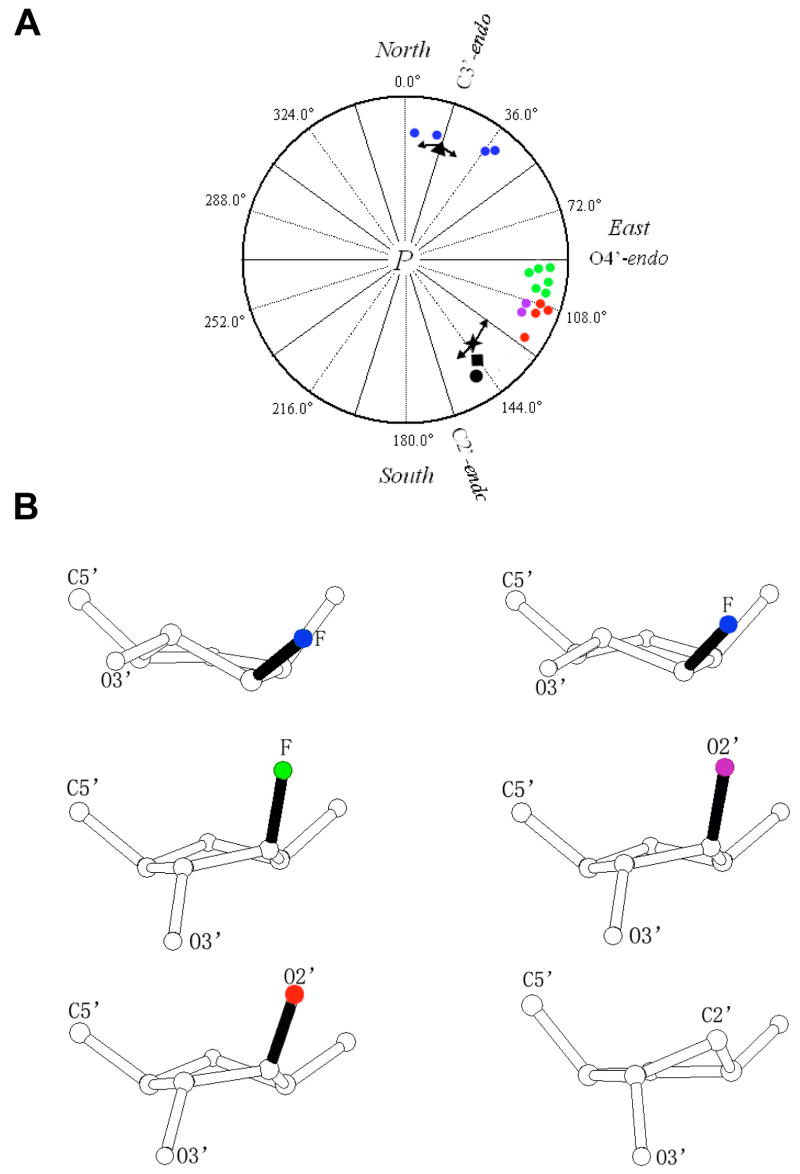
Arabinoses in the B-ANA1 and B-ANA2 duplexes exhibit pseudorotation phase angles between 107° and 123° (Figure 4). Thus, the typical pucker adopted by an arabinose in a B-form environment appears to be C1′-exo, although the 107° angle found for residue aU19 in B-ANA1 falls into the O4′-endo range (72–108°; Figure 4A). This indicates a more ‘Southern’ trend in the conformation of ANA compared with FANA residues inside B-DNA (19). The C1′-exo conformation is well tolerated in B-DNA and is commonly observed with 2′-deoxyribonucleosides. Similarly, an O4′-endo conformation of individual nucleosides does not result in drastic local distortions of a B-form duplex (19).
FIGURE 4.
Conformational properties of ANA and FANA nucleosides. (A) Pseudorotation phase angles P for FANA and ANA sugars in the A-form and B-form duplexes with reference values for DNA:RNA hybrids and canonical A-RNA and B-DNA duplexes. The phase angle is defined as tan P = [(ν4-ν1)–(ν3+ν0)]/[2ν2(sin36° + sin72°)], where ν0 to ν4 are the endocyclic torsion angles of the pentofuranose (ν0 O4′-C1′, ν1 C1′-C2′, ν2 C2′-C3′, ν3 C3′-C4′, ν4 C4′-O4′) (60). The color code is as follows: A-FANA1 and A-FANA2, blue circles; B-FANA, green circles (19); A-ANA, purple circles; B-ANA1 and B-ANA2, red circles; P angle ranges in the native A-DNA decamer (all residues) and B-DNA dodecamer duplexes (thymidines) are shown with arrows and average values are marked by a black triangle and cross, respectively (for a recent overview of P angle ranges in A- and B-form duplexes see (66)); average puckers of DNA residues in the crystals of complexes between DNA:RNA hybrids and HIV-I RT (33) and bacterial RNase H (36), black circle and square, respectively. (B) Examples of sugar conformations observed in the crystal structures of FANA-and ANA-modified DNA duplexes. The color code for 2′-substituents matches that in panel A. Top left: C3′-endo FANA (A-FANA1 residue faT6); top right: C4′-exo FANA (A-FANA2 residue faT36); middle left: O4′-endo B-FANA (19); middle right: C1′-exo A-ANA (residue aU6); bottom left: C1′-exo B-ANA1 (residue aU7); bottom right: DNA C2′-endo (reference).
The sugar conformation of ANA residues in the A-ANA duplex is also C1′-exo (Figure 4). However, unlike the same pucker type in the B-form environment, the conformational preferences by aU6 and aU16 lead to local perturbations of the A-form geometry. The 2′-O-methyl-As preceding the ANA residues adopt C3′-endo pucker and the relatively short P…P distances between residues 5 and 6 (5.52 Å) and 15 and 16 (5.50 Å) are consistent with an A-form geometry (Figure 3A). The South-Eastern pucker of aU6 and aU16 elongates the backbone locally and also appears to force residues A7 and A17, respectively, into C2′-endo conformations. The corresponding P…P distances are 6.66 Å (aU6-A7), 6.65 Å (A7-C8), 6.58 Å (aU16-A17) and 6.67 Å (A17-C18). Thus, they are about 1 Å longer than the distance between adjacent phosphate groups typical for A-form duplexes. 3′-Adjacent to the above adenosines, the backbone snaps back into a C4′-exo North-Eastern type conformation. The C1′-exo pucker of arabinoses seen both in the B-ANA and A-ANA duplexes is consistent with a gauche effect between O2′ and O4′ (Figure 4B).
The Southern-type pucker of A7 and A17 appears not to be chiefly a consequence of steric strain caused by the arabinose 2′-hydroxyl group. The distances between the 2′-oxygens of aU6 and aU16 and atoms from the respective 3′-adjacent residues are all longer than the expected sums of van der Waals radii (O-O, O-N and O-C; Figure 5A). For example, the distances between O2′ and O4′ are 5.18 Å (aU6…A7) and 5.16 Å (aU16…A17). And the distances between O2′ and C8 are 3.51 Å (aU6…A7) and 3.43 Å (aU16…A17). Thus, the 2′-hydroxyl groups are not hydrogen bonded to 4′-oxygen atoms from adjacent residues. In any case, the C1′-exo pucker of arabinonucleosides causes a conformational discontinuity in the backbones of the A-ANA duplex that is consistent with geometrical distortions (i.e. kink) observed at the global level.
FIGURE 5.
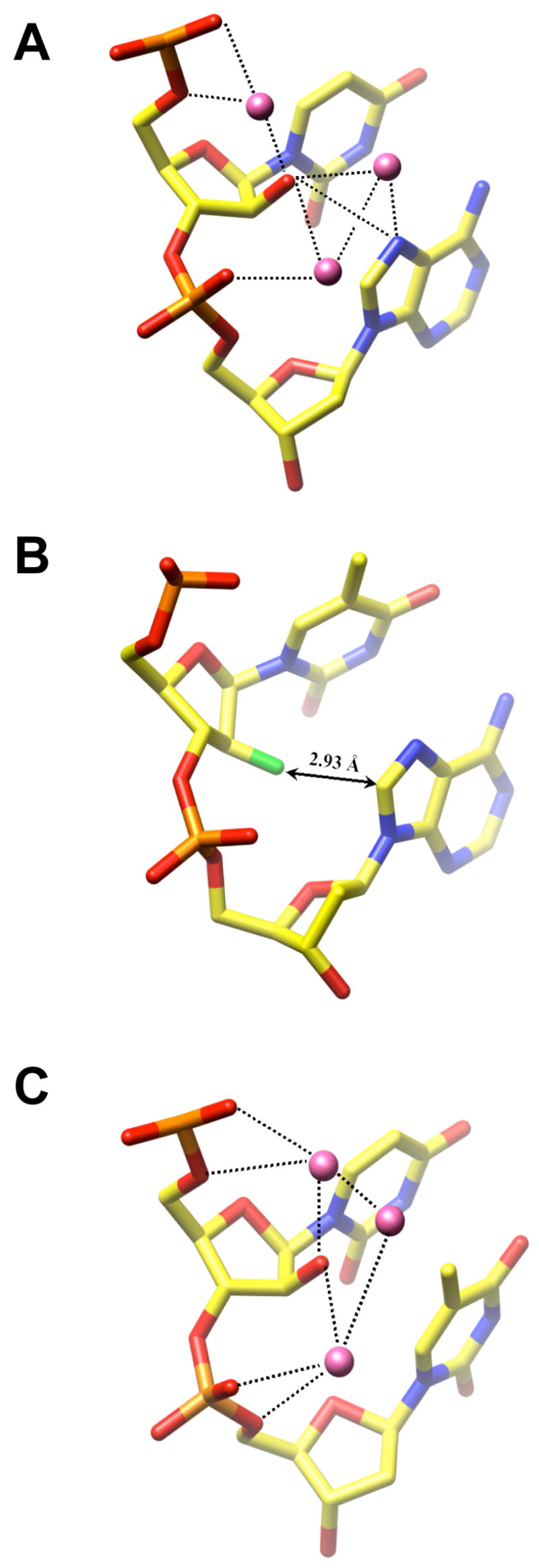
Hydration of ANA and FANA nucleotides. (A) A-ANA structure: Residues aU16 and A17. Water molecules are magenta spheres and hydrogen bonds are dashed lines. (B) A-FANA2 structure: Residues faT36 and A37. The shortest distance between the 2′-fluorine atom (green) and an atom of the 3′-adjacent A (C8) is indicated.
In the A-FANA1 duplex, residues faT6 and faT16 both adopt C3′-endo pucker. However, in the A-FANA2 duplex, the sugar conformation of these two residues is slightly different (C4′-exo; Figure 4). Interestingly, just as in the case of the A-ANA duplex, the 3′-adjacent adenosines in the A-FANA duplexes also adopt C2′-endo pucker (C3′-exo, P=181° for A7 in F-ANA1). The distances between P(2′OMeA) and P(faT) in all four strands are short (5.31 Å – 5.95 Å) whereas the spacings between P(faT) and the next two phosphates are enlarged. However, this coincidence appears to have different origins in the A-ANA and A-FANA duplexes. In the FANA duplexes, the glycosidic angles of 2′-fluoro-arabinonucleosides exhibit values (−151° to −160°) that are close to the average angle seen in the A-form DNAs (−160°). Adjacent adenosines display drastically different χ angles (avg. −85° in A-FANA1 and avg. −97° in A-FANA2), leading to a disruption in stacking between faT residues and 3′-adjacent As (Figures 3E, 5B). Unlike the situation in the A-ANA duplex, it appears that the primary reason for the Southern pucker of the residue 3′-adjacent to FANA-Ts is steric in nature. In F-ANA1 and F-ANA2, the average distance between F2′ and the 4′-oxygen of the adjacent A is 3.28 Å. The average distance between C2′ and the 4′-oxygen from adjacent residues is 4.19 Å, similar to the 4.30 Å seen in the A-ANA duplex. In A-form RNA duplexes, the ribose 2′-hydroxyl group is typically located at a distance of ca. 3.2 to 3.5 Å from the 4′-oxygen of the 3′-adjacent residue (18, 49). The above distance of 3.28 Å between F2′ (n) and O4′ (n+1) is the result of a C2′-endo pucker in residue A(n+1). Clearly, adoption of C3′-endo puckers by both the FANA-T and the following A residues would lead to a clash between fluorine and the sugar moiety of the 3′-adjacent residue. Thus, to avoid close contacts between F2′ and sugar, base and probably also the backbone portion (O5′) of the adjacent nucleotide, the latter is pushed away, requiring it to flip into a C2′-endo conformation. The existence of short orthogonal interactions between fluorine and electrophilic centers (i.e. nitrile or amide carbon) has been pointed out (reviewed in ref. 61) and they can be expected to contribute favorably to stability. However, we believe it to be unlikely that the above close contact between F2′ and C8 (shorter than 3 Å in three of four cases) adds much in terms of stability.
In summary, ANA residues adopt similar South-Eastern puckers in A- and B-form DNAs (Figure 4A), whereas FANA residues clearly favor the NorthEastern and Northern regions of the pseudorotation phase cycle. The conformational preferences of FANA residues in A- and B-form duplexes are different as FANA-thymidines in the B-DNA dodecamer were previously found to uniformly adopt the O4′-endo pucker (Figure 4A). Apparently, the A-form geometry allows for a slight variation in the sugar conformation of 2′-fluoro-arabinonucleosides (C3′-endo in A-FANA1 and C4′-exo in A-FANA2). The conformational deviations at the nucleoside level result in subtle differences between the overall duplex geometries as described in the previous section.
Hydration and Ion Coordination
ANA and FANA residues do not just exhibit clear conformational differences; they also differ fundamentally in terms of their hydration. The arabinose 2′-hydroxyl group acts as a hydrogen bond acceptor and donor in a DNA backbone otherwise devoid of donors. Thus, it provides a bridgehead for hydration networks linking phosphate groups and nucleobase atoms in the major groove (Figure 5). In A-ANA, water molecules link the 2′-OH to adjacent phosphate groups and N7 of A at both the aU6pA7 and aU16pA17 base steps (Figure 5A). As described above, the 2′-hydroxyl group is not properly positioned to hydrogen bond with the 4′-oxygen of the 3′-adjacent adenosine. Instead, it is engaged in a direct, but relatively weak hydrogen bond to N7 of A at the edge of the major groove (3.59 Å, aU6; 3.45 Å, aU16; Figure 5A). This interaction mode deviates somewhat from the one observed at aUpT steps in B-ANA1. The methyl group of thymine prevents a water-mediated hydrogen bond between O2′ and O4 of T (Figure 5C). However, at the aU8pC9 and aU20pC21 steps in B-ANA2, tandem water bridges link O2′ (aU) and N4(C). The hydration networks involving the arabinose 2′-hydroxyl group can be expected to have a stabilizing effect on the backbone conformation.
By comparison, the groove regions in the immediate vicinity of fluorine atoms in the A-FANA duplexes are dry as organic fluorine hardly ever makes hydrogen bonds (19, 62). The fluorine atoms occupy a very different position at the major groove edge (Figure 5B) compared with the 2′-hydroxyl groups of arabinoses in either the A- (Figure 5A) or B-DNA duplexes (Figure 5C). 2′-Fluorines are wedged between the unstacked thymine and adenine planes such they are located perpendicularly above the edges of the latter. Since fluorine is electron-rich, this arrangement may to some extent compensate for the loss of base stacking at the faTpA steps (Figures 3E, 5B).
The extent of bending into the major (deep) groove in the A-ANA and A-FANA duplexes correlates nicely with the number of positive charges as a result of cobalt hexamine coordination inside the groove to offset the build-up of negative electrostatic potential. The inner major groove of the A-ANA duplex features two cobalt hexamines but also a chloride anion that sits between the cations and edges of A:T pairs and is coordinated to N6 amino groups of adenines (positive charge +5; Figure 6A). In the A-FANA2 duplex with a somewhat stronger bend and therefore more closely spaced phosphate groups from opposite strands, two cobalt hexamines are bound (positive charge +6). And in the case of the A-FANA1 duplex that has the strongest bend three cobalt hexamines are bound inside the major groove (positive charge +9). In all three duplexes, an additional cobalt ion bridges tightly spaced phosphate groups from opposite strands at the groove periphery. These observations serve to reiterate the differences between the physical-chemical properties and conformational preferences of ANA and FANA residues, with consequences for local and global helical geometry as well as hydration and ion coordination.
FIGURE 6.
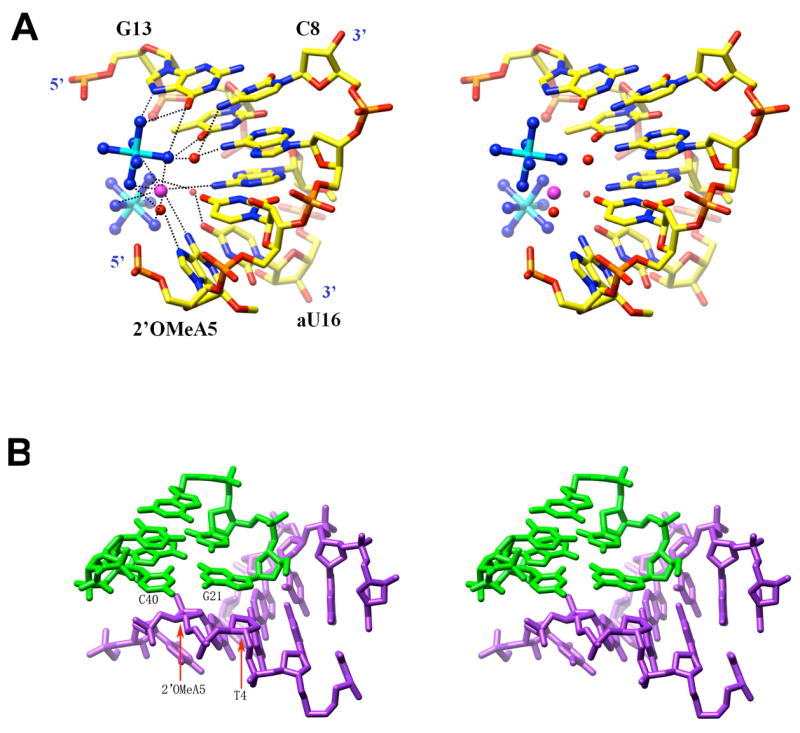
Ion coordination and packing interactions. (A) Coordination of cobalt hexamine ions (cyan and blue for Co3+ and N, respectively) and a chloride anion (large sphere in magenta) in the major groove of the A-ANA duplex. Selected residues are labeled, water molecules are red spheres and hydrogen bonds are dashed lines. (B) Example of a packing interaction in the A-ANA and A-FANA structures, involving terminal base pairs of a duplex (green bonds) and the sugar-phosphate backbone of a neighboring one (blue bonds). In the example shown here, the sugar moieties of residues T4 and 2′OMeA5 from an A-FANA1 decamer form a pseudo-base pair that stacks against the terminal C:G base pair from an A-FANA2 duplex.
In the B-ANA1 and B-ANA2 crystals a single magnesium hexahydrate ion coordinates to guanines from opposite strands near one end of duplexes. This binding mode is conserved in crystals of the Dickerson-Drew DNA dodecamer and occurs at the site of a facultative bend into the major groove (52, 63). Therefore cation binding in these crystals is not in any way linked to the presence of arabinonucleosides in the central A-tract of dodecamers.
Conserved Crystal Packing Interactions
The packing modes of duplexes in A-ANA and A-FANA crystals are nearly identical despite the higher symmetry of the former (one duplex per asymmetric unit in A-ANA and two duplexes in A-FANA; Table 1). Terminal base pairs of duplexes stack against the sugar-phosphate backbone from a symmetry mate (A-ANA). In A-FANA crystals terminal base pairs of A-FANA2 duplexes interact with the backbones from A-FANA1 duplexes and vice versa (Figure 6B). This somewhat unusual arrangement is also observed in orthorhombic crystals with different unit cell constants of DNA decamers of the same sequence but modified with single ribonucleosides or 2′-O-modified residues (see, for example ref. 49). However, the packing is considerably tighter in such crystals compared with A-ANA or A-FANA. The sugar moieties of nucleotides T4 and 2′OMeA5 (T14 and 2′OMeA15 in the second strand) stack onto terminal guanine and cytosine bases, respectively (Figure 6B). Thus these sugars form quasi-base pairs that extend the decamer duplexes at either end. In place of dispersive forces, several C-H…π interactions are observed. In addition, 4′-oxygen atoms of 2′-O-methylated residues are spaced at ca. 3.5 Å from the best planes through terminal guanines, consistent with a potentially stabilizing lone-pair…π* interaction (64). Most importantly, ANA and FANA residues are not directly involved in packing interactions, leading us to conclude that the observed conformations are intrinsically preferred rather than dictated by crystal packing forces.
Origins of FANA’s Higher RNA Affinity Compared to ANA
Our crystal structures provide an indication that the favorable RNA affinity of FANA relative to ANA is largely based on stereoelectronic effects involving fluorine that drive the conformational preorganization of the FANA strand for the C4′-exo (A-FANA2 duplex) and C3′-endo puckers (A-FANA1 duplex; Figure 4A). Northern and North-Eastern conformations of 2′-fluoro-arabinonucleosides lead to considerable local and global distortions of an A-form duplex and yet seem to be preferred over the Eastern and South-Eastern puckers of ANA residues in A-form DNA that cause significantly less disruption of local base stacking. Apparently the distortions in the A-form geometry as a result of a C3′-endo conformation of FANA residues are not too punishing energetically and the O4′-endo pucker seen with FANA nucleosides in B-form DNA (19) and paired with RNA (20) is disfavored. Thus, a 2′-fluorine substituent, either in the ribo (2′F-RNA) or arabino configuration (FANA), promotes conformational properties that result in significantly higher RNA affinities of 2′-fluoro analogues compared to the corresponding oligonucleotides with a 2′-OH substituent (RNA and ANA, resp.). The higher stabilities of heteroduplexes between RNA and either FANA or 2′F-RNA are afforded in the absence of hydrogen bonds by the 2′-substituent because fluorine is a poor acceptor (Figure 5). Extensive water networks around the 2′-OH groups of RNA (65) and ANA (i.e. this work) are apparently no match for the stereoelectronic effects mediated by fluorine in the sugar moiety of nucleic acids.
Conformations of ANA and FANA and Cleavage of Their Hybrids with RNA by RNase H
FANA:RNA duplexes are not only thermodynamically more stable than ANA:RNA duplexes (13), they also constitute better substrates for RNase H (12). It had been reasoned that the lower susceptibility of ANA:RNA heteroduplexes to cleavage by RNase H is related to the lower thermodynamic stability of such duplexes (fewer duplex molecules available for processing by the enzyme (12)). Obviously such a rationalization ignores potential conformational differences between FANA and ANA and the resulting differences in the geometry of their heteroduplexes with RNA (Figure 2). We show here that FANA and ANA differ in their conformational boundaries (Figure 4). Arabinonucleosides display sugar puckers in the Southern and South-Eastern region of the pseudorotation phase cycle, irrespective of whether they are located in an A- or a B-form duplex. Conversely, 2′-fluoro-arabinonucleoside puckering is limited to the Northern and Eastern regions (A- and B-form parent duplexes, resp.). Thus, it appears that Southern and even South-Eastern puckers are avoided by FANA strands. FANA paired to RNA was also found to adopt the O4′-endo (East) sugar conformation (20).
DNA:RNA hybrids can adopt a range of conformations, including pure A-form (49) and an intermediate geometry with RNA in the A-form and DNA either in between the A- and B-forms (32) or pure B-form. Recent crystal structures of complexes between RNase H and DNA:RNA hybrids are consistent with Southern (C2′-endo) or South-Eastern (C1′-exo) sugar conformations in the DNA (33, 36). The hybrid duplexes in both crystal structures manifest minor grooves that are narrower than in a canonical A-form duplex. Clearly, this feature and the precise spacing of intra- and inter-strand phosphates groups (the enzyme establishes several contacts to the DNA strand in the structures of complexes) are important for substrate recognition by RNase H. In addition, the unique conformational flexibility of DNA may also have a bearing on the differential rates of RNase H cleavage (21, 37): DNA:RNA > FANA:RNA ≫ ANA:RNA (12). Unlike 2′-deoxyribose in A- and B-form duplexes (66), neither FANA nor ANA sugars can fully cover the Eastern half of the pseudorotation wheel according to our data. The apparent contradiction that FANA:RNA duplexes are substrates of RNase H although FANA sugars are unable to adopt South-Eastern or Southern sugar puckers can be resolved in two ways. Either binding by the enzyme shifts the FANA sugar conformation toward the South-East (induced fit), or the enzyme tolerates a range of conformations in the strand paired to RNA that includes Southern, South-Eastern and Eastern puckers of sugars.
CONCLUSIONS
By analyzing the crystal structures of A- and B-DNA duplexes with incorporated ANA and FANA (A-form; for B-form see ref. 19) nucleosides at high resolution, we have shown that the two analogues have different conformational preferences. Sugar puckers of arabinonucleosides show a South-Eastern trend whereas the puckers of 2′-fluoro-arabinonucleosides fall into the 90° range of the phase angle between North and East (Figure 4A). The conformations of ANA nucleosides in the A- and B-DNA environments are virtually the same. On the other hand 2′-fluoro-arabinothymidines adopt different conformations in A- and B-form duplexes. The crystallographic data are supportive of the conclusion drawn based on earlier CD spectra in solution that FANA:RNA and ANA:RNA duplexes display subtle conformational differences (13) and point to stereoelectronic origins of the higher RNA affinity of FANA relative to ANA.
Although it is known that RNase H does not process RNA:RNA duplexes, the question whether some conformational variation in the strand opposite RNA is tolerated by the enzyme remains open. No structures of complexes for the most thoroughly investigated RNase H enzymes (from E. coli and Human) with substrate or inhibitor duplexes are available at present. Moreover, the details of how RNase H recognizes its substrates are still not completely understood. RNase H cleaves RNA in DNA:RNA and FANA:RNA duplexes with similar efficiency (12). Yet, the work presented here and the accumulated structural data regarding FANA show that this analogue is unlikely to adopt a Southern-type sugar conformation. Our structural investigation in the absence of enzyme of the ANA and FANA analogues whose heteroduplexes with RNA form substrates for RNase H indicates that the enzyme may tolerate a range of conformations in the strand paired to RNA and that the different cleavage efficiencies seen for ANA:RNA and FANA:RNA duplexes are also related to different conformational preferences of ANA and FANA. More conclusive answers will have to await the determination of crystal structures of complexes between RNase H and RNA paired to FANA and ANA.
Footnotes
Financial support by the US National Institutes of Health is gratefully acknowledged (GM55237 to ME). Use of the Advanced Photon Source was supported by the U.S. Department of Energy, Basic Energy Sciences, Office of Science, under Contract No. W-31-109-Eng-38. The DuPont-Northwestern-Dow Collaborative Access Team (DND-CAT) Synchrotron Research Center at the Advanced Photon Source (Sector 5) is supported by E. I. DuPont de Nemours & Co., The Dow Chemical Company, the National Science Foundation, and the State of Illinois.
Coordinates and structure factors and for all four structures have been deposited in the Protein Data Bank (http://www.rcsb.org): PDB ID codes 2FIJ (A-ANA), 2FIL (A-FANA), 2FIH (B-ANA1) and 2FII (B-ANA2).
References
- 1.Crooke ST. Basic principles of antisense therapeutics. In: Crooke ST, editor. Antisense Research and Application. Vol. 131. Springer; Berlin: 1998. pp. 1–50. [Google Scholar]
- 2.Lima WF, Crooke ST. Binding affinity and specificity of Escherichia coli RNase H1: impact on the kinetics of catalysis of antisense oligonucleotide-RNA hybrids. Biochemistry. 1997;36:390–398. doi: 10.1021/bi962230p. [DOI] [PubMed] [Google Scholar]
- 3.Walder RT, Walder JA. Role of RNase H in hybrid-arrested translation by antisense oligonucleotides. Proc Natl Acad Sci USA. 1988;85:5011–5015. doi: 10.1073/pnas.85.14.5011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Crooke ST. Phosphorothioate oligonucleotides. In: Crooke ST, editor. Therapeutic Applications of Oligonucleotides. R G Landes; Austin: 1995. pp. 63–79. [Google Scholar]
- 5.Tamm I, Dörken B, Hartmann G. Antisense therapy in oncology: new hope for an old idea? Lancet. 2001;358:489–497. doi: 10.1016/S0140-6736(01)05629-X. [DOI] [PubMed] [Google Scholar]
- 6.Jansen B, Zangemeister-Wittke U. Antisense therapy for cancer – the time of truth. Lancet Oncology. 2002;3:672–683. doi: 10.1016/s1470-2045(02)00903-8. [DOI] [PubMed] [Google Scholar]
- 7.Dove A. Antisense and sensibility. Nat Biotech. 2002;20:121–124. doi: 10.1038/nbt0202-121. [DOI] [PubMed] [Google Scholar]
- 8.Filmore D. Assessing antisense. Modern Drug Discovery. 2004 June;:49–50. [Google Scholar]
- 9.Levin AA. A review of issues in the pharmacokinetics and toxicology of phosphorothioate antisense oligonucleotides. Biochim Biophys Acta. 1999;1489:69–84. doi: 10.1016/s0167-4781(99)00140-2. [DOI] [PubMed] [Google Scholar]
- 10.Manoharan M. 2′-Carbohydrate modifications in antisense oligonucleotide therapy: importance of conformation, configuration and conjugation. Biochim Biophys Acta. 1999;1489:117–130. doi: 10.1016/s0167-4781(99)00138-4. [DOI] [PubMed] [Google Scholar]
- 11.Toulmé JJ. New candidates for true antisense. Nat Biotech. 2001;19:17–18. doi: 10.1038/83451. [DOI] [PubMed] [Google Scholar]
- 12.Damha MJ, Wilds CJ, Noronha A, Brukner I, Borkow G, Arion D, Parniak MA. Hybrids of RNA and arabinonucleic acids (ANA and 2′F-ANA) are substrates of ribonuclease H. J Am Chem Soc. 1998;120:12976–12977. [Google Scholar]
- 13.Wilds CJ, Damha MJ. 2′-Deoxy-2′-fluoro-β-D-arabinonucleosides and oligonucleotides (2′F-ANA): synthesis and physicochemical studies. Nucleic Acids Res. 2000;28:3625–3635. doi: 10.1093/nar/28.18.3625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Noronha A, Wilds CJ, Lok CN, Viazovkina K, Arion D, Parniak MA, Damha MJ. Synthesis and biophysical properties of arabinonucleic acids (ANA): CD spectra, melting temperatures, and RNase H susceptibility of ANA/RNA hybrids. Biochemistry. 2000;39:7050–7062. doi: 10.1021/bi000280v. [DOI] [PubMed] [Google Scholar]
- 15.Lok C-N, Viazovkina E, Min K-L, Nagy E, Wilds CJ, Damha MJ, Parniak MA. Potent gene-specific inhibitory properties of mixed-backbone antisense oligonucleotides comprised of 2′-deoxy-2′-fluoro-D-arabinose and 2′-deoxyribose nucleotides. Biochemistry. 2002;41:3457–3467. doi: 10.1021/bi0115075. [DOI] [PubMed] [Google Scholar]
- 16.Min KL, Viazovkina E, Galarneau A, Parniak MA, Damha MJ. Oligonucleotides comprised of alternating 2′-deoxy-2′-fluoro-β-D-arabinonucleosides and D-2′-deoxyribonucleosides (2′F-ANA/DNA ‘altimers’) induce efficient RNA cleavage mediated by RNase H. Bioorg Med Chem Lett. 2002;12:2651–2654. doi: 10.1016/s0960-894x(02)00439-0. [DOI] [PubMed] [Google Scholar]
- 17.Mangos MM, Min KL, Viazovkina E, Galarneau A, Elzagheid MI, Parniak MA, Damha MJ. Efficient RNase H-directed cleavage of RNA promoted by antisense DNA or 2′F-ANA constructs containing acyclic nucleotide inserts. J Am Chem Soc. 2003;125:654–661. doi: 10.1021/ja025557o. [DOI] [PubMed] [Google Scholar]
- 18.Saenger W. Principles of Nucleic Acid Structure. Springer-Verlag; New York, NY: 1984. [Google Scholar]
- 19.Berger I, Tereshko V, Ikeda H, Marquez VE, Egli M. Crystal structures of B-DNA with incorporated 2′-deoxy-2′-fluoro-arabino-furanosyl thymines: implications of conformational preorganization for duplex stability. Nucleic Acids Res. 1998;26:2473–2480. doi: 10.1093/nar/26.10.2473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Trempe JF, Wilds CJ, Denisov AY, Pon RT, Damha MJ, Gehring K. NMR solution structure of an oligonucleotide hairpin with a 2′F-ANA/RNA stem: implications for RNase H specificity toward DNA/RNA hybrid duplexes. J Am Chem Soc. 2001;123:4896–4903. doi: 10.1021/ja003859p. [DOI] [PubMed] [Google Scholar]
- 21.Denisov AY, Noronha AM, Wilds CJ, Trempe JF, Pon RT, Gehring K, Damha MJ. Solution structure of an arabinonucleic acid (ANA)/RNA duplex in a chimeric hairpin: comparison with 2′-fluoro-ANA/RNA and DNA/RNA hybrids. Nucleic Acids Res. 2001;29:4284–4293. doi: 10.1093/nar/29.21.4284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Venkateswarlu D, Ferguson DM. Effect of C2′-substitution on arabinonucleic acid structure and conformation. J Am Chem Soc. 1999;121:3009–3010. [Google Scholar]
- 23.Pieters JM, de Vroom E, van der Marel GA, van Boom JH, Koning TMG, Kaptein R, Altona C. Hairpin structures in DNA containing arabinofuranosylcytosine. A combination of nuclear magnetic resonance and molecular dynamics. Biochemistry. 1990;29:788–799. doi: 10.1021/bi00455a029. [DOI] [PubMed] [Google Scholar]
- 24.Godfredsen CH, Spielmann HP, Wengel J, Jacobsen JP. Structure of a DNA duplex containing a single 2′-O-methyl-β-D-araT: combined use of NMR, restrained molecular dynamics, and full relaxation matrix refinement. Bioconjug Chem. 1996;7:680–688. doi: 10.1021/bc960061f. [DOI] [PubMed] [Google Scholar]
- 25.Gmeiner WH, Konerding D, James TL. Effect of cytarabine on the NMR structure of a model Okazaki fragment from the SV40 genome. Biochemistry. 1999;38:1166–1175. doi: 10.1021/bi981702s. [DOI] [PubMed] [Google Scholar]
- 26.Gao Y-G, van der Marel GA, van Boom JH, Wang AH-J. Molecular structure of a DNA decamer containing an anticancer nucleoside arabinosylcytosine: conformational perturbation by arabinosylcytosine in B-DNA. Biochemistry. 1991;30:9922–9931. doi: 10.1021/bi00105a016. [DOI] [PubMed] [Google Scholar]
- 27.Schweitzer BI, Mikita T, Kellogg GW, Gardner KH, Beardsley GP. Solution structure of a DNA dodecamer containing the anti-neoplastic agent arabinocytosine: combined use of NMR, restrained molecular dynamics and full relaxation matrix refinement. Biochemistry. 1994;33:11460–11475. [PubMed] [Google Scholar]
- 28.Teng MK, Liaw YC, van der Marel GA, van Boom JH, Wang AH-J. Effects of the O2′ hydroxyl group on Z-DNA conformation: structure of Z-RNA and (araC)-[Z-DNA] Biochemistry. 1989;28:4923–4928. doi: 10.1021/bi00438a001. [DOI] [PubMed] [Google Scholar]
- 29.Zhang H, van der Marel GA, van Boom JH, Wang AH-J. Conformational perturbation of the anticancer nucleotide arabinosylcytosine on Z-DNA: molecular structure of (araC-dG)3 at 1.3 Å resolution. Biopolymers. 1992;32:1559–1569. doi: 10.1002/bip.360321113. [DOI] [PubMed] [Google Scholar]
- 30.Lima WF, Crooke ST. Binding affinity and specificity of Escherichia coli RNase H1: impact on the kinetics of catalysis of antisense oligonucleotide-RNA hybrids. Biochemistry. 1997;36:390–398. doi: 10.1021/bi962230p. [DOI] [PubMed] [Google Scholar]
- 31.Nakamura H, Oda Y, Iwai S, Inoue H, Ohtsuka E, Kanaya S, Kimura S, Katsuda C, Katayanagi K, Morikawa K, Miyashiro H, Ikehara M. How does RNase H recognize a DNA. RNA hybrid? Proc Natl Acad Sci USA. 1991;88:11535–11539. doi: 10.1073/pnas.88.24.11535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fedoroff OY, Salazar M, Reid BR. Structure of a DNA:RNA hybrid duplex. Why RNase H does not cleave pure RNA. J Mol Biol. 1993;233:509–523. doi: 10.1006/jmbi.1993.1528. [DOI] [PubMed] [Google Scholar]
- 33.Sarafianos SG, Das K, Tantillo C, Clark AD, Jr, Ding J, Whitcomb JM, Boyer PL, Hughes SH, Arnold E. Crystal structure of HIV-1 reverse transcriptase in complex with a polypurine tract RNA:DNA. EMBO J. 2001;20:1449–1461. doi: 10.1093/emboj/20.6.1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yazbeck DR, Min KL, Dahma MJ. Molecular requirements for degradation of a modified sense RNA strand by Escherichia coli ribonuclease H1. Nucleic Acids Res. 2002;30:3015–3025. doi: 10.1093/nar/gkf429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Minasov G, Teplova M, Nielsen P, Wengel J, Egli M. Structural basis of cleavage by RNase H of hybrids of arabinonucleic acids and RNA. Biochemistry. 2000;39:3525–3532. doi: 10.1021/bi992792j. [DOI] [PubMed] [Google Scholar]
- 36.Nowotny M, Gaidamakov SA, Crouch RJ, Yang W. Crystal structures of RNase H bound to an RNA/DNA hybrid: substrate specificity and metal-dependent catalysis. Cell. 2005;121:1005–1016. doi: 10.1016/j.cell.2005.04.024. [DOI] [PubMed] [Google Scholar]
- 37.Lima WF, Nichols JG, Wu H, Prakash TP, Migawa MT, Wyrzykiewicz TK, Bhat B, Crooke ST. Structural requirements at the catalytic site of the heteroduplex substrate for human RNase H1 catalysis. J Biol Chem. 2004;279:36317–36326. doi: 10.1074/jbc.M405035200. [DOI] [PubMed] [Google Scholar]
- 38.Berger I, Kang CH, Sinha N, Wolters M, Rich A. A highly efficient 24-condition matrix for the crystallization of nucleic acid fragments. Acta Cryst D. 1996;52:465–468. doi: 10.1107/s0907444995013564. [DOI] [PubMed] [Google Scholar]
- 39.Kabsch W. Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. J Appl Cryst. 1993;26:795–800. [Google Scholar]
- 40.Brünger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. Crystallography & NMR System: a new software suite for macromolecular structure determination. Acta Cryst D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 41.Egli M, Tereshko V, Teplova M, Minasov G, Joachimiak A, Sanishvili R, Weeks CM, Miller R, Maier MA, An H, Cook PD, Manoharan M. X-ray crystallographic analysis of the hydration of A-and B-form DNA at atomic resolution. Biopolymers (Nucleic Acid Sciences) 2000;48:234–252. doi: 10.1002/(SICI)1097-0282(1998)48:4<234::AID-BIP4>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 42.Kissinger CR, Gehlhaar DK, Fogel DB. Rapid automated molecular replacement by evolutionary search. Acta Cryst D. 1999;55:484–491. doi: 10.1107/s0907444998012517. [DOI] [PubMed] [Google Scholar]
- 43.Sheldrick GM, Schneider TR. SHELXL: high-resolution refinement. Meth Enzymol. 1997;277:319–343. [PubMed] [Google Scholar]
- 44.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Cryst D. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 45.Murshudov GN, Vagin AA, Lebedev A, Wilson KS, Dodson EJ. Efficient anisotropic refinement of macromolecular structures using FFT. Acta Cryst D. 1999;55:247–255. doi: 10.1107/S090744499801405X. [DOI] [PubMed] [Google Scholar]
- 46.CCP4. Collaborative computing project number 4. The CCP4 suite: programs for protein crystallography. Acta Cryst D. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 47.Cambillau C, Roussel A. Turbo Frodo, Version OpenGL1. Université Aix-Marseille II; Marseille, France: 1997. [Google Scholar]
- 48.Brünger AT. Free R value: a novel statistical quantity for assessing the accuracy of crystal structures. Nature. 1992;355:472–475. doi: 10.1038/355472a0. [DOI] [PubMed] [Google Scholar]
- 49.Egli M, Usman N, Rich A. Conformational influence of the ribose 2′-hydroxyl group: crystal structures of DNA-RNA chimeric duplexes. Biochemistry. 1993;32:3221–3237. [PubMed] [Google Scholar]
- 50.Egli M. Towards the structure-based design of nucleic acid therapeutics. In: Weber G, editor. Advan Enzyme Regul. Vol. 38. Elsevier Science Ltd.; Oxford, UK: 1998. pp. 181–203. [DOI] [PubMed] [Google Scholar]
- 51.Ban C, Ramakrishnan B, Sundaralingam M. A single 2′-hydroxyl group converts B-DNA to A-DNA. Crystal structure of the DNA-RNA chimeric decamer d(CCGGC)r(G)d(CCGG) with a novel intermolecular G•C base-paired quadruplet. J Mol Biol. 1994;236:275–285. doi: 10.1006/jmbi.1994.1134. [DOI] [PubMed] [Google Scholar]
- 52.Minasov G, Tereshko V, Egli M. Atomic-resolution crystal structures of B-DNA reveal specific influences of divalent metal ions on conformation and packing. J Mol Biol. 1999;291:83–99. doi: 10.1006/jmbi.1999.2934. [DOI] [PubMed] [Google Scholar]
- 53.Lubini P, Zürcher W, Egli M. Crystal structure of an oligodeoxynucleotide duplex containing 2′-O-methylated adenosines. Chem Biol. 1994;1:39–45. doi: 10.1016/1074-5521(94)90039-6. [DOI] [PubMed] [Google Scholar]
- 54.Tereshko V, Portmann S, Tay EC, Martin P, Natt F, Altmann K-H, Egli M. Correlating structure and stability of DNA duplexes with incorporated 2′-O-modified RNA analogues. Biochemistry. 1998;37:10626–10634. doi: 10.1021/bi980392a. [DOI] [PubMed] [Google Scholar]
- 55.Tereshko V, Wilds CJ, Minasov G, Prakash TP, Maier MA, Howard A, Wawrzak Z, Manoharan M, Egli M. Detection of alkali metal ions in DNA crystals using state-of-the-art X-ray diffraction experiments. Nucleic Acids Res. 2001;29:1208–1215. doi: 10.1093/nar/29.5.1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pattanayek R, Sethaphong L, Pan C, Prhavc M, Prakash TP, Manoharan M, Egli M. Structural rationalization of a large difference in RNA affinity despite a small difference in chemistry between two 2′-O-modified nucleic acid analogs. J Am Chem Soc. 2004;126:15006–15007. doi: 10.1021/ja044637k. [DOI] [PubMed] [Google Scholar]
- 57.Egli M, Minasov G, Tereshko V, Pallan PS, Teplova M, Inamati GB, Lesnik EA, Owens SR, Ross BS, Prakash TP, Manoharan M. Comprehensive analysis of the RNA affinity, nuclease resistance and crystal structure of ten 2′-O-ribonucleic acid modifications. Biochemistry. 2005;44:9045–9057. doi: 10.1021/bi050574m. [DOI] [PubMed] [Google Scholar]
- 58.Diop-Frimpong B, Prakash TP, Rajeev KG, Manoharan M, Egli M. Crystal structure of a DNA duplex with 2′-O-[2-methoxy)ethyl]-2-thiothymidines: stabilizing contributions of the sulfur-modified nucleotides. Nucleic Acids Res. 2005;33:5297–5307. doi: 10.1093/nar/gki823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lavery R, Sklenar H. Defining the structure of irregular nucleic acids: conventions and principles. J Biomol Struct Dyn. 1989;6:655–667. doi: 10.1080/07391102.1989.10507728. [DOI] [PubMed] [Google Scholar]
- 60.Altona C, Sundaralingam M. Conformational analysis of the sugar ring in nucleosides and nucleotides. New description using the concept of pseudorotation. J Am Chem Soc. 1972;94:8205–8212. doi: 10.1021/ja00778a043. [DOI] [PubMed] [Google Scholar]
- 61.Paulini R, Müller K, Diederich F. Orthogonal multipolar interactions in structural chemistry and biology. Angew Chem Int Ed. 2005;44:1788–1805. doi: 10.1002/anie.200462213. [DOI] [PubMed] [Google Scholar]
- 62.Dunitz JD. Organic fluorine: odd man out. Chem Biochem. 2004;5:614–621. doi: 10.1002/cbic.200300801. [DOI] [PubMed] [Google Scholar]
- 63.Egli M. DNA-cation interactions: quo vadis? Chem Biol. 2002;9:277–286. doi: 10.1016/s1074-5521(02)00116-3. [DOI] [PubMed] [Google Scholar]
- 64.Egli M, Gessner RV. Stereoelectronic effects of deoxyribose O4′ on DNA conformation. Proc Natl Acad Sci USA. 1995;92:180–185. doi: 10.1073/pnas.92.1.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Egli M, Portmann S, Usman N. RNA hydration: a detailed look. Biochemistry. 1996;35:8489–8494. doi: 10.1021/bi9607214. [DOI] [PubMed] [Google Scholar]
- 66.Sun G, Voigt JH, Fillippov IV, Marquez VE, Nicklaus MC. PROSIT: Pseudo-rotational online service and interactive tool, applied to a conformational survey of nucleosides and nucleotides. J Chem Inf Comput Sci. 2004;44:1752–1762. doi: 10.1021/ci049881+. [DOI] [PubMed] [Google Scholar]