Abstract
Hypothalamic GnRH neurons are essential for initiation and regulation of reproductive function. In addition to pituitary gonadotrope stimulation, activity of GnRH through its receptor (GnRHR) has been suggested to include autocrine regulation of the GnRH neuron. Two hypogonadal mouse strains, the Gnrh1 mutant (hpg) mice and Gnrhr mutant mice were used to investigate the potential role of GnRH signaling in the proper development and maintenance of GnRH neurons. Immunocytochemical analysis of heterozygous hpg mice revealed a GnRH neuron population that was normal in size and distribution, indicating no effect from reduced Gnrh1 gene dosage on the neurons themselves. To visualize GnRH neurons in homozygous GnRH-deficient hpg mice, heterozygous hpg mice were crossed with GnRH-green fluorescent protein (GFP) transgenic mice with targeted expression of the GFP reporter gene in GnRH neurons. Analysis of forebrains of homozygous hpg/GFP-positive mice immunostained for GFP revealed a normal population size and appropriate distribution of GnRH neurons in hpg mice, with immunoreactive neuronal processes present at the median eminence. Similarly, adult mice deficient in functional GnRHR possessed a full complement of GnRH neurons in the basal forebrain that was indistinguishable from the distribution of GnRH neurons in their wild-type counterparts. Moreover, hpg/GFP neurons retained the ability to generate spontaneous bursts of action potential firing activity, suggesting that GnRH peptide is not required for this function. These data establish that autocrine-paracrine GnRH-signaling is not a prerequisite for the developmental migration of GnRH neurons into the brain or for the projection of GnRH neurosecretory axons.
GnRH-1 DECAPEPTIDE IS A key central regulator of reproductive function. GnRH neurons develop outside the central nervous system in the embryonic nasal placodes and undergo a series of distinctive developmental events before their ultimate role in pituitary gonadotrope regulation (1,2). The migration of GnRH neurons from the nasal placode into the basal forebrain followed by the projection of neurosecretory axons to the median eminence (ME), where the hormone is released, are critical processes necessary to ensure proper central regulation of reproduction (3). GnRH neuronal development relies upon cooperative signaling from surrounding tissues during a short developmental window. Regulation of GnRH neuron development is dependent on factors within the nasal placode environment (e.g. activator protein-2α and fibroblast growth factors) (4,5) that first locally affect fate specification of GnRH neurons, and then on combinations of factors [e.g. nasal embryonic LHRH factor (NELF), γ-amino butyric acid (GABA), netrins] that directly affect GnRH neuron migration and axon targeting of the ME (6,7,8,9).
A potential autocrine role of GnRH itself in the development and function of GnRH neurons has been proposed from studies of the GnRH-deficient hypogonadal (hpg) mouse (10). A deletion of the Gnrh1 gene results in hpg mice that do not synthesize GnRH decapeptide (10). Previous examination of the distribution of GnRH neurons in the GnRH-deficient hpg mouse model was dependent upon detection of a short 5′ segment of GnRH mRNA because the deletion prevented transcription of the complete coding sequence and the production of detectable levels of peptide (10,11). GnRH mRNA levels as measured by in situ hybridization in adult hpg mouse brains were greatly reduced, detecting only 16–20% of the normal GnRH neuronal population size (10,11). The reduction in GnRH neuron number in hpg mice was attributed to either of two explanations. One hypothesis was that the truncated GnRH mRNA produced in the presence of the gene deletion was rendered unstable, resulting in accelerated degradation of the transcript with reduced cellular levels such that 80% of hpg GnRH neurons escaped detection by in situ hybridization (10,11). An alternative possibility was that a GnRH peptide is necessary during development for GnRH neuron proliferation, migration, or survival, resulting in the loss of 80% of the population in the absence of GnRH. This potential reduction of GnRH neurons may explain the results of our recent study using targeted viral vectors that identified only a small number of hpg GnRH neurons (12). The embryonic expression of the ligand and GnRH receptor (GnRHR) is consistent with a developmental role for GnRH (1,2,13); however, the role of GnRH as a candidate trophic factor on GnRH neurons in vivo remains unproven.
GnRH stimulation of pituitary gonadotropes activates the GnRHR, a G protein-coupled receptor, to release LH and FSH in large part through Gqα and G11α activation, effecting calcium mobilization to induce gonadotropin secretion (14,15,16). GnRHR activation of alternative signaling pathways in other nonpituitary tissues has been described, thus broadening the potential physiological roles of GnRH. These include evidence of a neuromodulatory role in amphibians to increase arterial blood pressure via catecholamine release (17), in sensory transmission in the visual system (18), and in chemosensory reception in both the vomeronasal and the olfactory systems (19,20), implicating the ability of GnRH to directly regulate other neurons. Autocrine roles of GnRH in the ovary, a tissue that expresses both GnRH and GnRHR, have been demonstrated because GnRH has antiproliferative and apoptosis-inducing effects in human ovarian surface epithelium and ovarian cancer, as well as in gynecological cancers (21,22,23,24). Recently, GnRH stimulation of slice cultures of pituitary tissue has demonstrated the ability to induce mobilization and reorganization of the gonadotrope cell cytoarchitecture (25). These broad examples suggest that GnRH can have potent and variable effects on development in addition its role in regulating pituitary gonadotropin secretion.
In mice, the earliest developmental expression of GnRH begins at embryonic d 10.5 in the nasal placode (26). The transcriptional activity of the Gnrh1 gene has been used to define the GnRH neuron because no other specific and unique marker to identify these cells has been demonstrated. The migration phase of development coincides with GnRHR expression on GnRH neurons (27), suggesting that GnRH may act as a regulatory factor during this period. Activation of the GnRHR in GnRH neuronal cell lines, primary GnRH neurons, and adult GnRH neurons in hypothalamic slice preparations has been shown to stimulate physiological responses, although trophic actions mediated by GnRHR have yet to be demonstrated in GnRH neurons in vivo (27,28).
To determine whether GnRH indeed plays a critical role in the establishment or the maintenance of GnRH neurons in vivo, GnRH neurons were studied in different hypogonadal mouse models: GnRH-deficient hpg mice (10) and GnRHR-mutant mice (Gnrhrmut/mut) (29). These GnRH signaling-deficient models were examined for GnRH neuron numbers, distribution, and electrophysiological firing activity. Because detection of GnRH cannot be used to identify GnRH neurons in hpg mice, mice heterozygous for the hpg mutation were crossed with transgenic mice with targeted green fluorescent protein (GFP) expression in GnRH neurons (30). Mice resulting from this cross allowed the visualization of hypothalamic GnRH neurons with GnRH-GFP expression and activity, thus facilitating analysis of the mutant GnRH neuronal population and distribution even in the absence of GnRH.
Materials and Methods
Animal husbandry
All mice were maintained in a 12-h light, 12-h dark cycle, with food and water available ad libitum. Humane animal care and welfare were in accordance to guidelines established by the Harvard Medical Area Standing Committee on Animals in the Harvard Medical School Center for Animal Resources and Comparative Medicine, approved by the Institutional Animal Protocol and Care Committee of Baylor College of Medicine, or approved by the Animal Care and Use Committee of the University of Virginia.
GnRH-deficient hpg mice
Mice heterozygous for the Gnrh1 gene deletion (Gnrh1+/ −; HET) in the C3H/HeHx101/H background strain were obtained from The Jackson Laboratory (Bar Harbor, ME). HET mice are fertile and generated wild-type (Gnrh1+/+; WT), HET, and homozygous (Gnrh1−/−; hpg) mice in Mendelian ratios. Genotypes of offspring were determined by PCR of genomic tail DNA with three primers: primer 1: 5′-TATGGCTTACAGTTCCAGCG (sense, intron two, upstream of the deletion); primer 2: 5′-AGGCTTGGAGAGCTGTAAGG (antisense, intron two, within the deleted region); primer 3: 5′-GTTTCAGTGCATCCTCTCAGG (antisense, downstream of the deleted region). Primers 1 and 2 are expected to generate a PCR product of 613 bp in size from the WT allele, and primers 1 and 3 are expected to generate a 536-bp product from the mutant allele.
GnRH-deficient GnRH-GFP mice
To facilitate the visualization of GnRH neurons in the brains of hpg mice devoid of immunoreactive GnRH, HET mice were crossed with transgenic mice engineered to specifically express enhanced GFP as a targeted reporter in GnRH neurons (30). A breeding pair [heterozygous for the Gnrh1 mutation and positive for the GnRH-GFP transgene; (HET/GFP)] was generated and used as founders to generate all subsequent GnRH-GFP reporter mice used for this study. PCR genotyping of genomic tail DNA was performed for the Gnrh1 gene as described above. The presence of the GnRH-GFP transgene was confirmed by PCR with GFP primers (forward, 5′-GACGTAAACGGCCAAAGTT; reverse, 5′-AAGTCGTGCTGCTTCATGTG).
Gnrhr mutant mice
Homozygous Gnrhrmut/mut mice and their WT littermates were generated by crossing heterozygotes in a breeding colony maintained at Baylor College of Medicine. Genotypes were determined as previously reported (29) using PCR and primers in the flanking genomic sequence (forward, 5′-CTCCACTCTTGAAGCCTGTCC; reverse, 5′-TCACCATGTTCACACAAATTC) followed by DdeI digestion of the amplified products. The mice were maintained in a 129S6/SvEv background strain.
Immunocytochemistry and analysis of GnRH and GnRH-GFP neurons
Adult male mice (8 wk old) were anesthetized, transcardially perfused with 10–20 ml saline followed by 10–20 ml of 4% paraformaldehyde in 0.1 m PBS (PBS; pH 7.4) using a 26-gauge needle and hand-held syringe. Testes were removed and imaged. Brains were dissected, post-fixed in 4% paraformaldehyde, cryoprotected in 20% sucrose solution, 0.01% sodium azide, and cryosectioned (Microm International GmbH, Walldorf, Germany) in the coronal plane from the accessory olfactory bulbs to the ME. For quantitative bright-field microscopy, 40-μm floating sections, or 20-μm thaw-mounted sections, were pretreated with 1% hydrogen peroxide in PBS containing 0.4% Triton X-100 (PBST) for 10 min to remove endogenous peroxidase activity, then incubated with either polyclonal rabbit anti-GnRH antibody (ImmunoStar, Hudson, WI; 1:32,000 dilution) or rabbit anti-GFP (Invitrogen, Carlsbad, CA; 1:10,000) in 10% normal horse serum, 4% normal donkey serum in PBST for three nights at 4 C. Sections were then incubated with a donkey antirabbit IgG biotinylated secondary antibody (Jackson ImmunoResearch, West Grove, PA) at 1:500 dilution using Vectastain Elite ABC Kit (Vector Laboratories, Burlingame, CA), and visualized with 3,3′-diaminobenzidine tetrahydrochloride (DAB; Pierce, Rockford, IL). Floating brain sections were serially aligned and mounted on glass microscope slides in a rostral to caudal order. After dehydration of sections, the slides were cleared, sealed, and coverslipped with Permount (Fisher Scientific, Pittsburgh, PA).
For immunofluorescence microscopy, 18-μm coronal brain sections were thaw-mounted onto poly-l-lysine coated slides. Slides were incubated in polyclonal goat anti-GFP (Genetex, Inc., San Antonio, TX; 1:5000). The subsequent secondary antibody, donkey antigoat was directly conjugated to Alexa-488 (Molecular Probes, Inc., Eugene, OR; 1:1000) to visualize GFP (green). The slides were mounted, cleared and coverslipped with Vectashield (Vector Laboratories).
Before bright-field and epifluorescence analysis of the numbers and distribution of GnRH and GnRH-GFP neurons, slides were coded to obscure sample identification. GnRH or GnRH-GFP neurons were counted in each section, and imaged at the organum vasculosum of the lamina terminalis (OVLT) and ME. Data for total neuron numbers and neurons counted in the rostral to caudal serial sections aligned at the OVLT as a common anatomical landmark between samples were graphed using Prism statistics software (GraphPad Software, Inc., San Diego, CA).
Immunocytochemistry of Gnrhr mutant mice
Eight-week-old male and female Gnrhrmut/mut mice, previously generated by N-ethyl-N-nitrosourea (ENU)-mutagenesis bear a T to C transition resulting in the missense L117P mutation (29), were transcardially perfused as above but without post-processing brain tissue in sucrose. Brains from Gnrhrmut/mut and WT animals were embedded in 5% agarose and cut coronally at 50 μm using a vibrating microtome (Leica VT1000S, Wetzlar, Germany). Sections were placed free-floating in containers with Nitex (Wildlife Supply Co., Buffalo, NY) mesh bottoms in 0.05 m PBS, pH 7.4. Each brain was cut into three sequential containers so that each antiserum/container used provided a full representation of sections from one third of the adult brain. Sections were pretreated at 4 C as follows: 0.1 m glycine in PBS for 30 min, 0.5% sodium borohydride in PBS for 15 min, and 5% normal goat serum (NGS) with 0.5% Triton X-100 and 1% hydrogen peroxide in PBS for 30 min. Adjacent sections were then incubated in different antisera directed toward GnRH, either LR-1 rabbit antiserum (1:40,000; a generous gift from Dr. Robert Benoit), or a commercial GnRH rabbit antiserum (1:1,000; PA121 obtained from Affinity Bioreagents, Golden, CO) in 1% BSA with 0.5% Tx for 3 nights at 4 C. After primary incubation, sections were washed at room temperature four times for 15 min each in 1% NGS with 0.02% Tx and then incubated with donkey antirabbit IgG biotinylated secondary antibody (Jackson ImmunoResearch) at 1:500 dilution in 1% NGS with 0.32% Tx for 2 h at room temperature. Sections were subsequently rinsed four times in 0.02% Tx-PBS followed by incubation in peroxidase-conjugated streptavidin (Jackson ImmunoResearch; diluted 1:2,500) for 1 h, washed for 1 h in Tris-buffered saline (TBS), and developed with 0.025% 3,3′-diaminobenzidine in TBS with 0.2% nickel ammonium and 0.02% hydrogen peroxide for 5 min. After immunocytochemistry, sections were washed in TBS, mounted on gelatin-subbed slides, dehydrated, and coverslipped with Permount (Fisher Scientific). GnRH-immunoreactive neurons were counted from two containers from each brain immunoreacted for one of two GnRH antisera giving a spread of 150 μm between each analyzed section within either antiserum. GnRH neurons were counted on a BH-2 photomicroscope (Olympus America, Inc., Center Valley, PA) and analyzed as a function of their rostral to caudal distribution centered and aligned at the OVLT.
Electrophysiological recordings of hpg/GFP neurons
To eliminate the effect of steroid milieu on GnRH neurons during electrophysiological recordings, adult (>2 months) female hpg/GFP mice were ovariectomized under isoflurane (Burns Veterinary Supply, Westbury, NY) anesthesia. Postoperative analgesia was provided by a long-acting local anesthetic (bupivacaine, 0.25%, 7.5 μl per site; Abbott Laboratories, North Chicago, IL). Recordings were performed 5–9 d after surgery. Brain slices from WT/GFP, n = 4) and hpg/GFP, n = 7) mice were prepared as previously described (31). Briefly, all solutions were bubbled with a 95% O2/5% CO2 mixture throughout the experiments and for at least 15 min before exposure to the tissue. Mice were decapitated, and the brain was rapidly removed and placed in ice-cold, high-sucrose saline solution containing (in mm) 250 sucrose, 3.5 KCl, 26 NaHCO3, 10 glucose, 1.25 Na2HPO4, 1.2 MgSO4, and 2.5 MgCl2. Coronal 300-μm brain slices were cut with a Vibratome 3000 (Technical Products, International, Inc., St. Louis, MO). Slices were incubated for 30 min at 30–32 C in a solution of 50% high-sucrose saline and 50% normal saline (NS) containing (in mm) 135 NaCl, 3.5 KCl, 10 glucose, 1.3 Na2HPO4, 1.2 MgSO4, and 2.5 CaCl2 and then were transferred to a solution of 100% NS at room temperature and kept at least 60 min and no more than 8 h before recording.
Targeted extracellular recordings were used for this study (32). Brain slices are placed in a recording chamber continuously superfused with oxygenated NS solution and kept at 30–32 C and mounted on an Olympus BX51WI upright fluorescent microscope with infrared differential interference contrast optics (Opelco, Dulles, VA). Recording pipettes (2–3 mΩ) were filled with normal HEPES-buffered solution containing (in mm) 150 NaCl, 10 HEPES, 10 glucose, 2.5 CaCl2, 1.3 MgCl2, and 3.5 KCl. Pipettes were placed in contact with the GnRH neurons using an MP-225 micromanipulator (Sutter Instruments, Novato, CA). No tight seal is formed (seals <50 MΩ); low resistance seals minimize the influence of the pipette potential on the cell’s membrane potential (33). The duration of recordings ranged from 6 to 60 min and was not different between groups (16.1 ± 3.2 vs. 15.1 ± 1.0 min, P > 0.10). Up to four cells per animal were recorded. If no activity was observed for 5 min, 15 mm KCl was added to the bath to check cell viability and recording integrity. If the cell did not respond to KCl, the data set was truncated at the time of last firing. If it fired in response to KCl, the data set was truncated for analysis at the time of adding KCl.
Extracellular recording data collection and analysis
Data were recorded using MultiClamp 700B amplifier (Axon Instruments, Foster City, CA) running Clampex 9.2 software (Axon Instruments) in voltage-clamp mode with a pipette holding potential of 0 mV. Data were filtered at 10 kHz and digitized with a Digidata 1332A (Axon Instruments). Extracellular firing activity (events) was analyzed with the MiniAnalysis program (Synaptosoft Inc., Decatur, GA).
Data are reported as mean ± sem. Events were counted and binned at 10-sec intervals, and were analyzed for mean firing rate. Firing rate (Hz) was determined in two ways: total events detected during 6 min (duration of the shortest recording) from each cell divided by 360 sec, and total number of events detected from each cell divided by the duration of the recording. No differences arose from these two approaches and the latter is reported.
Statistics
When appropriate, parametric statistics were used with data that satisfied Bartlett tests for equal variances. Otherwise, nonparametric statistics were used. Differences in number of GnRH neurons were analyzed by two-tailed Student’s t test or Mann-Whitney U test. Data for total neuron numbers and neurons counted in the rostral to caudal serial sections were analyzed using Prism statistics software (GraphPad Software, Inc., San Diego, CA). In electrophysiology experiments, groups were compared with Student’s t test. Differences were considered significant when P < 0.05.
Results
HET mice possess a normal complement of GnRH neurons
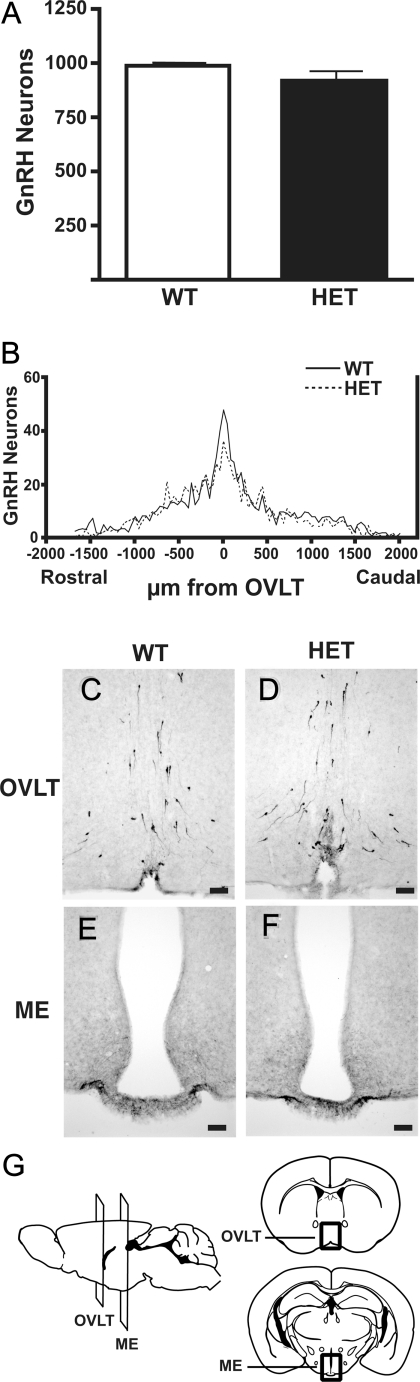
In addition to the complete lack of detectable GnRH in the homozygous hpg mice, the HET mice have been reported to contain only 20% of the hypothalamic GnRH content of their wild-type counterparts, yet they seemed phenotypically normal and had normal fertility (10). Therefore, to fully characterize the GnRH neuronal population in this mouse model, studies were first carried out to compare the GnRH neuronal population in the brains of WT mice with HET littermates, heterozygous for the Gnrh1 gene deletion. The size and distribution of the GnRH neuronal population were compared between these two genotypes to determine whether a dosage effect of the Gnrh1 gene was manifest at the level of the GnRH neurons. GnRH neurons were immunostained and counted in serial coronal sections of adult male WT and HET mice. GnRH immunoreactive neurons were distributed bilaterally through the medial ventral forebrain in similar patterns in both the WT and HET hpg mouse brains. The total number of hypothalamic GnRH neurons counted in each section from the septal preoptic area to the ME was not different between WT and HET (Fig. 1A). The rostral to caudal distribution of all GnRH neurons was compared by aligning the serial sections with the OVLT as a neuroanatomical reference point. The mean number of GnRH neurons in each section rostral and each section caudal to this reference point were plotted as a histogram of GnRH neurons per section in the coronal series to compare the distribution in WT and HET mice (Fig. 1B). GnRH neuron distributions were similar, with both WT and HET mice having the highest concentration of GnRH neurons located at the OVLT and smaller numbers of GnRH neurons present as far caudally as the ME (Fig. 1, C–G). The similar patterns of rostral to caudal distribution of GnRH neurons in the WT and HET mice suggest that similar GnRH neuronal migration patterns likely occurred developmentally between these two genotypes.
Figure 1.
Total number and distribution of GnRH neurons in the brain in WT and HET mice. GnRH neurons were immunostained and counted in coronal sections of adult male WT and HET mice (n = 4). A, Total hypothalamic GnRH neurons in WT and HET forebrains. B, Rostral to caudal distribution of GnRH neurons aligned at the OVLT and plotted as GnRH neurons per section in coronal series of 40-μm sections. No significant differences in GnRH neuron distribution were found. C–F, Immunocytochemistry of GnRH neurons at the OVLT in WT (C) and HET (D) and GnRH immunoreactive nerve terminals at the median eminence (ME) in WT (E) and HET (F). Bar, 100 μm. G, Illustrations of views of mouse brain to indicate planes of section of OVLT and ME and details of coronal sections presented in C–F (dark boxes). [Images redrawn with permission from G. Paxinos and K. B. J. Franklin: The Mouse Brain in Sterotaxic Coordinates, Elsevier, St. Louis, 2001 (55).]
HET mice bred with GnRH-GFP transgenic mice allow detection of hpg GnRH neurons
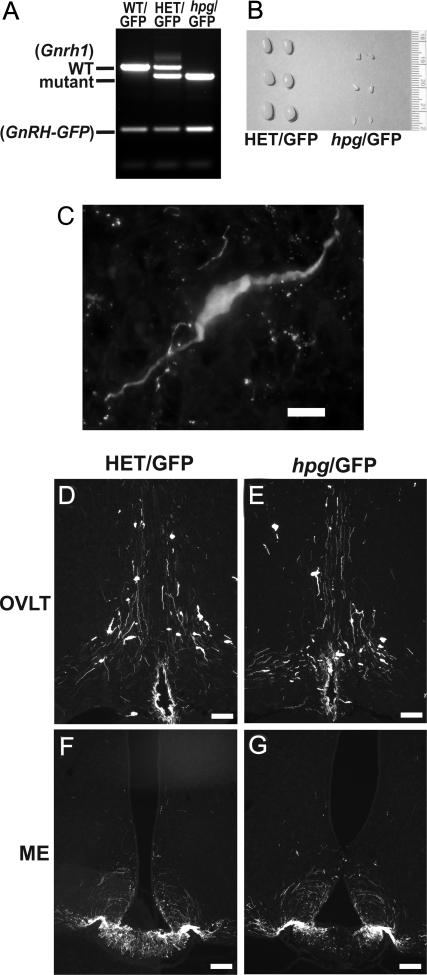
To facilitate the visualization of GnRH neurons in the brains of hpg mice devoid of immunoreactive GnRH, HET mice were crossed with transgenic mice engineered to specifically express enhanced-GFP as a targeted reporter in GnRH neurons (30). Breeding of HET mice with GnRH-GFP mice generated a GnRH-GFP transgene-positive Gnrh1-mutant mouse line as shown by PCR genotyping (Fig. 2A). The progeny of the resulting HET/GFP breeding pair were used for this study. hpg/GFP male mice were easily identified by their underdeveloped external genitalia and decreased testis size phenotype (Fig. 2B). Immunocytochemistry (ICC) for GFP in both HET/GFP and hpg/GFP males identified a hypothalamic population of neurons consistent in distribution and morphology with that of GnRH neurons (Fig. 2C). GFP immunostaining patterns were similar in the OVLT and ME of both HET/GFP and hpg/GFP male mice (Fig. 2, D–G).
Figure 2.
Detection of GFP-positive neurons in hpg mice crossed with transgenic GnRH-GFP mice. Crossing HET mice with GnRH-GFP mice generated a GnRH-GFP transgene-positive hpg mouse line. A, PCR genotyping data of wild-type (WT/GFP), heterozygous (HET/GFP), and GnRH-deficient (hpg/GFP) littermates with transgenic GnRH-GFP expression. B, Comparison of testes sizes from HET/GFP and hpg/GFP mice at four weeks of age. C, High resolution image of an hpg/GFP neuron (bar = 10 μm). D–G, GFP immunocytochemistry of HET/GFP mice (D and E) and hpg/GFP male mice (F and G) both depicting normal GnRH neuronal distribution at the OVLT and the presence of immunoreactive projections at the ME (bars, 100 μm).
hpg/GFP mice reveal an intact GnRH neuronal population size and distribution
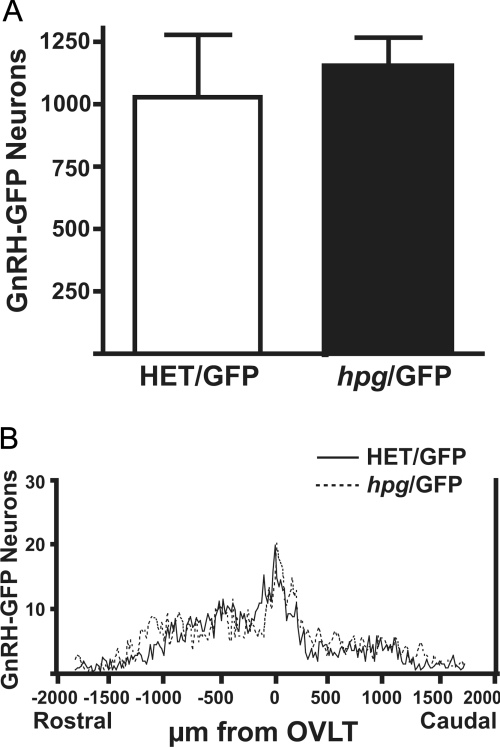
The total number of GnRH-GFP neurons detected in hpg/GFP male mice was not different than in the HET/GFP (Fig. 3A). In addition, there were no significant differences in the rostral to caudal distribution of GnRH-GFP neurons in brains of in hpg/GFP and HET/GFP male mice (n = 3) (Fig. 3B). The similar patterns of distribution of GnRH-GFP neurons suggest that GnRH neuronal differentiation, proliferation, migration patterns, and survival occur normally despite the absence of GnRH.
Figure 3.
Total number and distribution of GnRH neurons in GnRH-deficient hpg/GFP mice. Brain sections from adult male hpg/GFP mice were immunostained for GFP. A, Total number of GnRH-GFP neurons in the HET/GFP and hpg/GFP mice; B, Rostral to caudal distribution of GnRH-GFP neurons aligned at the OVLT and plotted as GnRH neurons per 20-μm section in the coronal series from brains of HET/GFP and hpg/GFP mice (n = 3).
GnRH receptor mutant mice possess an intact GnRH neuronal population
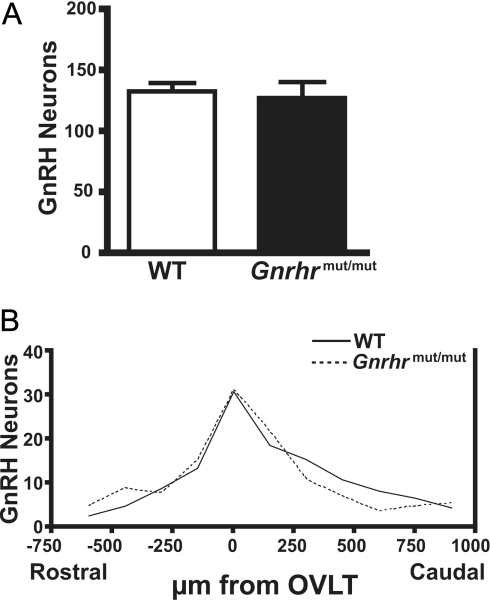
As an alternate hypogonadal mouse model, GnRH neurons in the ENU-generated Gnrhrmut/mut mice were compared with GnRH neurons in WT littermates. Homozygous Gnrhrmut/mut mice possess a GnRH receptor with a single amino acid change within the third transmembrane domain that renders the receptor nonfunctional (29). Detection of GnRH neurons using anti-GnRH PA121 demonstrated that the number of GnRH neurons found in the brains of Gnrhrmut/mut mice was not significantly different from the total GnRH neurons found in WT (Fig. 4A). Analysis of the rostral to caudal distribution of GnRH neurons counted in sections aligned at the OVLT showed near identical patterns between Gnrhrmut/mut mice and WT (Fig. 4B). GnRH neurons in mice possessing the nonfunctional GnRHR are thus present in normal numbers after an apparently normal developmental migration into the brain.
Figure 4.
Total number and distribution of GnRH neurons in WT and GnRHR mutant mice. Coronal forebrain sections of WT and Gnrhrmut/mut littermates (n = 5 for each genotype) immunostained with anti-GnRH antibody PA121. A, Total number of GnRH neurons in one-third of brain sections from WT and Gnrhrmut/mut mutant mice. B, Histogram of rostral to caudal distribution of GnRH neurons counted in 50-μm brain sections of either WT and Gnrhrmut/mut mutant mice.
To validate the use of the recently available PA121 antibody, the comparison of total GnRH neuron numbers and their distribution in Gnrhrmut/mut and WT mice were repeated using the well-characterized anti-GnRH LR1 antibody. In adjacent sections analyzed from the same brain region, the number of WT GnRH neurons was not different regardless of the anti-GnRH polyclonal antibody used (PA121, 132 ± 7 neurons; LR1, 121 ± 11 neurons; n = 5; P = N.S.). Similarly, both antibodies detected similar numbers of Gnrhrmut/mut GnRH neurons (PA121, 127 ± 13 neurons; LR1, 123 ± 16 neurons; n = 5) to reveal these antibodies had equivalent sensitivity to detect GnRH neurons.
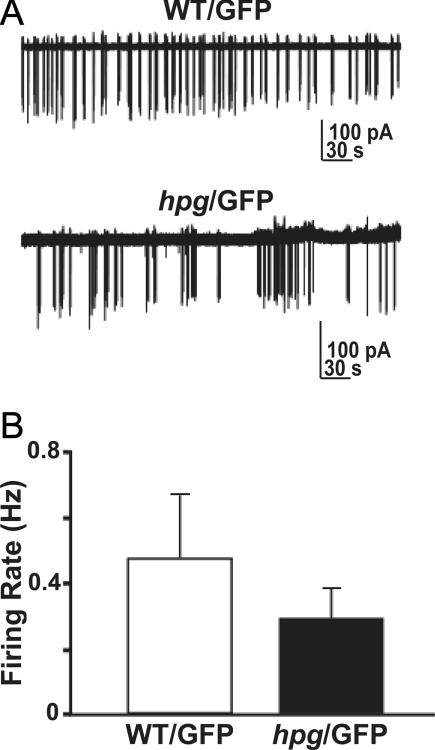
Electrophysiology of hpg/GFP neurons
The normal migration and position of GnRH neurons in hpg/GFP mice does not mandate their normal function. To begin to examine the effects of endogenous GnRH on GnRH neuronal activity, we used targeted extracellular recording to study firing activity of GnRH neurons in hpg/GFP mice. Representative examples of firing activity from WT/GFP and hpg/GFP mice are shown in Figure 5A. There were no significant differences in mean firing rate between groups (Fig. 5B; P = 0.38). Furthermore, GnRH neurons fired in small bursts of activity as previously described (32,34,35), suggesting that the lack of GnRH peptide does not preclude spontaneous generation of bursts of action potentials in GnRH neurons.
Figure 5.
GnRH neurons generate spontaneous bursts of neural activity in ovariectomized hpg/GFP female mice. A, Representative firing pattern over a 6-min period from WT/GFP and hpg/GFP mice (hpg). B, Mean ± sem firing rate of GnRH neurons from WT/GFP (n = 15 cells from 4 animals) and hpg/GFP (n = 17 cells from 7 animals) littermates. s, Seconds.
Discussion
Developmental organization of the GnRH neuronal network is a critical prerequisite for sexual differentiation, puberty, and fertility, yet a comprehensive understanding of the regulatory signals that establish this neuronal population is lacking (36). Here we report the characterization of adult GnRH neurons in two models of hypogonadal mice with primary deficiencies that abolished GnRH signaling. In contrast to the pro-migratory role of GnRH previously reported (37,38), the migration of hypothalamic GnRH neurons into the brain occurred normally both in mice deficient in GnRH (hpg), and mice with an inactivating Gnrhr mutation. In both hypogonadal strains used in this study, the sizes of the GnRH neuronal population and their distributions throughout the basal forebrain were normal in the ages used in this study, with axonal processes appropriately projecting to the ME. These findings provide evidence that the GnRH neuropeptide and its signaling through the cognate receptor are not critical for the development or maintenance of the hypothalamic GnRH neuronal population or for the formation of the neuroendocrine afferents.
The hypothesis that GnRH may developmentally regulate GnRH neurons originated from the distinctive expression of GnRH at times and in a place not associated with GnRH endocrine function—the embryonic nasal compartment (1,2). To date, however, a function of Gnrh1 expression coincident with the migration of GnRH neurons toward the brain has not yet been demonstrated. Embryonic GnRH expression and peptide processing have been measured (both directly and indirectly) to increase, paralleling maturational events of GnRH neurons, including cytoskeletal changes associated with migration (36,39,40,41). Therefore, a plausible role for GnRH in GnRH neuronal development has been suggested and supporting evidence has been described in different models.
In culture, the addition of exogenous GnRH to FNC-B4, GnRH-secreting cells derived from human olfactory epithelium, induced GnRHR-dependent changes consistent with neuronal differentiation, and included increased GnRH expression combined with down-regulation of nestin expression (37). Furthermore, changes in GnRH-stimulated FNC-B4 cells included axon growth, actin cytoskeleton remodeling and 3- to 4-fold increases in cell motility in Boyden chamber experiments (37). These findings in FNC-B4 cells strongly suggested an autocrine role for GnRH in the differentiation and migration of GnRH neurons. In vivo, a recent study in zebrafish that disrupted GnRH translation early in development by morpholino-modified antisense oligonucleotides altered the pattern of GnRH neuronal migration (38). This observation is a strikingly divergent from our findings in mice, where no deviations in the size or distribution of the adult GnRH neuron population were found despite the biosynthetic deficiency of GnRH or the loss of function of its specific receptor. This may represent a class difference of GnRH neuron development in fishes. Alternatively, the anomalous GnRH neuron development may represent the occasional non-target-related phenotype that can occur in morpholino knockdown experiments that may have simultaneously silenced an essential developmental gene for GnRH neurons (42).
It also may be possible that a part of the Gnrh1 gene or gene product spared in the deletion of exon 2 in hpg mice enables GnRH neuronal migration. This gene product would need to function independently of normal GnRHR signaling because GnRH neuronal migration occurred in Gnrhrmut/mut mice. Furthermore, a second form of GnRH (GnRH2) and a type II GnRH receptor (GnRHR2) exists in most species, and it could be argued that normal development may be mediated through an up-regulation of this second system in the absence of GnRH1 or GnRHR1. The compensatory GnRH2/GnRHR2 hypothesis is unlikely, however, in our model, because this system appears to be absent in mice (43).
We have provided further observations that the heterozygous deletion in the Gnrhr gene in adult male HET mice resulted in the identical reproductive phenotype as their WT counterparts, confirming previous reports (10). Moreover, we can now extend these observations to indicate that a comparable number of hypothalamic GnRH neurons is identified by ICC in the forebrain of HET mice as is found in their WT male littermates. This stands in contrast to an earlier description of an 80% reduction in hypothalamic GnRH content in hpg HET (10), a discrepancy that may reflect differences in methodology. Our observation in 2- to 3-month-old HET male mice suggests little to no effect of haploinsufficiency of the Gnrh1 gene on the GnRH neuronal network or on reproductive capacity.
To circumvent the reliance on potentially unstable Gnrh1 mRNA to identify GnRH neurons in homozygous hpg mice, transgenic GnRH-GFP mice were used, providing a reporter gene to identify the hpg GnRH neurons by crossing the two mouse strains. In these mice, genetically targeted GFP in GnRH neurons identifies 99.5% of GnRH neurons with little ectopic GFP fluorescence (30). GFP-labeled neurons were observed in neuroanatomical regions consistent with appropriate GnRH neuronal distribution in the caudal olfactory bulb, medial septal area, and medial preoptic area, with GFP-positive fibers found in the ME (30). The transgenic GnRH promoter/enhancer activity in hpg/GFP mice effectively directed GFP expression to identify hypothalamic neurons that are morphologically and neuroanatomically similar to GnRH neurons. It should be noted that other cells were observed with low levels of immunodetectable GFP in the brains of all GnRH-GFP mice; these cells appeared consistent with multipolar cells located dorsal and lateral to the normal distribution of GnRH neurons identified in previous reports (44,45). These cells were detected in all GnRH-GFP genotypes and were distinct from populations of hypothalamic GnRH neurons, thus easily avoided during cell count analysis by their location, size, morphology, and reduced intensity of GFP expression.
We substantiated that GnRH neurons can develop normally independent of GnRH signaling with the corollary experiment that examined the content and distribution of GnRH neurons in the Gnrhrmut/mut mouse generated by ENU mutagenesis (29). The Gnrhrmut/mut mouse has a hypogonadal phenotype similar to the hpg and hpg/GFP mice due to the inability of pituitary gonadotropes to respond to GnRH. These GnRH-insensitive mice provided the next logical avenue to determine whether a GnRHR signaling mechanism was necessary to support the development, differentiation, and maintenance of GnRH neurons. The similar distribution of GnRH neurons in homozygous Gnrhrmut/mut mice and WT control littermates supported the conclusion from the hpg GnRH neuron analysis that developmental regulation and maintenance of adult GnRH neurons is independent of GnRH activation of the GnRHR.
In the present study, the absence of any deficit in the size or the rostral to caudal distribution of the GnRH neuronal population implies that GnRH is not necessary for GnRH neurons to properly differentiate, migrate into the appropriate areas of the brain, extend axons to the ME, or survive into adulthood. The possibility remains that GnRH may act cooperatively to facilitate the activity of other factors. However, unlike the genetic removal or inhibition of other regulators (e.g. FGFR signaling, GABA, and cholecystokinin) which resulted in significant (20–30%) reductions in size of the GnRH neuronal population or disruption of its organization (46,47,48), the absence of GnRH signaling did not indicate any partial contribution to GnRH neuronal development. Rather, the completion of GnRH neuron development in the absence of GnRH narrows a potential role of GnRH to post-migratory neurosecretory phases of GnRH neuron differentiation and function.
Beyond effects on migration, autocrine or paracrine GnRH regulation of hypothalamic GnRH neuron function has been proposed to contribute to the regulation of pulsatile GnRH secretion through autocrine or paracrine feedback signaling (49). GnRH is postulated to regulate GnRH neuronal intrinsic pulsatility in vivo, in a GnRH neuronal cell line, in primary GnRH neurons, and in brain slices (13,28,50,51,52). Models of GnRHR-mediated GnRH autoregulation of pulsatile secretion have been proposed to be both inhibitory or stimulatory (28). Here we show hpg/GFP neurons are capable of firing spontaneous bursts of action potentials, a fundamental component of GnRH neuron function (53). The mean firing rate in the adult hpg/GFP neurons was not significantly different from WT/GFP neurons. hpg/GFP neurons further exhibited individual variability in burst firing similar to wild-type GnRH neurons in previous reports (32,34,35). Although GnRH peptide has been reported to alter the activity of GnRH neurons in both stimulatory and inhibitory directions (28,51,54), the decapeptide itself is apparently not critical for the genesis of electrical activity by these cells.
Interpretation of the electrophysiological data obtained in ovariectomized female hpg mice, whose male counterparts were used in the ICC studies, indicated that spontaneous neuronal activity occurred in GnRH-deficient neurons. We do not anticipate that this is a gender-specific event, and we expect that male hpg/GFP neurons would be similarly capable of spontaneous firing. Nonetheless, it is possible that sexual dimorphism may exist in WT GnRH neurons and future experiments will explore these questions in hpg GnRH neurons. Moreover, the investigation of electrophysiological responses of hpg/GFP neurons to relevant neuropeptide and hormonal stimuli may reveal as yet undetected effects of GnRH peptide on the electrophysiological properties of GnRH neurons.
In summary, normal GnRH neuronal development occurs in two strains of hypogonadal mice lacking the GnRH decapeptide or expressing inactive GnRHR, respectively. These results indicate that in the absence of GnRH autocrine or paracrine actions, the normal complement of GnRH neurons, become specified in the nasal placode and these neurons migrate normally into the medial basal forebrain to establish appropriate neurosecretory contacts with the ME.
Acknowledgments
We extend gratitude for the collaborative assistance from Monica Justice at the Department of Molecular and Human Genetics, Baylor College of Medicine, Houston, TX. Also, our appreciation is extended to Rona Carroll for the constructive comments and critical review during the manuscript preparation.
Footnotes
This work was supported by National Institutes of Health (NIH) Grants R21 HD050412 (to U.B.K.), R01 HD19938 (to U.B.K.), NIH MH63954 (to S.M.), NIH MH57442 (to S.M.), NIH R01 HD 34860 (to S.M.M.), NIH T32 HD07382 (P.C.), NIH F32 HD056765 (to P.C.) and NIH DC009034 (to S.T. and G.A.S.).
Disclosure Statement: The authors have nothing to disclose.
First Published Online May 22, 2008
Abbreviations: ENU, N-ethyl-N-nitrosourea; GFP, green fluorescent protein; GnRHR, GnRH receptor; HET, heterozygous for the Gnrh1 mutation; hpg, hypogonadal; ICC, immunocytochemistry; ME, median eminence; NGS, normal goat serum; NS, normal saline; OVLT, organum vasculosum of the lamina terminalis; TBS, Tris-buffered saline; WT, wild type.
References
- Schwanzel-Fukuda M, Pfaff DW 1989 Origin of luteinizing hormone-releasing hormone neurons. Nature 338:161–164 [DOI] [PubMed] [Google Scholar]
- Wray S, Nieburgs A, Elkabes S 1989 Spatiotemporal cell expression of luteinizing hormone-releasing hormone in the prenatal mouse: evidence for an embryonic origin in the olfactory placode. Brain Res Dev Brain Res 46:309–318 [DOI] [PubMed] [Google Scholar]
- Schwanzel-Fukuda M, Bick D, Pfaff DW 1989 Luteinizing hormone-releasing hormone (LHRH)-expressing cells do not migrate normally in an inherited hypogonadal (Kallmann) syndrome. Brain Res Mol Brain Res 6:311–326 [DOI] [PubMed] [Google Scholar]
- Kramer PR, Guerrero G, Krishnamurthy R, Mitchell PJ, Wray S 2000 Ectopic expression of luteinizing hormone-releasing hormone and peripherin in the respiratory epithelium of mice lacking transcription factor AP-2α. Mech Dev 94:79–94 [DOI] [PubMed] [Google Scholar]
- Gill JC, Moenter SM, Tsai PS 2004 Developmental regulation of gonadotropin-releasing hormone neurons by fibroblast growth factor signaling. Endocrinology 145:3830–3839 [DOI] [PubMed] [Google Scholar]
- Kramer PR, Wray S 2000 Novel gene expressed in nasal region influences outgrowth of olfactory axons and migration of luteinizing hormone-releasing hormone (LHRH) neurons. Genes Dev 14:1824–1834 [PMC free article] [PubMed] [Google Scholar]
- Tobet SA, Chickering TW, King JC, Stopa EG, Kim K, Kuo-Leblank V, Schwarting GA 1996 Expression of γ-aminobutyric acid and gonadotropin-releasing hormone during neuronal migration through the olfactory system. Endocrinology 137:5415–5420 [DOI] [PubMed] [Google Scholar]
- Fueshko SM, Key S, Wray S 1998 GABA inhibits migration of luteinizing hormone-releasing hormone neurons in embryonic olfactory explants. J Neurosci 18:2560–2569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarting GA, Kostek C, Bless EP, Ahmad N, Tobet SA 2001 Deleted in colorectal cancer (DCC) regulates the migration of luteinizing hormone-releasing hormone neurons to the basal forebrain. J Neurosci 21:911–919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason AJ, Hayflick JS, Zoeller RT, Young 3rd WS, Phillips HS, Nikolics K, Seeburg PH 1986 A deletion truncating the gonadotropin-releasing hormone gene is responsible for hypogonadism in the hpg mouse. Science 234:1366–1371 [DOI] [PubMed] [Google Scholar]
- Livne I, Gibson MJ, Silverman AJ 1993 Gonadotropin-releasing hormone (GnRH) neurons in the hypogonadal mouse elaborate normal projections despite their biosynthetic deficiency. Neurosci Lett 151:229–233 [DOI] [PubMed] [Google Scholar]
- Jeong KH, Bakowska JC, Song IO, Fu N, Breakefield XO, Kaiser UB2007 Improvement in reproductive parameters in hypogonadal mice: regulated gene replacement therapy in the central nervous system. Gene Ther 14:1092–1101 [DOI] [PubMed] [Google Scholar]
- Krsmanovic LZ, Martinez-Fuentes AJ, Arora KK, Mores N, Navarro CE, Chen HC, Stojilkovic SS, Catt KJ 1999 Autocrine regulation of gonadotropin-releasing hormone secretion in cultured hypothalamic neurons. Endocrinology 140:1423–1431 [DOI] [PubMed] [Google Scholar]
- Stanislaus D, Janovick JA, Brothers S, Conn PM 1997 Regulation of G(q/11)α by the gonadotropin-releasing hormone receptor. Mol Endocrinol 11:738–746 [DOI] [PubMed] [Google Scholar]
- Grosse R, Schmid A, Schoneberg T, Herrlich A, Muhn P, Schultz G, Gudermann T 2000 Gonadotropin-releasing hormone receptor initiates multiple signaling pathways by exclusively coupling to G(q/11) proteins. J Biol Chem 275:9193–9200 [DOI] [PubMed] [Google Scholar]
- Hansen JR, McArdle CA, Conn PM 1987 Relative roles of calcium derived from intra- and extracellular sources in dynamic luteinizing hormone release from perifused pituitary cells. Mol Endocrinol 1:808–815 [DOI] [PubMed] [Google Scholar]
- Wilson JX, Van Vliet BN, West NH 1984 Gonadotropin-releasing hormone increases plasma catecholamines and blood pressure in toads. Neuroendocrinology 39:437–441 [DOI] [PubMed] [Google Scholar]
- Maruska KP, Tricas TC 2007 Gonadotropin-releasing hormone and receptor distributions in the visual processing regions of four coral reef fishes. Brain Behav Evol 70:40–56 [DOI] [PubMed] [Google Scholar]
- Eisthen HL, Delay RJ, Wirsig-Wiechmann CR, Dionne VE 2000 Neuromodulatory effects of gonadotropin releasing hormone on olfactory receptor neurons. J Neurosci 20:3947–3955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirsig-Wiechmann CR, Wiechmann AF 2001 The prairie vole vomeronasal organ is a target for gonadotropin-releasing hormone. Chem Senses 26:1193–1202 [DOI] [PubMed] [Google Scholar]
- Savino L, Baldini B, Susini T, Pulli F, Antignani L, Massi GB 1992 GnRH analogs in gynecological oncology: a review. J Chemother 4:312–320 [DOI] [PubMed] [Google Scholar]
- Schally AV, Comaru-Schally AM, Nagy A, Kovacs M, Szepeshazi K, Plonowski A, Varga JL, Halmos G 2001 Hypothalamic hormones and cancer. Front Neuroendocrinol 22:248–291 [DOI] [PubMed] [Google Scholar]
- Grundker C, Emons G 2003 Role of gonadotropin-releasing hormone (GnRH) in ovarian cancer. Reprod Biol Endocrinol 1:65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang SK, Choi KC, Yang HS, Leung PC 2003 Potential role of gonadotrophin-releasing hormone (GnRH)-I and GnRH-II in the ovary and ovarian cancer. Endocr Relat Cancer 10:169–177 [DOI] [PubMed] [Google Scholar]
- Navratil AM, Knoll JG, Whitesell JD, Tobet SA, Clay CM 2007 Neuroendocrine plasticity in the anterior pituitary: gonadotropin-releasing hormone-mediated movement in vitro and in vivo. Endocrinology 148:1736–1744 [DOI] [PubMed] [Google Scholar]
- Wray S, Grant P, Gainer H 1989 Evidence that cells expressing luteinizing hormone-releasing hormone mRNA in the mouse are derived from progenitor cells in the olfactory placode. Proc Natl Acad Sci USA 86:8132–8136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Fuentes AJ, Hu L, Krsmanovic LZ, Catt KJ 2004 Gonadotropin-releasing hormone (GnRH) receptor expression and membrane signaling in early embryonic GnRH neurons: role in pulsatile neurosecretion. Mol Endocrinol 18:1808–1817 [DOI] [PubMed] [Google Scholar]
- Xu C, Xu XZ, Nunemaker CS, Moenter SM 2004 Dose-dependent switch in response of gonadotropin-releasing hormone (GnRH) neurons to GnRH mediated through the type I GnRH receptor. Endocrinology 145:728–735 [DOI] [PubMed] [Google Scholar]
- Pask AJ, Kanasaki H, Kaiser UB, Conn PM, Janovick JA, Stockton DW, Hess DL, Justice MJ, Behringer RR 2005 A novel mouse model of hypogonadotrophic hypogonadism: N-ethyl-N-nitrosourea-induced gonadotropin-releasing hormone receptor gene mutation. Mol Endocrinol 19:972–981 [DOI] [PubMed] [Google Scholar]
- Suter KJ, Song WJ, Sampson TL, Wuarin JP, Saunders JT, Dudek FE, Moenter SM 2000 Genetic targeting of green fluorescent protein to gonadotropin-releasing hormone neurons: characterization of whole-cell electrophysiological properties and morphology. Endocrinology 141:412–419 [DOI] [PubMed] [Google Scholar]
- Chu Z, Moenter SM 2005 Endogenous activation of metabotropic glutamate receptors modulates GABAergic transmission to gonadotropin-releasing hormone neurons and alters their firing rate: a possible local feedback circuit. J Neurosci 25:5740–5749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunemaker CS, DeFazio RA, Moenter SM 2002 Estradiol-sensitive afferents modulate long-term episodic firing patterns of GnRH neurons. Endocrinology 143:2284–2292 [DOI] [PubMed] [Google Scholar]
- Nunemaker CS, DeFazio RA, Moenter SM 2003 A targeted extracellular approach for recording long-term firing patterns of excitable cells: a practical guide. Biol Proced Online 5:53–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abe H, Terasawa E 2005 Firing pattern and rapid modulation of activity by estrogen in primate luteinizing hormone releasing hormone-1 neurons. Endocrinology 146:4312–4320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuehl-Kovarik MC, Pouliot WA, Halterman GL, Handa RJ, Dudek FE, Partin KM 2002 Episodic bursting activity and response to excitatory amino acids in acutely dissociated gonadotropin-releasing hormone neurons genetically targeted with green fluorescent protein. J Neurosci 22:2313–2322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tobet SA, Schwarting GA 2006 Minireview: recent progress in gonadotropin-releasing hormone neuronal migration. Endocrinology 147:1159–1165 [DOI] [PubMed] [Google Scholar]
- Romanelli RG, Barni T, Maggi M, Luconi M, Failli P, Pezzatini A, Pelo E, Torricelli F, Crescioli C, Ferruzzi P, Salerno R, Marini M, Rotella CM, Vannelli GB 2004 Expression and function of gonadotropin-releasing hormone (GnRH) receptor in human olfactory GnRH-secreting neurons: an autocrine GnRH loop underlies neuronal migration. J Biol Chem 279:117–126 [DOI] [PubMed] [Google Scholar]
- Abraham E, Palevitch O, Ijiri S, Du SJ, Gothilf Y, Zohar Y 2008 Early development of gonadotrophin-releasing hormone-I neurones and the role of GnRH-I as an autocrine migration factor. J Neuroendocrinol 20:394–405 [DOI] [PubMed] [Google Scholar]
- Livne I, Gibson MJ, Silverman AJ 1993 Biochemical differentiation and intercellular interactions of migratory gonadotropin-releasing hormone (GnRH) cells in the mouse. Dev Biol 159:643–656 [DOI] [PubMed] [Google Scholar]
- Moore Jr JP, Wray S 2000 Luteinizing hormone-releasing hormone (LHRH) biosynthesis and secretion in embryonic LHRH. Endocrinology 141:4486–4495 [DOI] [PubMed] [Google Scholar]
- Simonian SX, Herbison AE 2001 Regulation of gonadotropin-releasing hormone (GnRH) gene expression during GnRH neuron migration in the mouse. Neuroendocrinology 73:149–156 [DOI] [PubMed] [Google Scholar]
- Ekker SC, Larson JD 2001 Morphant technology in model developmental systems. Genesis 30:89–93 [DOI] [PubMed] [Google Scholar]
- Morgan K, Millar RP 2004 Evolution of GnRH ligand precursors and GnRH receptors in protochordate and vertebrate species. Gen Comp Endocrinol 139:191–197 [DOI] [PubMed] [Google Scholar]
- Skynner MJ, Slater R, Sim JA, Allen ND, Herbison AE 1999 Promoter transgenics reveal multiple gonadotropin-releasing hormone-I-expressing cell populations of different embryological origin in mouse brain. J Neurosci 19:5955–5966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spergel DJ, Kruth U, Shimshek DR, Sprengel R, Seeburg PH 2001 Using reporter genes to label selected neuronal populations in transgenic mice for gene promoter, anatomical, and physiological studies. Prog Neurobiol 63:673–686 [DOI] [PubMed] [Google Scholar]
- Tsai PS, Moenter SM, Postigo HR, El Majdoubi M, Pak TR, Gill JC, Paruthiyil S, Werner S, Weiner RI 2005 Targeted expression of a dominant-negative fibroblast growth factor (FGF) receptor in gonadotropin-releasing hormone (GnRH) neurons reduces FGF responsiveness and the size of GnRH neuronal population. Mol Endocrinol 19:225–236 [DOI] [PubMed] [Google Scholar]
- Bless EP, Westaway WA, Schwarting GA, Tobet SA 2000 Effects of γ-aminobutyric acid (A) receptor manipulation on migrating gonadotropin-releasing hormone neurons through the entire migratory route in vivo and in vitro. Endocrinology 141:1254–1262 [DOI] [PubMed] [Google Scholar]
- Giacobini P, Kopin AS, Beart PM, Mercer LD, Fasolo A, Wray S 2004 Cholecystokinin modulates migration of gonadotropin-releasing hormone-1 neurons. J Neurosci 24:4737–4748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valenca MM, Johnston CA, Ching M, Negro-Vilar A 1987 Evidence for a negative ultrashort loop feedback mechanism operating on the luteinizing hormone-releasing hormone neuronal system. Endocrinology 121:2256–2259 [DOI] [PubMed] [Google Scholar]
- Krsmanovic LZ, Stojilkovic SS, Mertz LM, Tomic M, Catt KJ 1993 Expression of gonadotropin-releasing hormone receptors and autocrine regulation of neuropeptide release in immortalized hypothalamic neurons. Proc Natl Acad Sci USA 90:3908–3912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todman MG, Han SK, Herbison AE 2005 Profiling neurotransmitter receptor expression in mouse gonadotropin-releasing hormone neurons using green fluorescent protein-promoter transgenics and microarrays. Neuroscience 132:703–712 [DOI] [PubMed] [Google Scholar]
- DePaolo LV, King RA, Carrillo AJ 1987 In vivo and in vitro examination of an autoregulatory mechanism for luteinizing hormone-releasing hormone. Endocrinology 120:272–279 [DOI] [PubMed] [Google Scholar]
- Nunemaker CS, Straume M, DeFazio RA, Moenter SM 2003 Gonadotropin-releasing hormone neurons generate interacting rhythms in multiple time domains. Endocrinology 144:823–831 [DOI] [PubMed] [Google Scholar]
- Xu C, Roepke TA, Zhang C, Ronnekleiv OK, Kelly MJ 2008 GnRH activates the M-current in GnRH neurons: an autoregulatory negative feedback mechanism? Endocrinology 149:2459–2466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paxinos G, Franklin KBJ 2001 The mouse brain in sterotaxic coordinates. 2nd ed. St. Louis: Elsevier (figures 26, 48, and 102) [Google Scholar]