Abstract
Connective tissue growth factor (CTGF), a member of the CCN family of proteins, is expressed in skeletal cells, and the ctgf null mutation leads to neonatal lethality due to defects in skeletal development. To define the function of CTGF in the postnatal skeleton, we created transgenic mice overexpressing CTGF under the control of the human osteocalcin promoter. CTGF transgenic female and male mice exhibited a significant decrease in bone mineral density, compared with wild-type littermate controls. Bone histomorphometry revealed that CTGF overexpression caused decreased trabecular bone volume due to impaired osteoblastic activity because mineral apposition and bone formation rates were decreased. Osteoblast and osteoclast number and bone resorption were not altered. Calvarial osteoblasts and stromal cells from CTGF transgenics displayed decreased alkaline phosphatase and osteocalcin mRNA levels and reduced bone morphogenetic protein (BMP) signaling mothers against decapentaplegic, Wnt/β-catenin, and IGF-I/Akt signaling. In conclusion, CTGF overexpression in vivo causes osteopenia, secondary to decreased bone formation, possibly by antagonizing BMP, Wnt, and IGF-I signaling and activity.
PRECURSOR MESENCHYMAL cells can differentiate into cells of various lineages, including osteoblasts, chondrocytes, and adipocytes (1). The fate of mesenchymal cells and their differentiation toward cells of the osteoblastic lineage is tightly controlled by extracellular and intracellular signals. Bone morphogenetic proteins (BMPs) are important determinants of cell fate, and play a central role in the regulation of osteoblastogenesis and endochondral bone formation (2). BMPs, in conjunction with Wnt, induce the differentiation of mesenchymal cells toward the osteoblastic lineage and enhance the pool of mature osteoblasts (3,4,5). The effects of BMPs and Wnt are controlled by a large group of secreted polypeptides that prevent BMP or Wnt signaling by binding to BMPs or Wnt, or to their receptors/coreceptors, precluding ligand-receptor interactions (2,5,6,7). IGFs do not direct the differentiation of immature cells toward cells of the osteoblastic lineage, but enhance the function of the mature osteoblast and increase bone formation (8).
Members of the CCN family of cysteine-rich (CR) secreted proteins include cysteine-rich 61 (Cyr 61), connective tissue growth factor (CTGF), nephroblastoma overexpressed (Nov), and Wnt-inducible secreted proteins 1, 2, and 3 (9,10). CCN proteins are highly conserved and share four distinct modules: an IGF-binding domain, a von Willebrand type C domain containing the CR domain, a thrombospondin-1 domain, and a C-terminal domain, important for protein-protein interactions (9,10). CCN proteins are structurally related to certain BMP antagonists, such as twisted gastrulation and chordin, and can have important interactions with regulators of osteoblast cell growth and differentiation (11).
CTGF is expressed in a variety of tissues, including bone and cartilage. In osteoblasts, CTGF expression is induced by BMP, TGFβ, Wnt, and cortisol, suggesting a possible role in the activity of these agents in bone cell function (12,13,14). CTGF regulates different cellular functions including adhesion, proliferation, migration and differentiation. The function of CTGF in skeletal cells is not well understood, and in vitro studies have yielded controversial results (13,15). By mechanisms that would resemble the activity of certain BMP or Wnt antagonists, CTGF binds to BMP-2 and -4 through its CR domain, and to Wnt coreceptors through its C-terminal domain, and inhibits osteoblastic differentiation (15,16). In vivo studies indicate that CTGF is necessary for endochondral bone formation, and deletion of ctgf in mice results in newborn lethality and skeletal abnormalities (17). Overexpression of CTGF in chondrocytes under the control of type XI collagen promoter has suggested that CTGF in excess can lead to osteopenia (18). However, the function of CTGF in the adult skeleton is not known.
The intent of this study was to investigate the direct effect of CTGF on bone remodeling and the mechanisms involved. For this purpose, we created transgenic mice overexpressing CTGF under the control of the osteoblast-specific osteocalcin promoter, and determined their skeletal phenotype. Cultures of osteoblastic and stromal cells from CTGF transgenics were performed to establish mechanisms responsible for the phenotype.
Materials and Methods
Osteocalcin/CTGF construct and generation of transgenic mice
After introduction of the Kozak consensus sequence upstream of the translation initiation codon, a 1046-bp fragment coding for murine ctgf (R.P. Ryseck, Princeton, NJ) was cloned downstream of a 182-bp artificial intron and a 3.8-kb fragment of the human osteocalcin promoter (E. Gardiner; Sydney, Australia), and upstream of polyadenylation sequences and a 3.5-kb fragment of the 3′-untranslated region and flanking DNA of the osteocalcin gene (19). Nucleotide sequence analysis confirmed the absence of mutations and the correct orientation of the construct. Microinjection of linearized DNA into pronuclei of fertilized oocytes from FVB (for tropism to Friend Leukemia Virus Strain B) inbred mice, and transfer of microinjected embryos into pseudopregnant FVB mice were carried out by the transgenic facility at the University of Connecticut Health Center (Farmington, CT). Positive founders were identified by Southern blot analysis of tail DNA (20). Founder mice were bred to wild-type FVB mice to generate transgenic lines. Intermatings of heterozygous transgenics were used to create a homozygous offspring. All animal experiments were approved by the Animal Care and Use Committee of Saint Francis Hospital and Medical Center.
X-ray analysis and bone mineral density (BMD)
Radiography was performed on mice anesthetized with tribromoethanol (Sigma Chemical Co., St. Louis, MO) on a Faxitron x-ray system (model MX 20; Faxitron X-Ray Corp., Wheeling, IL). The x-rays were performed at an intensity of 35 kW for 25 sec. Total bone mineral content (BMC; grams), skeletal area (cm2) and bone mineral density (BMD; grams per cm2) were measured on anesthetized mice using the PIXImus small animal DEXA system (GE Medical System/LUNAR, Madison, WI) (21). Calibrations were performed with a phantom of a defined value, and quality assurance measurements were performed before each use. The coefficient of variation for total BMD is less than 1% (n = 9).
Bone histomorphometric analysis
Static and dynamic histomorphometry was carried out on transgenic mice and wild-type littermate controls at one month of age. Mice were injected with calcein, 20 mg/kg, and demeclocycline, 50 mg/kg, at an interval of 2 d, and killed by CO2 inhalation 2 d after the demeclocycline injection. Femurs were dissected, fixed in 70% ethanol, dehydrated and embedded undecalcified in methyl methacrylate. Longitudinal sections, 5 μm thick, were cut on a microtome (Microm, Richards-Allan Scientific, Kalamazoo, MI) and stained with toluidine blue or Von Kossa. Static parameters of bone formation and resorption were measured in a defined area between 181 μm and 1086 μm from the growth plate, using an OsteoMeasure morphometry system (Osteometrics, Atlanta, GA) (22). For dynamic histomorphometry, mineralizing surface per bone surface and mineral apposition rate were measured in unstained sections under UV light, using a B-2A set long pass filter, and bone formation rate was calculated. The terminology and units used are those recommended by the Histomorphometry Nomenclature Committee of the American Society for Bone and Mineral Research (23).
Serum C-terminal cross-linked telopeptide of type I collagen (CTX)
The serum bone remodeling marker CTX was measured by ELISA using RatLaps ELISA kits (Nordic Bioscience Diagnostics, Herlev, Denmark), according to manufacturer’s instructions.
Primary osteoblast and bone marrow stromal cell cultures
Osteoblastic cells were isolated from parietal bones of 3-d-old CTGF heterozygous transgenic and wild-type control littermate mice, following the mating of CTGF heterozygous and wild-type FVB mice. Cells were obtained by five sequential digestions of the parietal bones using bacterial collagenase (CLS II, Worthington Biochemical, Freehold, NJ) (24). Cell populations harvested from the third to the fifth digestions were cultured as a pool, and were previously shown to have osteoblast characteristics (24). Osteoblastic cells were cultured in DMEM (Life Technologies, Inc., Grand Island, NY) supplemented with nonessential amino acids, 20 mm HEPES, 100 μg/ml ascorbic acid, and 10% fetal bovine serum (FBS, Atlanta Biologicals, Lawrenceville, GA), at 37 C in a humidified 5% CO2 incubator. Subconfluent cells were trypsinized and these first passage cells were used for subsequent experiments. To test for effects on BMP or Wnt signaling, cells were plated at a density of 25,000 cells/cm2, cultured to subconfluence (∼35,000 cells/cm2) and transfected with BMP/Signaling mothers against decapentaplegic (Smad) or Wnt/β-catenin reporter constructs and treated with BMP-2 (Wyeth Research, Collegeville, PA) or Wnt 3a (R&D Systems, Minneapolis, MN), as described below. To assess for effects on the phosphorylation of Smad or serine/threonine protein kinase B (Akt), or on cytosolic β-catenin levels, cells were plated at a density of 40,000 cells/cm2 and cultured to confluence (∼50,000 cells/cm2) or for 10 d after confluence, in the presence of 100 μg/ml ascorbic acid and 5 mm β-glycerophosphate (Sigma-Aldrich, St. Louis, MO), serum-deprived overnight and exposed to vehicle, BMP-2 for 20 min, Wnt 3a for 6 h or IGF-I (PeproTech Inc., Rocky Hill, NJ) for 15 min, as indicated in text and legends. Alternatively, cells cultured for 10 d after confluence were serum deprived overnight and analyzed for gene expression.
Bone marrow stromal cells were isolated from femurs of 4-wk-old CTGF heterozygous transgenic and wild-type littermate control mice, by bone marrow flushing, after CO2 asphyxiation, as described (22). Cells were plated at a density of 5 × 106 cells/cm2 and cultured in α-MEM (Invitrogen, Carlsbad, CA) containing 15% FBS at 37 C in a 5% CO2 incubator. All adherent cells without further cell isolation were grown to confluence (∼105/cm2; 6–7 d of culture), the medium was changed to α-MEM supplemented with 10% FBS, 50 μg/ml ascorbic acid, and 5 mm β-glycerophosphate. Cells were cultured for an additional 10-d period, serum deprived overnight, and analyzed for gene expression. To estimate the number of viable cells, mitochondrial dehydrogenase activity was measured using the Cell Titer 96 Aqueous One cell proliferation assay in accordance with the manufacturer’s instructions (Promega, Madison, WI). Metabolically active cells were estimated by their ability to reduce the tetrazolium compound 3-(4,5-dimethyl-thiazol-2-yl)-5(-3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium inner salt to a formazan product, which was quantified at an absorbance of 490 nm.
Real-time RT-PCR
Total RNA was extracted from calvariae or from cell cultures, and CTGF, alkaline phosphatase, osteocalcin, bone sialoprotein (BSP), and type I collagen mRNA levels determined by real-time RT-PCR (25,26). For this purpose, 1–10 μg of RNA were reverse-transcribed using SuperScript III Platinum Two-Step qRT-PCR kit (Invitrogen), according to manufacturer’s instructions and amplified in the presence of primers specific for these genes or primers for glyceraldehyde-3-phosphate dehydrogenase (GAPDH) or ribosomal protein L38 (RPL38) (Table 1), and Platinum Quantitative PCR SuperMix-UDG (Invitrogen) at 54–60 C for 45 cycles. Gene copy number was estimated by comparison with a standard curve constructed using CTGF (R. P. Ryseck), alkaline phosphatase, osteocalcin or BSP (all from American Type Culture Collection [ATCC (Manassas, VA)] and corrected for RPL38 (ATCC) copy number. CTGF copy number from calvarial extracts was normalized for GAPDH (R. Wu, Cornell University, Ithaca, NY) expression (27,28). Reactions were conducted in a 96-well spectrofluorometric thermal iCycler (Bio-Rad, Hercules, CA), and fluorescence was monitored during every PCR cycle at the annealing step.
Table 1.
Primers used for real-time RT-PCR amplification
| Gene | Accession no. | Strand | 3′ Locus | Sequence (5′–3′) |
|---|---|---|---|---|
| Ctgf | NM_010217 | F | 772 | CACTCCGGGAAATGCTGCGAGGAG[FAM]G |
| R | 840 | GTTGGGTCTGGGCCAAATGT | ||
| Alkaline phosphatase | BC065175 | F | 371 | CGGTTAGGGCGTCTCCACAGTAAC[FAM]G |
| R | 444 | CTTGGAGAGGGCCACAAAGG | ||
| Osteocalcin | L24429 | F | 496 | CACTTACGGCGCTACCTTGGGTAAGT[FAM]G |
| R | 523 | CCCAGCACAACTCCTCCCTA | ||
| Collagen type 1 | NM_007742 | F | 920 | GACCAGTGCCAAAGGAGATGCTGGT[FAM]C |
| R | 990 | GTCCAGGGCGACCTCTCTCA | ||
| Bone sialoprotein | BC045143 | F | 408 | CGAGACCACACTTTCCACACTCT[FAM]G |
| R | 469 | CTTCTTGGGCAGTTGGAGTGC | ||
| Rpl38 | BC055346 | F | 160 | CGAACCGGATAATGTGAAGTTCAAGGTT[FAM]G |
| R | 203 | CTGCTTCAGCTTCTCTGCCTTT | ||
| Gapdh | NM_017008 | F | 1425 | CACGCTCTGGAAAGCTGTGGCG[FAM]G |
| R | 1507 | AGCTTCCCGTTCAGCTCTGG |
Transient transfections
To determine changes in BMP-2 signaling, a construct containing 12 copies of a Smad 1/5 consensus sequence linked to an osteocalcin minimal promoter and a luciferase reporter gene (12xSBE-Oc-pGL3; M. Zhao, San Antonio, TX) was tested in transient transfection experiments (29). To determine changes in Wnt/β-catenin transactivating activity, a construct containing 16 copies of the lymphoid enhancer binding factor/T-cell-specific factor (Lef1/Tcf-4) recognition sequence, cloned upstream of a minimal thymidine kinase promoter and a luciferase reporter gene (16xTCF-Luc; J. Billiard, Wyeth Research) was tested in transient transfection experiments (30). To determine changes in osteocalcin promoter activity, a 1080-bp fragment of the rat osteocalcin promoter, cloned into pGL3 upstream of the luciferase reporter gene (I. Kalajzic, Farmington, CT), was tested in transient transfection experiments. Osteoblast cells were cultured to 70% confluence and transiently transfected with the indicated constructs using FuGENE6 (3 μl FuGENE/2 μg DNA), according to manufacturer’s instructions (Roche, Indianapolis, IN). A cytomegalovirus (CMV) directed β-galactosidase expression construct (Clontech, San Jose, CA) was used to control for transfection efficiency. Cells were exposed to the FuGENE-DNA mix for 16 h and transferred to serum-free medium for 8 h, treated with BMP-2, Wnt 3a or vehicle for 24 h, as indicated in the text and legends, and harvested. Luciferase and β-galactosidase activities were measured using an Optocomp luminometer (MGM Instruments, Hamden, CT). Luciferase activity was corrected for β-galactosidase activity.
Western immunoblot analysis
To determine the level of the phosphorylation of Smad 1/5/8 and Akt, the cell layer of control and CTGF overexpressing cells was washed with cold PBS and extracted in cell lysis buffer [20 mm Tris (pH 7.4), 150 mm NaCl, 1% Triton, and 1 mm EDTA] (Cell Signaling Technology, Beverly, MA) in the presence of protease and phosphatase inhibitors, as described (22,31). Protein concentration was determined by DC protein assay (Bio-Rad), and 10–20 μg of total cellular protein were fractionated by gel electrophoresis in 12.5% polyacrylamide gels under reducing conditions, and transferred to Immobilon P membranes, which were blocked with 3% BSA in PBS. For Smad 1/5/8, membranes were exposed to a rabbit polyclonal antibody, which recognizes Smads 1, 5, and 8 phosphorylated at Ser 463/465 (Smad 1 and 5), and Ser 426/428 (Smad 8) (Cell Signaling Technology), or exposed to a rabbit polyclonal antibody to unphosphorylated Smad 1 (Santa Cruz Biotechnology, Santa Cruz, CA) (32,33). For Akt phosphorylation, membranes were exposed to a rabbit polyclonal antibody, which recognizes Akt phosphorylated at Ser 473, or a polyclonal antibody to unphosphorylated Akt (both, Cell Signaling Technology). To determine cytosolic β-catenin levels, the cell layer was extracted in 10 mm Tris, 140 mm NaCl, 5 mm EDTA, and 2 mm dithiothreitol buffer at pH 7.6, in the presence of protease inhibitors, and the cytosolic fraction was separated by ultracentrifugation, as described (34). Twenty micrograms of protein were fractionated by gel electrophoresis in 7.5% polyacrylamide gels and transferred to Immobilon P membranes. The membranes were blocked with 3% BSA in PBS and exposed to a monoclonal antibody to unphosphorylated β-catenin or a polyclonal antibody to human actin (both from Santa Cruz Biotechnology). All blots were exposed to antirabbit or antimouse IgG antiserum conjugated to horseradish peroxidase and developed with a chemiluminescence detection reagent (PerkinElmer Life Sciences, Boston, MA).
Statistical analysis
Data are expressed as means ± sem. Statistical differences were determined by Student’s t test or ANOVA.
Results
Characterization of CTGF transgenic mice
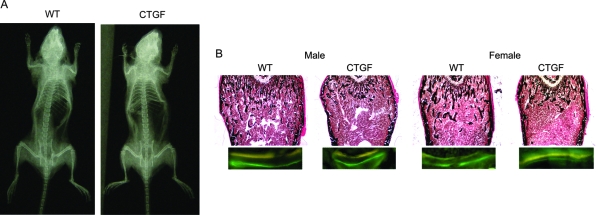
Two transgenic lines overexpressing CTGF under the control of the osteocalcin promoter were established. Their initial identification by Southern blot analysis revealed approximately 7 and more than 100 copies of ctgf transgene in heterozygous from line 1 and 2, respectively. Overexpression of CTGF transcripts in transgenics was confirmed by real-time RT-PCR in calvarial extracts; the ctgf/gapdh copy number in homozygous transgenic males from line 2 was 170 ± 59 compared with 3.7 ± 1.3 in wild-type littermate controls, and in homozygous transgenic females was 82 ± 27 compared with 1.9 ± 0.4 in controls (both 45-fold greater than control, both P < 0.05). CTGF transgenic mice were compared with wild-type littermate controls of identical sex, at the age of 4 wk, a time of high activity of the osteocalcin promoter (35). Line 2 was studied in detail, and 1-month-old homozygous CTGF transgenic male mice displayed reduced weight by 24% (Table 2). In contrast, only minor changes in body weight were observed in homozygous CTGF transgenic female mice, relative to their wild-type controls (Table 2) and no changes in body weight were noted in either male or female mice of line 1 (not shown). Both male and female CTGF overexpressing mice from line 2 displayed a 15–28% decrease in BMC and 8% decrease in BMD, when compared with control wild-type mice (Table 2). Contact radiography did not reveal obvious skeletal abnormalities (Fig. 1).
Table 2.
Weight, total BMC, bone area, and BMD of 1-month- old CTGF homozygous transgenics and wild-type control mice
| Weight | BMC | Bone Area | BMD | |
|---|---|---|---|---|
| Male | ||||
| WT | 15.7 ± 0.6 | 223 ± 11 | 6.4 ± 0.2 | 345 ± 7 |
| CTGF | 11.9 ± 0.7a | 162 ± 13a | 5.2 ± 0.4a | 314 ± 1a |
| Female | ||||
| WT | 15.2 ± 0.3 | 215 ± 7 | 6.1 ± 0.1 | 353 ± 5 |
| CTGF | 14.1 ± 0.6 | 184 ± 5a | 5.6 ± 0.1a | 329 ± 3a |
Weight (g), BMC (g × 104), bone area (cm2), and BMD (g/cm2 × 104) were obtained from one month old CTGF homozygous transgenics from line 2 and wild-type (WT) controls. Values are means ± sem (n = 5–8).
Significantly different from wild-type controls, P < 0.05.
Figure 1.
A, Skeletal radiograph of a 1-month-old homozygous male CTGF transgenic (line 2) and wild-type (WT) control. B, Representative femoral sections from 1-month-old male and female CTGF transgenic homozygous (line 2) and wild-type (WT) controls stained with von Kossa (upper panel) and visualization of calcein and demeclocycline labels (lower panel) by fluorescence microscopy at a final magnification of ×40 and ×400, respectively.
Bone histomorphometric analysis of distal femurs from 1-month-old female heterozygous and homozygous CTGF transgenics from line 2 revealed a 50–70% decrease in trabecular bone volume, due to a reduced number of trabeculae (Table 3). A 40 to 70% decrease in bone volume was observed in homozygous and heterozygous male transgenics from line 2 (Table 4). The decrease in bone volume observed was not associated with changes in osteoblast number. Although the number of osteoblasts per area was decreased because of the lesser number of trabeculae, the number of osteoblasts per perimeter and osteoblast surface were not different from controls. Fluorescence microscopy revealed a decrease in mineralizing surface, mineral apposition rate, and in bone formation rate, which was statistically significant in homozygous and heterozygous female mice, indicating that the decreased bone volume in CTGF transgenics was secondary to a decrease in osteoblast function (Tables 3 and 5 and Fig. 1). Changes in trabecular bone volume were not associated with changes in bone resorption because eroded surface was not different between transgenics and controls. Moreover, the serum levels of CTX did not differ between control and transgenic mice, indicating normal bone remodeling in CTGF transgenics. Serum concentrations of CTX (means ± sem; n = 4) were 111 ± 19 ng/ml in control male mice and 110 ± 32 ng/ml in CTGF homozygous transgenics and 139 ± 24 ng/ml in control female mice and 101 ± 19 ng/ml in CTGF homozygous transgenics. There was no difference between female (Table 3) and male (Table 4) phenotypes, except for the fact that the reduction in bone formation in male transgenics was less pronounced and a 30% decrease in bone formation observed in homozygous male transgenics did not reach statistical significance.
Table 3.
Femoral static and dynamic bone histomorphometry of 1-month-old CTGF transgenics and wild-type control females, line 2
| Line 2 | WT | CTGF+/− | CTGF+/+ |
|---|---|---|---|
| Bone volume/tissue volume (%) | 10.6 ± 0.7 | 5.4 ± 0.7a | 3.5 ± 0.7a |
| Trabecular separation (μm) | 110 ± 8 | 237 ± 36a | 400 ± 84a |
| Trabecular number (mm−1) | 8.5 ± 0.7 | 4.4 ± 0.6a | 2.9 ± 0.5a |
| Osteoblasts/bone perimeter (mm−1) | 38.0 ± 1.1 | 36.7 ± 1.5 | 38.8 ± 2.8 |
| Osteoblasts/tissue area (mm−2) | 506 ± 35 | 250 ± 26a | 179 ± 39a |
| Osteoclasts/bone perimeter (mm−1) | 7.0 ± 0.3 | 7.1 ± 0.4 | 8.6 ± 1.1 |
| Eroded surface/bone surface (%) | 21.0 ± 0.8 | 20.7 ± 0.9 | 23.0 ± 1.8 |
| Mineralizing surface/bone surface (%) | 6.7 ± 0.4 | 5.2 ± 0.3a | 5.2 ± 0.6a |
| Mineral apposition rate (μm/d) | 0.94 ± 0.02 | 0.79 ± 0.03a | 0.79 ± 0.02a |
| Bone formation rate/bone surface (μm3/μm2/d) | 0.063 ± 0.003 | 0.041 ± 0.002a | 0.041 ± 0.004a |
Bone histomorphometry was performed on femurs from 1-month-old female CTGF homozygous (CTGF+/+), CTGF heterozygous (CTGF+/−) and wild-type (WT) controls for line 2. For static histomorphometry, sections were stained with toluidine blue, and for dynamic histomorphometry unstained sections were analyzed by fluorescence microscopy. Values are means ± sem; n = 6 to 9.
Significantly different from wild type controls, P < 0.05.
Table 4.
Femoral static and dynamic bone histomorphometry of 1-month-old CTGF transgenics and wild-type control males, line 2
| WT | CTGF+/− | WT | CTGF+/+ | |
|---|---|---|---|---|
| Bone volume/tissue volume (%) | 10.7 ± 0.5 | 6.2 ± 0.5a | 12.4 ± 0.96 | 3.8 ± 0.6a |
| Trabecular separation (μm) | 92 ± 5 | 175 ± 15a | 88 ± 7 | 308 ± 50a |
| Trabecular number (mm−1) | 9.8 ± 0.5 | 5.6 ± 0.4a | 10.3 ± 0.6 | 3.5 ± 0.6a |
| Osteoblasts/bone perimeter (mm−1) | 58.0 ± 7.6 | 52.4 ± 3.3 | 33.3 ± 1.6 | 34.2 ± 2.8 |
| Osteoblasts/tissue area (mm−2) | 889 ± 105 | 458 ± 43a | 531 ± 23 | 192 ± 38a |
| Osteoclasts/bone perimeter (mm−1) | 12.1 ± 0.8 | 13.3 ± 0.6 | 6.5 ± 0.4 | 7.1 ± 0.4 |
| Eroded surface/bone surface (%) | 23.6 ± 1.5 | 25.6 ± 1.3 | 18.8 ± 1.2 | 18.0 ± 1.7 |
| Mineralizing surface/bone surface (%) | 4.1 ± 0.6 | 4.8 ± 0.6 | 5.08 ± 0.47 | 3.74 ± 0.5 |
| Mineral apposition rate (μm/d) | 0.87 ± 0.06 | 0.90 ± 0.04 | 0.812 ± 0.009 | 0.723 ± 0.057 |
| Bone formation rate/bone surface (μm3/μm2/d) | 0.036 ± 0.006 | 0.042 ± 0.004 | 0.041 ± 0.004 | 0.028 ± 0.006 |
Bone histomorphometry was performed on femurs from 1-month-old male CTGF heterozygous (CTGF+/−) and CTGF homozygous (CTGF+/+), of line 2 and their respective wild-type (WT) controls. For static histomorphometry, sections were stained with toluidine blue, and for dynamic histomorphometry unstained sections were analyzed by fluorescence microscopy. Values are means ± sem; n = 5 to 8 for static histomorphometry, and n = 3 to 6 for dynamic histomorphometry.
Significantly different from wild-type controls, P < 0.05.
Table 5.
Femoral static and dynamic bone histomorphometry of 1-month-old CTGF transgenics and wild-type control females, line 1
| Line 1 | WT | CTGF+/+ |
|---|---|---|
| Bone volume/tissue volume (%) | 13.2 ± 1.1 | 9.6 ± 0.9a |
| Trabecular separation (μm) | 85 ± 7 | 116 ± 6a |
| Trabecular number (mm−1) | 10.6 ± 0.7 | 7.9 ± 0.4a |
| Osteoblasts/bone perimeter (mm−1) | 46.9 ± 3.6 | 48.7 ± 2.3 |
| Osteoblasts/tissue area (mm−2) | 766 ± 52 | 605 ± 38a |
| Osteoclasts/bone perimeter (mm−1) | 10.6 ± 0.5 | 10.5 ± 0.8 |
| Eroded surface/bone surface (%) | 25.3 ± 1.5 | 24.1 ± 1.3 |
| Mineralizing surface/bone surface (%) | 3.4 ± 0.7 | 2.1 ± 0.6 |
| Mineral apposition rate (μm/d) | 1.10 ± 0.07 | 1.11 ± 0.05 |
| Bone formation rate/bone surface (μm3/μm2/d) | 0.040 ± 0.011 | 0.023 ± 0.006 |
Bone histomorphometry was performed on femurs from 1-month- old female CTGF homozygous (CTGF+/+), and wild-type (WT) controls for line 1. For static histomorphometry, sections were stained with toluidine blue, and for dynamic histomorphometry unstained sections were analyzed by fluorescence microscopy. Values are means ± sem; n = 6 to 9, except for dynamic histomorphometry, where n = 4 to 6.
Significantly different from wild-type controls, P < 0.05.
Both transgenic lines studied displayed analogous skeletal phenotypes, although the phenotype was less pronounced in line 1, probably because of the lesser degree of ctgf transgene expression. In accordance, one month old female CTGF homozygous transgenics from line 1 expressed an approximately 20-fold increase in CTGF transcripts in calvarial extracts and exhibited a modest decrease in trabecular bone volume with an approximately 40% decrease in bone formation (P > 0.05) (Tables 5). One-month-old male CTGF homozygous transgenics from line 1 also were osteopenic, and trabecular bone volume/tissue volume was 9.2 ± 0.8 in controls and 7.3 ± 0.5 in CTGF transgenics (P < 0.05) due to a decreased trabecular number, but bone formation was not suppressed significantly. CTGF heterozygous from line 1 did not exhibit a phenotype possibly due to the low level of transgene expression (not shown). Mice from line 1 showed only a 5 to 9% decrease in BMC (P > 0.05) without a significant change in BMD.
Primary osteoblast and stromal cell cultures
To investigate the impact of CTGF on osteoblastic cell function, calvarial osteoblastic cells from heterozygous transgenics (line 2) and wild-type controls were cultured. In accordance with the phenotype observed in CTGF transgenics in vivo, CTGF did not have an effect on cell number at confluence, and up to 14 d after confluence (data not shown). After 10 d of culture, alkaline phosphatase and osteocalcin mRNA levels were suppressed by about 50% in osteoblastic cells from transgenics when compared with control cells, confirming an inhibitory effect of CTGF on osteoblast function (Table 6). BSP expression was decreased by 40% (P < 0.08) and only a modest, approximately 20%, decrease in type I collagen was observed in cells from CTGF transgenics (P > 0.05, not shown). The activity of a transfected osteocalcin promoter construct was decreased from 14.9 ± 1.4 luciferase/β-galactosidase in control wild-type cells to 10.2 ± 0.3 luciferase/β-galactosidase in transgenic cells (P < 0.05).
Table 6.
Expression of osteoblast gene markers in osteoblastic cells from CTGF transgenics and wild-type controls
| Calvarial osteoblasts 10 d after confluence | WT | CTGF |
|---|---|---|
| Alkaline phosphatase | 2.7 ± 0.5 | 1.2 ± 0.2a |
| Osteocalcin | 1.1 ± 0.1 | 0.6 ± 0.1a |
| Bsp | 5.1 ± 0.7 | 3.1 ± 0.7 (P < 0.08) |
| Ctgf | 0.39 ± 0.03 | 0.8 ± 0.1a |
Calvarial osteoblastic cells from CTGF heterozygous transgenics (line 2) and wild type (WT) controls were cultured for 10 d after confluence. Total RNA was reverse-transcribed, and amplified by real time RT-PCR in the presence of specific primers. Data are expressed as alkaline phosphatase, osteocalcin, bsp and ctgf copy number, determined by real time RT-PCR, corrected for rpl38 expression. Values are means ± sem, n = 4.
Significantly different from wild-type controls, P < 0.05.
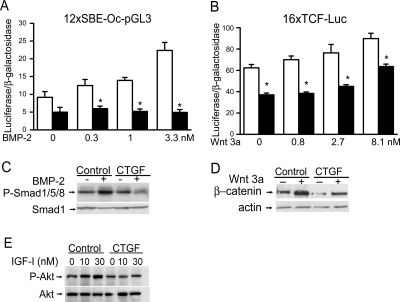
To examine possible mechanisms underlying the osteopenia and decreased bone formation of CTGF transgenics, we examined whether CTGF overexpression affected BMP/Smad, Wnt/β-catenin or IGF-I signaling in cells of the osteoblastic lineage. Confirming previous results, BMP-2 induced the transactivation of the 12xSBE-Oc-pGL3 reporter construct, where 12 Smad binding sites direct luciferase expression, and enhanced Smad1/5/8 phosphorylation (Fig. 2) (29,36,36). CTGF transgenic cells displayed decreased response to BMP-2 on the transactivation of the transfected 12xSBE-Oc-pGL3 reporter construct and on Smad 1/5/8 phosphorylation (Fig. 2, A and C). Wnt 3a caused a stimulatory effect on the transactivation of a 16xTCF-Luc reporter, where 16 TCF/Lef binding sites direct luciferase expression. The effect was modest, possibly because osteoblast-enriched cell cultures and not cultures of early osteoblast precursor cells were studied. Wnt 3a also increased the levels of cytosolic β-catenin (Fig. 2). The effect of Wnt 3a on the transactivation of the 16xTCF-Luc reporter and on β-catenin levels was decreased in cells from CTGF transgenics (Fig. 2, B and D).
Figure 2.
Effect of CTGF overexpression on BMP-2, Wnt 3a and IGF-I signaling in calvarial osteoblastic cells from CTGF heterozygous transgenics (line 2) and wild-type controls. A and B, Calvarial osteoblasts were cultured to subconfluence, and transfected with 12xSBE-Oc-pGL3 or 16xTCF-Luc, and a CMV/β-galactosidase expression vector. After 16 h, cells were switched to serum-free DMEM and treated or not with BMP-2 or Wnt 3a at the indicated doses for 24 h. Data shown represent luciferase/β-galactosidase activity for wild-type control cells (white bars) or CTGF overexpressing cells (black bars). Bars represent means ± sem for six observations. *, Significantly different from wild-type controls, P < 0.05. C–E, Osteoblasts were cultured to confluence and for 10 d after confluence, serum-deprived overnight and treated with BMP-2 at 3.3 nm for 20 min, Wnt 3a at 2.7 nm for 6 h, or IGF-I at 10 and 30 nm for 15 min for the determination of Smad 1/5/8 phosphorylation (C), β-catenin levels (D) or Akt phosphorylation (E). Total cell lysates (C and E) or cytosolic extracts (D) were resolved by gel electrophoresis and transferred to Immobilon P membranes, which were incubated with antibodies to phosphorylated Smad 1/5/8 or unphosphorylated Smad 1, with antibodies to β-catenin or actin, or antibodies to phosphorylated or unphosphorylated Akt.
To explore additional mechanisms involved in the inhibitory effect of CTGF on osteoblast function, we tested whether CTGF modified IGF-I signaling. IGF-I signals through the phosphatidyl-inositol-3 kinase, resulting in the activation of Akt. IGF-I at 10 and 30 nm increased phosphorylation of Akt in osteoblastic cells from control mice, but not in cells from CTGF transgenics, indicating an inhibitory effect of CTGF on IGF signaling (Fig. 2E).
In accordance with the results obtained in calvarial osteoblastic cells, CTGF overexpression did not modify cell number (not shown), and tempered the rise in alkaline phosphatase and osteocalcin mRNA levels observed in control marrow stromal cells cultured in the presence of differentiation medium for 10 d (Table 7). BSP expression was decreased in cells from CTGF transgenics at confluence and to a lesser extent 10 d after confluence. CTGF overexpression tempered the increase in type I collagen expression so that after 14 d of culture type I collagen mRNA levels were 40% lower in cells from transgenics than in control cells (P < 0.05).
Table 7.
Expression of osteoblast gene markers in stromal cells from CTGF transgenics and wild-type controls
| Confluence
|
10 d after confluence
|
|||
|---|---|---|---|---|
| Stromal cells | WT | CTGF | WT | CTGF |
| Alkaline phosphatase | 6.3 ± 1.7 | 2.8 ± 0.1 | 8.6 ± 1.0 | 4.0 ± 0.4a |
| Osteocalcin | 2.1 ± 0.4 | 1.9 ± 0.1 | 21.0 ± 3.0 | 7.7 ± 1.4a |
| Bsp | 2.7 ± 0.4 | 0.9 ± 0.4a | 3.7 ± 1.6 | 2.1 ± 0.8 |
| Ctgf | 1.3 ± 0.1 | 2.1 ± 0.1a | 0.8 ± 0.1 | 1.8 ± 0.1a |
Bone marrow stromal cells from CTGF heterozygous transgenics (line 2) and wild type (WT) controls were cultured for 10 d after confluence. Total RNA was reverse-transcribed, and amplified by real-time RT-PCR in the presence of specific primers. Data are expressed as alkaline phosphatase, osteocalcin, bsp and ctgf copy number, determined by real-time RT-PCR, corrected for rpl38 expression. Values are means ± sem, n = 4.
Significantly different from wild-type controls, P < 0.05.
Discussion
Our findings demonstrate that transgenic mice overexpressing CTGF develop decreased trabecular bone volume and osteopenia. The degree of osteopenia appeared to be related to the level of transgene expression, and heterozygous from a line expressing low transgene levels (line 1) did not exhibit a phenotype. There were no sex-dependent differences in the skeletal phenotype, although males from one transgenic line had decreased body weight, whereas females did not. The reason for this difference remains unexplained, but could involve skeletal and nonskeletal effects of CTGF because it is a secreted protein. The decrease in trabecular bone volume in CTGF transgenic mice appears to be secondary to decreased osteoblast function because there was a decrease in mineral apposition and bone formation rate, whereas osteoblast number was not affected. There was no apparent evidence of increased bone resorption. The lack of an effect of CTGF on osteoblast number may be a reflection of the osteocalcin promoter used in the study because it becomes active after osteoblast cell maturation. Consequently, CTGF is not overexpressed before the formation of mature osteoblasts. However, CTGF is a secreted soluble factor and could have affected neighboring undifferentiated cells of the osteoblastic lineage.
The in vivo skeletal phenotype of CTGF transgenics is consistent with previous studies indicating that transgenic mice overexpressing CTGF in chondrocytes, under the control of the type-XI collagen promoter, showed decreased bone density (18), suggesting that CTGF in excess leads to osteopenia. On the other hand, ctgf null mice have impaired bone formation and mineralization due to defects in cartilage and vascular development, indicating that CTGF is necessary for endochondral bone formation (17).
The in vivo findings were confirmed by in vitro experiments in osteoblastic cells from CTGF transgenic mice. CTGF overexpression caused a decrease in osteoblast gene marker expression in osteoblasts and stromal cells, indicating an inhibition of osteoblast cell function. The mechanism of the impaired osteoblastic function in CTGF transgenics could involve the binding and sequestration of BMPs by CTGF in the bone environment because osteoblasts from CTGF transgenics exhibited a decreased responsiveness to BMP-2 on Smad signaling. In fact, previous studies have shown direct interactions between CTGF and BMP-2, and as a consequence an inhibitory effect on osteoblastogenesis (15,16). Although, BMPs have been shown to induce the differentiation of mesenchymal cells toward cells of the osteoblastic lineage, they also enhance the function of the osteoblasts, and our results in CTGF transgenics suggest a more pronounced inhibitory effect on osteoblastic function than number, caused possibly by the expression of the osteocalcin promoter in mature cells (37). CTGF overexpression also has an inhibitory effect on Wnt/β-catenin signaling and activity. This could be explained by direct interactions between CTGF with Wnt coreceptors, although the suppression of BMP signaling can contribute to decreased Wnt activity (38,39). This dual inhibitory effect of CTGF is shared by BMP antagonists, which also have been shown to decrease BMP and Wnt activity. Similarly, Wnt antagonists, such as Dickkopf 1 and sclerostin oppose BMP actions (38,39). In addition, CTGF has been shown to bind IGF-I, although with low affinity, and as a consequence to reduce IGF-I effects on phosphatidyl-inositol-3 kinase (8). Because IGF-I enhances osteoblastic function, its inhibition by CTGF may contribute to the osteoblastic phenotype exhibited by CTGF transgenic mice.
The BMP, Wnt, and IGF-I antagonistic activity of CTGF would support our findings of its inhibitory role in osteoblastic function and possibly osteoblastogenesis. However, down-regulation of CTGF in vitro using RNA interference in C3H10T ½ cells has demonstrated that CTGF is necessary, under selected experimental conditions, for osteoblastogenesis (13,40,41). This dual permissive and inhibitory activity is not limited to CTGF, and it is shared by BMP antagonists of the twisted gastrulation/chordin families, which are structurally related to CCN proteins (11,42,43,44,45,46). Furthermore, under certain in vitro experimental conditions, forced overexpression of CTGF can favor osteoblastogenesis by mechanisms independent of BMP and Wnt signaling (47,48,49,50,51). Recently, we found that the constitutive expression of CTGF can induce hairy and enhancer of split 1 transcription and the transactivation of nuclear factor of activated T-cells (NFAT) in ST-2 stromal cells, and these two signals may have a stimulatory effect on osteoblastogenesis (47,48,49,50). Furthermore, the inhibitory effect of CTGF on BMP signaling shown in the present studies was not observed in ST-2 cells under conditions of forced overexpression of CTGF (47,48,49,50). It is apparent that the signals activated in vitro were not consistently operational in vivo, but they may have been active at selected times in certain mice. Although there were no changes in bone resorption, and the number of osteoclasts/ perimeter were not changed in CTGF transgenics as a whole, we noted that sporadic CTGF transgenics exhibited increased osteoclast surface. The mechanism could have involved induction of NFAT activity in these mice, and as a result an increase in osteoclastogenesis (51,52).
CTGF is expressed by endothelial cells, and has complex effects on angiogenesis. CTGF induces endothelial cell replication and survival and is required for angiogenesis in cartilage, but not in classic models of angiogenesis (53,54,55). The expression of CTGF is induced by hypoxia-inducible factor (HIF) acting either directly by increasing CTGF, or indirectly by enhancing the expression of vascular endothelial growth factor (VEGF), which in turn can stimulate CTGF expression (56,57). It is of interest that both HIF and VEGF act by posttranscriptional mechanisms to regulate CTGF expression. A feedback regulatory mechanism also plays a regulatory function because CTGF binds VEGF and suppresses HIF expression, and as a consequence decreases VEGF expression (58,59). It is of interest that CTGF can induce matrix metalloproteinases (MMP) 2 and 9, and MMP-2 can free VEGF from CTGF, and as a result favor angiogenesis (60). Consequently, the impact of CTGF on angiogenesis in a specific tissue depends on its regulation and association with proteases and other factors present in the cellular environment. CTGF may have an important function in angiogenesis, under basal or hypoxia-induced conditions, in the skeletal microenvironment. However, immunohistochemistry of femoral sections of CTGF transgenics failed to reveal an increase in either VEGF or platelet endothelial cell adhesion molecule (PECAM-1, CD31), which is expressed on the surface of endothelial cells and plays a role in angiogenesis (data not shown) (61,62). Additional studies in mice misexpressing ctgf are needed to define its role in skeletal angiogenesis.
In conclusion, our studies reveal that CTGF overexpression in vivo causes osteopenia and impaired osteoblast i.e. function, possibly by inhibiting BMP, Wnt, and IGF-I signaling and activity.
Acknowledgments
The authors thank Drs. M. Zhao for 12xSBE-Oc-pGL3 construct, E. Gardiner for human osteocalcin promoter construct, I. Kalajzic for osteocalcin promoter construct, R. P. Rysek for CTGF cDNA, R. Wu for GAPDH cDNA; Wyeth Research for BMP-2 and 16xTCF-Luc construct; T. X. Le and M. Burton for technical help, and M. Yurczak for secretarial assistance.
Footnotes
This work was supported by Grant AR021707 from the National Institute of Arthritis and Musculoskeletal and Skin Diseases, and Grants DK042424 and DK045227 from the National Institute of Diabetes and Digestive and Kidney Diseases.
Disclosure Statement: The authors have nothing to disclose.
First Published Online June 5, 2008
Abbreviations: Akt, Serine/threonine protein kinase B; ATCC, American Type Culture Collection; BMPs, bone morphogenetic proteins; BSP, bone sialoprotein; CMV, cytomegalovirus; CR, cysteine-rich; CTGF, connective tissue growth factor; CTX, C-terminal cross-link telopeptide of collagen type I; Cyr 61, cysteine-rich 61; FBS, fetal bovine serum; FVB, Friend Leukemia Virus Strain B; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; HIF, hypoxia-inducible factor; LEF, lymphoid enhancer binding factors; MMP, matrix metalloproteinases; NFAT, nuclear factor of activated T cells; Nov, nephroblastoma overexpressed; RPL38, ribosome protein L38; SBE, Smad binding element; Smad, Signaling mothers against decapentaplegic; TCF, T-cell-specific factor; VEGF, vascular endothelial growth factor.
References
- Bianco P, Gehron RP 2000 Marrow stromal stem cells. J Clin Invest 105: 1663–1668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canalis E, Economides AN, Gazzerro E 2003 Bone morphogenetic proteins, their antagonists, and the skeleton. Endocr Rev 24:218–235 [DOI] [PubMed] [Google Scholar]
- Thies RS, Bauduy M, Ashton BA, Kurtzberg L, Wozney JM, Rosen V 1992 Recombinant human bone morphogenetic protein-2 induces osteoblastic differentiation in W-20–17 stromal cells. Endocrinology 130:1318–1324 [DOI] [PubMed] [Google Scholar]
- Ghosh-Choudhury N, Harris MA, Feng JQ, Mundy GR, Harris SE 1994 Expression of the BMP 2 gene during bone cell differentiation. Crit Rev Eukaryot Gene Expr 4:345–355 [DOI] [PubMed] [Google Scholar]
- Westendorf JJ, Kahler RA, Schroeder TM 2004 Wnt signaling in osteoblasts and bone diseases. Gene 341:19–39 [DOI] [PubMed] [Google Scholar]
- Gazzerro E, Canalis E 2006 Bone morphogenetic proteins and their antagonists. Rev Endocr Metab Disord 7:51–65 [DOI] [PubMed] [Google Scholar]
- Kawano Y, Kypta R 2003 Secreted antagonists of the Wnt signalling pathway. J Cell Sci 116:2627–2634 [DOI] [PubMed] [Google Scholar]
- Gazzerro E, Canalis E 2006 Skeletal actions of insulin-like growth factors. Expert Rev Endocrinol Metab 1:47–56 [DOI] [PubMed] [Google Scholar]
- Brigstock DR 2003 The CCN family: a new stimulus package. J Endocrinol 178:169–175 [DOI] [PubMed] [Google Scholar]
- Brigstock DR, Goldschmeding R, Katsube KI, Lam SC, Lau LF, Lyons K, Naus C, Perbal B, Riser B, Takigawa M, Yeger H 2003 Proposal for a unified CCN nomenclature. Mol Pathol 56:127–128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia AJ, Coffinier C, Larrain J, Oelgeschlager M, De Robertis EM 2002 Chordin-like CR domains and the regulation of evolutionarily conserved extracellular signaling systems. Gene 287:39–47 [DOI] [PubMed] [Google Scholar]
- Parisi MS, Gazzerro E, Rydziel S, Canalis E 2006 Expression and regulation of CCN genes in murine osteoblasts. Bone 38:671–677 [DOI] [PubMed] [Google Scholar]
- Luo Q, Kang Q, Si W, Jiang W, Park JK, Peng Y, Li X, Luu HH, Luo J, Montag AG, Haydon RC, He TC 2004 Connective tissue growth factor (CTGF) is regulated by Wnt and bone morphogenetic proteins signaling in osteoblast differentiation of mesenchymal stem cells. J Biol Chem 279:55958–55968 [DOI] [PubMed] [Google Scholar]
- Pereira RC, Durant D, Canalis E 2000 Transcriptional regulation of connective tissue growth factor by cortisol in osteoblasts. Am J Physiol Endocrinol Metab 279:E570–E576 [DOI] [PubMed] [Google Scholar]
- Abreu JG, Ketpura NI, Reversade B, De Robertis EM 2002 Connective-tissue growth factor (CTGF) modulates cell signalling by BMP and TGF-β. Nat Cell Biol 4:599–604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercurio S, Latinkic B, Itasaki N, Krumlauf R, Smith JC 2004 Connective-tissue growth factor modulates WNT signalling and interacts with the WNT receptor complex. Development 131:2137–2147 [DOI] [PubMed] [Google Scholar]
- Ivkovic S, Yoon BS, Popoff SN, Safadi FF, Libuda DE, Stephenson RC, Daluiski A, Lyons KM 2003 Connective tissue growth factor coordinates chondrogenesis and angiogenesis during skeletal development. Development 130:2779–2791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakanishi T, Yamaai T, Asano M, Nawachi K, Suzuki M, Sugimoto T, Takigawa M 2001 Overexpression of connective tissue growth factor/hypertrophic chondrocyte-specific gene product 24 decreases bone density in adult mice and induces dwarfism. Biochem Biophys Res Commun 281:678–681 [DOI] [PubMed] [Google Scholar]
- Sims NA, White CP, Sunn KL, Thomas GP, Drummond ML, Morrison NA, Eisman JA, Gardiner EM 1997 Human and murine osteocalcin gene expression: conserved tissue restricted expression and divergent responses to 1,25-dihydroxyvitamin D3 in vivo. Mol Endocrinol 11:1695–1708 [DOI] [PubMed] [Google Scholar]
- Irwin N 1989 Molecular Cloning. In: Femt J, ed. Sambrook. New York: Cold Spring Harbor Laboratory Press; 9.32–9.36 [Google Scholar]
- Nagy TR, Prince CW, Li J 2001 Validation of peripheral dual-energy x-ray absorptiometry for the measurement of bone mineral in intact and excised long bones of rats. J Bone Miner Res 16:1682–1687 [DOI] [PubMed] [Google Scholar]
- Gazzerro E, Pereira RC, Jorgetti V, Olson S, Economides AN, Canalis E 2005 Skeletal overexpression of gremlin impairs bone formation and causes osteopenia. Endocrinology 146:655–665 [DOI] [PubMed] [Google Scholar]
- Parfitt AM, Drezner MK, Glorieux FH, Kanis JA, Malluche H, Meunier PJ, Ott SM, Recker RR 1987 Bone histomorphometry: standardization of nomenclature, symbols, and units. Report of the ASBMR Histomorphometry Nomenclature Committee. J Bone Miner Res 2:595–610 [DOI] [PubMed] [Google Scholar]
- McCarthy TL, Centrella M, Canalis E 1988 Further biochemical and molecular characterization of primary rat parietal bone cell cultures. J Bone Miner Res 3:401–408 [DOI] [PubMed] [Google Scholar]
- Nazarenko I, Lowe B, Darfler M, Ikonomi P, Schuster D, Rashtchian 2002 A Multiplex quantitative PCR using self-quenched primers labeled with a single fluorophore. Nucleic Acids Res 30:e37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nazarenko I, Pires R, Lowe B, Obaidy M, Rashtchian A 2002 Effect of primary and secondary structure of oligodeoxyribonucleotides on the fluorescent properties of conjugated dyes. Nucleic Acids Res 30:2089–2195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lian J, Stewart C, Puchacz E, Mackowiak S, Shalhoub V, Collart D, Zambetti G, Stein G 1989 Structure of the rat osteocalcin gene and regulation of vitamin D-dependent expression. Proc Natl Acad Sci USA 86:1143–1147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tso JY, Sun XH, Kao TH, Reece KS, Wu R 1985 Isolation and characterization of rat and human glyceraldehyde-3-phosphate dehydrogenase cDNAs: genomic complexity and molecular evolution of the gene. Nucleic Acids Res 13:2485–2502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao M, Qiao M, Harris SE, Oyajobi BO, Mundy GR, Chen D 2004 Smurf1 inhibits osteoblast differentiation and bone formation in vitro and in vivo. J Biol Chem 279:12854–12859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Billiard J, Way DS, Seestaller-Wehr LM, Moran RA, Mangine A, Bodine PV 2005 The orphan receptor tyrosine kinase Ror2 modulates canonical Wnt signaling in osteoblastic cells. Mol Endocrinol 19:90–101 [DOI] [PubMed] [Google Scholar]
- Schreiber E, Matthias P, Muller MM, Schaffner W 1989 Rapid detection of octamer binding proteins with ‘mini-extracts,’ prepared from a small number of cells. Nucleic Acids Res 17:6419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persson U, Izumi H, Souchelnytskyi S, Itoh S, Grimsby S, Engstrom U, Heldin CH, Funa K, ten Dijke P 1998 The L45 loop in type I receptors for TGF-β family members is a critical determinant in specifying Smad isoform activation. FEBS Lett 434:83–87 [DOI] [PubMed] [Google Scholar]
- Ishisaki A, Yamato K, Hashimoto S, Nakao A, Tamaki K, Nonaka K, ten Dijke P, Sugino H, Nishihara T 1999 Differential inhibition of Smad6 and Smad7 on bone morphogenetic protein- and activin-mediated growth arrest and apoptosis in B cells. J Biol Chem 274:13637–13642 [DOI] [PubMed] [Google Scholar]
- Young CS, Kitamura M, Hardy S, Kitajewski J 1998 Wnt-1 induces growth, cytosolic β-catenin, and Tcf/Lef transcriptional activation in Rat-1 fibroblasts. Mol Cell Biol 18:2474–2485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frenkel B, Capparelli C, Van Auken M, Baran D, Bryan J, Stein JL, Stein GS, Lian JB 1997 Activity of the osteocalcin promoter in skeletal sites of transgenic mice and during osteoblast differentiation in bone marrow-derived stromal cell cultures: effects of age and sex. Endocrinology 138:2109–2116 [DOI] [PubMed] [Google Scholar]
- Zhao M, Qiao M, Oyajobi BO, Mundy GR, Chen D 2003 E3 ubiquitin ligase Smurf1 mediates core-binding factor α1/Runx2 degradation and plays a specific role in osteoblast differentiation. J Biol Chem 278:27939–27944 [DOI] [PubMed] [Google Scholar]
- Spinella-Jaegle S, Roman-Roman S, Faucheu C, Dunn FW, Kawai S, Gallea S, Stiot V, Blanchet AM, Courtois B, Baron R, Rawadi G 2001 Opposite effects of bone morphogenetic protein-2 and transforming growth factor-β1 on osteoblast differentiation. Bone 29:323–330 [DOI] [PubMed] [Google Scholar]
- Winkler DG, Sutherland MS, Ojala E, Turcott E, Geoghegan JC, Shpektor D, Skonier JE, Yu C, Latham JA 2005 Sclerostin inhibition of Wnt-3a-induced C3H10T1/2 cell differentiation is indirect and mediated by bone morphogenetic proteins. J Biol Chem 280:2498–2502 [DOI] [PubMed] [Google Scholar]
- Morvan F, Boulukos K, Clement-Lacroix P, Roman RS, Suc-Royer I, Vayssiere B, Ammann P, Martin P, Pinho S, Pognonec P, Mollat P, Niehrs C, Baron R, Rawadi G 2006 Deletion of a single allele of the Dkk1 gene leads to an increase in bone formation and bone mass. J Bone Miner Res 21:934–945 [DOI] [PubMed] [Google Scholar]
- Nishida T, Nakanishi T, Asano M, Shimo T, Takigawa M 2000 Effects of CTGF/Hcs24, a hypertrophic chondrocyte-specific gene product, on the proliferation and differentiation of osteoblastic cells in vitro. J Cell Physiol 184:197–206 [DOI] [PubMed] [Google Scholar]
- Safadi FF, Xu J, Smock SL, Kanaan RA, Selim AH, Odgren PR, Marks Jr SC, Owen TA, Popoff SN 2003 Expression of connective tissue growth factor in bone: its role in osteoblast proliferation and differentiation in vitro and bone formation in vivo. J Cell Physiol 196:51–62 [DOI] [PubMed] [Google Scholar]
- Gazzerro E, Deregowski V, Vaira S, Canalis E 2005 Overexpression of twisted gastrulation inhibits bone morphogenetic protein action and prevents osteoblast cell differentiation in vitro. Endocrinology 146:3875–3882 [DOI] [PubMed] [Google Scholar]
- Gazzerro E, Deregowski V, Stadmeyer L, Gale NW, Economides AN, Canalis E 2006 Twisted gastrulation, a bone morphogenetic protein agonist/antagonist, is not required for post-natal skeletal function. Bone 39:1252–1260 [DOI] [PubMed] [Google Scholar]
- Ross JJ, Shimmi O, Vilmos P, Petryk A, Kim H, Gaudenz K, Hermanson S, Ekker SC, O'Connor MB, Marsh JL 2001 Twisted gastrulation is a conserved extracellular BMP antagonist. Nature 410:479–483 [DOI] [PubMed] [Google Scholar]
- Chang C, Holtzman DA, Chau S, Chickering T, Woolf EA, Holmgren LM, Bodorova J, Gearing DP, Holmes WE, Brivanlou AH 2001 Twisted gastrulation can function as a BMP antagonist. Nature 410:483–487 [DOI] [PubMed] [Google Scholar]
- Nosaka T, Morita S, Kitamura H, Nakajima H, Shibata F, Morikawa Y, Kataoka Y, Ebihara Y, Kawashima T, Itoh T, Ozaki K, Senba E, Tsuji K, Makishima F, Yoshida N, Kitamura T 2003 Mammalian twisted gastrulation is essential for skeleto-lymphogenesis. Mol Cell Biol 23:2969–2980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smerdel-Ramoya A, Zanotti S, Deregowski V, Canalis E Connective tissue growth factor (CTGF) enhances osteoblastogenesis in vitro. J Biol Chem, in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koga T, Matsui Y, Asagiri M, Kodama T, de Crombrugghe B, Nakashima K, Takayanagi H 2005 NFAT and Osterix cooperatively regulate bone formation. Nat Med 11:880–885 [DOI] [PubMed] [Google Scholar]
- Sun L, Blair HC, Peng Y, Zaidi N, Adebanjo OA, Wu XB, Wu XY, Iqbal J, Epstein S, Abe E, Moonga BS, Zaidi M 2005 Calcineurin regulates bone formation by the osteoblast. Proc Natl Acad Sci USA 102:17130–17135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mammucari C, Tommasi d, V, Sharov AA, Neilson J, Havrda MC, Roop DR, Botchkarev VA, Crabtree GR, Dotto GP 2005 Integration of Notch 1 and calcineurin/NFAT signaling pathways in keratinocyte growth and differentiation control. Dev Cell 8:665–676 [DOI] [PubMed] [Google Scholar]
- Winslow MM, Pan M, Starbuck M, Gallo EM, Deng L, Karsenty G, Crabtree GR 2006 Calcineurin/NFAT signaling in osteoblasts regulates bone mass. Dev Cell 10:771–782 [DOI] [PubMed] [Google Scholar]
- Ikeda F, Nishimura R, Matsubara T, Hata K, Reddy SV, Yoneda T 2006 Activation of NFAT signal in vivo leads to osteopenia associated with increased osteoclastogenesis and bone-resorbing activity. J Immunol 177: 2384–2390 [DOI] [PubMed] [Google Scholar]
- Babic AM, Chen CC, Lau LF 1999 Fisp12/mouse connective tissue growth factor mediates endothelial cell adhesion and migration through integrin alphavbeta3, promotes endothelial cell survival, and induces angiogenesis in vivo. Mol Cell Biol 19:2958–2966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimo T, Nakanishi T, Nishida T, Asano M, Kanyama M, Kuboki T, Tamatani T, Tezuka K, Takemura M, Matsumura T, Takigawa M 1999 Connective tissue growth factor induces the proliferation, migration, and tube formation of vascular endothelial cells in vitro, and angiogenesis in vivo. J Biochem 126:137–145 [DOI] [PubMed] [Google Scholar]
- Kuiper EJ, Roestenberg P, Ehlken C, Lambert V, van Treslong-de Groot HB, Lyons KM, Agostini HJ, Rakic JM, Klaassen I, Van Noorden CJ, Goldschmeding R, Schlingemann RO 2007 Angiogenesis is not impaired in connective tissue growth factor (CTGF) knock-out mice. J Histochem Cytochem 55:1139–1147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo S, Tanaka N, Kubota S, Mukudai Y, Yosimichi G, Sugahara T, Takigawa M 2006 Novel angiogenic inhibitor DN-9693 that inhibits post-transcriptional induction of connective tissue growth factor (CTGF/CCN2) by vascular endothelial growth factor in human endothelial cells. Mol Cancer Ther 5:129–137 [DOI] [PubMed] [Google Scholar]
- Kondo S, Kubota S, Shimo T, Nishida T, Yosimichi G, Eguchi T, Sugahara T, Takigawa M 2002 Connective tissue growth factor increased by hypoxia may initiate angiogenesis in collaboration with matrix metalloproteinases. Carcinogenesis 23:769–776 [DOI] [PubMed] [Google Scholar]
- Inoki I, Shiomi T, Hashimoto G, Enomoto H, Nakamura H, Makino K, Ikeda E, Takata S, Kobayashi K, Okada Y 2002 Connective tissue growth factor binds vascular endothelial growth factor (VEGF) and inhibits VEGF-induced angiogenesis. FASEB J 16:219–221 [DOI] [PubMed] [Google Scholar]
- Chang CC, Lin MT, Lin BR, Jeng YM, Chen ST, Chu CY, Chen RJ, Chang KJ, Yang PC, Kuo ML 2006 Effect of connective tissue growth factor on hypoxia-inducible factor 1α degradation and tumor angiogenesis. J Natl Cancer Inst 98:984–995 [DOI] [PubMed] [Google Scholar]
- Dean RA, Butler GS, Hamma-Kourbali Y, Delbe J, Brigstock DR, Courty J, Overall CM 2007 Identification of candidate angiogenic inhibitors processed by matrix metalloproteinase 2 (MMP-2) in cell-based proteomic screens: disruption of vascular endothelial growth factor (VEGF)/heparin affin regulatory peptide (pleiotrophin) and VEGF/Connective tissue growth factor angiogenic inhibitory complexes by MMP-2 proteolysis. Mol Cell Biol 27:8454–8465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimaio TA, Wang S, Huang Q, Scheef EA, Sorenson CM, Sheibani N 2008 Attenuation of retinal vascular development and neovascularization in PECAM-1-deficient mice. Dev Biol 315:72–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman PJ 1997 The biology of PECAM-1. J Clin Invest 100:S25–S29 [PubMed] [Google Scholar]