Abstract
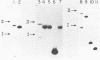
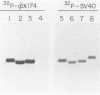
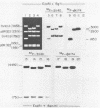
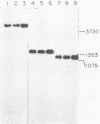
Three simian virus (SV40)-phi X174 recombinant genomes were isolated from single BSC-1 monkey cells cotransfected with SV40 and phi X174 RF1 DNAs. The individual cell progenies were amplified, cloned, and mapped by a combination of restriction endonuclease and heteroduplex analyses. In each case, the 600 to 1,000 base pairs of phi X174 DNA (derived from different regions of the phi X174 genome) were present as single inserts, located in either the early or late SV40 regions; the deletion of SV40 DNA was greater than the size of the insert; and the remaining portions of the hybrid genome were indistinguishable from wild-type SV40 DNA, as judged by both mapping and biological tests. Hence, apart from the deletion which accommodates the phi X174 DNA insert, no other rearrangements of SV40 DNA were detected. The restriction map of a SV40-phi X174 recombinant DNA isolate before molecular cloning was indistinguishable from those of two separate cloned derivatives of that isolate, indicating that the species cloned was the major amplifiable recombinant structure generated by a single recombinant-producing cell. The relative simplicity of the SV40-phi X174 recombinant DNA examined is consistent with the notion that most recombinant-producing BSC-1 cells support single recombination events generating only one amplifiable recombinant structure.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Brack C. DNA electron microscopy. CRC Crit Rev Biochem. 1981;10(2):113–169. doi: 10.3109/10409238109114551. [DOI] [PubMed] [Google Scholar]
- Dorsett D. L., Keshet I., Winocour E. Quantitation of a simian virus 40 nonhomologous recombination pathway. J Virol. 1983 Oct;48(1):218–228. doi: 10.1128/jvi.48.1.218-228.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gluzman Y. SV40-transformed simian cells support the replication of early SV40 mutants. Cell. 1981 Jan;23(1):175–182. doi: 10.1016/0092-8674(81)90282-8. [DOI] [PubMed] [Google Scholar]
- Kelly T. J., Jr, Nathans D. The genome of simian virus 40. Adv Virus Res. 1977;21:85–173. doi: 10.1016/s0065-3527(08)60762-9. [DOI] [PubMed] [Google Scholar]
- Oren M., Kuff E. L., Winocour E. The presence of common host sequences in different populations of substituted SV40 DNA. Virology. 1976 Sep;73(2):419–430. doi: 10.1016/0042-6822(76)90403-7. [DOI] [PubMed] [Google Scholar]
- Oren M., Lavi S., Winocour E. The structure of a cloned substituted SV40 genome. Virology. 1978 Apr;85(2):404–421. doi: 10.1016/0042-6822(78)90448-8. [DOI] [PubMed] [Google Scholar]
- Rigby P. W., Dieckmann M., Rhodes C., Berg P. Labeling deoxyribonucleic acid to high specific activity in vitro by nick translation with DNA polymerase I. J Mol Biol. 1977 Jun 15;113(1):237–251. doi: 10.1016/0022-2836(77)90052-3. [DOI] [PubMed] [Google Scholar]
- Smith G. E., Summers M. D. The bidirectional transfer of DNA and RNA to nitrocellulose or diazobenzyloxymethyl-paper. Anal Biochem. 1980 Nov 15;109(1):123–129. doi: 10.1016/0003-2697(80)90019-6. [DOI] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Villarreal L. P., Berg P. Hybridization in situ of SV40 plaques: detection of recombinant SV40 virus carrying specific sequences of nonviral DNA. Science. 1977 Apr 8;196(4286):183–185. doi: 10.1126/science.191907. [DOI] [PubMed] [Google Scholar]
- Vogelstein B., Gillespie D. Preparative and analytical purification of DNA from agarose. Proc Natl Acad Sci U S A. 1979 Feb;76(2):615–619. doi: 10.1073/pnas.76.2.615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winocour E., Keshet I. Indiscriminate recombination in simian virus 40-infected monkey cells. Proc Natl Acad Sci U S A. 1980 Aug;77(8):4861–4865. doi: 10.1073/pnas.77.8.4861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winocour E., Singer M., Kuff E. Rapid detection, isolation, and amplification of host-substituted SV40 variants. Cold Spring Harb Symp Quant Biol. 1980;44(Pt 1):621–628. doi: 10.1101/sqb.1980.044.01.065. [DOI] [PubMed] [Google Scholar]