Abstract
In an effort to identify novel endo-α-N-acetylgalact- osaminidases (endo-α-GalNAcases), four potential genes were cloned. Three of the expressed proteins EngEF from Enterococcus faecalis, EngPA from Propionibacterium acnes, and EngCP from Clostridium perfringens were purified and characterized. Their substrate specificity was investigated and compared to the commercially available endo-α-GalNAcases from Streptococcus pneumoniae (EngSP) and Alcaligenes sp. (EngAL). All enzymes were incubated with various synthetic substrates, and natural glycoproteins and the released sugars were detected by colorimetric assay and thin layer chromatography analysis. The Core 1 disaccharide Galβ1,3GalNAcα1pNP was the most rapidly hydrolyzed substrate by all enzymes tested. EngEF exhibited the highest kcat for this substrate. EngEF and EngPA were also able to fully hydrolyze the Core 3 disaccharide GlcNAcβ1,3GalNAcα1pNP. This is the first report of endo-α-GalNAcases EngEF and EngPA acting on Core 3 in addition to Core 1 O-glycans. Interestingly, there were no significant differences in transglycosylation activities when Galβ1,3GalNAcα1pNP or GlcNAcβ1,3GalNAcα1pNP was incubated with various 1-alkanols in the presence of the endo-α-GalNAcases tested in this work.
Keywords: deglycosylation, endo-α-N-acetylgalactosaminidases, O-glycosylation
Introduction
Glycosylation is a common posttranslational modification of proteins. Glycans are implicated in a wide range of biological events such as cell–cell interactions and recognition, inflammation, and autoimmune diseases (Varki 1993; Ohtsubo and Marth 2006). Detailed knowledge of the glycan structure helps to correlate them to their respective function. To do so, tools are required for highly sensitive analysis of glycan chains. For structural analysis of asparagine-linked carbohydrates (N-linked glycans), sugars are released from the protein backbone by enzymes such as PNGase F (Tarentino and Plummer 1994). The O-linked glycans are most commonly attached to serine or threonine residues through the GalNAc residue at the reducing end. Presently, there is no enzymatic way of releasing of O-glycans intact. This is achieved by chemical methods, typically by β-elimination with mild alkali (Kakehi et al. 1994) or mild hydrazinolysis (Royle et al. 2002). These chemical methods are limited because after this treatment the protein is no longer functional.
Endo-α-N-acetylgalactosaminidase (endo-α-GalNAcase, EC 3.2.1.97) catalyzes the hydrolysis of an O-glycosidic α-linkage between galactosyl β1,3 N-acetyl-D-galactosamine (Galβ1,3GalNAc) and the serine or threonine residue in mucins and mucin-type glycoproteins from various animal sources. This O-linked disaccharide (Core 1 type O-glycan) is one of the most abundant core structures found in mucin glycoproteins. It is known as the Thomsen– Friedenreich antigen (T antigen) immunodeterminant group and is used as a specific marker of carcinoma (Varki 1993; Ohtsubo and Marth 2006).
Endo-α-GalNAcases have been purified from Clostridium perfringens (Huang and Aminoff 1972), Streptococcus pneumoniae (Glasgow et al. 1977; Umemoto et al. 1977; Brooks and Savage 1997), Alcaligenes sp. (Fan et al. 1990), Bacillus sp. (Ashida et al. 2000), and Bifidobacterium longum (Fujita et al. 2005). All of these enzymes have a strict substrate specificity, acting only on the α-linked disaccharide, Galβ1,3GalNAc. An enzyme isolated from Streptomyces sp., has been reported to release longer sugar chains than the disaccharide from porcine mucin (Ishii-Karakasa et al. 1992, 1997) although further studies are needed to confirm this activity. These endo-α-GalNAcases liberate O-linked oligosaccharides from glycoproteins without damaging the protein backbone. This makes these enzymes powerful tools for the investigation of the structure and function of O-glycans.
The first gene encoding endo-α-GalNAcase was isolated from B. longum JCM1217 (engBF) (Fujita et al. 2005). In an effort to identify endo-α-GalNAcases with broader substrate specificity a BLAST search was done (Altschul et al. 1997) using the EngBF protein sequence as a template. Four potential endo-α-GalNAcases were selected from the BLAST results and their genes were cloned; the expressed proteins were purified and characterized. Two of these enzymes can release Core 1 and Core 3 type O-glycans. This is the first report of these enzymes exhibiting this broader substrate specificity.
Results
Identification and selection of the putative endo-α-N-acetylgalactosaminidases
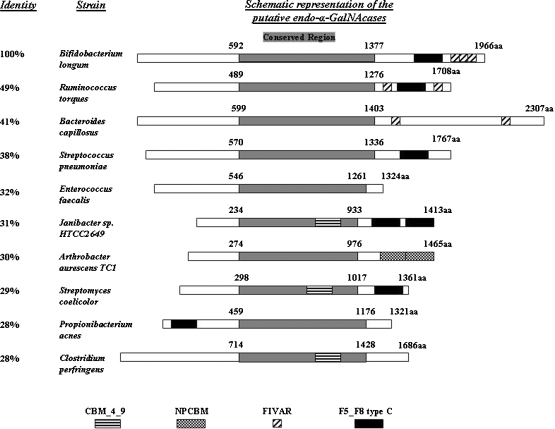
In order to identify putative endo-α-GalNAcases, a BLAST (Altschul et al. 1997) search was run using the endo-α-GalNAcase protein sequence EngBF from B. longum JCM1217. The top nine hits were from bacteria and had 28% and higher overall sequence identity with EngBF (Figure 1). The predicted molecular mass of EngSP corresponds to that previously characterized from S. pneumoniae (Brooks and Savage 1997; Glasgow et al. 1977; Umemoto et al. 1977). All of the putative endo-α-GalNAcases had a centrally located conserved region of 700–800 amino acid residues (Figure 1). Using the Pfam database (Finn et al. 2006) to analyze the protein sequences, all sequences with the exception of EngEF from Enterococcus faecalis contained predicted domains that are presumably involved with the identification or binding of sugars (sugar binding domain (SBD)). To determine if the existence or the positioning of the SBDs could play a role on the substrate specificity of this family of enzymes, it was decided to clone and characterize four of these proteins each with a different organization. EngEF has no predicted SBD. EngPA from Propionibacterium acnes has an F5_F8 type C domain (a member of the galactose-binding domain-like superfamily) close to the N-terminus. EngAA from Arthrobacter aurescens has two novel putative carbohydrate binding module (NPCBM) domains at the C-terminus. EngCP from C. perfringens has a CBM 4_9 domain (carbohydrate binding module) inside the conserved region. All of the other potential endo-α-GalNacases as well as the already characterized enzymes EngBF and EngSP have SBDs at the C-terminus or inside the conserved region (Figure 1).
Fig. 1.
Schematic representation of the putative endo-α-GalNAcases. Alignment of the putative endo-α-GalNAcases that were revealed by BLAST search against the protein sequence of EngBF and the identity results are shown. The gray shaded boxes correspond to the centrally located conserved region, while the black and patterned boxes indicate different types of SBDs. CBM_4_9, carbohydrate binding module (Pfam 02018); NPCBM, novel putative carbohydrate binding module (Pfam 08305); FIVAR, uncharacterized sugar-binding domain (Pfam 07554); F5-F8 type C, member of the galactose-binding domain-like superfamily (Pfam 00754). The protein accession numbers are B. longum (ABY836679), R. torques (ZP_01966813), B. capillosus (ZP_02035456), S. pneumoniae (YP_873926.1), E. faecalis (NP_815498.1), Janibacter sp. (ZP_00993766.1), A. aurescens (YP_947239.1), S. coelicolor (NP_630440.1), P. acnes (YP_056270.1), and C. perfrigens (YP_695137.1).
Cloning end expression of the target endo-α-GalNAcase genes
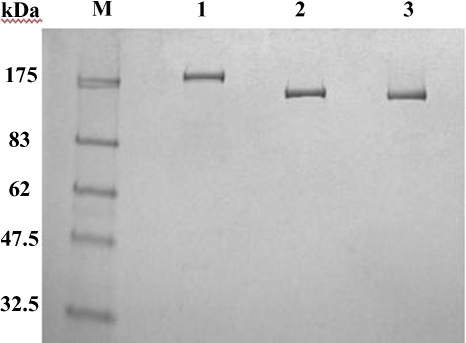
Oligonucliotide primers (Table I) were designed to amplify the genes engEF, engCP, and engAA from the genomic DNA of E. faecalis ATCC 700802, C. perfringens ATCC 13124, and A. aurescens TCI. Following polymerase chain reaction (PCR), the amplified genes were cloned in frame into the pET21a expression vector. Unlike engEF and engCP, expression of engAA was very low. The very weak endo-α-GalNAcase activity of EngAA could be detected using Galβ1,3GalNAcα1pNP as a substrate but only after an extended incubation. For this reason, EngAA was dropped from this study. The gene coding the putative endo-α-GalNAcase protein engPA from P. acnes could not be amplified by PCR from the genomic DNA of P. acnes ATCC25746. To clone the ORF coding for protein EngPA, it was necessary to chemically synthesize the gene using codons optimized for gene expression in Escherichia coli. Once synthesized the gene was cloned into the expression vector pNEB206A. All three recombinant proteins were purified to apparent homogeneity by the chromatographic steps described in the experimental procedures section. The purified proteins migrated as single bands and the apparent molecular weights were in agreement with the predicted molecular masses (188,000 Da for EngCP, 147,000 Da for EngEF, 142,000 Da for EngPA) (Figure 2).
Table I.
Primers used for the cloning of the endo-α- N-acetylgalactosaminidase genes
| Primer | Sequence |
|---|---|
| AAfor-NdeI | 5′-CCCATATGCCCCGCTTGTCATCCC-3′ |
| AArev-HindIII | 5′-CCAAGCTTTGCGCAGCTGAACTTCGCG-3′ |
| EFfor-NdeI | 5′-CCCATATGAAACATGGAAAAATAAAACG- |
| ATTTAGTAC-3′ | |
| EFrev-XhoI | 5′-CCCTCGAGTTTTTTTGATTCCCCACTGTG- |
| ACCGTAAAG-3′ | |
| CPfor-NdeI | 5′-CCCATATGGGTAGAAAATGCATGAATAA- |
| GAAGATTG-3′ | |
| CPrev-XhoI | 5′-CCCTCGAGTCTAGCAGTTCTAACAGTTATT- |
| GATTCCTTAG-3′ | |
| PAfor | 5′-GGAGACAUCCATATGAGTCGCACCC-3′ |
| PArev | 5′- GGGAAAGUTTAACGACCTTGACGTGAAAC-3′ |
Underlined are the restriction sites, while in bold are the uracil residues that are excised from the amplified DNA product by using the USERTM Friendly Cloning Kit (Bitinaite et al. 2007).
Fig. 2.
Purified endo-α-GalNAcases. SDS–PAGE analysis using 10–20% polyacrylamide gel and stained by Coomassie Brilliant Blue R-250. Lane M, molecular mass standards; lane 1, EngCP; lane 2, EngEF; and lane 3, EngPA.
Substrate specificity
The substrate specificities of the purified enzymes EngCP, EngEF, and EngPA and the commercially available EngSP and EngAL were determined. Each enzyme was incubated with various synthetic substrates. The released sugars were detected by colorimetric assay and thin layer chromatography (TLC) analysis (Table II, Figure 3). Galβ1,3GalNAcα1pNP was the most rapidly hydrolyzed substrate by all the enzymes tested. After 16 h of incubation, only EngEF and EngPA were capable of fully hydrolyzing the Core 3 disaccharide (GlcNAcβ1, 3GalNAcα1pNP). EngAL could only partially hydrolyze the Core 3 disaccharide (27%) after 16 h of incubation (Table II). EngAL could also partially release GalNAc while the rest of the enzymes released only traces of the monosaccharide (Table II). None of the enzymes could act on Galβ1,3GlcNAcα1pNP and low or no activity was detected when Core 2 trisaccharide (Galβ1,3(GlcNAcβ1,6)GalNAcα1pNP) was used as a substrate (Table II).
Table II.
Substrate specificity using pNP substrates
| EngCP | EngEF | EngPA | EngSP | EngAL | |
|---|---|---|---|---|---|
| Product released (%) | |||||
| Galβ1,3GalNAcα1pNP (Core 1) | 100 | 100 | 100 | 100 | 100 |
| Galβ1,3(GlcNAcβ1,6)GalNAcα1pNP (Core 2) | 2.5 | 2 | 0 | 0.6 | 0 |
| GlcNAcβ1,3GalNAcα1pNP (Core 3) | 6 | 100 | 100 | 3 | 27 |
| Galβ1,3GlcNAcα1pNP | 0 | 0 | 0 | 0 | 0 |
| GalNAcα1pNP | 4.4 | 2.2 | 1.8 | 1.2 | 30 |
Reaction mixtures were incubated with different endo-α-GalNAcases at 25°C for 16 h. Product release was measured at 405 nm.
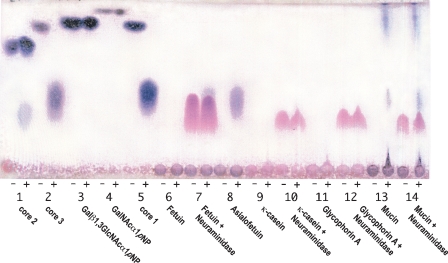
Fig. 3.
TLC analysis of the reaction products using pNP substrates and natural glycoproteins. Reaction mixtures were incubated with (lane +) and without (lane −) EngEF at 25°C for 16 h. Lane 1, Core 2 trisaccharide Galβ1,3(GlcNAcβ1,6)GalNAcα1pNP; lane 2, Core 3 disaccharide GlcNAcβ1,3GalNAcα1pNP; lane 3; Galβ1,3GlcNAcα1pNP disaccharide; lane 4, GalNAcα1pNP monosaccharide; lane 5, Core 1 disaccharide Galβ1,3GalNAcα1pNP; lane 6, Fetuin; lane 7, Fetuin + Neuraminidase; lane 8, Asialofetuin; lane 9, κ-casein glycopeptides; lane 10, κ-casein glycopeptides + Neuraminidase; lane 11, Glycophorin A; lane 12, Glycophorin A + Neuraminidase; lane 13, Mucin glycopeptides; and lane 14, Mucin glycopeptides + Neuraminidase.
The enzymes were also tested for their ability to release sugars from natural glycoproteins. Calf κ-casein, human glycophorin A, porcine mucin, calf fetuin, and calf asialofetuin were incubated with different endo-α-GalNAcases. Based on the TLC analysis, sugars were only released when asialofetuin and mucin where used as substrates (Figure 3). Coincubating the substrates with neuraminidase enabled the enzymes to release the core sugars. The released sugars migrated at the same Rf on the TLC plate as the Core 1 and 3 disaccharides (Figure 3). As expected from the pNP data, when EngAL was incubated with mucin an extra spot at the same Rf as GalNAc was observed (data not shown).
The kcat of all enzymes was determined on Gal β1,3GalNAc- α1pNP and GlcNAcβ1,3GalNAcα1pNP (Table III). When Galβ1,3GalNAcα1pNP was used as a substrate EngEF exhibited the highest kcat. EngPA had the lowest activity on that substrate with a kcat about 25 times lower than EngEF. In the case of GlcNAcβ1,3GalNAcα1pNP the kinetic parameters were determined only for EngPA and EngEF. Interestingly, EngPA had a 3-fold higher kcat than EngEF.
Table III.
Kinetic parameters of endo-α-GalNAcases using Galβ1,3GalNAcα1pNP (Core 1) and GlcNAcβ1,3GalNAcα1pNP (Core 3) as substrates
| kcat (1/s) | Km (μM) | |
|---|---|---|
| Core 1 | ||
| EngCP | 19.9 | 70.93 |
| EngEF | 51.17 | 47.85 |
| EngPA | 2.009 | 3.781 |
| EngSP | 10.51 | 40.37 |
| EngAL | 25.89 | 33.87 |
| EngBFa | 17.8 | 21.8 |
| Core 3 | ||
| EngEF | 9.434 | 20.03 |
| EngPA | 28.9 | 11.15 |
aData from Fujita et al. (2005).
Transglycosylation activity
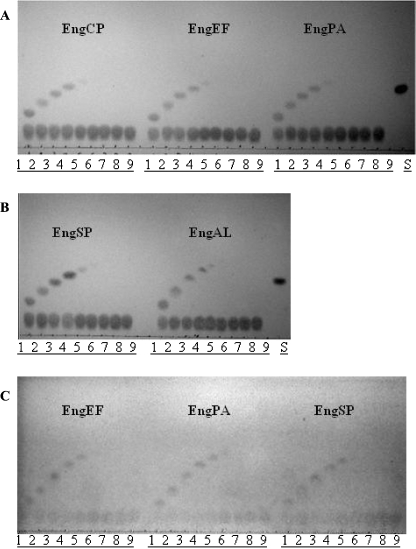
Several endoglycosidases have transglycosylation activity in addition to hydrolysis activity. To test for this activity, Galβ1,3GalNAcα1pNP was incubated with various 1-alkanols in the presence of EngCP, EngEF, EngPA, EngSP, and EngAL. Reaction products were analyzed on TLC plates (Figure 4). All of the enzymes tested exhibited similar transglycosylation activity. The longest 1-alkanol successfully incorporated in these transglycosylation reactions was 1-pentanol though the level of product was very low (Figure 4A and B). EngEF, EngPA, and EngSP were also tested for transglycosylation activity using the disaccharide GlcNAcβ1,3GalNAcα1pNP as the donor and 1-alkanols again as the acceptors. Since only EngEF and EngPA were capable of fully hydrolyzing GlcNAcβ1,3GalNAcα1pNP compared to the rest of the endo-α-GalNAcases which had very low activity on this substrate (Table II), it was surprising to find no significant difference in the amount of transglycosylation products produced by all three enzymes when observed on a TLC (Figure 4C).
Fig. 4.
TLC analysis of the transglycosylation reactions. Transglycosylation of disaccharide Galβ1,3GalNAcα1pNP (A, B) or GlcNAcβ1,3GalNAcα1pNP (C) to various 1-alkanols by endo-α-GalNAcases. Lane 1, methanol; lane 2, ethanol; lane 3, 1-propanol; lane 4, 1-butanol; lane 5, 1-pentanol; lane 6, 1-hexanol; lane 7, 1-heptanol; lane 8, 1-octanol; lane 9, 1-nonalol; and lane S, Galβ1,3GalNAcα1pNP.
Discussion
This is the first report presenting data about the substrate specificity of all available endo-α-GalNAcases. Three novel endo-α-GalNAcases: EngEF from E. faecalis, EngPA from P. acnes, and EngCP from C. perfringens have been cloned, purified, and characterized. The commercially available endo-α-GalNAcases from S. pneumoniae (EngSP) and Alcaligenes sp. (EngAL) have also been tested. Data from a previous report on EngBF from B. longum (Fujita et al. 2005) have also been included. All six of these enzymes were able to completely hydrolyze Core 1 disaccharide Galβ1,3GalNAcα1pNP. None of them could act on Galβ1,3GlcNAcα1pNP showing that there is a strict preference for GalNAc as the sugar participating in the O-glycosidic linkage. EngEF exhibited the highest kcat on Galβ1,3GalNAcα1pNP while EngPA had the lowest.
EngEF and EngPA were the only enzymes in this study able to fully hydrolyze the Core 3 disaccharide (GlcNAcβ1,3GalNAcα1pNP). EngAL could partially hydrolyze it (Table II) while EngCP, EngSP, and EngBL had even lower activity on this substrate (less than 6%). Interestingly, EngPA was more active on the Core 3 substrate than EngEF contrary to the results on the Core 1 disaccharide (Table III). A detailed mutagenesis study will help in identifying the functional residues responsible for the ability of EngEF and EngPA to act on Core 3 disaccharides.
Unfortunately, none of the enzymes could act on natural glycoproteins prior to treatment with neuraminidase. A major challenge would be to design an endo-α-GalNAcase that could act on sialyated O-glycans.
All the enzymes were tested for their transglycosylation activity. When the enzymes were incubated in the presence of Galβ1,3GalNAcα1pNP and several 1-alkanols, we observed no major differences in the amount of transglycosylation products produced (Figure 4A and B). EngEF, EngPA, and EngSP were also tested for transglycosylation activity using GlcNAcβ1,3GalNAcα1pNP as a donor. Interestingly, the same transglycosylation activity profile for these three enzymes was observed with this donor (Figure 4C) even though only EngEF and EngPA are capable of fully hydrolyzing GlcNAcβ1,3GalNAcα1pNP in contrast to EngSP which is significantly less active on this substrate.
After submission of this manuscript, a characterization of EngBF from B. longum and EngCP from C. perfringens was published (Ashida et al. 2008). Their results for the substrate specificity of EngCP agreed closely with the substrate specificity we had observed for EngCP.
In this study, three endo-α-GalNAcases were cloned, purified, and characterized. This is the first report of these endo-α-GalNAcases EngEF and EngPA acting on Core 3 in addition to Core 1 O-glycans. This property could make these enzymes a powerful tool for the release of O-glycan sugars from glycoproteins. They can also be used as templates in future protein engineering experiments toward the creation of endo-α-GalNAcases capable of acting on O-linked glycans regardless of their sugar composition.
Materials and methods
Cloning and expression of the endo-α-GalNAcase genes
Based on the DNA sequence of AAO81568 from E. faecalis, YP_695137.1 from C. perfringens, and YP_947239.1 from A. aurescens, oligonucleotide primer pairs EFfor-NdeI/EFrev-XhoI, CPfor-NdeI/CPrev-XhoI, and AAfor-NdeI/AArev-HindIII were designed (Table I). The putative endo-α-GalNAcase genes were amplified by PCR using these primer pairs from the genomic DNA of E. faecalis ATCC 700802, C. perfringens ATCC 13124, and A. aurescens TC1, respectively. The amplified genes (engEF, engCP, and engAA) were digested with the appropriate restriction enzymes (New England Biolabs, Inc., Ipswich, MA) and inserted into the corresponding sites of pET-21a (Novagen, Madison, WI). The resulting plasmids (pET21a-engEF, pET21a-engCP, and pET21a-engAA) were transformed into E. coli T7 Express lysY (New England Biolabs, Inc., Ipswich, MA). The transformed cells were grown overnight at 30°C in a 20 mL LB medium containing 100 μg/mL ampicillin. These transformations were used to inoculate 1000 mL of fresh medium and antibiotics and grown at 25°C. After these cultures reached an A600 of 0.6–0.7, 0.3 mM isopropyl thio-β-D-galactopyranoside was added and the cultures were shifted to 20°C. Incubation was continued for 12–14 h. The cells were harvested by centrifugation. The cell pellet was suspended in a final volume of 15 mL of 20 mM Tris–HCl, pH 7.6, 200 mM NaCl, 1 mM dithiothreitol. The resuspended cells were sonicated for eight 20-s bursts at 50% duty cycle using a Sonicator Ultrasonic processor model-375 (Misonix, Farmingdale, NY). Samples were subsequently centrifuged and the supernatant was collected for further purification. All purification steps were carried out at 4°C. All columns used for protein purification were purchased from GE Healthcare (Piscataway, NJ).
Based on the DNA sequence of YP_056270.1 from P. acnes, the putative endo-α-GalNAcase gene was chemically synthesized. The codons were optimized for gene expression in E. coli using DNAWorks software (Hoover and Lubkowski 2002). The optimized sequence was divided into six building blocks and synthesized (Hoover and Lubkowski 2002). After the sequence of each block was verified, the full sized gene was assembled using the USER method (Bitinaite et al. 2007). Once assembled the synthesized engPA gene was amplified by PCR using the primer pair PAfor/PArev (Table I) and inserted into the corresponding sites of pNEB206A using the USERTM Friendly Cloning Kit (New England Biolabs, Inc., Ipswich, MA) (Bitinaite et al. 2007). The resulting plasmid pNEB206A-engPA was transformed, expressed, and lysed as previously described for other endo-α-GalNAcase genes.
EngEF purification
The cell extract was diluted 3-fold with a buffer containing 20 mM Tris–HCl, pH 7.6 (buffer A) and loaded onto a HiTrap Q HP column that had been previously equilibrated in buffer A. After the column was washed with five column volumes of buffer A, the enzyme was eluted with a linear gradient of 0–1 M NaCl in buffer A. Fractions with the enzyme eluted from 0.2 to 0.75 M NaCl. This was pooled and loaded onto a HisTrap HP column previously equilibrated with buffer B (20 mM Tris–HCl, pH 7.6, 500 mM NaCl). After the column was washed with five column volumes of buffer B, the enzyme was eluted with a linear gradient of 0–0.5 M imidazole in buffer B. The peak of enzyme activity eluted at an imidazole concentration range of 0.06–0.27 M. These fractions were combined, dialyzed overnight against buffer A, and loaded onto a Source 15Q column equilibrated in buffer A. After the column was washed with five column volumes of buffer A, the proteins were eluted with a linear gradient of 0.1–0.23 M NaCl in buffer A. Active EngEF eluted from 0.14 to 0.18 M NaCl. These fractions were combined and concentrated with a Centricon Concentrator 10 (Millipore, Billerica, MA) to about 5 mL. Concentrated samples were loaded onto a Superdex 75 column previously equilibrated with buffer C (20 mM Tris–HCl, pH 7.6, 200 mM NaCl) and washed with buffer C. Fractions with EngEF activity were pooled and concentrated. After the addition of glycerol at 50%, the purified enzyme preparation was stored at −20°C. Enzyme purity was judged by gradient polyacrylamide gel electrophoresis under denaturing conditions (Laemmli 1970). Protein concentration was determined using Bradford's dye binding assay (Bio-Rad, Hercules, CA) (Bradford 1976). Highly purified bovine serum albumin (Thermo Fisher Scientific Inc., Pittsburgh, PA) was used as the protein standard.
EngCP purification
The cell extract was diluted 3-fold with buffer A and loaded onto a Source 15Q column previously equilibrated in the same buffer. After the column was washed with five column volumes of buffer A, the proteins were eluted with a linear gradient of 0–1 M NaCl in buffer A. The enzyme activity eluted from 0.3 to 0.45 M NaCl. These fractions were loaded onto a HisTrap HP column previously equilibrated with buffer B. After the column was washed with five column volumes of buffer B, the proteins were eluted with a linear gradient of 0–0.5 M imidazole in buffer B. The enzyme eluted from column at a concentration range of 0.12–0.19 M imidazole. These fractions were combined, dialyzed overnight against buffer D (20 mM sodium acetate, pH 6.0), and subsequently loaded onto a Source 15S column previously equilibrated in buffer D. After the column was washed with five column volumes of buffer D, the proteins were eluted with a linear gradient of 0–1 M NaCl in buffer D. Active EngCP fractions (eluting from 0.45 to 0.60 M NaCl) were combined and concentrated with a Centricon Concentrator 10 (Millipore, Billerica, MA) to about 5 mL. Concentrated samples were loaded onto a Superdex 75 column previously equilibrated with buffer E (20 mM sodium acetate, pH 6.0, 200 mM NaCl) and washed with buffer E. Fractions with EngCP activity were pooled and concentrated. After the addition of glycerol at 50%, the purified enzyme preparation was stored at −20°C.
EngPA purification
The cell extract was diluted 3-fold with buffer A and loaded onto a Source 15Q column previously equilibrated in the same buffer. After the column was washed with five column volumes of buffer A, the majority of the EngPA activity was detected in the flow through. This was dialyzed against buffer D and loaded onto a Source 15S column previously equilibrated in buffer D. The column was washed with five column volumes of buffer D and EngPA activity was eluted with a linear gradient of 0–1 M NaCl in buffer D. Active EngPA fractions (eluting from 0.31 to 0.36 M NaCl) were combined, concentrated (5 mL), and loaded onto a Superdex 75 column previously equilibrated with buffer E. The column was washed with buffer E and fractions with EngPA activity were pooled and concentrated. After the addition of glycerol at 50%, purified enzyme preparation was stored at −20°C.
Assay of endo-α-GalNAcase activity
The hydrolytic activity of the enzymes was assayed using Galβ1,3GalNAcα1pNP as a substrate. A 100 μL standard reaction mixture contained a 50 mM sodium phosphate buffer, pH 7.5, 5 mM MgCl2, and 0.25 mM substrate. The time-dependent release of p-nitrophenol (pNP) at room temperature was measured by reading the absorbance at 405 nm using a 96-well plate reader (SpectraMax M5, Molecular Devices, Inc., Sunnyvale, CA). Pure protein samples were used for the kinetic measurements. Endo-α-GalNAcases from S. pneumoniae (EngSP) and Alcaligenes sp. (EngAL) were purchased from Roche (Basel, Switzerland) and Seikagaku Corporation (Tokyo, Japan), respectively. The substrate specificity was determined using various pNP glycosides and natural glycoproteins. The substrates Galβ1,3GalNAcα1pNP (CAS 59837-14-8), GlcNAcβ1,3GalNAcα1pNP (CAS 125455-64-3), Galβ1,3GlcNAcα1pNP (CAS 57467-13-7), GalNAcα1pNP (CAS 23646-68-6), calf κ-casein, human glycophorin A, porcine mucin, calf fetuin, and calf asialofetuin were purchased from Sigma. Galβ1,3(GlcNAcβ1,6)GalNAcα1pNP (CAS 139459-55-5) was from Toronto Research Chemicals Inc. (North York, Ontario). For TLC analysis, a Silica Gel 60 plate (Merck, Whitehouse Station, NJ) was developed in a solvent system of chloroform/methanol/water, 3/3/1 (v/v/v), and the sugars were visualized by spraying a diphenylamine/aniline/phosphate reagent (Bailey and Bourne 1960).
Transglycosylation assays
The 15 μL transglycosylation reaction mixture contained a 50 mM sodium phosphate buffer, pH 7.5, 5 mM MgCl2, 0.8 mM GlcNAcβ1,3GalNAcα1pNP or 1.6 mM Galβ1,3GalNAcα1pNP as donors, 0.8 μg of endo-α-GalNAcase and various 1-alcanols as acceptors (13%, v/v). The reactions were incubated at room temperature for 16 h. Methanol, ethanol, 1-propanol, 1-butanol, 1-pentanol, 1-hexanol, 1-heptanol, 1-octanol, and 1-nonalol were purchased from Sigma (St. Louis, MO). The transglycosylation reaction mixtures were analyzed on a Silica Gel 60 TLC plate using chloroform/methanol/water 65/35/8 as the developing solvent and the sugars were visualized by spraying a diphenylamine/aniline/phosphate reagent.
Optimum pH
The pH optimum for each enzyme was determined in a pH range of 2.0–9.0 using the following buffers (50 mM): glycine–HCl (2.0–4.0), sodium acetate (3.5–6.0), sodium phosphate (5.5–8.0), and Tris–HCl (7.0–9.0).
Enzyme kinetics
Steady-state enzyme kinetics were performed at 25°C. The program HYPER v 1.01 was used to determine Vmax and Km values. The kcat values were calculated from Vmax using a molecular mass of 188,000 Da for EngCP, 147,000 Da for EngEF, 142,000 Da for EngPA, 190,000 Da for EngSP, and 160,000 Da for EngAL. Reported values are the average of three measurements. The standard deviations do not exceed 5%.
Funding
This work was supported by New England Biolabs, Inc.
Acknowledgments
The authors wish to thank Dr. Romas Vaisvila for his guidance and assistance in the gene synthesis of the EngPA and Dr. Paula Magnelli and Elizabeth McLeod for careful reading of this manuscript. Special thanks should be given to Dr. Donald Comb for support of this project.
Conflict of interest statement
This work was supported by New England Biolabs, Inc., a company that may profit from the sale of enzymes described herein. The authors are (DL, EPG) or were (DK) employees of New England Biolabs, Inc.
Abbreviations
- Endo-α-GalNAcase
endo-α-N-acetylgalactosaminidase
- Gal β1,3GalNAc
d-galactopyranosyl-β1,3N-acetyl-d-galactos- amine pyranoside
- GlcNAcβ1,3GalNAc
N-acetyl-d-glucos- aminepyranosyl-β1,3N-acetyl-d-galactosamine pyranoside
- ORF
open reading frame
- PCR
polymerase chain reaction
- pNP
p-nitrophenol
- SBD
sugar binding domain
- TLC
thin layer chromatography
References
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashida H, Maki R, Ozawa H, Tani Y, Kiyohara M, Fujita M, Imamura A, Ishida H, Kiso M, Yamamoto K. Characterization of two different endo-alpha-N-acetylgalactosaminidases from probiotic and pathognic enterobacteria, Bifidobacterium longum and Clostridium perfringens. Glycobiology. 2008 doi: 10.1093/glycob/cwn053. Advance Access, published on June 17, 2008, doi:10.1093/glycob/cwn053. [DOI] [PubMed] [Google Scholar]
- Ashida H, Yamamoto K, Murata T, Usui T, Kumagai H. Characterization of endo-alpha-N-acetylgalactosaminidase from Bacillus sp. and syntheses of neo-oligosaccharides using its transglycosylation activity. Arch Biochem Biophys. 2000;373:394–400. doi: 10.1006/abbi.1999.1565. [DOI] [PubMed] [Google Scholar]
- Bailey R, Bourne E. Colour reactions given by sugars and dipenylamine-aniline spray reagents on paper chromatograms. J Chromatogr A. 1960;4:206–213. [Google Scholar]
- Bitinaite J, Rubino M, Varma KH, Schildkraut I, Vaisvila R, Vaiskunaite R. USER friendly DNA engineering and cloning method by uracil excision. Nucleic Acids Res. 2007;35:1992–2002. doi: 10.1093/nar/gkm041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Brooks MM, Savage AV. The substrate specificity of the enzyme endo-alpha-N-acetyl-d-galactosaminidase from Diplococcus pneumonia. Glycoconj J. 1997;14:183–190. doi: 10.1023/a:1018585604073. [DOI] [PubMed] [Google Scholar]
- Fan JQ, Yamamoto K, Kumagai H, Tochikura T. Induction and efficient purification of endo-alpha-N-acetylgalactosaminidase from Alcaligenes sp. Agric Biol Chem. 1990;54:233–234. [PubMed] [Google Scholar]
- Finn RD, Mistry J, Schuster-Bockler B, Griffiths-Jones S, Hollich V, Lassmann T, Moxon S, Marshall M, Khanna A, Durbin R, Eddy SR, Sonnhammer EL, Bateman A. Pfam: Clans, web tools and services. Nucleic Acids Res. 2006;34:D247- D251. doi: 10.1093/nar/gkj149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita K, Oura F, Nagamine N, Katayama T, Hiratake J, Sakata K, Kumagai H, Yamamoto K. Identification and molecular cloning of a novel glycoside hydrolase family of core 1 type O-glycan-specific endo-alpha- N-acetylgalactosaminidase from Bifidobacterium longum. J Biol Chem. 2005;280:37415–37422. doi: 10.1074/jbc.M506874200. [DOI] [PubMed] [Google Scholar]
- Glasgow LR, Paulson JC, Hill RL. Systematic purification of five glycosidases from Streptococcus (Diplococcus) pneumoniae. J Biol Chem. 1977;252:8615–8623. [PubMed] [Google Scholar]
- Hoover DM, Lubkowski J. DNAWorks: An automated method for designing oligonucleotides for PCR-based gene synthesis. Nucleic Acids Res. 2002;30:e43. doi: 10.1093/nar/30.10.e43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang CC, Aminoff D. Enzymes that destroy blood group specificity: V. The oligosaccharase of Clostridium perfringens. J Biol Chem. 1972;247:6737–6742. [PubMed] [Google Scholar]
- Ishii-Karakasa I, Iwase H, Hotta K. Partial purification and characterization of an endo-alpha-N-acetylgalactosaminidase from the culture medium of Streptomyces sp. OH-11242. Biochem J. 1992;288(Pt 2):475–482. doi: 10.1042/bj2880475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii-Karakasa I, Iwase H, Hotta K, Tanaka Y, Omura S. Structural determination of the O-linked sialyl oligosaccharides liberated from fetuin with endo-alpha-N-acetylgalactosaminidase-S by HPLC analysis and 600-MHz 1H-NMR spectroscopy. Eur J Biochem. 1997;247:709–715. doi: 10.1111/j.1432-1033.1997.00709.x. [DOI] [PubMed] [Google Scholar]
- Kakehi K, Susami A, Taga A, Suzuki S, Honda S. High-performance capillary electrophoresis of O-glycosidically linked sialic acid-containing oligosaccharides in glycoproteins as their alditol derivatives with low-wavelength UV monitoring. J Chromatogr A. 1994;680:209–215. doi: 10.1016/0021-9673(94)80069-3. [DOI] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Ohtsubo K, Marth JD. Glycosylation in cellular mechanisms of health and disease. Cell. 2006;126:855–867. doi: 10.1016/j.cell.2006.08.019. [DOI] [PubMed] [Google Scholar]
- Royle L, Mattu TS, Hart E, Langridge JI, Merry AH, Murphy N, Harvey DJ, Dwek RA, Rudd PM. An analytical and structural database provides a strategy for sequencing O-glycans from microgram quantities of glycoproteins. Anal Biochem. 2002;304:70–90. doi: 10.1006/abio.2002.5619. [DOI] [PubMed] [Google Scholar]
- Tarentino AL, Plummer TH., Jr Enzymatic deglycosylation of asparagine-linked glycans: Purification, properties, and specificity of oligosaccharide-cleaving enzymes from Flavobacterium meningosepticum. Methods Enzymol. 1994;230:44–57. doi: 10.1016/0076-6879(94)30006-2. [DOI] [PubMed] [Google Scholar]
- Umemoto J, Bhavanandan VP, Davidson EA. Purification and properties of an endo-alpha-N-acetyl-d-galactosaminidase from Diplococcus pneumoniae. J Biol Chem. 1977;252:8609–8614. [PubMed] [Google Scholar]
- Varki A. Biological roles of oligosaccharides: All of the theories are correct. Glycobiology. 1993;3:97–130. doi: 10.1093/glycob/3.2.97. [DOI] [PMC free article] [PubMed] [Google Scholar]