Abstract
Motivation: Predicting interactions between small molecules and proteins is a crucial step to decipher many biological processes, and plays a critical role in drug discovery. When no detailed 3D structure of the protein target is available, ligand-based virtual screening allows the construction of predictive models by learning to discriminate known ligands from non-ligands. However, the accuracy of ligand-based models quickly degrades when the number of known ligands decreases, and in particular the approach is not applicable for orphan receptors with no known ligand.
Results: We propose a systematic method to predict ligand–protein interactions, even for targets with no known 3D structure and few or no known ligands. Following the recent chemogenomics trend, we adopt a cross-target view and attempt to screen the chemical space against whole families of proteins simultaneously. The lack of known ligand for a given target can then be compensated by the availability of known ligands for similar targets. We test this strategy on three important classes of drug targets, namely enzymes, G-protein-coupled receptors (GPCR) and ion channels, and report dramatic improvements in prediction accuracy over classical ligand-based virtual screening, in particular for targets with few or no known ligands.
Availability: All data and algorithms are available as Supplementary Material.
Contact: laurent.jacob@ensmp.fr
Supplementary information: Supplementary data are available at Bioinformatics online.
1 INTRODUCTION
Predicting interactions between small molecules and proteins is a key element in the drug discovery process. In particular, several classes of proteins such as G-protein-coupled receptors (GPCR), enzymes and ion channels represent a large fraction of current drug targets and important targets for new drug development (Hopkins and Groom, 2002). Understanding and predicting the interactions between small molecules and such proteins could therefore help in the discovery of new lead compounds.
Various approaches have already been developed and have proved very useful to address this in silico prediction issue (Manly et al., 2001). The classical paradigm is to predict the modulators of a given target, considering each target independently from other proteins. Usual methods are classified into ligand-based and structure-based or docking approaches. Ligand-based approaches compare a candidate ligand to the known ligands of the target to make their prediction, typically using machine learning algorithms (Butina et al., 2002; Byvatov et al., 2003) whereas structure-based approaches use the 3D-structure of the target to determine how well each candidate binds the target (Halperin et al., 2002).
Ligand-based approaches require the knowledge of sufficient ligands of a given target with respect to the complexity of the ligand/non-ligand separation to produce accurate predictors. If few or no ligands are known for a target, one is compelled to use docking approaches, which in turn require the 3D structure of the target and are very time consuming. If for a given target with unavailable 3D structure no ligand is known, none of the classical approaches can be applied. This is the case for many GPCR as very few structures have been crystallized so far (Ballesteros and Palczewski, 2001) and many of these receptors, referred to as orphan GPCR, have no known ligand.
An interesting idea to overcome this issue is to stop considering each protein target independently from other proteins, and rather take the point of view of chemogenomics (Jaroch and Weinmann, 2006; Kubinyi et al., 2004). Roughly speaking, chemogenomics aims at mining the entire chemical space, which corresponds to the set of all small molecules, for interactions with the biological space, i.e. the set of all proteins or at least protein families, in particular drug targets. A salient motivation of the chemogenomics approach is the realization that some classes of molecules can bind ‘similar’ proteins, suggesting that the knowledge of some ligands for a target can be helpful to determine ligands for similar targets. Besides, this type of method allows for a more rational approach to design drugs since controlling a whole ligand's selectivity profile is crucial to make sure that no side effect occurs and that the compound is compatible with therapeutical usage.
Recent reviews (Jaroch and Weinmann, 2006; Kubinyi et al., 2004; Klabunde, 2007; Rognan, 2007) describe several chemogenomic approaches to predict interactions between compounds and targets. A first class of approaches, called ligand-based chemogenomics by Rognan (2007), pool together targets at the level of families (such as GPCR) or subfamilies (such as purinergic GPCR) and learn a model for ligands at the level of the family (Balakin et al., 2002; Klabunde, 2006). Other approaches, termed target-based chemogenomic approaches by Rognan (2007), cluster receptors based on ligand binding site similarity and again pool together known ligands for each cluster to infer shared ligands (Frimurer et al., 2005). Finally, a third strategy termed target-ligand approach by Rognan (2007) attempts to predict ligands for a given target by leveraging binding information for other targets in a single step, that is, without first attempting to define a particular set of similar receptors. For example, Bock and Gough (2005) merge descriptors of ligands and targets to describe putative ligand–receptor complexes, and use machine learning methods to discriminate real complexes from ligand–receptor pairs that do not form complexes. Erhan et al. (2006) show how the same idea can be casted in the framework of neural networks and support vector machines (SVM). In particular, they show that a given set of receptor descriptors can be combined with a given set of ligand descriptors in a computationally efficient framework, offering in principle a large flexibility in the choice of the receptor and ligand descriptors.
In this article, we go one step further in this direction and investigate various kinds of receptor and ligand descriptors that can be combined for in silico chemogenomics screening with SVM, building on recent development in the field of kernel methods for bio- and chemoinformatics. In particular, we propose a new kernel for receptors, based on a priori defined hierarchies of receptors. We test the different methods for the prediction of ligands for three major classes of therapeutic targets, namely enzymes, GPCR and ion channels. We show that the choice of representation has a strong influence on the accuracy of the model estimated, and in particular that the new hierarchy kernel systematically outperforms other descriptors used in multitask learning or involving receptor sequences. We show that the chemogenomics approach is, particularly, relevant for targets with few known ligands. In particular we estimate that, for orphan receptors with no known ligands, our method reaches a normalized accuracy of 86.2%, 77.6% and 80.5% on the enzymes, GPCR and ion channels, respectively, well above the 50% accuracy of a random predictor that would be trained in a classical ligand-based virtual screening framework with no training example.
2 METHOD
We formulate the typical in silico chemogenomics problem as the following learning problem: given a collection of n target/molecule pairs (t1, c1), …,(tn, cn) known to form complexes or not, estimate a function f(t, c) that would predict whether any chemical c binds to any target t. In this section, we propose a rigorous and general framework to solve this problems building on recent developments of kernel methods in bio- and chemoinformatics. This approach is similar to the approaches proposed in the context of MHC-I-peptide binding prediction (Jacob and Vert, 2008) and in (Erhan et al., 2006).
2.1 From single-target screening to chemogenomics
Much effort in chemoinformatics has been devoted to the more restricted problem of mining the chemical space for interaction with a single target t, using a training set of molecules c1,…, cn known to interact or not with the target. Machine learning approaches, such as artificial neural networks (ANN) or SVM, often provide competitive models for such problems. The simplest linear models start by representing each molecule c by a vector representation Φ(c), before estimating a linear function ft(c)=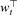 Φ(c) whose sign (positive or negative) is used to predict whether or not the small molecule c is a ligand of the target t. The weight vector wt is typically estimated based on its ability to correctly predict the classes of molecules in the training set.
Φ(c) whose sign (positive or negative) is used to predict whether or not the small molecule c is a ligand of the target t. The weight vector wt is typically estimated based on its ability to correctly predict the classes of molecules in the training set.
The in silico chemogenomics problem is more general because data involving interactions with different targets are available to train a model which must be able to predict interactions between any molecule and any protein. In order to extend the previous machine learning approaches to this setting, we need to represent a pair (t, c) of target t and chemicals c by a vector Φ(t, c), then estimate a linear function f(t, c)=w⊤Φ(t, c) whose sign is used to predict whether or not c can bind to t. As before the vector w can be estimated from the training set of interacting and non-interacting pairs, using any linear machine learning algorithm.
To summarize, we propose to cast the in silico chemogenomics problem as a learning problem in the ligand–target space thus making it suitable to any classical linear machine learning approach as soon as a vector representation Φ(t, c) is chosen for protein/ligand pairs. We propose in the next sections a systematic way to design such a representation.
2.2 Vector representation of target/ligand pairs
A large literature in chemoinformatics has been devoted to the problem of representing a molecule c by a vector Φlig(c)∈ℝdc, e.g. using various molecular descriptors (Todeschini and Consonni, 2002). These descriptors encode several features related to the physicochemical and structural properties of the molecules, and are widely used to model interactions between the small molecules and a single target using linear models described in the previous section (Gasteiger and Engel, 2003). Similarly, much work in computational biology has been devoted to the construction of descriptors for genes and proteins, in order to represent a given protein t by a vector Φtar(t)∈ℝdt. The descriptors typically capture properties of the sequence or structure of the protein, and can be used to infer models to predict, e.g. the structural or functional class of a protein.
For our in silico chemogenomics problem, we need to represent each pair (c, t) of small molecule and protein by a single vector Φ(c, t). In order to capture interactions between features of the molecule and of the protein that may be useful predictors for the interaction between c and t, we propose to consider features for the pair (c, t) obtained by multiplying a descriptor of c with a descriptor of t. Intuitively, if for example, the descriptors are binary indicators of specific structural features in each small molecule and proteins, then the product of two such features indicates that both the small molecule and the target carry specific features, which may be strongly correlated with the fact that they interact. More generally, if a molecule c is represented by a vector of descriptors Φlig(c)∈ℝdc and a target protein by a vector of descriptors Φtar(t)∈ℝdt, this suggests to represent the pair (c, t) by the set of all possible products of features of c and t, i.e. by the tensor product:
| (1) |
Remember that the tensor product in (1) is a dc × dt vector whose (i, j)-th entry is exactly the product of the i-th entry of Φlig(c) by the j-th entry of Φtar(t). This representation can be used to combine in an algorithmic way any vector representation of small molecules with any vector representation of proteins, for the purpose of in silico chemogenomics or any other task involving pairs of molecules/protein. A potential issue with this approach, however, is that the size of the vector representation for a pair may be prohibitively large for practical computation and storage. For example, using a vector of molecular descriptors of size 1024 for molecules and representing a protein by the vector of counts of all 2mers of amino acids in its sequence (dt=20 × 20=400) results in more than 400 k dimensions for the representation of a pair. In order to circumvent this issue we now show how kernel methods such as SVM can efficiently work in such large spaces.
2.3 Kernels for target/ligand pairs
SVM is an algorithm to estimate linear binary classifiers from a training set of patterns with known class (Boser et al., 1992; Vapnik, 1998). A salient feature of SVM, often referred to as the kernel trick, is its ability to process large- or even infinite-dimensional patterns as soon as the inner product between any two patterns can be efficiently computed. This property is shared by a large number of popular linear algorithms, collectively referred to as kernel methods, including for example, algorithms for regression, clustering or outlier detection (Schölkopf and Smola, 2002; Shawe-Taylor and Cristianini, 2004).
In order to apply kernel methods such as SVM for in silico chemogenomics, we therefore need to show how to efficiently compute the inner product between the vector representations of two molecule/protein pairs. Interestingly, a classical property of tensor products allows us to factorize the inner product between two tensor product vectors as follows:
| (2) |
This factorization dramatically reduces the burden of working with tensor products in large dimensions. For example, in our previous example where the dimensions of the small molecule and proteins are vectors of respective dimensions 1024 and 400, the inner product in>400 k dimensions between tensor products is simply obtained from (2) by computing two inner products, respectively in dimensions 1024 and 400, before taking their product.
Even more interestingly, this reasoning extends to the case where inner products between vector representations of small molecules and proteins can themselves be efficiently computed with the help of positive definite kernels (Vapnik, 1998), as explained in the next sections. Positive definite kernels are linked to inner products by a fundamental result (Aronszajn, 1950): the kernel between two points is equivalent to an inner product between the points mapped to a Hilbert space uniquely defined by the kernel. Now by denoting
we obtain the inner product between tensor products by:
| (3) |
In summary, as soon as two kernels Kligand and Ktarget corresponding to two implicit embeddings of the chemical and biological spaces in two Hilbert spaces are chosen, we can solve the in silico chemogenomics problem with an SVM (or any other relevant kernel method) using the product kernel (3) between pairs. The particular kernels Kligand and Ktarget should ideally encode properties related to the ability of similar molecules to bind similar targets or ligands, respectively. We review in the next two sections possible choices for such kernels.
2.4 Kernels for ligands
Recent years have witnessed impressive advances in the use of SVM in chemoinformatics (Ivanciuc, 2007). In particular, much work has focused on the development of kernels for small molecules for the purpose of single-target virtual screening and prediction of pharmacokinetics and toxicity. For example, simple inner products between vectors of classical molecular descriptors have been widely investigated, including physicochemical properties of molecules or 2D and 3D fingerprints (Azencott et al., 2007; Todeschini and Consonni, 2002). Other kernels have been designed directly from the comparison of 2D and 3D structures of molecules, including kernels based on the detection of common substructures in the 2D structures of molecules seen as graphs (Borgwardt and Kriegel, 2005; Gärtner et al., 2003; Horváth et al., 2004; Kashima, 2003; Kashima et al., 2004; Mahé and Vert, 2006; Mahé et al., 2005; Ralaivola et al., 2005) or on the encoding of various properties of the 3D structure of molecules (Azencott et al., 2007; Mahé et al., 2006).
While any of these kernels could be used to model the similarities of small molecules and be plugged into (3), we restrict ourselves in our experiment to a particular kernel proposed by Ralaivola et al. (2005) called the Tanimoto kernel, a classical choice that usually gives state-of-the-art performances in molecule classification tasks. It is defined as:
 |
(4) |
where Φlig(c) is a binary vector whose bits indicate the presence or absence of all linear path of length l or less as subgraph of the 2D structure of c. We chose l=8 in our experiment, i.e. characterize the molecules by the occurrences of linear subgraphs of length 8 or less, a value previously observed to give good results in several virtual screening task (Mahé et al., 2005). We used the freely and publicly available ChemCPP1 software to compute this kernel in the experiments.
2.5 Kernels for targets
SVM and kernel methods are also widely used in bioinformatics (Schölkopf et al., 2004), and a variety of approaches have been proposed to design kernels between proteins, ranging from kernels based on the amino-acid sequence of a protein (Cuturi and Vert, 2005; Jaakkola et al., 2000; Kuang et al., 2005; Leslie et al., 2002, 2004; Tsuda et al., 2002; Vert et al., 2004) to kernels based on the 3D structures of proteins (Borgwardt et al., 2005; Dobson and Doig, 2005; Qiu et al., 2007) or the pattern of occurrences of proteins in multiple sequenced genomes (Vert, 2002). These kernels have been used in conjunction with SVM or other kernel methods for various tasks related to structural or functional classification of proteins. While any of these kernels can theoretically be used as a target kernel in (3), we investigate in this article a restricted list of specific kernels described below, aimed at illustrating the flexibility of our framework and test various hypothesis.
- The Dirac kernel between two targets t, t′ is:
This basic kernel simply represents different targets as orthonormal vectors. From (3) we see that orthogonality between two proteins t and t′ implies orthogonality between all pairs (c, t) and (c′, t′) for any two small molecules c and c′. This means that a linear classifier for pairs (c, t) with this kernel decomposes as a set of independent linear classifiers for interactions between molecules and each target protein, which are trained without sharing any information of known ligands between different targets. In other words, using Dirac kernel for proteins amounts to performing classical learning independently for each target, which is our baseline approach.
(5) - The multitask kernel between two targets t, t′ is defined as:
This kernel, originally proposed in the context of multitask learning (Evgeniou et al., 2005), removes the orthogonality of different proteins to allow sharing of information. As explained in Evgeniou et al. (2005), plugging Kmultitask in (3) amounts to decomposing the linear function used to predict interactions as a sum of a linear function common to all targets and of a linear function specific to each target: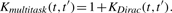
A consequence is that only data related to the target t are used to estimate the specific vector wt, while all data are used to estimate the common vector wgeneral. In our framework this classifier is therefore the combination of a target-specific part accounting for target-specific properties of the ligands and a global part accounting for general properties of the ligands across the targets. The latter term allows to share information during the learning process, while the former ensures that specificities of the ligands for each target are not lost. While the multitask kernel provides a basic framework to share information across proteins, it does not allow to weigh differently how known interactions with a protein t should contribute to predict interactions with a target t′. Empirical observations underlying chemogenomics, on the other hand, suggest that molecules binding a ligand t are only likely to bind ligand t′ similar to t in terms of structure or evolutionary history. In terms of kernels this suggest to plug into (3) a kernel for proteins that quantifies this notion of similarity between proteins, which can, for example, be detected by comparing the sequences of proteins. In order to test this approach, we therefore tested two commonly used kernels between protein sequences: the mismatch kernel (Leslie et al., 2004), which compares proteins in terms of common short sequences of amino acids up to some mismatches, and the local alignment kernel (Vert et al., 2004) which measures the similarity between proteins as an alignment score between their primary sequences. In our experiments involving the mismatch kernel, we use the classical choice of 3-mers with a maximum of one mismatch, and for the datasets where some sequences were not available in the database, we added KDirac(t, t′) to the kernel (and normalized to one on the diagonal) in order to keep it valid.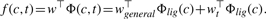
(6) -
Alternatively, we propose a new kernel aimed at encoding the similarity of proteins with respect to the ligands they bind. Indeed, for most major classes of drug targets such as the ones investigated in this study (GPCR, enzymes and ion channels), proteins have been organized into hierarchies that typically describe the precise functions of the proteins within each family. Enzymes are labeled with Enzyme Commission numbers (EC numbers) defined in (International Union of Biochemistry and Molecular Biology (1992), that classify the chemical reaction they catalyze, forming a four-level hierarchy encoded into four numbers. For example, EC;1 includes oxidoreductases, EC;1.2 includes oxidoreductases that act on the aldehyde or oxo group of donors, EC;1.2.2 is a subclass of EC;1.2 with NAD+ or NADP+ as acceptor and EC;1.2.2.1 is a subgroup of enzymes catalyzing the oxidation of formate to bicarbonate. These number define a natural and very informative hierarchy on enzymes: one can expect that enzymes that are closer in the hierarchy will tend to have more similar ligands. Similarly, GPCRs are grouped into four classes based on sequence homology and functional similarity: the rhodopsin family (class A), the secretin family (class B), the metabotropic family (class C) and a last class regrouping more diverse receptors (class D). The KEGG database (Kanehisa et al., 2002) subdivides the large rhodopsin family in three subgroups (amine receptors, peptide receptors and other receptors) and adds a second level of classification based on the type of ligands or known subdivisions. For example, the rhodopsin family with amine receptors is subdivided into cholinergic receptors, adrenergic receptors, etc. This also defines a natural hierarchy that we could use to compare GPCRs. Finally, KEGG also provides a classification of ion channels. Classification of ion channels is a less simple task since some of them can be classified according to different criteria like voltage dependence or ligand gating. The classification proposed by KEGG includes Cys-loop superfamily, glutamate-gated cation channels, epithelial and related Na+ channels, voltage-gated cation channels, related to voltage-gated cation channels, related to inward rectifier K+ channels, chloride channels and related to ATPase-linked transporters and each of these classes is further subdivided according, for example to the type of ligands (e.g. glutamate receptor) or to the type of ion passing through the channel (e.g. Na+ channel). Here again, this hierarchy can be used to define a meaningful similarity in terms of interaction behavior.
For each of the three target families, we define the hierarchy kernel between two targets of the family as the number of common ancestors in the corresponding hierarchy plus one, that is,
where Φh(t) contains as many features as there are nodes in the hierarchy, each being set to 1 if the corresponding node is part of t's hierarchy and 0 otherwise, plus one feature constantly set to one that accounts for the ‘plus one’ term of the kernel. One might not expect the EC classification to be a good similarity measure in terms of binding, since it does not closely reflect evolutionary or mechanistic similarities except for the case of identical subclasses with different serial numbers. However, using the full hierarchy gave a better accuracy in our experiments. Even if the hierarchy itself is not fully relevant in this case, the improvement can be explained, on the one hand, by the multitask effect, i.e. by the fact that we use the data from the target and the data from other targets with a smaller weight, and on the other hand, by the fact that we give more weight to the enzymes with the same serial number than to the other enzymes.
3 DATA
We extracted compound interaction data from the KEGG BRITE Database (Kanehisa et al., 2002, 2004) concerning enzyme, GPCR and ion channel, three target classes particularly relevant for novel drug development.
For each family, the database provides a list of known compounds for each target. Depending on the target families, various categories of compounds are defined to indicate the type of interaction between each target and each compound. These are, for example, inhibitor, cofactor and effector for enzyme ligands, antagonist or (full/partial) agonist for GPCR and pore blocker, (positive/negative) allosteric modulator, agonist or antagonist for ion channels. The list is not exhaustive for the latter since numerous categories exist. Although different types of interactions on a given target might correspond to different binding sites, it is theoretically possible for a non-linear classifier like SVM with non-linear kernels to learn classes consisting of several disconnected sets. Therefore, for the sake of clarity of our analysis, we do not differentiate between the categories of compounds.
For each target class, we retained only one protein by element of the hierarchy. In particular, we did not take into account the different orthologs of the targets, and the different enzymes corresponding to the same EC number. We then eliminated all compounds for which no molecular descriptor was available (principally peptide compounds), and all the targets for which no compound was known. For each target, we generated as many negative ligand–target pairs as we had known ligands forming positive pairs by combining the target with a ligand randomly chosen among the other targets' ligands (excluding those that were known to interact with the given target). This protocol generates false negative data since some ligands could actually interact with the target although they have not been experimentally tested, and our method could benefit from experimentally confirmed negative pairs.
This resulted in 2436 data points for enzymes (1218 known enzyme–ligand pairs and 1218 generated negative points) representing interactions between 675 enzymes and 524 compounds, 798 training data points for GPCRs representing interactions between 100 receptors and 219 compounds and 2330 ion channel data points representing interactions between 114 channels and 462 compounds. Besides, Figure 1 shows the distribution of the number of known ligands per target for each dataset and illustrates the fact that for most of them, few compounds are known.
Fig. 1.
Distribution of the number of training points for a target for the enzymes, GPCR and ion channel datasets. Each bar indicates the proportion of targets in the family for which a given (x-axis) number of data points is available.
For each target t in each family, we carried out two experiments. First, all data points corresponding to other targets in the family were used for training only and the nt points corresponding to t were k-folded with k=min(nt, 10). That is, for each fold, an SVM classifier was trained on all points involving other targets of the family plus a fraction of the points involving t, then the performances of the classifier were tested on the remaining fraction of data points for t. This protocol is intended to assess the incidence of using ligands from other targets on the accuracy of the learned classifier for a given target. Second, for each target t we trained an SVM classifier using only interactions that did not involve t and tested on the points that involved t. This is intended to simulate the behavior of our framework when making predictions for orphan targets, i.e. for targets for which no ligand is known.
For both experiments, we used the area under the ROC curve (AUC) as a performance measure. The ROC curve was computed for each target using the test points pooled from all the folds. For the first protocol, since training an SVM with only one training point does not really make sense and can lead to ‘anti-learning’ less than 0.5 performances, we set all results r involving the Dirac target kernel on targets with only one known ligand to max(r, 0.5). This is to avoid any artifactual penalization of the Dirac approach and make sure we measure the actual improvement brought by sharing information across targets.
4 RESULTS
We first discuss the results obtained on the three datasets for the first experiment, assessing how using training points from other targets of the family improves prediction accuracy with respect to individual (Dirac-based) learning. Table 1 shows the mean AUC across the family targets for an SVM with a product kernel using the Tanimoto kernel for ligands and various kernels for proteins. For the enzymes and ion channels datasets, we observe significant improvements when the multitask kernel is used in place of the Dirac kernel, on the one hand, and when the hierarchy kernel replaces the multitask kernel, on the other hand. For example, the Dirac kernel only performs at an average AUC of 77% for the ion channel dataset, while the multitask kernel increases the AUC to87.3% and the hierarchy kernel brings it to 92.5%. For the enzymes, a global improvement of 30.9% is observed between the Dirac and the hierarchy approaches. This clearly demonstrates the benefits of sharing information among known ligands of different targets, on the one hand, and the relevance of incorporating prior information into the kernels, on the other hand.
Table 1.
AUC for the first protocol on each dataset with various target kernels
| Ktar \ Target | Enzymes | GPCR | Channels |
|---|---|---|---|
| Dirac | 0.646±0.009 | 0.750±0.023 | 0.770±0.020 |
| Multitask | 0.931±0.006 | 0.749±0.022 | 0.873±0.015 |
| Hierarchy | 0.955±0.005 | 0.926±0.015 | 0.925±0.012 |
| Mismatch | 0.725±0.009 | 0.805±0.023 | 0.875±0.015 |
| Local alignment | 0.676±0.009 | 0.824±0.021 | 0.901±0.013 |
On the GPCR dataset though, the multitask kernel performs slightly worse than the Dirac kernel, probably because some targets in different subclasses show very different binding behavior, which results in adding more noise than information when sharing naively with this kernel. However, a more careful handling of the similarities between GPCRs through the hierarchy kernel results in significant improvement over the Dirac kernel (from 75% to 92.6%), again demonstrating the relevance of the approach.
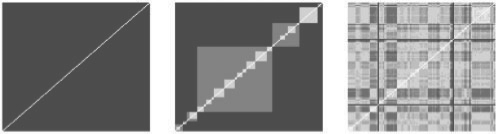
Sequence-based target kernels do not achieve the same performance as the hierarchy kernel, although they perform relatively well for the ion channel dataset, and give better results than the multitask kernel for both GPCR and ion channel datasets. In the case of enzymes, it can be explained by the diversity of the proteins in the family and for the GPCR, by the well-known fact that the receptors do not share overall sequence homology (Gether, 2000). Figure 2 shows three of the tested target kernels for the ion channel dataset. The hierarchy kernel adds some structure information with respect to the multitask kernel, which explains the increase in AUC. The local alignment sequence-based kernels fail to precisely rebuild this structure but retain some substructures. In the cases of GPCR and enzymes, almost no structure is found by the sequence kernels, which, as alluded to above, was expected and suggests that more subtle comparison of the sequences would be required to exploit the information they contain.
Fig. 2.
Target kernel Gram matrices (Ktar) for ion channels with multitask, hierarchy and local alignment kernels.
Figure 3 illustrates the influence of the number of training points for a target on the improvement brought by using information from similar targets. As one could expect, the improvement is very strong when few ligands are known and decreases when enough training points become available. After a certain point (around 30 training points), using similar targets can even impair the performances. This suggests that the method could be globally improved by learning for each target independently how much information should be shared, for example, through kernel learning approaches (Lanckriet et al., 2004).
Fig. 3.
Relative improvement of the hierarchy kernel against the Dirac kernel as a function of the number of known ligands for enzymes, GPCR and ion channel datasets. Each point indicates the mean performance ratio between individual and hierarchy approaches across the targets of the family for which a given (x-axis) number of training points was available.
The second experiment aims at pushing this remark to its limit by assessing how each strategy is able to predict ligands for proteins with no known ligand. Table 2 shows the results in that case. As expected, the classifiers using Dirac kernels show random behavior in this case since using a Dirac kernel with no data for the target amounts to learning with no training data at all. In particular, in the SVM implementation that we used, the classifier learned with no data from the task gave constant scores to all the test points, hence the 0.500 ± 0.000 AUC on the test data. On the other hand, we note that it is still possible to obtain reasonable results using adequate target kernels. In particular, the hierarchy kernel loses only 7.2% of AUC for the ion channel dataset, 5.1% for the GPCR dataset and 1.7% for the enzymes compared to the first experiment where known ligands were used, suggesting that if a target with no known compound is placed in the hierarchy, e.g. in the case of GPCR homology detection with known members of the family using specific GPCR alignment algorithms (Kratochwil et al., 2005) or fingerprint analysis (Attwood et al., 2003), it is possible to predict some of its ligands almost as accurately as if some of them were already available.
Table 2.
AUC for the second protocol on each dataset with various target kernels
| Ktar \ Target | Enzymes | GPCR | Channels |
|---|---|---|---|
| Dirac | 0.500±0.000 | 0.500±0.000 | 0.500±0.000 |
| Multitask | 0.902±0.008 | 0.576±0.026 | 0.704±0.026 |
| Hierarchy | 0.938±0.006 | 0.875±0.020 | 0.853±0.019 |
| Mismatch | 0.602±0.008 | 0.703±0.027 | 0.729±0.024 |
| Local alignment | 0.535±0.005 | 0.751±0.025 | 0.772±0.023 |
In this second setting, our approach when using the hierarchy kernel on the targets is closely related to annotation transfer. Indeed, the learned predictor in this case will predict a molecule to be a ligand of a given target if the molecule is similar to the known ligands of close targets in the hierarchy. In particular, it will predict that the ligands of the target's direct neighbors are ligands of the target (which is an intuitive and natural way to choose new candidate binders). A major difference, however, is that a candidate molecule which is very similar to ligands of a close target, but not a ligand itself, will not be be predicted to be a ligand by the annotation transfer approach. In particular, if the candidate molecule is not present anywhere else in the ligand database, it will never be predicted to be a ligand. Exemples can be found in each of the considered target classes. The 4-aminopyridine is a blocker of the ion channel KCJN5, a potassium inwardly rectifying channel. Although this molecule is a known blocker of other channels (in particular, many potassium channels), it is not a known ligand of any other channel of KCJN5's superfamily. However, the most similar molecule in the database, in the sense of the Tanimoto kernel, is the Pinacidil, which happens to be a known ligand of two direct neighbors of KCJN5. This allows our method to predict 4-aminopyridine as a ligand for this target. Similarly, N-acetyl-d-glucosamine 1,6-bisphosphate is the only known effector of phosphoacetylglucosamine mutase, an enzyme of the isomerase family. This molecule is not a known ligand of any other enzyme in the database, so a direct annotation transfer approach would never predict it as a ligand. Our method, on the other hand, predicts it correctly, taking advantage of the fact that very similar molecules like d-ribose 1,5-bisphosphate or α-d-glucose 1,6-bisphosphate are known ligands of direct neighbors. The same observation can be made for several GPCRs, including the prostaglandin F receptor whose three known ligands are not ligands of any other GPCR but whose direct neighbors have similar ligands.
5 DISCUSSION
We propose a general method to combine the chemical and the biological space in an algorithmic way and predict interaction between any small molecule and any target, which makes it a very valuable tool for drug discovery. The method allows one to represent systematically a ligand–target pair, including information on the interaction between the ligand and the target. Prediction is then performed by any machine learning algorithm (an SVM in our case) in the joint space, which makes targets with few known ligands benefit from the data points of similar targets, and which allows one to make predictions for targets with no known ligand. Our information-sharing process is therefore simply based on a choice of description for the ligands, another one for the targets and on classical machine learning methods. Everything is done by casting the problem in a joint space and no explicit procedure to select which part of the information is shared is needed. Since it subdivides the representation problem into two subproblems, our approach makes use of previous work on kernels for molecular graphs and kernels for biological targets. For the same reason, it will automatically benefit from future improvements in both fields. This leaves plenty of room to increase the performance.
Results on experimental ligand datasets show that using target kernels allowing to share information across the targets considerably improve the prediction, especially in the case of targets with few known ligands. The improvement is particularly strong when the target kernel uses prior information on the structure between the targets, e.g. a hierarchy defined on a target class. Although the usage of a kernel based on the hierarchy is restricted to protein families where hierarchical classification schemes exist, it applies to the three main classes of proteins targeted by drugs, and others like cytochromes and abc transporters. Sequence kernels, on the other hand, did not give very good results in our experiments. However, we believe using the target sequence information could be an interesting alternative or complement to the hierarchy kernel. For example, Jacob et al. (2008) used a kernel based on the sequence of the GPCR that performed as well as the kernel based on the GPCR hierarchy. Further improvement could come from the use of kernel for structures in the cases where 3D structure information is available (e.g. for the enzymes, but not for the GPCR). Our method also shows good performances even when no ligand is known at all for a given target, which is excellent news since classical ligand-based approaches fail to predict ligand for these targets on the one hand, and docking approaches are computationally expensive and not feasible when the target 3D structure is unknown, which is the case of GPCR on the other hand.
In future work, it could be interesting to apply this framework to quantitative prediction of binding affinity using regression methods in the joint space. It would also be important to confirm predicted ligands experimentally or at least by docking approaches when the target 3D structure is available.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Pierre Mahé for his help with ChemCPP and kernels for molecules, and Véronique Stoven for insightful discussions on the biological aspects of the problem.
Conflict of Interest: none declared.
Footnotes
1Available at http://chemcpp.sourceforge.net.
REFERENCES
- Aronszajn N. Theory of reproducing kernels. Trans. Am. Math. Soc. 1950;68:337–404. [Google Scholar]
- Attwood TK, et al. Prints and its automatic supplement, preprints. Nucleic Acids Res. 2003;31:400–402. doi: 10.1093/nar/gkg030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azencott C-A, et al. One- to four-dimensional kernels for virtual screening and the prediction of physical, chemical, and biological properties. J. Chem. Inf. Model. 2007;47:965–974. doi: 10.1021/ci600397p. [DOI] [PubMed] [Google Scholar]
- Balakin KV, et al. Property-based design of GPCR-targeted library. J. Chem. Inf. Comput. Sci. 2002;42:1332–1342. doi: 10.1021/ci025538y. [DOI] [PubMed] [Google Scholar]
- Ballesteros J, Palczewski K. G protein-coupled receptor drug discovery: implications from the crystal structure of rhodopsin. Curr. Opin. Drug Discov. Devel. 2001;4:561–574. [PMC free article] [PubMed] [Google Scholar]
- Bock JR, Gough DA. Virtual screen for ligands of orphan g protein-coupled receptors. J. Chem. Inf. Model. 2005;45:1402–1414. doi: 10.1021/ci050006d. [DOI] [PubMed] [Google Scholar]
- Borgwardt K, et al. Protein function prediction via graph kernels. Bioinformatics. 2005;21(Suppl. 1):i47–i56. doi: 10.1093/bioinformatics/bti1007. [DOI] [PubMed] [Google Scholar]
- Borgwardt KM, Kriegel H-P. Proceedings of the Fifth International Conference on Data Mining. Washington, DC, USA: IEEE Computer Society; 2005. Shortest-path kernels on graphs; pp. 74–81. [Google Scholar]
- Boser BE, et al. Proceedings of the 5th Annual ACM Workshop on Computational Learning Theory. New York, USA: ACM Press; 1992. A training algorithm for optimal margin classifiers; pp. 144–152. [Google Scholar]
- Butina D, et al. Predicting ADME properties in silico: methods and models. Drug Discov. Today. 2002;7(Suppl. 11):S83–S88. doi: 10.1016/s1359-6446(02)02288-2. [DOI] [PubMed] [Google Scholar]
- Byvatov E, et al. Comparison of support vector machine and artificial neural network systems for drug/nondrug classification. J. Chem. Inf. Comput. Sci. 2003;43:1882–1889. doi: 10.1021/ci0341161. [DOI] [PubMed] [Google Scholar]
- Cuturi M, Vert J-P. The context-tree kernel for strings. Neural Netw. 2005;18:1111–1123. doi: 10.1016/j.neunet.2005.07.010. [DOI] [PubMed] [Google Scholar]
- Dobson P, Doig A. Predicting enzyme class from protein structure without alignments. J. Mol. Biol. 2005;345:187–199. doi: 10.1016/j.jmb.2004.10.024. [DOI] [PubMed] [Google Scholar]
- Erhan D, et al. Collaborative filtering on a family of biological targets. J. Chem. Inf. Model. 2006;46:626–635. doi: 10.1021/ci050367t. [DOI] [PubMed] [Google Scholar]
- Evgeniou T, et al. Learning multiple tasks with kernel methods. J. Mach. Learn. Res. 2005;6:615–637. [Google Scholar]
- Frimurer TM, et al. A physicogenetic method to assign ligand-binding relationships between 7tm receptors. Bioorg. Med. Chem. Lett. 2005;15:3707–3712. doi: 10.1016/j.bmcl.2005.05.102. [DOI] [PubMed] [Google Scholar]
- Gärtner T, et al. On graph kernels: hardness results and efficient alternatives. In: Schölkopf B, Warmuth M, editors. Proceedings of the Sixteenth Annual Conference on Computational Learning Theory and the Seventh Annual Workshop on Kernel Machines. Vol. 2777 of Lecture Notes in Computer Science. Heidelberg: Springer; 2003. pp. 129–143. [Google Scholar]
- Gasteiger J, Engel T. Chemoinformatics : a Textbook. Wiley; 2003. [Google Scholar]
- Gether U. Uncovering molecular mechanisms involved in activation of g protein-coupled receptors. Endocr. Rev. 2000;21:90–113. doi: 10.1210/edrv.21.1.0390. [DOI] [PubMed] [Google Scholar]
- Halperin I, et al. Principles of docking: an overview of search algorithms and a guide to scoring functions. Proteins. 2002;47:409–443. doi: 10.1002/prot.10115. [DOI] [PubMed] [Google Scholar]
- Hopkins AL, Groom CR. The druggable genome. Nat. Rev. Drug Discov. 2002;1:727–730. doi: 10.1038/nrd892. [DOI] [PubMed] [Google Scholar]
- Horváth T, et al. Proceedings of the tenth ACM SIGKDD international conference on Knowledge discovery and data mining. New York, NY: ACM Press; 2004. Cyclic pattern kernels for predictive graph mining; pp. 158–167. [Google Scholar]
- International Union of Biochemistry and Molecular Biology. Enzyme Nomenclature 1992. California, USA: Academic Press; 1992. [Google Scholar]
- Ivanciuc O. Applications of support vector machines in chemistry. In: Lipkowitz KB, Cundari TR, editors. Reviews in Computational Chemistry. Vol. 23. Weiheim: Wiley-VCH; 2007. pp. 291–400. [Google Scholar]
- Jaakkola T, et al. A discriminative framework for detecting remote protein homologies. J. Comput. Biol. 2000;7:95–114. doi: 10.1089/10665270050081405. [DOI] [PubMed] [Google Scholar]
- Jacob L, Vert J-P. Efficient peptide-MHC-I binding prediction for alleles with few known binders. Bioinformatics. 2008;24:358–366. doi: 10.1093/bioinformatics/btm611. [DOI] [PubMed] [Google Scholar]
- Jacob L, et al. BMC Bioinformatics. 2008. Virtual screening of GPCRs: anin silicochemogenomics approach. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaroch SE, Weinmann H. Ernst Schering Research Foundation Workshop. Berlin: Springer; 2006. Chemical Genomics: Small Molecule Probes to Study Cellular Function. [Google Scholar]
- Kanehisa M, et al. The KEGG databases at Genome Net. Nucleic Acids Res. 2002;30:42–46. doi: 10.1093/nar/30.1.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M, et al. The KEGG resource for deciphering the genome. Nucleic Acids Res. 2004;32(Database issue):D277–D280. doi: 10.1093/nar/gkh063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashima H, et al. Marginalized kernels between labeled graphs. In: Faucett T, Mishra N, editors. Proceedings of the Twentieth International Conference on Machine Learning. AAAI Press; 2003. pp. 321–328. [Google Scholar]
- Kashima H, et al. Kernels for graphs. In: Schölkopf B, editor. Kernel Methods in Computational Biology. MIT Press; 2004. pp. 155–170. [Google Scholar]
- Klabunde T. Ligand Design for G Protein-coupled Receptors. Wiley-VCH, Great Britain; 2006. Chemogenomics approaches to ligand design; pp. 115–135. Ch. 7. [Google Scholar]
- Klabunde T. Chemogenomic approaches to drug discovery: similar receptors bind similar ligands. Br. J. Pharmacol. 2007;152:5–7. doi: 10.1038/sj.bjp.0707308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kratochwil NA, et al. An automated system for the analysis of g protein-coupled receptor transmembrane binding pockets: alignment, receptor-based pharmacophores, and their application. J. Chem. Inf. Model. 2005;45:1324–1336. doi: 10.1021/ci050221u. [DOI] [PubMed] [Google Scholar]
- Kuang R, et al. Profile-based string kernels for remote homology detection and motif extraction. J. Bioinform. Comput. Biol. 2005;3:527–550. doi: 10.1142/s021972000500120x. [DOI] [PubMed] [Google Scholar]
- Kubinyi H, et al. Methods and Principles in Medicinal Chemistry. New York: Wiley-VCH; 2004. Chemo-Genomics in Drug Discovery: A Medicinal Chemistry Perspective. [Google Scholar]
- Lanckriet GRG, et al. A statistical framework for genomic data fusion. Bioinformatics. 2004;20:2626–2635. doi: 10.1093/bioinformatics/bth294. [DOI] [PubMed] [Google Scholar]
- Leslie C, et al. The spectrum kernel: a string kernel for SVM protein classification. In: Altman RB, editor. Proceedings of the Pacific Symposium on Biocomputing 2002. Singapore: World Scientific; 2002. pp. 564–575. [PubMed] [Google Scholar]
- Leslie CS, et al. Mismatch string kernels for discriminative protein classification. Bioinformatics. 2004;20:467–476. doi: 10.1093/bioinformatics/btg431. [DOI] [PubMed] [Google Scholar]
- Mahé P, Vert J-P. Graph kernels based on tree patterns for molecules. Technical Report ccsd-00095488. 2006 HAL. [Google Scholar]
- Mahé P, et al. Graph kernels for molecular structure-activity relationship analysis with support vector machines. J. Chem. Inf. Model. 2005;45:939–951. doi: 10.1021/ci050039t. [DOI] [PubMed] [Google Scholar]
- Mahé P, et al. The pharmacophore kernel for virtual screening with support vector machines. J. Chem. Inf. Model. 2006;46:2003–2014. doi: 10.1021/ci060138m. [DOI] [PubMed] [Google Scholar]
- Manly C, et al. The impact of informatics and computational chemistry on synthesis and screening. Drug Discov. Today. 2001;6:1101–1110. doi: 10.1016/s1359-6446(01)01990-0. [DOI] [PubMed] [Google Scholar]
- Qiu J, et al. A structural alignment kernel for protein structures. Bioinformatics. 2007;23:1090–1098. doi: 10.1093/bioinformatics/btl642. [DOI] [PubMed] [Google Scholar]
- Ralaivola L, et al. Graph kernels for chemical informatics. Neural Netw. 2005;18:1093–1110. doi: 10.1016/j.neunet.2005.07.009. [DOI] [PubMed] [Google Scholar]
- Rognan D. Chemogenomic approaches to rational drug design. Br. J. Pharmacol. 2007;152:38–52. doi: 10.1038/sj.bjp.0707307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schölkopf B, Smola AJ. Learning with Kernels: Support Vector Machines, Regularization, Optimization, and Beyond. Cambridge, MA: MIT Press; 2002. [Google Scholar]
- Schölkopf B, et al. Kernel Methods in Computational Biology. Cambridge, Massachussetts: MIT Press; 2004. [Google Scholar]
- Shawe-Taylor J, Cristianini N. Kernel Methods for Pattern Analysis. New York, USA: Cambridge University Press; 2004. [Google Scholar]
- Todeschini R, Consonni V. Handbook of Molecular Descriptors. New York, USA: Wiley-VCH; 2002. [Google Scholar]
- Tsuda K, et al. Marginalized kernels for biological sequences. Bioinformatics. 2002;18:S268–S275. doi: 10.1093/bioinformatics/18.suppl_1.s268. [DOI] [PubMed] [Google Scholar]
- Vapnik VN. Statistical Learning Theory. New York: Wiley; 1998. [Google Scholar]
- Vert J-P. A tree kernel to analyze phylogenetic profiles. Bioinformatics. 2002;18:S276–S284. doi: 10.1093/bioinformatics/18.suppl_1.s276. [DOI] [PubMed] [Google Scholar]
- Vert J-P, et al. Local alignment kernels for biological sequences. In: Schölkopf B, editor. Kernel Methods in Computational Biology. Cambridge, Massachussetts: MIT Press; 2004. pp. 131–154. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.