Abstract
Paracoccidioides brasiliensis is a dimorphic fungus that is the causative agent of paracoccidioidomycosis, the most important prevalent systemic mycosis in Latin America. Recently, the existence of three genetically isolated groups in P. brasiliensis was demonstrated, enabling comparative studies of molecular evolution among P. brasiliensis lineages. Thirty-two gene sequences coding for putative virulence factors were analyzed to determine whether they were under positive selection. Our maximum likelihood–based approach yielded evidence for selection in 12 genes that are involved in different cellular processes. An in-depth analysis of four of these genes showed them to be either antigenic or involved in pathogenesis. Here, we present evidence indicating that several replacement mutations in gp43 are under positive balancing selection. The other three genes (fks, cdc42 and p27) show very little variation among the P. brasiliensis lineages and appear to be under positive directional selection. Our results are consistent with the more general observations that selective constraints are variable across the genome, and that even in the genes under positive selection, only a few sites are altered. We present our results within an evolutionary framework that may be applicable for studying adaptation and pathogenesis in P. brasiliensis and other pathogenic fungi.
Author Summary
The fungus Paracoccidioides brasiliensis is the causative agent of paracoccidioidomycosis, a severe pulmonary mycosis that is endemic to Latin America, where an estimated 10 million people are infected with the fungus. Despite the importance of this disease, we know little about the ecological and evolutionary history of this fungus. Here, we present a survey of genetic variation in putative virulence genes in P. brasiliensis in what constitutes the first systematic approach to understand the molecular evolution of the fungus. We used a population genetics approach to determine the role has natural selection played in the coding genes for proteins involved in pathogenesis. We found that nonsynonymous mutations are more common in genes that code for virulence factors than in housekeeping genes. Our results suggest that positive selection has played an important role in the evolution of virulence factors of P. brasiliensis and is therefore an important factor in the host–pathogen dynamics. Our results also have implications for the possible development of a vaccine against paracoccidioidomycosis, since gp43—the main vaccine candidate—has a high level of polymorphism maintained by natural selection.
Introduction
The neutral theory of evolution states that most evolutionary change at the molecular level is caused by the fixation of neutral alleles through random genetic drift [1]. Nonetheless, it is the impact of natural selection on genomic evolution that is of interest if we wish to understand patterns of adaptive evolution by distinguishing between selectively neutral and non-neutral evolutionary change, and relate this change to the biology and history of the organism. The arms race between hosts and their pathogens is a particularly useful system for relating potentially non-neutral evolutionary change to the biology and history of the organisms [2],[3] because of the role natural selection plays in maintaining or fixing different alleles in both host and pathogen populations [4].
Human-fungal interactions provide a privileged system to study the impact of natural selection on the genome of fungal pathogens. Paracoccidoides brasiliensis is the etiological agent of paracoccidioidomycosis (PCM), a human systemic mycosis of importance in Latin America [5]. It is endemic to an area extending from Mexico to Argentina, and infects an estimated 10 million people [6]. Recently, the existence of genetically distinct evolutionary lineages within P. brasiliensis was demonstrated through analysis of DNA sequence data for multiple genes [7],[8]. These groups are currently designated S1 (species 1), PS2 (phylogenetic species 2), PS3 (phylogenetic species 3) and Pb01. Additional support for these lineages comes from variation in virulence and expression levels of antigenic proteins previously found between P. brasiliensis isolates which are now known to belong to S1 and PS2 groups [9]. The recent publication of genomic sequences in the form of expressed sequence tag (EST) databases for several isolates of the different genetic groups of P. brasiliensis [10],[11],[12] and the closely-related species Histoplasma capsulatum (Ajellomyces capsulatum) (unpublished results) presents an opportunity to investigate the role that natural selection may have played in shaping the molecular evolution of the P. brasiliensis genome. Comparative studies between the P. brasiliensis genetic groups and H. capsulatum can be useful to understand host-pathogen evolution, especially in the genes encoding pathogenesis-related proteins which are likely to evolve in response to selective pressure from the host's immune system.
Detecting natural selection at the molecular level requires statistical tests that distinguish the genomic signature of selection from that of neutral mutation and genetic drift alone. Positive selection is inferred when ω [13] (the ratio of non-synonymous (dN) to synonymous (dS) mutations between species) exceeds 1. Positive directional selection occurs when successive amino acid changes make a protein better adapted in a particular biological context, and as a result the changes will tend to be fixed in future lineages. Positive diversifying selection occurs when multiple phenotypes in a population are favored, resulting in an overall increase of the genetic diversity within the species [14],[15]. Several likelihood methods have also been developed to detect deviations from neutral expectation. Under an infinite-sites model, the level of DNA polymorphism within a species is proportional to the amount of divergence at that locus among closely related species [16]. Deviations from this model form the basis for various tests of natural selection, such as the HKA test [17], and the M-K test [18]. Moreover, likelihood methods that allow ω to vary among the branches in a phylogeny, as well as between codons, have been proposed [19],[20],[21],[22],[23]. Using such methods, several genes involved in defense systems and immunity, as well as toxic protein genes, have been shown to be under diversifying or positive directional selection [24],[25],[26],[27].
In this study, we sought to understand the molecular evolution of candidate genes associated with P. brasilensis fungal pathogenesis, which are hypothesized as being under positive selection due to their role in the host-pathogen immune system interaction. Thirty-two putative virulence factors described in previous studies [9],[10],[11],[12],[28] were selected from two available EST databases [10],[11]. In addition, we randomly selected 32 putative housekeeping genes without known antigenic or virulence properties to be used as controls. Orthologous sequences from P. brasiliensis and H. capsulatum were tested for positive selection by means of the Nei and Gojobori method [29], which calculates the average ratio across all amino acid sites. For those genes that showed some evidence of positive selection we obtained sequences from the three lineages of P. brasilensis and used maximum likelihood methods to identify amino acid residues on which positive selection has acted [30]. Our results suggest that positive selection has indeed played an important role in the molecular evolution of virulence factors of P. brasiliensis.
Materials and Methods
P. brasiliensis isolates
The P. brasiliensis strains used in this study were described previously [7]. The sample included individuals from four biotypes recognized for P. brasiliensis: Pb01 (n = 1), S1 (n = 46), PS2 (n = 6) and PS3 (n = 23) and was representative of six endemic areas for paracoccidiodomycosis. We used sequences from GenBank under accession numbers DQ003724 to DQ003788 as well as new sequences obtained by methods previously described [7]. Briefly, total DNA was extracted from the yeast culture with protocols using glass beads [31] or maceration of frozen cells [32]. PCR primers and conditions were as previously reported [7]. The new sequences were deposited in GenBank under accession numbers EU283774 to EU283809.
Selection of putative virulence factors
Molecular genetic tools are still not fully developed for P. brasiliensis, hindering studies that seek to molecularly define genetic factors involved in P. brasiliensis pathogenesis. For the dimorphic fungi, a virulence factor has been functionally defined as a gene product that has an effect on the survival and growth of the organism in its mammalian host but is not essential for growth of the parasitic phase in vitro [33]. Nevertheless, the study of virulence genes sensu Rappleye and Goldman in isolation [33] does not provide full picture of their evolution, because the molecular basis of virulence involves complex networks that comprise many classes of genes. We focused on all the genes proposed to have an impact on the virulence of P. brasiliensis. Table S1 lists the genes that, following genomic analysis in P. brasiliensis, were considered as potential virulence factors and, as such, candidates for this survey [10],[11],[12]. For a gene to be included in this analysis, it had to fulfill three conditions: (i) to have been reported as a putative virulence factor in the previous literature [9],[10],[11],[12],[28], (ii) to be present in the three analyzed databases (two ESTs databases from P. brasiliensis and the genome of H. capsulatum), and (iii) have been demonstrated to be a virulence factor or be an ortholog of a proven virulence factor and have a high homology with it (<1E-10). Fifty percent (32 genes) of the 64 initial candidates fulfilled our requirements and were analyzed to detect positive selection.
Tests for positive selection: H. capsulatum vs. P. brasiliensis
Data retrieving and alignment
Gene sequences were obtained from the National Center for Biotechnology Information (NCBI). The genome sequences from H. capsulatum and the EST sequences of P. brasiliensis were obtained from the EMBL database (http://www.ebi.ac.uk/Databases/nucleotide.html) as of October 13, 2007. BLAST programs were obtained from the NCBI and run locally (Table S2). The two EST databases used in this study [10],[11] include genes expressed in the yeast phase of P. brasiliensis. These databases were compared with the Bastos EST database [12] and early versions of the genome sequence of P. brasiliensis Pb18 (unpublished results) to verify that we were working with high quality EST sequences. The sequences were visually checked and edited to avoid frame shift mutations. No false polymorphisms due to sequencing errors were found in the sequences. The orthology of the genes was assessed by using the preliminary version of the P. brasiliensis genome (http://www.broad.mit.edu/annotation/genome/paracoccidioides_brasiliensis/MultiHome.html).
Housekeeping genes were selected from the P. brasiliensis available sequences in the Gen Bank by using a PERL script, which randomly selected thirty-two genes that did not present any annotation related to virulence or antigenicity.
Alignments of the sequences of the putative virulence factors and housekeeping genes were generated with MUSCLE [34], and the quality of the alignment was assessed with MacClade [35].
dN/dS calculation and Z-tests
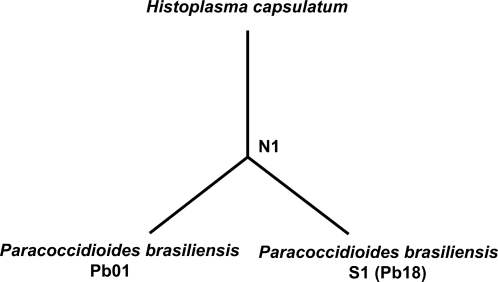
Using a distance-based Bayesian method, the ancestral sequences were reconstructed (i.e. the common ancestor of the three branches of the tree (N1 in Figure 1)), using the Ancestor software [36] for each gene in the dataset. The predicted sequence of each ancestral state was given a probability, with a 95% or higher cut-off. To test for positive selection we calculated the dN and dS values for each branch of the phylogeny (Figure 1) using the random effect likelihood method of Pond and Frost [37],[38], available in HyPhy [38]. The distance from the common ancestor of the last common ancestor of the two P. brasiliensis groups was calculated using an optimal model of nucleic acid selection. Similar results were obtained with other models (HKY85, TN93, and REV).
Figure 1. The phylogeny of H. capsulatum, P. brasiliensis Pb18, and P. brasiliensis Pb01 .N1 is the common ancestor of the three branches of the tree.
Additionally, we estimated the dS and dN variances: Var(dS) and Var(dN), respectively. With this information, we calculated dN/dS and tested the null hypothesis of no selection (H0: dN = dS) versus the positive selection hypothesis (H1: dN>dS) using the Z-test: Z = (dN−dS)/√(Var(dS)+Var(dN)). Z tests calculations were performed using the MEGA software [39],[40].
Mutational saturation dynamics
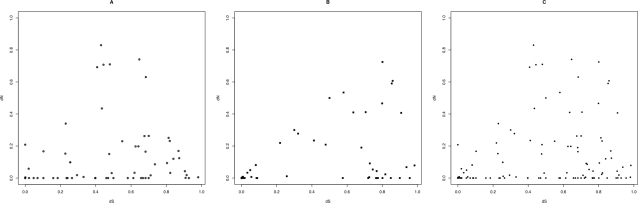
To examine the relative degree of mutational saturation in non-synonymous and synonymous substitutions in our dataset, we plotted the number of non-synonymous nucleotide differences between the two P. brasiliensis groups and the common ancestor against the number of synonymous nucleotide differences for both sets of genes (housekeeping and virulence factors) (Figure 2). Additionally, we fitted a linear model (with functional form dN = A(dS)+B) and a model involving a square term dN = (A(dS)2+BdS+C) to the data by the method of least squares [41]. All the statistical analyses were performed with R.
Figure 2. Observed nonsynonymous differences per site (dN) and synonymous differences per site (dS) in pairwise comparisons for three different partitions of genes.
A. Putative Virulence factors. B. Randomly selected controls. C. Both groups of genes analyzed altogether.
M-K tests
M-K tests [18] between the P. brasiliensis sensu lato and H. capsulatum, using the aligned regions previously sequenced as well as sequences retrieved from GenBank, were calculated using the DNASP analysis program [42].
Codon-Based Likelihood Analyses within P. brasiliensis
To validate our results, we selected a smaller subset of genes that had demonstrated to be under positive selection pressures and for which population datasets were available. The only genes that fulfilled these characteristics were gp43, p27, fks and cdc42. In this set of sequences we searched for evidence of positive selection using the CODEML program of the PAML package (version 4) [22],[30] by using several likelihood-based tests. For each test, equilibrium codon frequencies were estimated from the average nucleotide frequencies at each codon position, amino acid distances were assumed to be equal, and the transition/transversion ratio (κ) was estimated from the data. For all other parameters, we used the default settings described by Yang and Bielawski [30]. Given the observed intraspecific variability, the lack of homoplasy found in individual gene trees, and the phylogenetically recognized groups, we assumed linkage between colinear sites (i.e., there was no recombination within each data set).
To determine which model best fit the data, likelihood ratio tests (LRTs) were performed by comparing the differences in log-likelihood values (LRT = −2lnL) between two models using a χ2 distribution, with the number of degrees of freedom equal to the difference in the number of parameters between the models. We used six models implemented in PAML [13],[22],[30] to test for the presence of sites under positive selection (ω>1). The one-ratio model (M0) assumes one ω for all sites. The neutral model (M1) assumes two classes of sites in the protein: the conserved sites at which ω = 0, and the neutral sites that are defined by ω = 1. The beta model (M7) uses a β distribution of ω over sites: β (p,q), which, depending on parameters p and q, can take various shapes in the 0 to 1 interval. The other three models allow sites with ω>1 and can be considered as tests of positive selection. The selection model (M2) has an additional class of sites compared to the neutral model, in which ω is a free parameter and, as such, can change among residues. The discrete model (M3) uses a distribution with three site classes, with the proportions (p0, p1, and p2) and the ω ratios (ω0, ω1, and ω2) estimated from the data. The beta and ω model (M8) added an extra class of sites to the beta model, estimating the proportion of ω from the data. We used LRTs to make 3 comparisons: to find out whether positive selection has played a role in the molecular evolution of these genes the one-ratio model (M0) was compared with the discrete model (M3) and the neutral model (M1) was compared with the selection model (M2). A third comparison (the beta model (M7) vs. the beta and ω model, M8) [30] was used to identified particular sites in the genes that were likely to have evolved under positive selection by using the Bayesian Empirical Bayes (BEB) analysis previously proposed by Yang [13]. Bayes' theorem was used to estimate the posterior probability that a given site came from the class of positively selected sites [13],[30],[43]. In order to predict potential antigenic determinants for HLA recognition, we used the program SYPFETHI [44].
Estimation of the Time to the Most Recent Common Ancestor (TMRCA)
To determine whether any of the studied loci presented coalescence times within the P. brasiliensis clade (which were older than any other loci) we calculated the Time to the Most Recent Common Ancestor (TMRCA). TMRCAs for S1 and PS2 were estimated based on genetic variation at the eight nuclear loci using the program IM [45]. Estimates of TMRCA do not directly estimate the date of divergence; they provide the timing of coalescence of alleles within a taxon. TMRCA estimates can post- or pre-date the speciation event, and thus can indicate whether the polymorphism in any given gene is older or more recent than the polymorphism in the other genes.
Results
Tests for positive selection (dN/dS): H. capsulatum vs. P. brasiliensis
Thirty-two putative virulence factors fulfilled the requirements for inclusion in this analysis. All the virulence factors showed to be single-copy genes (data not shown, available upon request). To be considered as being under positive selection, these genes had to exhibit a dN/dS ratio larger than 1 and a p-value for the Z-test below 0.05. Table 1 shows the dN/dS ratios for the putative virulence factors and their p-values as determined by using the Z test. According to these criteria, 12 genes were determined to be under positive selection. The dN/dS ratio is correlated to the strength of selection, where values >1 indicate positive selection, and larger values indicate stronger selection. Thirty-two housekeeping genes were randomly selected from the P. brasiliensis available sequences by using a PERL script and their dN/dS (and associated Z values) were calculated and were used as source of comparison. None of these genes showed evidence of being under positive selection in the P. brasiliensis branches, as illustrated in Table 2.
Table 1. Ratio of nonsynonymous to synonomous mutation rate (dN/dS) values for putative virulence factors in the P. brasiliensis lineage.
| Gene | P. brasiliensis Pb18/N1 | P. brasiliensis Pb01/N1 | ||
| dN/dS | p-value | dN/dS | p-value | |
| ade2 | 0.175 | 1 | 0.374 | 1 |
| his1 | 2.318 | 0.01* | 1.482 | 0.06 |
| mls1 | 0 | 1 | 0.005 | 1 |
| icl1 | 0.375 | 0.47 | 0.053 | 1 |
| hem3 | 0.115 | 0.12 | 0.8 | 0.18 |
| chs3 | 0 | 1 | 0.142 | 0.53 |
| cst20 | 1.930 | 0.047* | 0.282 | 1 |
| cdc42 | 1.890 | 0.04* | 0.438 | 1 |
| R = asB | 0 | 1 | 0 | 1 |
| ags1 | 0 | 1 | 1.855 | 0.042* |
| cpn10 | 0.195 | 1 | 0 | 1 |
| groEL | 0.361 | 0.48 | 0.117 | 1 |
| fgsc A4 | 0.385 | 1 | 0.385 | 1 |
| ssc1 | 0 | 1 | 0.148 | 0.93 |
| hsp70 (mitochondrial) | 0 | 1 | 0.020 | 1 |
| hsp70 | 0 | 1 | 0 | 1 |
| hsp82 | 0 | 1 | 0 | 1 |
| hsp88 | 0.047 | 1 | 1.595 | 0.05* |
| hsp90 | 0 | 1 | 0.316 | 1 |
| mdj1 | 0.024 | 1 | 0 | 1 |
| ura3 | 0.039 | 0.51 | 3.215×10−11 | 1 |
| fas2b | 1.980 | 0.05* | 2.160 | 0.03* |
| sod1 | 0.060 | 0.27 | 2.654 | 0.02* |
| ure1b | 0 | 1 | 0.064 | 1 |
| tsa1 | 0.062 | 1 | 1.612 | 0.045* |
| gas1 | 3.980 | 0.02 | 0 | 1 |
| asp | 0/0 | 1 | 0/0 | 1 |
| mnn5 | 0 | 1 | 5.147 | 0.01* |
| tcp1 | 0.309 | 1 | 0.117 | 1 |
| fks | 0/0 | 1 | 23.041 | 0.001* |
| p27 | 0.388 | 1 | 1.699 | 0.043* |
| gp43 | 1.478 | 0.01* | 3.100 | 0.02* |
dN/dS values are shown for the branches that lead towards P. brasiliensis groups as showed in Figure 1. dN/dS ratios are correlated with the strength of selection, where values >1 indicate positive selection, and larger values indicate stronger selection. The P value associated to each dN/dS ratio represents the significance of the Z-test for each branch. Genes that had dN/dS value above 1 and its Z-value was significant (<0.05) were considered under positive selection are marked with *.
Table 2. dN/dS values for a set of randomly selected genes not related to pathogenesis in the P. brasiliensis lineage.
| Gene | P. brasiliensis Pb18/N1 | P. brasiliensis Pb01/N1 | ||
| dN/dS | p-value | dN/dS | p-value | |
| 14-3-3 | 0 | 1 | 0 | 1 |
| cyr1 | 0.007 | 1 | 0.005 | 1 |
| adh | 0 | 1 | 0 | 1 |
| pepN | 0.08 | 1 | 0.993 | 0.3 |
| atp-synt_B | 0 | 1 | 0 | 1 |
| erg6 | 0 | 1 | 0 | 1 |
| Calnexin | 0 | 1 | 0 | 1 |
| cts1 | 0 | 1 | 0 | 1 |
| CLPA | 0.57 | 0.25 | 0.436 | 0.26 |
| cox15 | 0.692 | 1 | 0 | 1 |
| cox17 | 0 | 1 | 0 | 1 |
| cox23 | 0 | 1 | 0 | 1 |
| cox8 | 0.521 | 0.28 | 0.92 | 0.18 |
| cox11 | 0 | 1 | 0 | 1 |
| RibH | 0 | 1 | 0 | 1 |
| Glycos_transf_2 | 0.629 | 0.18 | 0.281 | 0.17 |
| eno | 0.697 | 1 | 0.697 | 1 |
| Fer4 | 0 | 1 | 0 | 1 |
| FBP_aldolase_IIA | 0 | 1 | 0 | 1 |
| Gp_dh_N | 0 | 1 | 0 | 1 |
| hyd2 | 0 | 1 | 0 | 1 |
| l10 | 0 | 1 | 0 | 1 |
| L-Dopa | 0.583 | 1 | 0 | 1 |
| nag | 0 | 1 | 0 | 1 |
| oxa1 | 0.9 | 1 | 0 | 1 |
| pet100 | 0 | 1 | 0 | 1 |
| phb1 | 0 | 1 | 0.559 | 1 |
| pra | 0 | 1 | 0 | 1 |
| sco1 | 0 | 1 | 0.231 | 1 |
| sep1 | 0.125 | 1 | 0.46 | 1 |
| zip | 1 | 0.13 | 0.864 | 0.16 |
| tub1 | 0.011 | 1 | 0.642 | 0.42 |
Conventions are explained in Table 1.
Mutational saturation
A possible explanation for the high proportion of genes under positive selection is that the high proportion of virulence factors showing significantly higher dN/dS are partly artifacts caused by the methods used to estimate the number of non-synonymous and synonymous mutations [46]. Such an explanation would require saturation to occur faster in synonymous than in non-synonymous sites, i.e., the number of non-synonymous nucleotide differences should be a concave function of the number of synonymous nucleotide differences [41]. We plotted the number of non-synonymous nucleotide differences between the two groups of P. brasiliensis and their common ancestor, against the number of synonymous nucleotide differences (Figure 2). No differences were found between the linear and the quadratic models, neither for virulence factors (LRT = 2.134, p = 0.144), nor the housekeeping genes (LRT = 0.112, p = 0.7378), nor for the pooled data (LRT = 1.631; p = 0.2015) indicating that the lineal model is more appropriate to explain the relationship between dN and dS. Therefore, mutational saturation is not responsible for the elevated dN/dS ratios observed in the virulence factors. Similar comparisons were performed including H. capsulatum: one virulence factor (ags1) and housekeeping gene (Gp_dh_N) were found to be under positive selection in the branch that leads towards H. capsulatum (data not shown).
Another possibility is that sequencing errors had inflated dN values. Such errors could artificially increase the significance level of the dN/dS test because they would tend to elevate the number of non-synonymous mutations. However, sequencing errors should also elevate the proportion synonymous mutations and missense mutations. If sequencing errors had, indeed, increased dN, then a large proportion of points in Figure 2 should be located in the upper-left region of the plane. Because no such pattern is observed in Figure 2, we consider this explanation unlikely.
Detection of positive selection by several computing packages program is “reliable” but “conservative” [19],[30],[47] when few sequences are used. Increased accuracy and power are most easily gained with more sequences [19],[30]. Therefore, to further validate our methods and distinguish between directional and diversifying selection, we selected a subset of genes. We choose from among the 12 genes that showed both evidence for positive selection and had more than 25 sequences of P. brasiliensis in GenBank, then reapplying population genetics analysis to these genes. From the 12 genes listed in Table 1, four were selected to be analyzed more in-depth: gp43, p27, cdc42 and fks.
M-K tests
For the gp43 case, the M-K test yielded no significant results between H. capsulatum and P. brasiliensis (Fischer's exact test, P = 0.40, Table 3). M-K tests were significant for p27, cdc42 and fks (p27: P = 0.043594; cdc42: P = 0.000993; fks: P = 0.000017; Table 3) when H. capsulatum was used as an outgroup.
Table 3. McDonald-Kreitman tests of neutrality.
| Fixed between H.capsulatum and S1 | Polymorphic Within species | Fisher's exact test. P-value | |||
| Syn. | Non-Syn. | Syn. | Non-Syn. | ||
| cdc42 | 125 | 119 | 11 | 0 | <0.001 |
| fks | 40 | 90 | 14 | 2 | <0.001 |
| gp43 | 1 | 7 | 5 | 13 | 0.6279 |
| p27 | 35 | 22 | 8 | 0 | 0.0436 |
Codon-Based Likelihood Analyses within P. brasiliensis
gp43
DNA sequences were obtained from 77 P. brasiliensis individuals that yielded twenty-six unique alleles in the exon 2 region of gp43. A total of 29 polymorphic sites and 33 mutations, including 8 singleton and 21 parsimony informative sites, were found among the gp43 alleles (πS1 = 0.00571; πPS2 = 0.00206; πPS3 = 0.00031). Eight silent and twenty-five replacement substitutions were found, where the majority (75.7%) of non-synonymous differences occurred as singletons. No insertion-deletions were found.
Log-likelihood values and parameter estimates under each model are listed in Table 4. Selection models provided a significantly better fit to the data than the neutral models (Table 5); comparisons of M2 versus single-ratio and neutral models yielded LRT values of 18.106 (df = 2, P<0.0001) and 9.14 (df = 2, P = 0.0103), respectively. Likewise, tests between beta (M7) and ω (M8) models strongly supported positive selection (LRT = 18.64, P<0.0001). We found evidence of variation in ω among lineages, as well as substantial variation in ω between sites in the data set. The free-ratio model (M3) was compared with a model that assumes a constant ω across all lineages (M0) by performing LRTs. We could not reject M0 for any of the genes except gp43. Using the one-ratio model (M0), the average value of ω for the gp43 gene was 1.168 - significantly higher than for any of the housekeeping genes [48]. The values of the parameters under the discrete model (M3) indicated that 59.3% of the sites in the gp43 gene were under purifying selection (ω = 0), whereas 37.07% belonged to a site class with ω = 1.63, and 3.6% had an ω equal to 18.53, indicating that the two latter classes are under positive selection.
Table 4. Likelihood values, parameter estimates, and sites under positive selection as inferred under the six proposed models applied to each of the four loci.
| Model | lnL | Parameter Estimate | dN/dS | Selected sites | ||||
| GP43 | One-ratio (M0) | −971.307 | 1.168 | 1.168 | None | |||
| Neutral (M1) | −975.791 | p0 = 0.559996 w0 = 0.0 | 0.44 | Not allowed | ||||
| p1 = 0.44004 w1 = 1.00 | ||||||||
| Selection (M2) | −966.738 | p0 = 0.43329 w0 = 0.00 | 1.2303 | 231 V** | 241 S** | 266 I+ | 296 G+ | |
| p1 = 0.52265 w1 = 1.00 | 335 P* | 336 L+ | ||||||
| p2 = 0.04406 w2 = 16.20255 | ||||||||
| Free-ratio (M3) | −965.824 | p0 = 0.58630 w0 = 0.00 | 1.2628 | 218 S** | 225 E** | 226 D** | 229 H** | |
| p1 = 0.37715 w1 = 1.58443 | 231 V** | 241 S** | 248 P** | 251 T** | ||||
| p2 = 0.03655 w2 = 18.19716 | 260 T** | 265 Y** | 266 I** | 296 G** | ||||
| 335 P** | 330 S** | 348 K** | 360 K** | |||||
| 363 L** | 374 E** | 376 G** | ||||||
| Beta (M7) | −976.065 | p = 0.005 q = 0.00829 | 0.375 | Not allowed | ||||
| Beta+w (M8) | −966.745 | p0 = 0.95493 p = 0.03212 | 1.2303 | 231 V** | 241 S** | 266 I+ | 296 G+ | |
| q = 0.02627 | 335 P* | 336 L+ | ||||||
| p1 = 0.04507 w = 15.93096 | ||||||||
| FKS | One-ratio (M0) | −869.645 | 0.1136 | 0.1136 | None | |||
| Neutral (M1) | −869.645 | P0 = 1.00000 wo = 0.11334 | 0.1136 | Not allowed | ||||
| P1 = 0.00000 w1 = 1.00000 | ||||||||
| Selection (M2) | −869.645 | P0 = 1.00000 wo = 0.11324 | 0.1136 | |||||
| P1 = 0.00000 w1 = 1.00000 | ||||||||
| P2 = 0.00000 w2 = 3.00000 | ||||||||
| Free-ratio (M3) | −869.645 | p0 = 0.03490 wo = 0.11330 | 0.1136 | None | ||||
| p1 = 0.91236 w1 = 0.11324 | ||||||||
| p2 = 0.05274 w2 = 0.11328 | ||||||||
| Beta (M7) | −869.645 | p = 12.68894 q = 99.00000 | 0.1136 | Not allowed | ||||
| Beta+w (M8) | −869.645 | p0 = 1.00000 p = 12.73125 | ||||||
| q = 99.00000 | ||||||||
| (p1 = 0.00000) w = 1.00000 | ||||||||
| CDC42 | One-ratio (M0) | −1151.0403 | 0.1136 | 0.39594 | None | |||
| Neutral (M1) | −1151.429 | P0 = 0.6041 wo = 1.000 | 0.39594 | Not allowed | ||||
| P1 = 0.3959 w1 = 0.00000 | ||||||||
| Selection (M2) | −1150.091 | P0 = 0.7645 wo = 0.58454 | 3.12653 | |||||
| P1 = 0.2227 w1 = 1.00000 | ||||||||
| P2 = 0.01283 w2 = 46.95373 | ||||||||
| Free-ratio (M3) | −1150.3119 | p0 = 0.8300 wo = 0.46384 | 1.93988 | None | ||||
| p1 = 0.1464 w1 = 0.46384 | ||||||||
| p2 = 0.0246 w2 = 27.5156 | ||||||||
| Beta (M7) | −1179.7537 | p = 0.05 q = 0.604263 | 0.365995 | Not allowed | ||||
| Beta+w (M8) | −1178.436 | p0 = 1.00000 p = 12.73125 | 0.396092 | |||||
| q = 85 | ||||||||
| (p1 = 0.05) w = 1.00000 | ||||||||
| P27 | One-ratio (M0) | −1341.1883 | 0.1136 | 0.506987 | None | |||
| Neutral (M1) | −1341.1883 | P0 = 0.500 wo = 0.01397325 | 0.506987 | Not allowed | ||||
| P1 = 0.500 w1 = 1.000000 | ||||||||
| Selection (M2) | −1341.1883 | P0 = 0.3333 wo = 0.000 | 2.0047 | |||||
| P1 = 0.014195 w1 = 0.3333 | ||||||||
| P2 = 0.05 w2 = 5.0 | ||||||||
| Free-ratio (M3) | −1341.1883 | p0 = 0.3333 wo = 0.01711 | 0.05133 | None | ||||
| p1 = 0.3333 w1 = 0.03422 | ||||||||
| p2 = 0.3333 w2 = 0.10267 | ||||||||
| Beta (M7) | −1341.1883 | p = 1.000 q = 1.000 | 0.5 | Not allowed | ||||
| Beta+w (M8) | −1341.1883 | p0 = 0.6667 p = 1.000 | 0.555556 | |||||
| q = 2.00 w = 1.000 | ||||||||
| (p1 = 0.667) w = 1.000 | ||||||||
Amino acid sites inferred to be under positive selection with a probability. lnL: log-Likelihood
>99% are marked with a **, more than 95% with a * and more than 75% with a +.
Table 5. Likelihood ratio statistics of different models.
| Locus | Comparison | Df | lnL | X2 Critical value (1%) |
| GP43 | One-ratio (M0) vs. Discrete (M 3) | 4 | 10.966608 | 9.21 |
| Neutral (M 1) vs. Selection (M 2) | 2 | 18.10675 | 13.28 | |
| Beta (M 7) vs. Beta+w (M 8) | 2 | 18.641266 | 9.21 | |
| FKS | One-ratio (M0) vs. Discrete (M 3) | 4 | 0 | 9.21 |
| Neutral (M 1) vs. Selection (M 2) | 2 | 0 | 13.28 | |
| Beta (M 7) vs. Beta+w (M 8) | 2 | 0 | 9.21 | |
| CDC42 | One-ratio (M0) vs. Discrete (M 3) | 4 | 1.4568 | 9.21 |
| Neutral (M 1) vs. Selection (M 2) | 2 | 2.676 | 13.28 | |
| Beta (M 7) vs. Beta+w (M 8) | 2 | 2.6354 | 9.21 | |
| P27 | One-ratio (M0) vs. Discrete (M 3) | 4 | 0 | 9.21 |
| Neutral (M 1) vs. Selection (M 2) | 2 | 0 | 13.28 | |
| Beta (M 7) vs. Beta+w (M 8) | 2 | 0 | 9.21 |
Twice the difference in log likelihood ratio between a null model and an alternative model was compared with a χ2 distribution in order to test whether an alternative model fits the data better than the null model. Df: Degrees of Freedom; LRT: Likelihood Ratio Test.
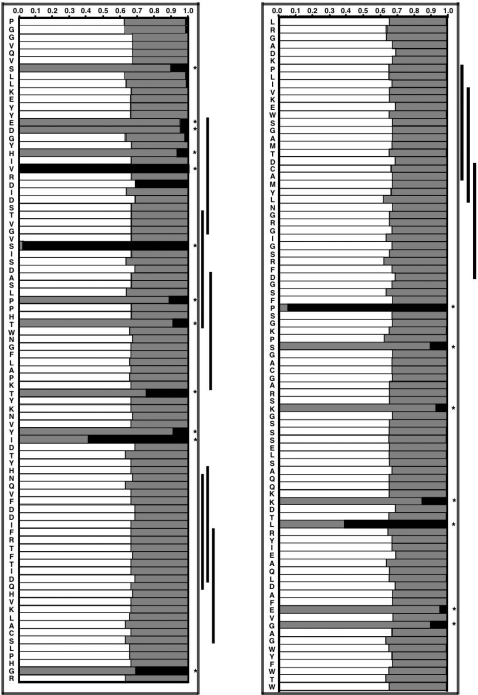
Models of positive selection (discrete, selection, beta and ω models) that allow for sites with ω greater than 1 fit the gp43 data significantly better than the corresponding neutral models (one-ratio, neutral and beta models) (Table 5). Posterior probabilities, as revealed by the discrete model, indicate that the gp43 codons belong to one of the three classes with different selective pressures, as indicated by the beta and ω model (Figure 3). Using the Bayesian Empirical Bayes (BEB) analysis, 19 sites with a posterior probability greater than 95% of having a greater than 1 value were identified. In order to predict potential antigenic determinants for HLA recognition, we used the program SYPFETHI [44]. As illustrated in Figure 3, seven of the sites under positive selection were located as potential epitopes as predicted with SYFPEITHI.
Figure 3. Posterior probabilities showed by each site in the exon 2 of the PBGP43 gene belonging to site classes with different selective pressures (of 18.20 [black], 1.58 [gray], and 0.00 [white bars]) under the free-ratio model.
The gp43 amino acid sequence is shown to the left. Sites with a posterior probability higher than 95% have a greater than 1 and are indicated by an asterisk (*). The underlined parts correspond to the regions that according to SYFPEITHI prediction are potential epitopes.
fks, p27 and cdc42
DNA sequences obtained from 15 individuals showed low levels of polymorphism in p27 and cdc42 (p27: πS1 = 0.00571; πPS2 = 0.00206; πPS3 = 0.00031; cdc42: πS1 = 0.00071; πPS2 = 0.00006; πPS3 = 0.000011). No insertion-deletions were found. In the fks case, most of the sequences were retrieved from the NCBI and the polymorphism level was low (fks: πS1 = 0.000001; πPS2 = 0.00006; πPS3 = 0.000013).
Estimation of Time to the Most Recent Common Ancestor (TMRCA)
The TMRCAs for S1 and PS2 were estimated based on genetic variation at the gp43 locus and seven other nuclear loci. The results showed that the TRMCA for the gp43 alleles is longer than for any other gene in P. brasiliensis (Table 6), indicating that the polymorphism in gp43 is significantly older than the polymorphism in the other genes (Signed rank test; P<0.01). This constitutes evidence for balancing selection [49],[50]. Additional evidence for the balancing selection hypothesis in gp43 comes from the haplotype network previously described for this gene, in which several high frequency haplotypes are separated by long branches [7].
Table 6. Maximum-Likelihood Estimates (MLE) and the 95% confidence intervals of Time to the Most Recent Common Ancestor (TMRCA).
| Gene | Mean | TRMCA | Confidence interval (95%) |
| Variance | |||
| p27 | 0.54 | 0.321 | 0.324–1.534 |
| cdc42 | 0.489 | 0.546 | 0.297–2.672 |
| gp43 | 2.238 | 7.118 | 1.5125–3.262 |
| arf | 0.815 | 0.836 | 0.5678–2.5875 |
| ord | 1.534 | 2.411 | 0.89–1.65 |
| tub | 1.188 | 1.865 | 0.8625–1.7625 |
| ord | 1.618 | 2.840 | 0.9375–3.5625 |
| fks | 0.611 | 0.471 | 0.4341–2.7225 |
Conversely, the TRMCAs for cdc42, p27 and fks were significantly lower than the other genes as is expected if a gene is under positive directional selection.
Discussion
Identification of putative virulence factors
Comparisons of DNA sequence differences within and between closely related species can give insights into the temporal scales of molecular evolutionary processes, and into selective pressures on different type of loci. In this study, evidence of different types of positive selection acting on the putative virulence factors was obtained from analysis of the ratio between non-synonymous and synonymous substitution rates in coding regions. A comparison of these virulence factors with housekeeping genes in P. brasiliensis showed that a higher proportion of virulence genes evolve under positive selection (37.5% vs. 0%), suggesting that at least some of these genes have an adaptive role. Substantial heterogeneity in the mode of evolution was found both among and within the genes investigated in this study. As predicted from previous studies of evolution of virulence factors in other organisms, the 12 putative virulence factors genes identified as having evolved under positive selection have a wide variety of functions (Table 1, Table S1 and Text S1) [27].
This analysis of positive selection using genomic data identified a set of genes that together with data derived from genetic, expression and biochemical essays, provides some insights into the evolution of P. brasiliensis virulence. Some of these genes are involved in the escape from immune recognition (tsa1, sod1). However, this is just one aspect of the ability of a pathogen to successfully invade and colonize its host, and other genes have proven to be important in pathogenesis, such as the case of heat shock genes that are connected to virulence [32]–[34]. Previous studies have suggested that although virulence factors sensu Rappleye and Goldman [33] are key factors in pathogenesis, their study as isolated entities does not provide a holistic picture of the evolutionary dynamics of virulence. The results of this study, and others, support the notion that many essential genes participate in complex networks that comprise the molecular basis of virulence, and that their history is shaped by natural selection.
For most of the genes found to be under positive selection (10 out of 12), biochemical and physiological characteristics are known. Only two genes (p27 and gp43) have unknown functions. All the others were classified in four different categories of genes according to their functions: metabolic related genes (fas2, his1), cell wall related genes (fks, mnn5, ags1), heat shock proteins, detoxification related genes (tsa1, sod1, hsp88) and signal transduction genes (cdc42, cst20). A detailed biochemical description and information related to these genes is presented in the Text S1.
M-K and codon analysis of p27, cdc42, fks and gp43
p27, cdc42 and fks are genes that are depauperate in genetic variation, as is expected for regions in which advantageous amino acid replacements have been fixed by positive selection. Judging by the significant results of the M-K tests, positive selection has played an important role in the history of these three genes and the depletion of genetic variation within P. brasiliensis (at these three loci) is a consequence of positive selection.
The M-K test was not significant for gp43. This test has proven to be robust because the sites in which synonymous and non-synonymous mutations occur are interspersed, so that they would be similarly affected by genetic drift and changes in geography [20],[45]. In gp43, the M-K test was not able to detect positive selected within the P. brasiliensis lineage due to the excess of non-synonymous substitutions within and across species. The persistence of non-synonymous intra- and trans-specific gp43 polymorphisms within and between lineages of the P. brasiliensis complex suggests they have been maintained by historical or contemporary selection [51].
Several recent studies have used the power of modern molecular selection analyses to design experiments based on the molecular evolutionary hypothesis [20]. An example of the importance of this kind of study is that immunization with gp43 epitopes from one isolate would not be expected to be effective against allthe species complex due to the high level of polymorphism in gp43. This has profound implications for the development of a gp43 vaccine and immunotherapy [52].
It is likely that the evolution of putative virulence factors of P. brasiliensis has been driven by the interaction between the pathogen and its extracellular environment. However, it remains unclear whether the positive pressure was derived from the environment when the fungus is in its free-living stages, or from the host's immune system. Determining the function and biochemical roles of the proteins encoded by the genes found to be under positive selection in P. brasiliensis should shed light on the corresponding selective pressures.
Conclusions
Molecular evolutionary analysis should facilitate the identification of biologically important genes through the comparison of nucleotide sequences. Although the methods for positive selection used here are not perfect [23], the identification of positively selected proteins offers a good approach for understanding human pathogenic fungi, in which transformation or production of mutants is difficult (McEwen, personal communication). Positive selection in virulence factors might have different outcomes, including: adaptation of a species to optimize the process of infection, to escape host immune response, inhabit different environmental niches, and also lead to functional diversification of members of multi-gene families.
We hope that identifying and cataloging these loci for this and other groups of fungi will provide others with an evolutionary framework for pursuing directed mutation experiments on the specific functional significance of these genes.
Supporting Information
P. brasiliensis genes assigned as putative virulence genes by genomic and proteomic studies (10,11). The table includes the biochemical role of the gene product and study that defined each gene as a putative virulence factor in P. brasiliensis and constitutes a more expanded version of Table 1.
(0.05 MB XLS)
Accession numbers of the nucleotide sequences of the virulence genes that were analyzed in this study.
(0.04 MB XLS)
Biochemical information related to these genes under positive selection.
(0.18 MB DOC)
Acknowledgments
This work constitutes a prime example of the Coyne's Law [53]. We would like to acknowledge L. Scordato, B. He and two anonymous reviewers for their insightful comments. We also would like to thank M. Sprigge and J. A. Coyne for reading the whole manuscript and editing it very conscientiously.
Footnotes
The authors have declared that no competing interests exist.
This work was supported by Comite de Investigaciones de la Universidad de Antioquia (Sostenibilidad 2005–2006) and financially by Corporación para Investigaciones Biológicas. D.R.M. thanks Fundación Sofia Perez de Soto. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Kimura M. The neutral theory of molecular evolution. Cambridge, UK: Cambridge University Press; 1983. [Google Scholar]
- 2.Price DA, Goulder PJ, Klenerman P, Sewell AK, Easterbrook PJ, et al. Positive selection of HIV-1 cytotoxic T lymphocyte escape variants during primary infection. Proc Natl Acad Sci U S A. 1997;94:1890–1895. doi: 10.1073/pnas.94.5.1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang P, Wang Q, Sims PF, Hyde JE. Rapid positive selection of stable integrants following transfection of Plasmodium falciparum. Mol Biochem Parasitol. 2002;123:1–10. doi: 10.1016/s0166-6851(02)00105-6. [DOI] [PubMed] [Google Scholar]
- 4.Escalante AA, Lal AA, Ayala FJ. Genetic polymorphism and natural selection in the malaria parasite Plasmodium falciparum. Genetics. 1998;149:189–202. doi: 10.1093/genetics/149.1.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Restrepo A. Paracoccidioidomycosis. In: Dismukes WE, Pappas PG, Sobel JD, editors. Clinical micology. New York, NY: Oxford University Press; 2003. [Google Scholar]
- 6.Brummer E, Castaneda E, Restrepo A. Paracoccidioidomycosis: an update. Clin Microbiol Rev. 1993;6:89–117. doi: 10.1128/cmr.6.2.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Matute DR, McEwen JG, Puccia R, Montes BA, San-Blas G, et al. Cryptic speciation and recombination in the fungus Paracoccidioides brasiliensis as revealed by gene genealogies. Mol Biol Evol. 2006;23:65–73. doi: 10.1093/molbev/msj008. [DOI] [PubMed] [Google Scholar]
- 8.Carrero LL, Niño-Vega G, Teixeira MM, Carvalho MJA, Soares CMA, et al. New Paracoccidioides brasiliensis isolate reveals unexpected genomic variability in this human pathogen. Fungal Genetics and Biology. 2008;45:605–612. doi: 10.1016/j.fgb.2008.02.002. [DOI] [PubMed] [Google Scholar]
- 9.Carvalho KC, Ganiko L, Batista WL, Morais FV, Marques ER, et al. Virulence of Paracoccidioides brasiliensis and gp43 expression in isolates bearing known PbGP43 genotype. Microbes Infect. 2005;7:55–65. doi: 10.1016/j.micinf.2004.09.008. [DOI] [PubMed] [Google Scholar]
- 10.Goldman GH, dos Reis Marques E, Duarte Ribeiro DC, de Souza Bernardes LA, Quiapin AC, et al. Expressed sequence tag analysis of the human pathogen Paracoccidioides brasiliensis yeast phase: identification of putative homologues of Candida albicans virulence and pathogenicity genes. Eukaryot Cell. 2003;2:34–48. doi: 10.1128/EC.2.1.34-48.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Felipe MS, Andrade RV, Petrofeza SS, Maranhao AQ, Torres FA, et al. Transcriptome characterization of the dimorphic and pathogenic fungus Paracoccidioides brasiliensis by EST analysis. Yeast. 2003;20:263–271. doi: 10.1002/yea.964. [DOI] [PubMed] [Google Scholar]
- 12.Bastos KP, Bailao AM, Borges CL, Faria FP, Felipe MS, et al. The transcriptome analysis of early morphogenesis in Paracoccidioides brasiliensis mycelium reveals novel and induced genes potentially associated to the dimorphic process. BMC Microbiol. 2007;7:29. doi: 10.1186/1471-2180-7-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nielsen R, Yang Z. Likelihood models for detecting positively selected amino acid sites and applications to the HIV-1 envelope gene. Genetics. 1998;148:929–936. doi: 10.1093/genetics/148.3.929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Storz JF. Using genome scans of DNA polymorphism to infer adaptive population divergence. Mol Ecol. 2005;14:671–688. doi: 10.1111/j.1365-294X.2005.02437.x. [DOI] [PubMed] [Google Scholar]
- 15.Vallender EJ, Lahn BT. Positive selection on the human genome. Hum Mol Genet 13 Spec No. 2004;2:R245–254. doi: 10.1093/hmg/ddh253. [DOI] [PubMed] [Google Scholar]
- 16.Nei M. Molecular evolutionary genetics. New York: Columbia University Press; 1987. [Google Scholar]
- 17.Hudson RR, Kreitman M, Aguade M. A test of neutral molecular evolution based on nucleotide data. Genetics. 1987;116:153–159. doi: 10.1093/genetics/116.1.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McDonald JH, Kreitman M. Adaptive protein evolution at the Adh locus in Drosophila. Nature. 1991;351:652–654. doi: 10.1038/351652a0. [DOI] [PubMed] [Google Scholar]
- 19.Yang Z. PAML 4: Phylogenetic Analysis by Maximum Likelihood. Mol Biol Evol. 2007;24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- 20.Nielsen R. Molecular signatures of natural selection. Annu Rev Genet. 2005;39:197–218. doi: 10.1146/annurev.genet.39.073003.112420. [DOI] [PubMed] [Google Scholar]
- 21.Anisimova M, Bielawski JP, Yang Z. Accuracy and power of bayes prediction of amino acid sites under positive selection. Mol Biol Evol. 2002;19:950–958. doi: 10.1093/oxfordjournals.molbev.a004152. [DOI] [PubMed] [Google Scholar]
- 22.Yang Z. PAML: a program package for phylogenetic analysis by maximum likelihood. Comput Appl Biosci. 1997;13:555–556. doi: 10.1093/bioinformatics/13.5.555. [DOI] [PubMed] [Google Scholar]
- 23.Hughes AL. Looking at Darwin in all the wrong places: the misguided quest for positive selection at the nucleotide sequence level. Heredity. 2007;99:364–373. doi: 10.1038/sj.hdy.6801031. [DOI] [PubMed] [Google Scholar]
- 24.Johannesson H, Vidal P, Guarro J, Herr RA, Cole GT, et al. Positive directional selection in the proline-rich antigen (PRA) gene among the human pathogenic fungi Coccidioides immitis, C. posadasii and their closest relatives. Mol Biol Evol. 2004;21:1134–1145. doi: 10.1093/molbev/msh124. [DOI] [PubMed] [Google Scholar]
- 25.Liu Z, Bos JI, Armstrong M, Whisson SC, da Cunha L, et al. Patterns of diversifying selection in the phytotoxin-like scr74 gene family of Phytophthora infestans. Mol Biol Evol. 2005;22:659–672. doi: 10.1093/molbev/msi049. [DOI] [PubMed] [Google Scholar]
- 26.Stahl EA, Bishop JG. Plant-pathogen arms races at the molecular level. Curr Opin Plant Biol. 2000;3:299–304. doi: 10.1016/s1369-5266(00)00083-2. [DOI] [PubMed] [Google Scholar]
- 27.Anisimova M BJ, Dunn K, Yang Z. Phylogenomic analysis of natural selection pressure in Streptococcus genomes. BMC Evol Biol. 2007;7:154. doi: 10.1186/1471-2148-7-154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ortiz BL, Garcia AM, Restrepo A, McEwen JG. Immunological characterization of a recombinant 27-kilodalton antigenic protein from Paracoccidioides brasiliensis. Clin Diagn Lab Immunol. 1996;3:239–241. doi: 10.1128/cdli.3.2.239-241.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nei M, Gojobori T. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol Biol Evol. 1986;3:418–426. doi: 10.1093/oxfordjournals.molbev.a040410. [DOI] [PubMed] [Google Scholar]
- 30.Yang Z, Bielawski JP. Statistical methods for detecting molecular adaptation. Trends In Ecology And Evolution. 2000;15:496–503. doi: 10.1016/S0169-5347(00)01994-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.van Burik JA, Schreckhise RW, White TC, Bowden RA, Myerson D. Comparison of six extraction techniques for isolation of DNA from filamentous fungi. Med Mycol. 1998;36:299–303. [PubMed] [Google Scholar]
- 32.Morais FV, Barros TF, Fukada MK, Cisalpino PS, Puccia R. Polymorphism in the gene coding for the immunodominant antigen gp43 from the pathogenic fungus Paracoccidioides brasiliensis. J Clin Microbiol. 2000;38:3960–3966. doi: 10.1128/jcm.38.11.3960-3966.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rappleye CA, Goldman WE. Defining Virulence Factors in the Dimorphic Fungi. Annual Reviewof Microbiology. 2006;60:281–303. doi: 10.1146/annurev.micro.59.030804.121055. [DOI] [PubMed] [Google Scholar]
- 34.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Maddison DR, Maddison WP. Sinauer Associates; 2005. MacClade. [Google Scholar]
- 36.Zhang JaN M. Accuracies of ancestral amino acid sequences inferred by the parsimony, likelihood, and distance methods. J Mol Evol. 1997;44:S139–S146. doi: 10.1007/pl00000067. [DOI] [PubMed] [Google Scholar]
- 37.Pond SL, Frost SD. A simple hierarchical approach to modeling distributions of substitution rates. Mol Biol Evol. 2005;22:223–234. doi: 10.1093/molbev/msi009. [DOI] [PubMed] [Google Scholar]
- 38.Pond SL, Frost SD, Muse SV. HyPhy: hypothesis testing using phylogenies. Bioinformatics. 2005;21:676–679. doi: 10.1093/bioinformatics/bti079. [DOI] [PubMed] [Google Scholar]
- 39.Kumar S, Nei M, Dudley J, Tamura K. MEGA: A biologist-centric software for evolutionary analysis of DNA and protein sequences. Brief Bioinform. 2008 doi: 10.1093/bib/bbn017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- 41.Nielsen R. The ratio of replacement to silent divergence and tests of neutrality. J Evol Biol. 1997;10:217–231. [Google Scholar]
- 42.Rozas J, Rozas R. DnaSP version 3: an integrated program for molecular population genetics and molecular evolution analysis. Bioinformatics. 1999;15:174–175. doi: 10.1093/bioinformatics/15.2.174. [DOI] [PubMed] [Google Scholar]
- 43.Yang Z, Wong WS, Nielsen R. Bayes empirical bayes inference of amino acid sites under positive selection. Mol Biol Evol. 2005;22:1107–1118. doi: 10.1093/molbev/msi097. [DOI] [PubMed] [Google Scholar]
- 44.Rammensee H, Bachmann J, Emmerich NP, Bachor OA, Stevanovic S. SYFPEITHI: database for MHC ligands and peptide motifs. Immunogenetics. 1999;50:213–219. doi: 10.1007/s002510050595. [DOI] [PubMed] [Google Scholar]
- 45.Nielsen R. Statistical tests of selective neutrality in the age of genomics. Heredity. 2001;86:641–647. doi: 10.1046/j.1365-2540.2001.00895.x. [DOI] [PubMed] [Google Scholar]
- 46.Hasegawa M, Cao Y, Yang Z. Preponderance of slightly deleterious polymorphism in mitochondrial DNA: nonsynonymous/synonymous rate ratio is much higher within species than between species. Mol Biol Evol. 1998;15:1499–1505. doi: 10.1093/oxfordjournals.molbev.a025877. [DOI] [PubMed] [Google Scholar]
- 47.Chen SL, Hung CS, Xu J, Reigstad CS, Magrini V, et al. Identification of genes subject to positive selection in uropathogenic strains of Escherichia coli: a comparative genomics approach. Proc Natl Acad Sci U S A. 2006;103:5977–5982. doi: 10.1073/pnas.0600938103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Matute DR, Torres IP, Salgado-Salazar C, Restrepo A, McEwen JG. Background selection at the chitin synthase II (chs2) locus in Paracoccidioides brasiliensis species complex. Fungal Genet Biol. 2007;44:357–367. doi: 10.1016/j.fgb.2007.01.006. [DOI] [PubMed] [Google Scholar]
- 49.Bamshad M, Wooding SP. Signatures of natural selection in the human genome. Nat Rev Genet. 2003;4:99–111. doi: 10.1038/nrg999. [DOI] [PubMed] [Google Scholar]
- 50.Kreitman M. Methods to detect selection in populations with applications to the human. Annual Review Of Genomics and Human Genetics. 2000;1:539–559. doi: 10.1146/annurev.genom.1.1.539. [DOI] [PubMed] [Google Scholar]
- 51.Canino MF, Bentzen P. Evidence for positive selection at the pantophysin (Pan I) locus in walleye pollock, Theragra chalcogramma. Mol Biol Evol. 2004;21:1391–1400. doi: 10.1093/molbev/msh141. [DOI] [PubMed] [Google Scholar]
- 52.Iwai LK, Yoshida M, Sidney J, Shikanai-Yasuda MA, Goldberg AC, et al. In silico prediction of peptides binding to multiple HLA-DR molecules accurately identifies immunodominant epitopes from gp43 of Paracoccidioides brasiliensis frequently recognized in primary peripheral blood mononuclear cell responses from sensitized individuals. Mol Med. 2003;9:209–219. [PMC free article] [PubMed] [Google Scholar]
- 53.Coyne J. Jerry Coyne. Curr Biol. 2004;14:R825–826. doi: 10.1016/j.cub.2004.09.031. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
P. brasiliensis genes assigned as putative virulence genes by genomic and proteomic studies (10,11). The table includes the biochemical role of the gene product and study that defined each gene as a putative virulence factor in P. brasiliensis and constitutes a more expanded version of Table 1.
(0.05 MB XLS)
Accession numbers of the nucleotide sequences of the virulence genes that were analyzed in this study.
(0.04 MB XLS)
Biochemical information related to these genes under positive selection.
(0.18 MB DOC)