Abstract
The enzyme ribonucleotide reductase, responsible for the synthesis of deoxyribonucleotides (dNTP), is upregulated in response to DNA damage in all organisms. In Saccharomyces cerevisiae, dNTP concentration increases ∼6- to 8-fold in response to DNA damage. This concentration increase is associated with improved tolerance of DNA damage, suggesting that translesion DNA synthesis is more efficient at elevated dNTP concentration. Here we show that in a yeast strain with all specialized translesion DNA polymerases deleted, 4-nitroquinoline oxide (4-NQO) treatment increases mutation frequency ∼3-fold, and that an increase in dNTP concentration significantly improves the tolerance of this strain to 4-NQO induced damage. In vitro, under single-hit conditions, the replicative DNA polymerase ε does not bypass 7,8-dihydro-8-oxoguanine lesion (8-oxoG, one of the lesions produced by 4-NQO) at S-phase dNTP concentration, but does bypass the same lesion with 19–27% efficiency at DNA-damage-state dNTP concentration. The nucleotide inserted opposite 8-oxoG is dATP. We propose that during DNA damage in S. cerevisiae increased dNTP concentration allows replicative DNA polymerases to bypass certain DNA lesions.
INTRODUCTION
Ribonucleotide reductases (RNRs) catalyze the formation of dNTPs by reducing the corresponding ribonucleotides, and are instrumental in controlling dNTP concentration (1). In eukaryotes and in some bacteria, RNR is composed of a large and a small subunit, both necessary for catalysis. RNR expression increases in response to DNA damage. In Escherichia coli, nrdA and nrdB (encoding the large and the small RNR subunits, respectively) are among the most potently induced lexA-independent genes following UV exposure (induced ∼20- and ∼7-fold, respectively, within 60 min of UV exposure) (2,3). In resting mammalian cells, DNA damage induces the p53R2 protein, an alternative small RNR subunit, about 4-fold in a p53-dependent manner (4–6). Similarly, Drosophila large RNR subunit, RnrL, is induced by ionizing radiation in wild-type, but not p53-deficient strains (7). In the yeasts S. cerevisiae and S. pombe, RNR genes are also among the most robustly induced genes following DNA damage (8–10). In addition to transcriptional regulation, RNR activity in both yeasts is controlled by Sml1 and Spd1, small proteins that bind to RNR and inhibit its activity (11–13). Sml1 and Spd1 are degraded upon entry into S phase and in response to DNA damage (12,14). In S. cerevisiae, the large subunit is encoded under normal growth conditions by the RNR1 gene. During DNA damage, the highly similar RNR3 gene is activated, which leads to increased levels of the large subunit (8). The small subunit, responsible for generation of the free tyrosyl radical important for catalysis, is a heterodimer encoded by the RNR2 and RNR4 genes (15). The Mec1/Rad53 DNA damage checkpoint is responsible both for activation of RNR2-4 genes transcription and Sml1 degradation (14,16).
RNR activity is also controlled allosterically. The enzyme's allosteric specificity sites, located in the large subunit, adjust the balance between the four individual dNTPs. The allosteric activity sites, also located in the large subunit control the overall concentration of dNTP: when the concentration of dNTP reaches a certain level, RNR activity is down-regulated by dATP feedback inhibition (17). Saccharomyces cerevisiae RNR has a relaxed dATP feedback inhibition, which allows at least a 6- to 8-fold increase of dNTP concentration in response to DNA damage, or at least an ∼3- to 5-fold increase above the dNTP concentration of an S-phase yeast cell (18). This increase in dNTP concentration correlates directly to DNA damage tolerance. In the rnr1-D57N mutant strain, in which the dATP feedback inhibition of RNR is non-functional, dNTP concentration increases ∼30-fold in response to DNA damage, ∼4 times more than in a wild-type strain under similar conditions. The ability of the rnr1-D57N mutant to increase dNTP concentration above wild-type levels in response to DNA damage is associated with higher tolerance of DNA damage induced by 4-NQO, methyl methane sulfonate (MMS) and UV-light (18). 4-NQO produces several types of quinoline adducts at guanine and adenine bases as well as 8-oxoG (19). Overexpression of the wild-type RNR1 gene in logarithmically growing yeast elevates dNTP concentration ∼10-fold and similarly leads to an increased DNA damage tolerance to 4-NQO (20). Deletion of Crt1/Rfx1, Rox1 or Mot3, transcriptional repressors of RNR2, RNR3 and RNR4 genes, also leads to 4-NQO resistance (21).
The improved DNA damage tolerance of S. cerevisiae in the presence of high dNTP concentration is associated with higher mutation frequency (18), and can be best explained by a more efficient translesion DNA synthesis (TLS). The specialized TLS polymerases Rev1, Polζ and Polη are believed to be responsible for the mutagenic bypass of DNA lesions and increased damage tolerance. To identify translesion polymerases that increase DNA damage tolerance in the dNTP concentration-dependent manner, we made deletions of REV1, RAD30 (Polη), REV3 (the catalytic subunit of Polζ), and POL4 (non-replicative DNA polymerase involved in DNA repair), and compared DNA damage tolerance of these deletion strains towards 4-NQO in the presence of normal and high dNTP concentrations. Deletion of REV1 or REV3, but not of RAD30 or POL4 resulted in sensitivity to 4-NQO. Interestingly, increased dNTP concentration significantly improved the 4-NQO tolerance in all TLS polymerase-deleted strains, including a strain with all non-replicative polymerases deleted. Mutation frequency in this strain increased ∼3-fold after treatment with 4-NQO. These observations indicate that replicative DNA polymerases are able to bypass certain DNA lesions when dNTP concentration is elevated after DNA damage. In support of this hypothesis we show that in vitro, under single-hit conditions, the replicative DNA polymerase ε (Polε) does not bypass 8-oxoG lesion at S-phase dNTP concentration, but does bypass the same lesion with 19–27% efficiency at DNA-damage-state dNTP concentration.
MATERIALS AND METHODS
Yeast strains
All yeast strains are derivatives of W4069-4C (MATa CAN1 ade2-1 his3-11,15 leu2-3,112 trp1-1) (18) used as wild type and were grown in YP media (1% yeast extract, 2% peptone) with 2% dextrose (YPD) or 2% galactose (YPGal). Construction of the pGAL-RNR1 strain was described before (20). TLS polymerase genes were deleted using cassettes polymerase chain reaction (PCR)-amplified from pFA6a-HIS3MX6 (for rev1Δ), pFA6a-kanMX6 (for rev1Δ, rev3Δ, rad30Δ) and pFA6a-TRP1(for pol4Δ) as previously described (22). REV3 was also deleted with LEU2 using pAM56 plasmid (23) kindly provided by Dr Alan Morrison. All deletion strains were back-crossed to wild-type and the correct insertion of a deletion cassette was confirmed by PCR. Construction of the rev1Δ::HIS3 rad30Δ::KanMX6 rev3Δ::LEU2 pol4Δ::TRP1 strain was done by crossing single TLS polymerase deletion strains with each other. Introduction of the pGAL-RNR1 into different strains was also done by crossing.
dNTP analysis
At a density from 0.5 × 107 to 1.5 × 107 cells/ml, ∼1 × 108 cells were harvested by filtration through 25 mm White AAWP nitrocellulose filters (0.8 µm, Millipore AB, Solna, Sweden). The filters were immersed in 500 µl of ice-cold extraction solution (12% w/v trichloroacetic acid, 15 mM MgCl2) in Eppendorf tubes. The following steps were carried out at 4°C. The tubes were vortexed for 30 s, incubated for 15 min and vortexed again for 30 s. The filters were removed and the supernatants were collected after centrifugation at 20 000g for 1 min and added to 800 µl of ice-cold Freon-trioctylamine mixture [10 ml of Freon (1,1,2-trichlorotrifluoroethane, Aldrich, Sigma-Aldrich Sweden AB, Stockholm, Sweden, 99%) and 2.8 ml of trioctylamine (Fluka, Sigma-Aldrich Sweden AB, Stockholm, Sweden, >99%)]. The samples were vortexed and centrifuged for 1 min at 20 000g. The aqueous phase was collected and added to 800 µl of ice-cold Freon-trioctylamine mixture. The mixture was vortexed and centrifuged as above. Twenty microliters of the aqueous phase containing dNTP and NTP was analyzed by HPLC on a Partisphere 5 SAX column (PolyLC Inc., Columbia, MD, USA) using a UV-2075 Plus detector (Jasco, Mölndal, Sweden). Nucleotides were isocratically eluted with 2.5% acetonitrile, 0.3 M potassium phosphate, pH 5.0 buffer.
Flow cytometry
At the density 0.5 × 107 to 1.5 × 107 cells/ml, ∼1 × 107 cells were harvested by filtration through 25 mm White AAWP nitrocellulose filters (0.8 µm, Millipore). The filters were immersed into 13 ml tubes with 1.5 ml H2O and vortexed to wash the cells off the filters. Total 3.5 ml of 99% ethanol was added dropwise with slow vortexing and cells were kept at 4°C overnight. The filters were removed; the cells were collected by centrifugation, resuspended in 700 µl of H2O, transferred to Eppendorf tubes and centrifuged. The cells were resuspended in RNAse solution (2 mg/ml RNAse in 50 mM Tris pH 8.0, boiled 15 min) and incubated 6–15 h at 37°C. Fifty microliters of 20 mg/ml proteinase K in H2O was added and the cells were incubated 1 h at 50°C. The cells were collected by centrifugation, resuspended in 0.5 ml 50 mM Tris pH 7.5. For analysis, 50 µl of cell suspension was placed into 1 ml of staining solution (SYBR®-Green I (Molecular Probes) diluted 10 000 times in 50 mM Tris, pH 7.5). Samples were sonicated at low output and analyzed on a Cytomics FC500 (Beckman Coulter Inc, Bromma, Sweden).
Primer extension assay
Polε was purified as described (24). Primer extension assays were performed as described (25), but with varying dNTP concentrations as indicated in Table 1 and with the following primer (5′-CTGACAGTGTAACCATTACACGGATTCGATAGTATCCTCTAAGGACGATTCGATCCTG-3′) annealed to the wild-type, 8-oxoG or MeG templates (5′-GATCGATCGTAACzTAGCAGGATCGAATCGTCCTTAGAGGATACTATCGAATCCGTGTAATGGTTACACTGTCAG-3′), where z indicates a G, an 8-oxoG or MeG. The reaction mixtures were separated on an 8% denaturing polyacrylamide gel and visualized with a Typhoon 9400 PhosphorImager (GE Healthcare Biosciences, Uppsala, Sweden). The intensities of the bands were quantified using ImageQuant software package supplied with the PhosphorImager.
Table 1.
dNTP concentrations used in primer extension assays shown in Figure 4
| Cell volume (μm3) | dNTP concentration | dNTP (μM) |
|||
|---|---|---|---|---|---|
| dCTP | dTTP | dATP | dGTP | ||
| 90 | Low | 10 | 16 | 5.5 | 3 |
| Normal | 19.5 | 33 | 11 | 5.5 | |
| High | 97.5 | 191.5 | 97 | 25 | |
| 45 | Low | 19.5 | 33 | 11 | 5.5 |
| Normal | 39 | 66 | 22 | 11 | |
| High | 195 | 383 | 194 | 49.5 | |
‘Normal’ is an estimated S-phase cell dNTP concentration; ‘Low’ is half of ‘Normal’ and is approximately an average concentration of a logarithmically growing yeast culture; ‘High’ is an approximated maximal dNTP concentration of a DNA-damaged cell (dCTP is 5-fold, dTTP is 5.8-fold, dATP is 8.8-fold and dGTP is 4.5-fold above ‘Normal’).
Analysis of base insertion opposite 8-oxoG
Biotinylated Acc65I overhang primer (5′-Biotin GTAGGTACCGATCTACGAGAGATACCATTACACGGATTCGATAGTATCCTCTAAGGACGATTCGATCCTG-3′) was annealed at a 1:1 molar ratio to a complementary template, EcoRI template (5′-AGATGGAATTCGTTTACACTGTCGCGTAACzTAGCAGGATCGAATCGTCCTTAGAGGATACTATCGAATCCGTGTAATGG-3′). The z on the template indicates the position of 8-oxoG. The primer-template (1 pmol) was elongated by wild-type Polε (0.2 μM) for 30 min at 30°C as described for primer extension reactions at DNA-damage-state dNTP concentrations for a 45 μm3 cell (Table 1). The reactions were stopped at 70°C for 1 h. The elongated product was bound to Dynabeads M-280 Streptavidin overnight and immobilized on the beads according to the manufacturer's protocol. Next, the product was washed twice with Washing buffer (Dynal Biotech, ASA, Oslo, Norway) and with water to remove all non-biotinylated components by utilizing a Dynal Magnetic Particle Concentrator (Dynal MPC). To remove the template containing the 8-oxoG, the primer-template was denaturated in 0.1 M NaOH for 5 min. After the denaturation the DNA was again bound to Dynal MPC to remove the template containing 8-oxoG, which was not biotinylated. The denaturation step was repeated once. The biotinylated primer was washed eight times with 0.1 M NaOH and two times with TE buffer pH 7.6 by utilizing the Dynal MPC. To amplify the biotinylated primer, PCR was run with Phusion high fidelity DNA polymerase (Finnzymes). The PCR product (129 bp) was purified and cleaved by Acc65I and EcoRI at 37°C. The PCR was carried out with upstream primer (5′-GTAGGTACCGATCTACGAGAG-3′) and downstream primer (5′-CTAGCAGATGATGTAACGCTTCTCAGATGGAATTCGTTTACACTGTCGC-3′). The vector pBluescript II SK+ was cleaved by Acc65I and EcoRI at 37°C. The cleaved products were purified, ligated and transformed into E. coli. Colonies were picked by blue/white screening. White colonies were purified and sent to Eurofins MWG operon (Germany) for sequencing.
RESULTS
Overexpression of RNR1 efficiently elevates dNTP concentration
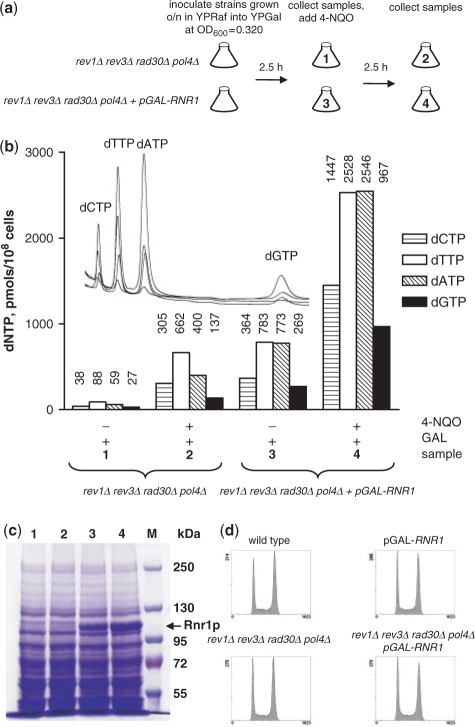
To establish strains, in which dNTP concentration could be experimentally controlled, we utilized the GAL1-driven wild-type RNR1 gene introduced into the URA3 locus of the yeast genome. We measured dNTP pools in the rev1Δ rad30Δ rev3Δ pol4Δ and rev1Δ rad30Δ rev3Δ pol4Δ pGAL-RNR1 strains grown in galactose-containing media before and after DNA damage induced by 4-NQO (Figure 1a). Induction of the RNR1 gene by galactose in the rev1Δ rad30Δ rev3Δ pol4Δ pGAL-RNR1 strain resulted in overexpression of the Rnr1 protein and a 9- to 13-fold elevation of dNTP concentration compared to rev1Δ rad30Δ rev3Δ pol4Δ strain (Figure 1b and c). Addition of 4-NQO to the rev1Δ rad30Δ rev3Δ pol4Δ pGAL-RNR1 strain induced by galactose further increased dNTP concentration 3- to 4-fold (Figure 1b). This further increase can be explained by the induction of the RNR2-4 genes, degradation of Sml1 and a decreased utilization of dNTP during DNA damage. Addition of 4-NQO to the rev1Δ rad30Δ rev3Δ pol4Δ strain elevated the dNTP concentration 5- to 8-fold (Figure 1b). The same fold increase in dNTP concentration occurs in wild-type yeast during DNA damage (18). Simultaneous deletion of all non-replicative polymerases had no effect on cell proliferation or cell division cycle under normal growth conditions (i.e. in the absence of 4-NQO) (Figure 1d). Overexpression of RNR1 in all strains did not affect proliferation rates and viability as judged by the number and the size of colonies (Figure 2a).
Figure 1.
Overexpression of RNR1 efficiently elevates dNTP concentration in yeast strains lacking TLS polymerases. (a) rev1Δ rad30Δ rev3Δ pol4Δ and rev1Δ rad30Δ rev3Δ pol4Δ pGAL-RNR1 strains were incubated in liquid YP media with 2% galactose and treated with 0.2 mg/L 4-NQO as shown in the diagram. (b) Samples (indicated by numbers 1–4) treated as outlined in (a) were used for determination of dNTP pools. The numbers above the bars indicate the amount of the individual dNTP expressed in pmols/108 cells. Four overlaid HPLC chromatograms (raw data, not normalized by the number of cells) are shown on the inset. (c) Samples (indicated by numbers 1–4) treated as outlined in (a) were used for analysis of Rnr1 protein levels by 6% SDS–PAGE. M indicates protein marker lane. (d) The cell cycle progression is not altered in the strains lacking TLS polymerases. wild-type, pGAL-RNR1, rev1Δ rad30Δ rev3Δ pol4Δ and rev1Δ rad30Δ rev3Δ pol4Δ pGAL-RNR1 strains were inoculated in liquid YPD and incubated overnight at 30°C. Next morning cultures were diluted in fresh YPD to an OD600 of 0.1 and grown at 30°C. Samples were collected after 4.5 h and prepared for flow-cytometric analysis.
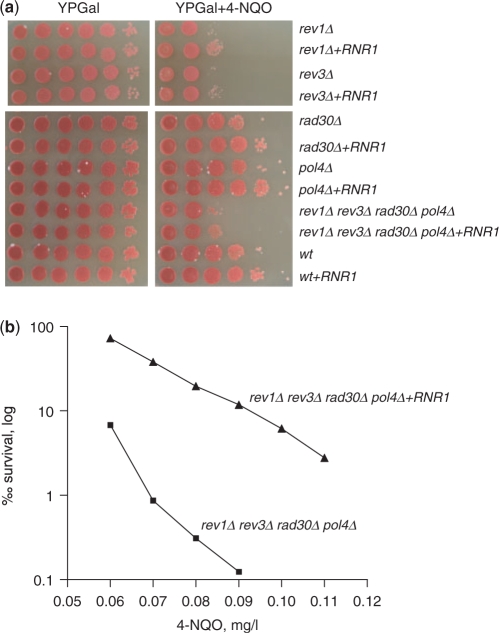
Figure 2.
Increased dNTP concentration improves DNA damage tolerance in the absence of TLS polymerases. (a) Stationary phase cultures grown in YPD were spotted at 10-fold serial dilutions on YPGal (control) and YPGal with 0.24 mg/l 4-NQO plates, and incubated for 4 days at 30°C. (b) rev1Δ rad30Δ rev3Δ pol4Δ and rev1Δ rad30Δ rev3Δ pol4Δ pGAL-RNR1 strains were grown overnight in YPD; appropriate dilutions were plated on YPGal plates containing indicated amounts of 4-NQO, and on YPGal plates to calculate the number of viable cells. Colonies were counted after 4 days of incubation at 30°C.
DNA damage tolerance of the TLS polymerase deletion strains increases in the presence of elevated dNTP concentration
If a certain TLS polymerase were responsible for the bypass of a 4-NQO lesion only at a high dNTP concentration, then deletion of this polymerase would result in a yeast strain equally sensitive to 4-NQO at normal and high dNTP concentrations. In all single polymerase deletion strains the elevation of dNTP concentration improved DNA damage tolerance (survival of DNA damage) (Figure 2a). Deletion of REV1 or REV3 resulted in sensitivity to 4-NQO, while the rad30Δ and pol4Δ strains were not 4-NQO sensitive (Figure 2a). Next, we tested the DNA damage tolerance of a strain with all non-replicative nuclear polymerases deleted (rev1Δ rad30Δ rev3Δ pol4Δ), with or without RNR1 overexpression. Strikingly, overexpression of RNR1 (resulting in a 3- to 4-fold higher dNTP concentration under these conditions, compare samples 2 and 4 in Figure 1b) improved the DNA damage tolerance of the rev1Δ rad30Δ rev3Δ pol4Δ strain to 4-NQO up to 100-fold (Figure 2b and Supplementary Figure 1). The elevation of dNTP concentration also improved the tolerance to 4-NQO in a wild-type strain with all polymerases present, as observed earlier (18,21).
4-NQO increases the mutation frequency 3-fold in a rev1Δ rad30Δ rev3Δ pol4Δ strain
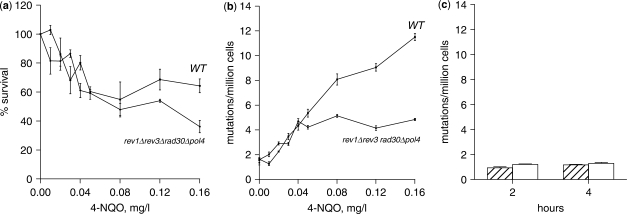
The increased DNA damage tolerance of the rev1Δ rad30Δ rev3Δ pol4Δ strain in the presence of elevated dNTP concentration suggests that the replicative DNA polymerases are able to bypass some lesions produced by 4-NQO. Alternatively, other DNA repair pathways, e.g. nucleotide excision repair (NER) or base excision repair (BER), are somehow stimulated by increased dNTP pools. However, these pathways do not involve a direct bypass of a lesion by a DNA polymerase and should not be mutagenic. Therefore, we measured the induced mutation frequencies in the rev1Δ rad30Δ rev3Δ pol4Δ and wild-type strains after 2 h incubation with increasing concentrations of 4-NQO. The initial increase in the induced mutation frequencies (about 3-fold) and the initial decrease in survival showed the same dynamics in both strains (Figure 3a and b). At 0.04 mg/l 4-NQO the induced mutation frequency in the rev1Δ rad30Δ rev3Δ pol4Δ strain reached a plateau, while the induced mutation frequency in the wild-type strain continued to increase. Since in both strains the treatment with 4-NQO leads to elevation of dNTP pools ∼8-fold (Figure 1b), it is possible that the observed initial increase in mutation frequencies is due to higher error rates of replicative polymerases in the presence of high dNTP concentration and not due to lesion bypass. However, mutation frequencies did not increase in the rev1Δ rad30Δ rev3Δ pol4Δ pGAL-RNR1 strain induced by galactose for 2 or 4 h (Figure 3c), even though the dNTP concentration increases ∼10-fold after the galactose induction in the absence of 4-NQO (Figure 1b). Thus, the increase in the 4-NQO-induced mutation frequency in the rev1Δ rad30Δ rev3Δ pol4Δ strain is most likely due to increased translesion synthesis by the replicative DNA polymerases.
Figure 3.
4-NQO increases mutation frequency in the rev1Δ rad30Δ rev3Δ pol4Δ strain. (a) wild-type and rev1Δ rad30Δ rev3Δ pol4Δ logarithmically growing in YPD were treated with increasing amounts of 4-NQO for 2 h and were after appropriate dilutions spread on YPD plates in triplicates to determine survival. (b) Yeast cells treated as in (a) were after appropriate dilutions spread on synthetic complete medium −arginine +L-canavanine to determine mutation frequencies in CAN1 gene by dividing the number of Can1r mutants by the average number of surviving cells (c) rev1Δ rad30Δ rev3Δ pol4Δ pGAL-RNR1 strain grown in YPRaf was divided into two cultures, one of which was induced by 2% galactose and mutation frequencies were determined after 2 and 4 h induction. Hatched bars: uninduced cells; open bars: galactose-induced cells.
Bypass of 8-oxoG by Polε at S-phase and DNA-damage-state dNTP concentrations
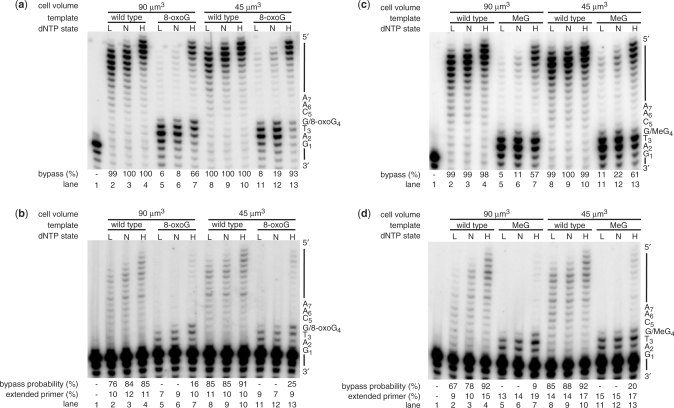
4-NQO produces several types of quinoline adducts to guanine and adenine bases as well as a common DNA lesion, 8-oxoG (19). The ratio between the quinoline-bound adducts and the 8-oxoG found in the DNA of Ehrlich ascites cells exposed to 4-NQO was estimated to be 4:1 (26). We assessed the ability of yeast replicative Polε to bypass 8-oxoG in vitro at the dNTP concentration found in vivo in wild-type cells during a normal S phase and during DNA damage. Polε is one of the three replicative yeast DNA polymerases and, together with Polδ, is responsible for the bulk of DNA synthesis (27,28). The intracellular dNTP concentrations were calculated using the published amount of dNTP per million of wild-type haploid yeast cells grown in YPD (11,18), and the reported wet (60 × 10–12 g) and dry (15 × 10–12 g) weight of a haploid yeast cell (29). Next, the dry weight was subtracted from the wet weight to estimate the volume of the soluble fraction of a haploid cell (45 × 10–12 g or ∼45 µm3). Because yeast cells increase in volume during the cell cycle arrest elicited by DNA damage, and because in some reports the volume of yeast cells is 70 µm3 and greater, we calculated dNTP concentration using two volumes: 45 and 90 µm3 (Table 1).
The ability of the wild-type, proofreading-proficient, Polε to bypass an 8-oxoG lesion increased dramatically at an elevated dNTP concentration approximating the DNA-damaged-state concentration (Table 1, ‘High’) as compared to S-phase dNTP concentration (Table 1, ‘Normal’). In the presence of excess Polε over template, the 8-oxoG lesion bypass increased from 19% at S-phase dNTP concentration to 93% at DNA-damaged-state dNTP concentration for a 45 µm3 cell (Figure 4a, compare lanes 12 and 13), or from 8% to 66% for a 90 µm3 cell (Figure 4a, compare lanes 6 and 7). Under single-hit conditions, when the reactions were performed with an excess of template over Polε to ensure that each product was formed from only one replication event, we observed no 8-oxoG bypass at low (Table 1, ‘Low’ and Figure 4b, lanes 5 and 11) or S-phase dNTP concentrations (Table 1, ‘Normal’ and Figure 4b, lanes 6 and 12), but 16 and 25% bypass probability at DNA-damaged-state dNTP concentrations for the 90 and 45 µm3 cell, respectively (Table 1, ‘High’ and Figure 4b, lanes 7 and 13). To calculate the bypass efficiency we divided the bypass probability of the damaged template with the bypass probability of the undamaged template (30). The bypass efficiency at ‘Low’ and S-phase dNTP concentrations was 0%, and at DNA-damage-state dNTP concentrations 19 and 27%, for the 90 and 45 µm3 cell, respectively. Therefore, approximately 20% of the time, Polε bypasses an 8-oxoG lesion at DNA-damaged-state dNTP concentration without dissociating from the template. We have identified dAMP as the major nucleotide inserted by Polε opposite 8-oxoG (Table 2) at dNTP concentrations present in vivo after DNA damage. Insertion of dAMP opposite 8-oxoG has also been observed for Polδ, although in the presence of equimolar dNTP concentrations (31). Another common lesion, O6-Methylguanine (MeG) is also bypassed by Polε under single-hit conditions at DNA-damaged-state dNTP concentrations, but not at S-phase dNTP concentrations (Figure 4c and d). The bypass efficiency at S-phase dNTP concentrations was 0% and at DNA-damage-state dNTP concentrations 10 and 22%, for the 90 and 45 µm3 cell, respectively.
Figure 4.
Polε bypasses an 8-oxoG and MeG lesions at DNA-damage-state, but not at normal S-phase-state, dNTP concentration. (a) Primer extension assays were performed with 4 nM Polε and 2 nM wild-type or 8-oxoG templates at low (L), normal S-phase (N) and DNA-damage-state (H) dNTP concentrations (see Table 1 for details) for 10 min at 30°C. The lesion bypass was calculated by dividing the sum of the products at position 5 (position after G/8-oxoG) or greater by the sum of the products at position 3 or greater. The sequence of the template and the positions of the nucleotides are indicated on the right. (b) Assays under single-hit conditions were performed with 0.17 nM Polε and 2 nM wild-type or 8-oxoG templates at low (L), normal S-phase (N) and DNA-damage-state (H) dNTP concentrations (see Table 1 for details) for 2 min at 30°C. The bypass probability was calculated by dividing the sum of the products at position 5 (position after G/8-oxoG) or greater by the sum of the products at position 3 or greater as previously described (30). The amount of extended primer is the intensity of all products greater than the primer divided by the intensity of the primer and all products greater than the primer. The total amount of primer extended in all reactions was far <20%, demonstrating that the conditions for single completed hits were reached (40). The sequence of the template and the positions of the nucleotides are indicated on the right. (c) Primer extension assays were performed with 4 nM Polε and 2 nM wild-type or MeG templates at low (L), normal S-phase (N) and DNA-damage-state (H) dNTP concentrations (see Table 1 for details) for 10 min at 30°C. The sequence of the template and the positions of the nucleotides are indicated on the right. (d) Assays under single-hit conditions were performed with 0.06 nM Polε and 2 nM wild-type or MeG templates at low (L), normal S-phase (N) and DNA-damage-state (H) dNTP concentrations for 2 min at 30°C. The sequence of the template and the positions of the nucleotides are indicated on the right.
Table 2.
Base insertion and deletion during bypass of 8-oxoG by Polε
| Number (%) | |
|---|---|
| A insertion | 22 (92) |
| C insertion | 1 (4) |
| Single deletion | 1 (4) |
DISCUSSION
Until the discovery of specialized TLS polymerases, it was generally believed that DNA lesions were bypassed by replicative DNA polymerases. In the last decade the focus has almost entirely shifted to investigation of DNA lesion bypass by TLS polymerases, while the role of replicative polymerases has received much less attention. Here, we address the involvement of yeast replicative and TLS DNA polymerases in the bypass of DNA lesions at normal and elevated dNTP concentrations.
In S. cerevisiae, the dNTP concentration increases 6- to 8-fold in response to DNA damage (18). The dNTP concentration increase is noticeable already at very low concentrations of mutagens (e.g. 0.01 mg/l NQO), long before cell proliferation is affected. An artificial elevation of dNTP concentration above normal levels in an rnr1 mutant strain results in increased DNA damage tolerance and higher mutation rates. It is important to note that the DNA damage checkpoint is not pre-activated by an artificial elevation of dNTP concentration (20), and thus cannot be accounted for higher DNA damage tolerance. Conversely, a decrease in RNR activity by deletion of the RNR4 gene leads to a smaller increase of dNTP concentration after DNA damage, lower induced mutation frequencies and higher DNA damage sensitivity (32–34). These observations indicate that an increase in dNTP concentration is important for the ability of DNA polymerases to bypass DNA lesions. Here, we demonstrate that an artificial increase in dNTP concentration in a strain with all non-replicative polymerases deleted increases 4-NQO tolerance up to 100-fold. Furthermore, we demonstrate that 4-NQO elevates mutation frequency ∼3-fold in a strain with all non-replicative polymerases deleted. This increase is not due to higher mutation rates of replicative DNA polymerases in the presence of elevated dNTP under these conditions, as an artificial increase in dNTP concentration (to the same levels as in the 4-NQO treated yeast) in the absence of DNA damage does not elevate the mutation frequency. These observations indicate that replicative DNA polymerases can bypass certain DNA lesions in the presence of increased dNTP concentration. It should be, however, noted that the involvement of replicative polymerases in lesion bypass in vivo has only been demonstrated when all non-replicative polymerases have been deleted, and that the involvement of replicative polymerases in TLS in normal cells has not yet been proven.
Yeast replicative DNA polymerases α and δ (mainly involved in the lagging strand synthesis) are able to insert nucleotides opposite DNA lesions when dNTP concentration is sufficiently high (35,36). For example, Polδ is able to partially bypass an 8-oxoG lesion at 100 µM, but not at 5 µM dNTP (35). However, as the dNTP concentration in vivo under the DNA damaging conditions has not been defined, the physiological relevance of these observations was not obvious. Here we show for the first time that nucleotides can be inserted opposite 8-oxoG and MeG lesions by the yeast leading strand polymerase, Polε, at dNTP concentrations and at correct dNTP pool bias present in vivo during DNA damage. These data underscore the importance of elevated dNTP concentration for unaided lesion bypass by replicative DNA polymerases. In addition, the insertion of nucleotides by replicative polymerases opposite lesions can provide a substrate for the TLS Polζ. This TLS polymerase is inefficient at inserting nucleotides opposite various DNA lesions including 8-oxoG, but is efficient at extending from nucleotides inserted opposite these lesions by the replicative DNA Polδ (35,37).
It should be pointed out that 8-oxoG and MeG are examples demonstrating the ability of replicative polymerases to directly bypass certain lesions at dNTP concentrations present during DNA damage in vivo. It might be, however, some other lesions and not necessarily the 8-oxoG lesion that are bypassed by the replicative polymerases in the experiments with 4-NQO and elevated dNTP concentration. We were not able to test the ability of Polε to bypass other known lesions induced by 4-NQO, e.g. those containing guanine bases with quinoline adducts, because of the difficulty to synthesize such templates. It is, however, less likely that Polε would be able to bypass such bulky lesions as quinoline adducts efficiently, considering that Polε can not bypass thymine dimer or (+)- and (–)-trans-anti-benzo[a]pyrene-7,8-dihydrodiol-9,10-epoxide-N2-dG DNA adducts (38,39). The inability of replicative polymerases to bypass bulky lesions even at very high dNTP concentration is in agreement with our observation that the increased dNTP concentration only partially rescues the sensitivity of the TLS polymerase deletion strain towards the 4-NQO-produced lesions (Figure 2).
Although the TLS polymerases clearly play a major role in the bypass of most DNA lesions, we propose a new pathway, in which the elevated dNTP concentration present in S. cerevisiae after DNA damage engages replicative DNA polymerases (directly or in cooperation with Polζ) in the bypass of certain, perhaps less bulky, DNA lesions. Recently, Lis et al. (34) have proposed a similar pathway that appears to induce mutations at damaged DNA in S. cerevisiae by up-regulating dNTP levels and facilitating translesion synthesis by the replicative Polδ. The DNA-damage-dependent upregulation of RNR transcription in all studied organisms ranging from bacteria to mammals suggests an important role for increased dNTP production during DNA damage. It will be interesting to explore whether replicative polymerases of other organisms are also involved in the bypass of lesions during DNA damage.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Swedish Research Council (A.C. and E.J.); the Swedish Cancer Society (A.C. and E.J.); the Swedish Foundation for Strategic Research (A.C.); Åke Wibergs stiftelse (A.C. and E.J.); Magnus Bergwalls stiftelse (E.J.); Svenska Smärtafonden (E.J.); Stiftelserna J.C. Kempe och Seth M Kempes Minne (A.C. and N.S.); Medical Faculty of Umeå University; Insamlingsstiftelsen for Medicinsk Forskning vid Umeå Universitet. Funding for open access charge: Swedish Research Council.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENT
We thank Thomas Kunkel for critical reading of the manuscript.
REFERENCES
- 1.Reichard P. Interactions between deoxyribonucleotide and DNA synthesis. Annu. Rev. Biochem. 1988;57:349–374. doi: 10.1146/annurev.bi.57.070188.002025. [DOI] [PubMed] [Google Scholar]
- 2.Gibert I, Calero S, Barbe J. Measurement of in vivo expression of nrdA and nrdB genes of Escherichia coli by using lacZ gene fusions. Mol. Gen. Genet. 1990;220:400–408. doi: 10.1007/BF00391745. [DOI] [PubMed] [Google Scholar]
- 3.Courcelle J, Khodursky A, Peter B, Brown PO, Hanawalt PC. Comparative gene expression profiles following UV exposure in wild-type and SOS-deficient Escherichia coli. Genetics. 2001;158:41–64. doi: 10.1093/genetics/158.1.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tanaka H, Arakawa H, Yamaguchi T, Shiraishi K, Fukuda S, Matsui K, Takei Y, Nakamura Y. A ribonucleotide reductase gene involved in a p53-dependent cell-cycle checkpoint for DNA damage. Nature. 2000;404:42–49. doi: 10.1038/35003506. [DOI] [PubMed] [Google Scholar]
- 5.Nakano K, Balint E, Ashcroft M, Vousden KH. A ribonucleotide reductase gene is a transcriptional target of p53 and p73. Oncogene. 2000;19:4283–4289. doi: 10.1038/sj.onc.1203774. [DOI] [PubMed] [Google Scholar]
- 6.Hakansson P, Hofer A, Thelander L. Regulation of mammalian ribonucleotide reduction and dNTP pools after DNA damage and in resting cells. J. Biol. Chem. 2006;281:7834–7841. doi: 10.1074/jbc.M512894200. [DOI] [PubMed] [Google Scholar]
- 7.Akdemir F, Christich A, Sogame N, Chapo J, Abrams JM. p53 directs focused genomic responses in Drosophila. Oncogene. 2007;26:5184–5193. doi: 10.1038/sj.onc.1210328. [DOI] [PubMed] [Google Scholar]
- 8.Elledge SJ, Davis RW. Two genes differentially regulated in the cell cycle and by DNA-damaging agents encode alternative regulatory subunits of ribonucleotide reductase. Genes Dev. 1990;4:740–751. doi: 10.1101/gad.4.5.740. [DOI] [PubMed] [Google Scholar]
- 9.Elledge SJ, Davis RW. Identification and isolation of the gene encoding the small subunit of ribonucleotide reductase from Saccharomyces cerevisiae: DNA damage-inducible gene required for mitotic viability. Mol. Cell Biol. 1987;7:2783–2793. doi: 10.1128/mcb.7.8.2783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fernandez Sarabia MJ, McInerny C, Harris P, Gordon C, Fantes P. The cell cycle genes cdc22+ and suc22+ of the fission yeast Schizosaccharomyces pombe encode the large and small subunits of ribonucleotide reductase. Mol. Gen. Genet. 1993;238:241–251. doi: 10.1007/BF00279553. [DOI] [PubMed] [Google Scholar]
- 11.Zhao X, Muller EG, Rothstein R. A suppressor of two essential checkpoint genes identifies a novel protein that negatively affects dNTP pools. Mol. Cell. 1998;2:329–340. doi: 10.1016/s1097-2765(00)80277-4. [DOI] [PubMed] [Google Scholar]
- 12.Liu C, Powell KA, Mundt K, Wu L, Carr AM, Caspari T. Cop9/signalosome subunits and Pcu4 regulate ribonucleotide reductase by both checkpoint-dependent and -independent mechanisms. Genes Dev. 2003;17:1130–1140. doi: 10.1101/gad.1090803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hakansson P, Dahl L, Chilkova O, Domkin V, Thelander L. The Schizosaccharomyces pombe replication inhibitor Spd1 regulates ribonucleotide reductase activity and dNTPs by binding to the large Cdc22 subunit. J. Biol. Chem. 2006;281:1778–1783. doi: 10.1074/jbc.M511716200. [DOI] [PubMed] [Google Scholar]
- 14.Zhao X, Chabes A, Domkin V, Thelander L, Rothstein R. The ribonucleotide reductase inhibitor Sml1 is a new target of the Mec1/Rad53 kinase cascade during growth and in response to DNA damage. EMBO J. 2001;20:3544–3553. doi: 10.1093/emboj/20.13.3544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chabes A, Domkin V, Larsson G, Liu A, Graslund A, Wijmenga S, Thelander L. Yeast ribonucleotide reductase has a heterodimeric iron-radical-containing subunit. Proc. Natl Acad. Sci. USA. 2000;97:2474–2479. doi: 10.1073/pnas.97.6.2474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang M, Zhou Z, Elledge SJ. The DNA replication and damage checkpoint pathways induce transcription by inhibition of the Crt1 repressor. Cell. 1998;94:595–605. doi: 10.1016/s0092-8674(00)81601-3. [DOI] [PubMed] [Google Scholar]
- 17.Thelander L, Reichard P. Reduction of ribonucleotides. Annu. Rev. Biochem. 1979;48:133–158. doi: 10.1146/annurev.bi.48.070179.001025. [DOI] [PubMed] [Google Scholar]
- 18.Chabes A, Georgieva B, Domkin V, Zhao X, Rothstein R, Thelander L. Survival of DNA damage in yeast directly depends on increased dNTP levels allowed by relaxed feedback inhibition of ribonucleotide reductase. Cell. 2003;112:391–401. doi: 10.1016/s0092-8674(03)00075-8. [DOI] [PubMed] [Google Scholar]
- 19.Friedberg EC, Walker GC, Siede W, Wood RD, Schultz RA, Ellenberger T. DNA Repair and Mutagenesis. Washington, DC: ASM press; 2005. [Google Scholar]
- 20.Chabes A, Stillman B. Constitutively high dNTP concentration inhibits cell cycle progression and the DNA damage checkpoint in yeast Saccharomyces cerevisiae. Proc. Natl Acad. Sci. USA. 2007;104:1183–1188. doi: 10.1073/pnas.0610585104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Klinkenberg LG, Webb T, Zitomer RS. Synergy among differentially regulated repressors of the ribonucleotide diphosphate reductase genes of Saccharomyces cerevisiae. Eukaryot.Cell. 2006;5:1007–1017. doi: 10.1128/EC.00045-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Longtine MS, McKenzie A, 3rd, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 23.Morrison A, Christensen RB, Alley J, Beck AK, Bernstine EG, Lemontt JF, Lawrence CW. REV3, a Saccharomyces cerevisiae gene whose function is required for induced mutagenesis, is predicted to encode a nonessential DNA polymerase. J. Bacteriol. 1989;171:5659–5667. doi: 10.1128/jb.171.10.5659-5667.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chilkova O, Jonsson BH, Johansson E. The quaternary structure of DNA polymerase epsilon from Saccharomyces cerevisiae. J. Biol. Chem. 2003;278:14082–14086. doi: 10.1074/jbc.M211818200. [DOI] [PubMed] [Google Scholar]
- 25.Asturias FJ, Cheung IK, Sabouri N, Chilkova O, Wepplo D, Johansson E. Structure of Saccharomyces cerevisiae DNA polymerase epsilon by cryo-electron microscopy. Nat. Struct. Mol. Biol. 2006;13:35–43. doi: 10.1038/nsmb1040. [DOI] [PubMed] [Google Scholar]
- 26.Kohda K, Tada M, Kasai H, Nishimura S, Kawazoe Y. Formation of 8-hydroxyguanine residues in cellular DNA exposed to the carcinogen 4-nitroquinoline 1-oxide. Biochem. Biophys. Res. Commun. 1986;139:626–632. doi: 10.1016/s0006-291x(86)80036-5. [DOI] [PubMed] [Google Scholar]
- 27.Pursell ZF, Isoz I, Lundström EB, Johansson E, Kunkel TA. Yeast DNA polymerase epsilon participates in leading strand DNA replication. Science. 2007;317:127–130. doi: 10.1126/science.1144067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nick McElhinny SA, Gordenin DA, Stith CM, Burgers PM, Kunkel TA. Division of labor at the eukaryotic replication fork. Mol. Cell. 2008;30:137–144. doi: 10.1016/j.molcel.2008.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guthrie C, Fink GR. Guide to Yeast Genetics and Molecular Biology. San Diego: Academic Press; 1991. [Google Scholar]
- 30.Kokoska RJ, McCulloch SD, Kunkel TA. The efficiency and specificity of apurinic/apyrimidinic site bypass by human DNA polymerase eta and Sulfolobus solfataricus Dpo4. J. Biol. Chem. 2003;278:50537–50545. doi: 10.1074/jbc.M308515200. [DOI] [PubMed] [Google Scholar]
- 31.Haracska L, Yu SL, Johnson RE, Prakash L, Prakash S. Efficient and accurate replication in the presence of 7,8-dihydro-8-oxoguanine by DNA polymerase eta. Nat. Genet. 2000;25:458–461. doi: 10.1038/78169. [DOI] [PubMed] [Google Scholar]
- 32.Wang PJ, Chabes A, Casagrande R, Tian XC, Thelander L, Huffaker TC. Rnr4p, a novel ribonucleotide reductase small-subunit protein. Mol. Cell Biol. 1997;17:6114–6121. doi: 10.1128/mcb.17.10.6114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Strauss M, Grey M, Henriques JA, Brendel M. RNR4 mutant alleles pso3-1 and rnr4Delta block induced mutation in Saccharomyces cerevisiae. Curr. Genet. 2007;51:221–231. doi: 10.1007/s00294-007-0120-7. [DOI] [PubMed] [Google Scholar]
- 34.Lis ET, O'Neill BM, Gil-Lamaignere C, Chin JK, Romesberg FE. Identification of pathways controlling DNA damage induced mutation in Saccharomyces cerevisiae. DNA Repair. 2008;7:801–810. doi: 10.1016/j.dnarep.2008.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Haracska L, Prakash S, Prakash L. Yeast DNA polymerase zeta is an efficient extender of primer ends opposite from 7,8-dihydro-8-Oxoguanine and O6-methylguanine. Mol. Cell Biol. 2003;23:1453–1459. doi: 10.1128/MCB.23.4.1453-1459.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Niimi A, Limsirichaikul S, Yoshida S, Iwai S, Masutani C, Hanaoka F, Kool ET, Nishiyama Y, Suzuki M. Palm mutants in DNA polymerases alpha and eta alter DNA replication fidelity and translesion activity. Mol. Cell Biol. 2004;24:2734–2746. doi: 10.1128/MCB.24.7.2734-2746.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Haracska L, Unk I, Johnson RE, Johansson E, Burgers PM, Prakash S, Prakash L. Roles of yeast DNA polymerases delta and zeta and of Rev1 in the bypass of abasic sites. Genes Dev. 2001;15:945–954. doi: 10.1101/gad.882301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McCulloch SD, Kokoska RJ, Chilkova O, Welch CM, Johansson E, Burgers PM, Kunkel TA. Enzymatic switching for efficient and accurate translesion DNA replication. Nucleic Acids Res. 2004;32:4665–4675. doi: 10.1093/nar/gkh777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang Y, Wu X, Guo D, Rechkoblit O, Wang Z. Activities of human DNA polymerase kappa in response to the major benzo[a]pyrene DNA adduct: error-free lesion bypass and extension synthesis from opposite the lesion. DNA Repair. 2002;1:559–569. doi: 10.1016/s1568-7864(02)00055-1. [DOI] [PubMed] [Google Scholar]
- 40.Creighton S, Bloom LB, Goodman MF. Gel fidelity assay measuring nucleotide misinsertion, exonucleolytic proofreading, and lesion bypass efficiencies. Methods Enzymol. 1995;262:232–256. doi: 10.1016/0076-6879(95)62021-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.