Abstract
Here, we report the first example that one enantiomer of a supramolecular cylinder can selectively stabilize human telomeric G-quadruplex DNA. The P-enantiomer of this cylinder has a strong preference for G-quadruplex over duplex DNA and, in the presence of sodium, can convert G-quadruplexes from an antiparallel to a hybrid structure. The compound's chiral selectivity and its ability to discriminate quadruplex DNA have been studied by DNA melting, circular dichroism, gel electrophoresis, fluorescence spectroscopy and S1 nuclease cleavage. The chiral supramolecular complex has both small molecular chemical features and the large size of a zinc-finger-like DNA-binding motif. The complex is also convenient to synthesize and separate enantiomers. These results provide new insights into the development of chiral anticancer agents for targeting G-quadruplex DNA.
INTRODUCTION
Human telomeres are essential structures composed of telomeric DNA and telomere-binding proteins located at the ends of every human chromosome. There exists a very strong relationship between maintaining telomeres and tumor progression. Telomere quadruplexes, telomere-specific proteins and associated enzymes are attractive drug targets for cancer chemotherapy and for modulation of gene transcription (1–8). A number of small molecules have been reported to efficiently stabilize G-quadruplex DNA, and recently some metal complexes as G-quadruplex DNA stabilizers were reported (4,9–12). However, to our knowledge, there is no report to show that one of the enantiomers of a chiral compound can selectively stabilize G-quadruplex. We report here that chiral metallo-supramolecular complexes can discriminate G-quadruplex DNA. Only one enantiomer can stabilize human telomeric G-quadruplex DNA and convert antiparallel G-quadruplex to hybrid structure in sodium. Chiral recognition of DNA has been considered important for rational drug design and for structural probes of DNA conformation, such as B–Z DNA transition (13,14). G-quadruplexes are important targets for drug design; therefore, the chiral supramolecular complex reported here may be a potential drug candidate targeting towards G-quadruplex DNA.
MATERIALS AND METHODS
Chemicals and reagents
DNA oligomers, 5′-AGGGTTAGGGTTAGGGTTAGGG-3′ (human telomeric G-quadruplex, ε260 = 228 500 M−1 cm−1/strand), its complementary strand 5′-CCCTAACCCTAACCCTAACCCT-3′ (i-motif, ε260 = 193 700 M−1 cm−1/strand) and fluorescent analogs 5′-Fluoro-AGGGTTAGGGTTAGGGTTAGGG-3′, 2-aminopurine (2-Ap)-labeled individual sequence on TTA loop in three different positions, 5′-TTTTGGGGTTTTGGGGTTTTGGGGTTTTGGGG-3′ (ε260 = 293 800 M−1 cm−1/strand), 5′-GGGGTTGGGGTTGGGGTTGGGG-3′ (ε260 = 213 400 M−1 cm−1/strand), 5′-TGGGGT-3′ (ε260 = 57 800 M−1 cm−1/strand) were purchased from Sangon (Shanghai, China) and used without further purification (7,8). Concentrations of these oligomers were determined by measuring the absorbance at 260 nm after melting (8,13). Extinction coefficients were estimated by the nearest neighbor method by using mononucleotide and dinucleotide values. All the experiments were carried out in 10 mM Tris buffer (100 mM NaCl, pH 7.2) unless stated otherwise. Calf thymus DNA (CT-DNA) was obtained from Sigma (St Louis, MO, USA) and purified as described earlier (8,13). The concentration was determined by ultraviolet absorbance measurements using the extinction coefficient (4): ε260 = 12 824 M−1 cm−1.
The metallo-supramolecular cylinders [M2L3](PF6)4 and [M2L3]Cl4 (M = Ni or Fe) were synthesized following literature methods (15–17). Electrospray ionization mass spectra were recorded on a Finnigan LCQ ion trap mass spectrometer (ThermoFinnigan, San Jose, CA, USA). Element analysis was carried out on Elementar Aanlysensysteme GmbH Vario EL (HAMAU, Germany). 1HNMR spectra were carried out on a Bruker Avance 600 MHz NMR Spectrometer (8). [Ni2L3](PF6)4, ESI-MS (CH3CN): m/z 311.7 ([Ni2L3]4+); elemental analysis: calculated percentage: C, 49.27; H, 3.29; N, 9.20; found percentage: C, 49.48; H, 3.32; N, 9.17. [Fe2L3](PF6)4, ESI-MS (CH3CN): m/z 310.4 ([Fe2L3]4+), 420.1 ([Fe2L3(F)]3+), 1675.5 ([Fe2L3(PF6)3]+); elemental analysis: calculated percentage: C, 49.0; H, 3.6; N, 8.9; found percentage: C, 49.0; H, 3.1; N, 8.7; 1HNMR (600 MHz, CD3CN, 25°C): δ = 8.96 (2H, s, Hi), 8.61 (2H, d, H3), 8.44(2H, t, H4), 7.81(2H, t, H5), 7.39(2H, d, H6), 7.00(4H, br s, Hph), 5.59 (4H, br s, Hph), 4.08 (2H, s, CH2 spacer). The racemic [M2L3]Cl4 was concentrated by lyophilization and enantiomerically pure material was obtained by using a cellulose (∼20 μ, Aldrich, Steinheim, Germany, lot: 03016MB-444) column and eluting with 20 mM NaCl aqueous solution (17). UV–Vis spectroscopy was used to determine the enantiomer concentration and CD spectra of the two enantiomers prepared at the same concentration were used to estimate their purity (13,17). The samples of purified M- and P-enantiomer were collected and freeze-dried, respectively for future use.
Absorbance and UV melting
Absorbance measurements and melting experiments were carried out on a Cary 300 UV/Vis spectrophotometer equipped with a Peltier temperature control accessory. All UV/Vis spectra were measured in 1.0-cm path-length cell with the same concentration of corresponding metal complex aqueous solution as the reference solution. Absorbance changes at either 260 nm or 295 nm versus temperature were collected (8,13) at a heating rate of 1°C·min−1.
Circular dichroism measurements
Circular dichroism (CD) spectra and CD melting experiments were carried out on a JASCO J-810 spectropolarimeter equipped with a temperature controlled water bath (7,8,13). The optical chamber of CD spectrometer was deoxygenated with dry purified nitrogen (99.99%) for 45 min before use and kept the nitrogen atmosphere during experiments. Three scans were accumulated and automatically averaged. The various concentration of M- or P-cylinder was scanned (8,13) as a control and subtracted from the spectra of metal cylinder/DNA mixture to eliminate its influence on DNA CD signal between 340 and 220 nm.
Fluorescence measurements
Fluorescence measurements were carried out on a JASCO FP-6500 spectrofluorometer at 20°C. Fluorescence spectra of 2-Ap-labeled G-quadruplex DNA were measured by using an excitation wavelength of 305 nm and recorded from 320 nm to 500 nm in the absence or presence of different amount of metal cylinder (8,13). The concentration of 2-Ap-labeled G-quadruplex DNA was fixed at 1 μM in strand.
Gel electrophoresis and S1 nuclease cleavage
Native gel electrophoresis was carried out on acrylamide gel (20%) and run at 4°C, 12.5 V cm−1 in 1 × TB buffer containing 10 mM NaCl or 10 mM KCl and was silver stained.
S1 nuclease digestion and PAGE using fluorescent oligonucleotide were carried out as described previously (8). Reaction mixtures contained S1 buffer and 3 U of S1 nuclease. The samples of 5′-fluorescein-labeled oligomer (5 μl × 10 μM), and the mixtures of 5′-fluorescein-labeled oligomer–metal complex were incubated before initiating digestion at 37°C by adding S1 nuclease and Zn2+. After 5 min, digestions were stopped by adding 4 μl of stop buffer (70% formamide/57 mM EDTA, pH 7.5) and then freezing. The frozen samples were treated with 1 μl of formamide, heated at 95°C for 3 min. They were loaded on a 20% denaturing gel electrophoresis containing 7 M urea and electrophoresed at room temperature at 20 V cm−1.
RESULTS AND DISCUSSION
Enantioselectivity and discrimination between quadruplex and duplex DNA
Supramolecular chemistry has been described as an information science (18,19). It provides an excellent methodology for designing large synthetic compounds targeting DNA major groove (20,21), because the compound can have similar size like DNA-binding protein recognition motifs (such as zinc fingers or α-helices) and multiple cationic charges favoring the noncovalent binding to anionic DNA. The chiral compound we used, [M2L3]4+ (M = Ni2+ or Fe2+), has a bimetallo triple helicate structure (15–17). Structures and CD spectra of the two pairs of enantiomers, M and P, are shown in Figure 1. Each enantiomer has a hydrophobic surface and a size (15–17) (length ∼18 Å, diameter ∼8 Å) compatible with G-quartet [length ∼14 Å, width (22) ∼11 Å], and the positive charged triple helical structure has the potential to interact with the loops and grooves of G-quadruplex, which has been proposed for zinc-finger protein and macrocyclic and helical oligoamides binding (6,23).
Figure 1.
(A) Structures of the M-enantiomer (left) and P-enantiomer (right) of [Ni2L3]4+ cation. Nickel: gray; nitrogen: yellow; carbon atoms in three ligand L are shown in red, green and blue, respectively. Hydrogen atoms are omitted for clarity. The crystal data of [Ni2L3]4+ are from the Cambridge Crystallographic Data Centre CCDC 182/570 (15). (B) Structures of the M-enantiomer (left) and P-enantiomer (right) of [Fe2L3]4+ cation. Iron atoms are in purple, other atoms are the same as in [Ni2L3]4+. The crystal data of [Fe2L3]4+ are from the Cambridge Crystallographic Data Centre CCDC 622770 (16). (C) CD spectra of the M-enantiomer (black) and P-enantiomer (red) of [Ni2L3]4+; (D) CD spectra of the M-enantiomer (black) and P-enantiomer (red) of [Fe2L3]4+. The CD spectra are measured at the concentration of 10 µM for each enantiomer in 100 mM NaCl, 10 mM Tris buffer (pH 7.2).
The chiral selectivity on stabilization of human telomeric G-quadruplex DNA by [Ni2L3]4+-M and [Ni2L3]4+-P enantiomer is remarkable (Figure 2). Nonfluorescent-labeled DNA UV melting studies (7,8) directly demonstrate that P-enantiomer can increase G-quadruplex melting temperature (Tm) 10°C at 1:1 ratio of [complex]/[DNA], but M cannot increase Tm even at higher ratio (Figure 2). For i-motif DNA and G-quadruplex/i-motif complementary duplex DNA (4) (Figure 2), the two enantiomers do not show chiral selectivity. Both of them destabilize i-motif DNA and do not influence G-quadruplex/i-motif duplex DNA stability.
Figure 2.
Plot of DNA stabilization temperature versus the concentration of [Ni2L3]4+-P (filled symbols) or [Ni2L3]4+-M (open symbols) binding to G-quadruplex (black squares), G-quadruplex/i-motif complementary duplex (red circles) and i-motif DNA (green diamonds). Insert: UV melting profiles of G-quadruplex DNA (1 µM/strand) in the absence (blue) or presence of 1 µM P-enantiomer (cyan) or M-enantiomer (magenta) in 10 mM Tris buffer containing 100 mM NaCl, pH 7.2.
[Fe2L3]4+-P enantiomer shows the same chiral selectivity on G-quadruplex as [Ni2L3]4+-P enantiomer (Table 1). This indicates that P-enantiomer has stronger selectivity to G-quadruplex than M and the different central metal ion, Fe2+ or Ni2+, does not alter their chiral selectivity. We also studied the effect of ligand L on DNA stability. As shown in Table 1, clearly, ligand L does not affect DNA stability (Table 1) under our experimental conditions. Both [Ni2L3]4+ and [Fe2L3]4+ enantiomers can slightly increase CT-DNA Tm, about 0.5°C for [Ni2L3]4+ and 1°C for [Fe2L3]4+ enantiomers in 100 mM NaCl, 10 mM (pH 7.2) Tris buffer (24), demonstrating that [Ni2L3]4+-P enantiomer has even stronger preference to G-quadruplex than [Fe2L3]4+-P enantiomer, and the difference may be due to their subtle structural differences (15–17). [Ni2L3]4+-P enantiomer has a more than 20-fold selectivity for G-quadruplex over duplex DNA (4), and the selectivity is general, better than BRACO-19 (4), a lead G-quadruplex-interactive molecule which can inhibit tumor growth in vivo and has been in clinical trials.
Table 1.
Stabilization temperature (ΔTm) of G-quadruplex DNA, i-motif DNA, dsDNA (G-quadruplex/i-motif complementary duplex DNA) and CT-DNA by ligand (L, 3 µM), M-enantiomer (M, 1 µM) and P-enantiomer (P, 1 µM) of [Ni2L3]4+ and [Fe2L3]4+, respectively, in 10 mM Tris buffer containing 100 mM NaCl at pH 7.2 or pH 5.5 (for i-motif DNA melting studies)
| Compound | ΔTm (°C) |
|||
|---|---|---|---|---|
| G-quadruplex | i-motif | dsDNA | CT-DNA | |
| L | 0 | −0.5 | 0 | 0 |
| [Ni2L3]4+-M | 0 | −2.5 | 0 | 0.7 |
| [Ni2L3]4+-P | 10.6 | −2.8 | 0 | 0.5 |
| [Fe2L3]4+-M | 0 | −2.2 | 0 | 1.3 |
| [Fe2L3]4+-P | 10.8 | −2.8 | 0 | 1.0 |
The DNA concentration was 1 µM/strand or in equivalent concentration for dsDNA.
The chiral selectivity of the two enantiomers binding to human telomeric G-quadruplex is further studied by gel electrophoresis (Figure 3). In the presence of P-enantiomer, a new band with mobility slower than that of G-quadruplex DNA alone was observed. The retarded migration is due to the molecular weight/charge contribution of supramolecule to supramolecule–quadruplex complex (25). While in the presence of M-enantiomer, no delayed band was observed (Figure 3). These results are in accordance with DNA melting data (Figure 2).
Figure 3.
Native gel electrophoretic analysis (20% PAGE) of human telomeric d[AG3(T2AG3)3] in the presence of various concentration of [Ni2L3]4+ (A and C) and [Fe2L3]4+ (B and D). The gels were run in TB buffer with 10 mM NaCl (A and B) or 10 mM KCl (C and D). Lane 1 was the DNA alone. Samples of lanes 2–4 were prepared as DNA with P-enantiomer at the ratios of 2:1, 1:1 and 2:3. Samples of lanes 5–7 were prepared as DNA with M-enantiomer at the ratios of 2:1, 1:1 and 2:3.
To our knowledge, supramolecular [Ni2L3]4+ complex binding to any DNA has not been reported. Hannon and co-workers (26) have shown that both [Fe2L3]4+-M and [Fe2L3]4+-P can increase CT-DNA Tm > 10°C at low-salt concentration (20 mM NaCl, 1 mM cacodylate buffer, pH 6.8) and M-enantiomer has even stronger effect (26,27). Their recent studies compellingly demonstrate that the same helicates recognize specific DNA three-way junctions (28,29), and also show anticancer activity (30). Inspired by these findings, we focus on [Ni2L3]4+ enantiomers binding to DNA and find their chiral selectivity to G-quadruplex. At physiological salt concentration as we used (100 mM NaCl, 10 mM Tris buffer, pH 7.2), M and P can only slightly increase CT-DNA Tm. It should be pointed out that this difference can be due to the different experimental conditions. We use higher ionic strength which can decrease the positively charged complex binding to duplex DNA. This indicates that the electrostatic effect is playing an important role for this complex binding to duplex DNA major groove (26), while the chiral selectivity is essential when the complex binding to G-quadruplex (Figure 2). According to G-quadruplex melting data measured under different ionic strength conditions (Figure 4), and the equation (31): 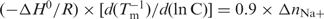 , where the values of ΔH0 are calculated (8) from the melting curves (ΔH0G-DNA, ΔH0G-DNA-M and ΔH0G-DNA-P, are 157.3 ± 0.2, −145.6 ± 0.6 and −135.7 ± 0.5 kJ mol−1 for G-quadruplex DNA alone, G-quadruplex DNA with M- or P-enantiomer, respectively), the number of sodium ion releasing (31) is calculated about 1.8 sodium no matter in the absence or presence of the enantiomer, consistent with G-quadruplex DNA NMR analysis (32). Therefore, M and P binding to G-quadruplex do not lead to more sodium ion release.
, where the values of ΔH0 are calculated (8) from the melting curves (ΔH0G-DNA, ΔH0G-DNA-M and ΔH0G-DNA-P, are 157.3 ± 0.2, −145.6 ± 0.6 and −135.7 ± 0.5 kJ mol−1 for G-quadruplex DNA alone, G-quadruplex DNA with M- or P-enantiomer, respectively), the number of sodium ion releasing (31) is calculated about 1.8 sodium no matter in the absence or presence of the enantiomer, consistent with G-quadruplex DNA NMR analysis (32). Therefore, M and P binding to G-quadruplex do not lead to more sodium ion release.
Figure 4.
Variation of the reciprocal melting temperature for G-quadruplex in the absence (black squares) or presence of [Ni2L3]4+-P (red circles) and [Ni2L3]4+-M (blue uptriangles) with the logarithm of NaCl concentration in 10 mM Tris buffer (pH 7.2).
Mode of binding
2-Ap fluorescence modified in different loops (Table 2) has been widely used to verify the mode of ligand binding to G-quadruplex (33) and i-motif DNA (8). Our results show that P-enantiomer can decrease 2-Ap fluorescence (8,33) and the decrease follows the order: A7 ≈ A19 > A13 (Figure 5A). This indicates that P-enantiomer may preferentially bind to the end of G-quartet by external stacking (2,33,34) and leads to the decrease of 2-Ap fluorescence (8) labeled in the two lateral loops (A7 and A19), which is consistent with a breakpoint observed at 1:1 binding ratio (Figure 6) in CD titrations (18). The interactions of the positive charged triple helical structure with the two lateral loops, grooves and DNA backbone (6,23,26) can further stabilize P-enantiomer binding. Nonlinear least-squares analysis of the fluorescence titration data (8) of A7 or A19 by P-enantiomer yielded an association constant of 2.6 ± 0.6 × 107 M−1 (Table S1). For M-enantiomer, its binding hardly decreases the 2-Ap fluorescence modified in any of the three loops (Figure 5A), and a breakpoint was observed at 1.67:1 binding ratio in CD titrations (Figure 6). These results indicate that M-enantiomer, like terbium–amino acid complex (34), may take electrostatic nonspecific binding to G-quadruplex which binding mode has been proposed by Hurley group and ours (3,34). The different binding mode of the two enantiomers is further supported by S1 nuclease digestion (8). Figure 5B shows the enzyme cleavage patterns of 5′-fluorescein-labeled quadruplex DNA after S1 nuclease digestion (8). Digestion by S1 nuclease resulted in three major cleavages (lane 2, DNA alone) occurred at three loops (8), consistent with previous studies on i-motif DNA (8) and c-myc promoter cleavage (35). In the presence of P-enantiomer, the amount of the cleavage at 5′-end (band 4) and at 3′-end (band 2) decreased dramatically while the cleavage at the diagonal loop (band 3) was hardly influenced. In the presence of M-enantiomer, the cleavage pattern was almost the same as that of DNA alone. This is in accordance with the 2-Ap fluorescence results and further supports that P-enantiomer may bind to the end of G-quartet by external stacking and contact with the two lateral loops.
Table 2.
Sequences of fluorescent analogs of human telomeric G-quadruplex DNA labeled by 2-Ap in three TTA loops
| Labeled position | Sequence |
|---|---|
| A7 | 5′-AGGGTT(2-Ap)GGGTTAGGGTTAGGG-3′ |
| A13 | 5′-AGGGTTAGGGTT(2-Ap)GGGTTAGGG-3′ |
| A19 | 5′-AGGGTTAGGGTTAGGGTT(2-Ap)GGG-3′ |
Figure 5.
Fluorescence changes and S1 digestion of human telomeric G-quadruplex in the presence of chiral metal complexes. (A) Plot of fluorescence intensity at 370 nm of 2-Ap individually labeled G-quadruplex versus binding ratio of complex/DNA in 100 mM NaCl, 10 mM Tris buffer (pH 7.2). Closed squares (black), [P-[Ni2L3]4+]/[A7]; closed circles (red), [P-[Ni2L3]4+]/A13; closed triangles (green), [P-[Ni2L3]4+]/[A19]; open squares (blue), [M-[Ni2L3]4+]/[A7]; open circles (cyan), [M-[Ni2L3]4+]/[A13]; open triangles (magenta), [M-[Ni2L3]4+]/[A19]. DNA concentration was fixed at 1 µM/strand. (B) Image of fluorescent denaturing PAGE (20%) after S1 digestion. Lane 1, untreated 5′-fluorescein-labeled 22-mer AG3(T2AG3)3; Lane 2, S1-treated DNA; Lanes 3–5, S1 treated the mixture of DNA with P-[Ni2L3]4+ at the ratio of 2:1, 1:1 and 2:3; Lanes 6–8, S1 treated the mixture of DNA with M-[Ni2L3]4+ at the ratio of 2:1, 1:1 and 2:3.
Figure 6.
(A) CD titration of d[AG3(T2AG3)3] with [Ni2L3]4+-P in 100 mM NaCl, 10 mM Tris buffer (pH 7.2) at 20°C. The concentration of [Ni2L3]4+-P was varied from 0 µM to 3 µM. (B) The change in ellipticity at 265 nm with increased concentration of [Ni2L3]4+-P derived from the CD titration, a breakpoint was observed at 1:1 ratio. DNA concentration was 1 µM/strand. (C) CD titration of d[AG3(T2AG3)3] with [Ni2L3]4+-M in 100 mM NaCl, 10 mM Tris buffer (pH 7.2) at 20°C. The concentration of [Ni2L3]4+-M was varied from 0 to 3 µM. (D) The change in ellipticity at 265 nm with increased concentration of [Ni2L3]4+-M derived from the CD titration, a breakpoint was observed at 1.67:1 ratio. DNA concentration was 1 µM/strand.
Discrimination between quadruplexes
We also studied another two distinct telomeric G-quadruplex DNA: tetrahymena G4(T2G4)3, a monomeric hybrid structure (37) and oxytricha telomeric DNA (T4G4)4, a monomeric antiparallel structure (38), and compared with human telomeric G-quadruplex DNA in sodium and in potassium (39,40) buffer (Table 3). Clearly, no matter in the presence of sodium or potassium, both M- and P-enantiomers destabilize these two telomeric DNA without chiral selectivity. The results of gel electrophoresis (Figure S1) show that there are no new bands emerged in the case of tetrahymena or oxytricha telomeric DNA with either M- or P-enantiomer indicating that the two enantiomers bind weakly to the two DNA. These results demonstrate that P-enantiomer is capable of discriminating between quadruplexes (Figure S1). We also examined the interactions of M- and P-enantiomer with a parallel-stranded tetramolecular quadruplex TG4T (41) by means of CD melting and gel electrophoresis. Both M- and P-enantiomer binding destabilized the quadruplex stability (Figure S2A), and no new band was observed in the gel (Figure S2B). The enantiomer binding to unfolded quadruplexes and the detailed destabilization mechanism are not clear yet and we are undertaking further studies. The results presented here indicate that the chiral selective stabilization of human telomeric G-quadruplex is not only related to G-quadruplex topology, but also related to the sequence and the loop constitution. Furthermore, we extend our work by investigating the effect of the 5′- or 3′-flanking sequence of human telomeric G-quadruplex (42) on the chiral selectivity of the enantiomers. As shown in Table 3, for DNA alone, its stability is decreased with longer flanking sequence. However, flanking sequence does not influence P-enantiomer chiral selectivity either in sodium or in potassium buffer (Table 3). This demonstrates that the 5′- or 3′-end capping sequence (42) does not influence P-enantiomer binding to the end of G-quartet (Scheme 1) by external stacking further supporting the 2-Ap fluorescence results (8,33) and S1 nuclease cleavage data.
Table 3.
Stabilization effect of [Ni2L3]4+-M (M) and [Ni2L3]4+-P (P) on different telomeric G-quadruplex DNA in 10 mM Tris, 100 mM NaCl (or 10 mM KCl), pH 7.2 buffer
| DNA sequence | In sodium buffer |
In potassium buffer |
||||||
|---|---|---|---|---|---|---|---|---|
| Structure | DNA Tm | DNA+M ΔTm (°C) | DNA+P ΔTm (°C) | Structure | DNA Tm | DNA+M ΔTm (°C) | DNA+P ΔTm (°C) | |
| 22mer-AG3(T2AG3)3 | Human Telomeric Repeat, Antiparallel36 | 59.6 | 0 | 10.6 | Human Telomeric Repeat, Hybrid | 54.6 | 11.7 | 19.8 |
| 22mer- G4(T2G4)3 | Tetrahymena Telomeric Repeat, Hybrid37 | 62.2 | −3.1 | −1.9 | Tetrahymena Telomeric Repeat, Hybrid39 | 77.6 | −4.4 | −5.0 |
| 32mer-(T4G4)4 | Oxytricha Telomeric Repeat, Antiparallel38 | 62.8 | −5.0 | −5.0 | Oxytricha Telomeric Repeat, Antiparallel40 | 86.9 | −6.0 | −5.7 |
| 26mer AA-AG3(T2AG3)3-AA | Antiparallel | 47.5 | −0.8 | 10.2 | Hybrid42 | 61.8 | 7.0 | 18.8 |
| 26mer TT-AG3(T2AG3)3-TT | Antiparallel | 52.6 | 0 | 9.8 | Hybrid42 | 59.8 | 5.2 | 14.1 |
| 24mer T-AG3(T2AG3)3-T | Antiparallel | 54.3 | −0.2 | 9.2 | Hybrid42 | 66.6 | 5.1 | 13.2 |
Meltings of human telomeric G-quadruplex DNA with different flanking sequences on the 5′ and 3′ terminus were measured in 10 mM Tris, 100 mM NaCl (or 100 mM KCl), pH 7.2 buffer. M- or P-enantiomer was fixed at 1 µM. DNA concentration was 1 µM/strand. The bold data serves as telomeric DNA reference.
Scheme 1.
Representative illustration of chiral supramolecular complex selective recognition of human telomeric G-quadruplex DNA.
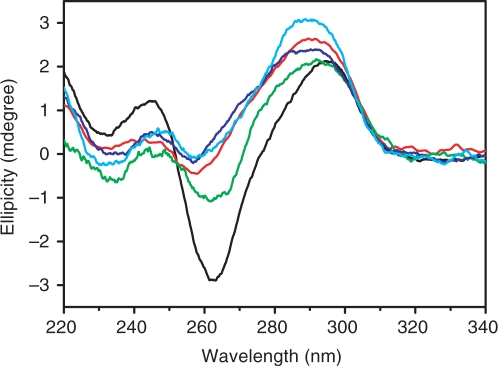
Human telomeric G-quadruplex DNA is polymorphic, which can form an antiparallel structure (7,36) in the presence of sodium or adopt a hybrid structure in the presence of potassium. With addition of DNA equimolar M- or P-enantiomer in sodium, unexpectedly, P-enantiomer binding can induce DNA positive CD band intensity significantly increased and shifted from 295 to 290 nm. The negative band intensity simultaneously decreased and shifted from 265 to 256 nm (Figure 7, red line). This phenomenon is similar to the structural transition of quadruplex alone occurred in Na+ buffer titrated (23) by K+ (Figure 7, cyan line). Besides, similar G-quadruplex CD spectra were observed upon P-enantiomer binding no matter in Na+ or in K+ buffer (Figure S3). It should be pointed out that the CD band of the enantiomers at longer wavelength does not change when binding to G-quadruplex. As for M-enantiomer, its binding can just decrease the negative band intensity at 265 nm without band shift and cannot increase the positive band intensity (Figure 7, green line). However, the 1:1 mixture (13) of M and P (Figure 7, blue line) shows the same effect as P-enantiomer does. These results indicate that only P-enantiomer can convert G-quadruplex (Scheme 1) from antiparallel to hybrid structure (42), similar to previous reports that Se2SAP and TMPYP4 prefer antiparallel/parallel hybrid structure (3), but in contrast with telomestatin and macrocylic and helical oligoamides (23), which favor the antiparallel structure. DNA UV melting studies in K+ buffer (Figure 8) further support that P-enantiomer prefers the hybrid structure over M because P-enantiomer can increase Tm 20°C and M-enantiomer can only increase Tm 12°C. The difference in Tm increase shows P-enantiomer chiral preference even in K+ buffer. This can be the reason why P-enantiomer binding can drive G-quadruplex from antiparallel to hybrid structural transition in Na+ while M binding cannot. Gel electrophoresis (Figure 3B) and 2-Ap fluorescence titration data (Figure S4) obtained in K+ buffer further demonstrate that P-enantiomer prefers hybrid structure over M no matter in Na+ or in K+ buffer. Our preliminary data in cancer cells indicate that the two enantiomers show telomerase inhibition (Figure S5), and influence telomere shortening, β-galactosidase activity and upregulation of cyclin-dependent kinase (CDK) inhibitors p16 and p21 (data not shown), further studies are undergoing and will be reported in due course.
Figure 7.
CD spectra of human telomeric DNA d[AG3(T2AG3)3] (1 µM/strand) in the absence (black) or presence of 1 µM P-enantiomer (red), 1 µM M-enantiomer (green), 1:1 mixer (blue) of M and P and 2 mM K+ (cyan) in 10 mM Tris, 10 mM NaCl, pH 7.2 buffer. CD spectra are obtained by individual background subtraction.
Figure 8.
The melting profiles of human telomeric G-quadruplex (1 µM/strand) in the absence (black) or presence of 1 µM M (red) or P-enantiomer (blue) of [Ni2L3]4+ (A) and [Fe2L3]4+ (B) in 10 mM KCl, 10 mM Tris buffer, pH 7.2.
CONCLUSIONS
In summary, we report here that one of the enantiomers of a chiral metal complex is capable of discriminating between quadruplexes. The P-enantiomer selectively stabilizes human telomeric G-quadruplex DNA and can convert G-quadruplex from antiparallel to hybrid structure in sodium showing remarkable chiral preference. These findings will prompt the rational design and discovery of chiral anticancer agents targeting towards G-quadruplex DNA.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
NSFC; CAS and Jilin Province. Funding for open access charge: National Natural Science Foundation of China and Chinese Academy of Sciences.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors are grateful for the referees’ helpful comments on the article. They would also like to thank Drs Y. Zheng and H. Zhang for their technical assistance.
REFERENCES
- 1.Blackburn EH. Structure and function of telomeres. Nature. 1991;350:569–573. doi: 10.1038/350569a0. [DOI] [PubMed] [Google Scholar]
- 2.Mergny JL, Hélène C. G-quadruplex DNA: a target for drug design. Nature. 1998;4:1366–1367. doi: 10.1038/3949. [DOI] [PubMed] [Google Scholar]
- 3.Rezler EM, Seenisamy J, Bashyam S, Kim M.-Y, White E, Wilson WD, Hurley LH. Telomestatin and diseleno sapphyrin bind selectively to two different forms of the human telomeric G-quadruplex structure. J. Am. Chem. Soc. 2005;127:9439–9447. doi: 10.1021/ja0505088. and cited references. [DOI] [PubMed] [Google Scholar]
- 4.Reed JE, Arnal AA, Neidle S, Vilar R. Stabilization of G-quadruplex DNA and inhibition of telomerase activity by square-planar nickel complexes. J. Am. Chem. Soc. 2006;128:5992–5993. doi: 10.1021/ja058509n. and cited references. [DOI] [PubMed] [Google Scholar]
- 5.Li W, Wu P, Ohmichi T, Sugimoto N. Characterization and thermodynamic properties of quadruplex/duplex competition. FEBS Lett. 2002;526:77–81. doi: 10.1016/s0014-5793(02)03118-6. [DOI] [PubMed] [Google Scholar]
- 6.Ladame S, Schouten JA, Roldan J, Redman JE, Neidle S, Balasubramanian S. Exploring the recognition of quadruplex DNA by an engineered Cys2-His2 zinc finger protein. Biochemistry. 2006;45:1393–1399. doi: 10.1021/bi050229x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ren J, Qu X, Trent JO, Chaires JB. Tiny telomere DNA. Nucleic Acids Res. 2002;30:2307–2315. doi: 10.1093/nar/30.11.2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li X, Peng Y, Ren J, Qu X. Carboxyl-modified single-walled carbon nanotubes selectively induce human telomeric i-motif formation. Proc. Natl Acad. Sci. USA. 2006;103:19658–19663. doi: 10.1073/pnas.0607245103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Reed JE, Neidle S, Vilar R. Stabilisation of human telomeric quadruplex DNA and inhibition of telomerase by a platinum-phenanthroline complex. Chem. Commun. 2007:4366–4368. doi: 10.1039/b709898g. [DOI] [PubMed] [Google Scholar]
- 10.Bertrand H, Monchaud D, De Cian A, Guillot R, Mergny JL, Teulade-Fichou MP. The importance of metal geometry in the recognition of G-quadruplex-DNA by metal-terpyridine complexes. Org. Biomol. Chem. 2007;5:2555–2559. doi: 10.1039/b708635k. [DOI] [PubMed] [Google Scholar]
- 11.Kieltyka R, Fakhoury J, Moitessier N, Sleiman HF. Platinum phenanthroimidazole complexes as G-quadruplex DNA selective binders. Chem. Eur. J. 2008;14:1145–1154. doi: 10.1002/chem.200700783. [DOI] [PubMed] [Google Scholar]
- 12.Bertrand H, Bombard S, Monchaud D, Teulade-Fichou MP. A platinum-quinacridine hybrid as a G-quadruplex ligand. J. Biol. Inorg. Chem. 2007;12:1003–1014. doi: 10.1007/s00775-007-0273-3. [DOI] [PubMed] [Google Scholar]
- 13.Qu X, Trent JO, Fokt I, Priebe W, Chaires JB. Allosteric, chiral-selective drug binding to DNA. Proc. Natl Acad. Sci. USA. 2000;97:12032–12037. doi: 10.1073/pnas.200221397. and cited references. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xu Y, Zhang Y, Sugiyama H, Umano T, Osuga H, Tanaka K. (P)-helicene displays chiral selection in binding to Z-DNA. J. Am. Chem. Soc. 2004;126:6566–6567. doi: 10.1021/ja0499748. [DOI] [PubMed] [Google Scholar]
- 15.Hannon MJ, Painting CL, Hamblin J, Jackson A, Errington W. An inexpensive approach to supramolecular architecture. Chem. Commun. 1997:1807–1808. [Google Scholar]
- 16.Kerckhoffs JMCA, Peberdy JC, Meistermann I, Childs LJ, Isaac CJ, Pearmund CR, Reudegger V, Khalid S, Alcock NW, Hannon MJ, et al. Enantiomeric resolution of superamolecular helicates with different surface topographies. Dalton Trans. 2007:734–742. doi: 10.1039/b614093a. [DOI] [PubMed] [Google Scholar]
- 17.Hannon MJ, Meistermann I, Isaac CJ, Blomme C, Aldrich-Wright J, Rodger A. Paper: a cheap yet effective chiral stationary phase for chromatographic resolution of metallo-supramolecular helicates. Chem. Commun. 2001:1078–1079. [Google Scholar]
- 18.Davis AV, Yeh RM, Raymond KN. Supramolecular assembly dynamics. Proc. Natl Acad. Sci. USA. 2002;99:4793–4796. doi: 10.1073/pnas.052018299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lehn J-M. From supramolecular chemistry towards constitutional dynamic chemistry and adaptive chemistry. Chem. Soc. Rev. 2007;36:151–160. doi: 10.1039/b616752g. [DOI] [PubMed] [Google Scholar]
- 20.Schoentjes B, Lehn J-M. Interaction of double-helical polynuclear copper(I) complexes with double-stranded DNA. Helv. Chim. Acta. 1995;78:1–12. [Google Scholar]
- 21.Hannon MJ. Supramolecular DNA recognition. Chem. Soc. Rev. 2007;36:280–295. doi: 10.1039/b606046n. [DOI] [PubMed] [Google Scholar]
- 22.De Cian A, Delemos E, Mergny JL, Teulade-Fichou MP, Monchaud D. Highly efficient G-quadruplex recognition by bisquinolinium compounds. J. Am. Chem. Soc. 2007;129:1856–1857. doi: 10.1021/ja067352b. [DOI] [PubMed] [Google Scholar]
- 23.Shirude PS, Gillies ER, Ladame S, Godde F, Shin-ya K, Huc I, Balasubramanian S. Macrocyclic and helical oligoamides as a new class of G-quadruplex ligands. J. Am. Chem. Soc. 2007;129:11890–11891. doi: 10.1021/ja073775h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang H, Yu H, Ren J, Qu X. Reversible B/Z-DNA transition under the low salt condition and Non-B-Form PolydApolydT selectivity by a cubane-like europium-L-aspartic acid complex. Biophys. J. 2006;90:3203–3207. doi: 10.1529/biophysj.105.078402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.De Cian A, Mergny JL. Quadruplex ligands may act as molecular chaperones for tetramolecular quadruplex formation. Nucleic Acids Res. 2007;35:2483–2493. doi: 10.1093/nar/gkm098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Meistermann I, Moreno V, Prieto MJ, Moldrheim E, Sletten E, Khalid S, Rodger PM, Peberdy JC, Isaac CJ, Rodger A, et al. Intramolecular DNA coiling mediated by metallosupramolecular cylinders: differential binding of P and M helical enantiomers. Proc. Natl Acad. Sci. USA. 2002;99:5069–5074. doi: 10.1073/pnas.062634499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Malina J, Hannon MJ, Brabec V. DNA binding of dinuclear iron(II) metallosupramolecular cylinders. DNA unwinding and sequence preference. Nucleic Acids Res. 2008;36:3630–3638. doi: 10.1093/nar/gkn244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Oleksy A, Blanco AG, Boer R, Usón I, Aymamí J, Rodger A, Hannon MJ, Coll M. Molecular recognition of a three-way DNA junction by a metallosupramolecular helicate. Angew. Chem. Int. Ed. 2006;45:1227–1231. doi: 10.1002/anie.200503822. [DOI] [PubMed] [Google Scholar]
- 29.Malina J, Hannon MJ, Brabec V. Recognition of DNA three-way junctions by metallosupramolecular cylinders: gel electrophoresis studies. Chemstry. 2007;13:3871–3877. doi: 10.1002/chem.200700159. [DOI] [PubMed] [Google Scholar]
- 30.Pascu GI, Hotze AC, Sanchez-Cano C, Kariuki BM, Hannon MJ. Dinuclear ruthenium(II) triple-stranded helicates: luminescent supramolecular cylinders that bind and coil DNA and exhibit activity against cancer cell lines. Angew. Chem. Int. Ed. 2007;46:4374–4378. doi: 10.1002/anie.200700656. and cited references. [DOI] [PubMed] [Google Scholar]
- 31.Spink CH, Chaires JB. Effects of hydration, ion release, and excluded volume on the melting of triplex and duplex DNA. Biochemistry. 1999;38:496–508. doi: 10.1021/bi9820154. [DOI] [PubMed] [Google Scholar]
- 32.Hud NV, Smith FW, Anet FAL, Feigon J. The Selectivity for K+ versus Na+ in DNA quadruplexes is dominated by relative free energies of hydration: a thermodynamic analysis by 1H NMR. Biochemistry. 1996;35:15383–15390. doi: 10.1021/bi9620565. [DOI] [PubMed] [Google Scholar]
- 33.Kimura T, Kawai K, Fujitsuka M, Majima T. Detection of the G-quadruplex-TMPyP4 complex by 2-aminopurine modified human telomeric DNA. Chem. Commun. 2006;401:402. doi: 10.1039/b514526k. [DOI] [PubMed] [Google Scholar]
- 34.Xu H, Zhang H, Qu X. Interactions of the human telomeric DNA with terbium–amino acid complexes. J. Inorg. Biochem. 2006;100:1646–1652. doi: 10.1016/j.jinorgbio.2006.05.015. [DOI] [PubMed] [Google Scholar]
- 35.Kumar P, Verma A, Maiti S, Gargallo R, Chowdhury S. Tetraplex DNA transtions within the human c-myc promoter detected by multivariate curve resolution of fluorescence resonance energy transfer. Biochemistry. 2005;44:16426–16434. doi: 10.1021/bi051452x. [DOI] [PubMed] [Google Scholar]
- 36.Wang Y, Patel DJ. Solution structure of the human telomeric repeat d[AG3(T2AG3)3] G-tetraplex. Structure. 1993;1:263–282. doi: 10.1016/0969-2126(93)90015-9. [DOI] [PubMed] [Google Scholar]
- 37.Wang Y, Patel DJ. Solution structure of the Tetrahymena telomeric repeat d(T2G4)4 G-tetraplex. Structure. 1994;2:1141–1156. doi: 10.1016/s0969-2126(94)00117-0. [DOI] [PubMed] [Google Scholar]
- 38.Wang Y, Patel DJ. Solution structure of the Oxytricha telomeric repeat d[G4(T4G4)3] G-tetraplex. J. Mol. Biol. 1995;251:76–94. doi: 10.1006/jmbi.1995.0417. [DOI] [PubMed] [Google Scholar]
- 39.Dapić V, Abdomerović V, Marrington R, Peberdy J, Rodger A, Trent JO, Bates PJ. Biophysical and biological properties of quadruplex oligodeoxyribonucleotides. Nucleic Acids Res. 2003;31:2097–2107. doi: 10.1093/nar/gkg316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Balagurumoorthy P, Brahmachari SK. Intra- and interloop interactions in the folded G quartet structure of Oxytricha telomeric sequence. Indian J. Biochem. Biophys. 1995;32:385–390. [PubMed] [Google Scholar]
- 41.Aboul-ela F, Murchie AIH, Lilley DMJ. NMR study of parallel-stranded tetraplex formation by the hexadeoxynucleotide d(TG4T) Nature. 1992;360:280–282. doi: 10.1038/360280a0. [DOI] [PubMed] [Google Scholar]
- 42.Dai J, Carver M, Punchihewa C, Jones RA, Yang D. Structure of the Hybrid-2 type intramolecular human telomeric G-quadruplex in K+ solution: insights into structure polymorphism of the human telomeric sequence. Nucleic Acids Res. 2007;35:4927–4940. doi: 10.1093/nar/gkm522. and cited references. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.