Abstract
In the field of phylogenetics and comparative genomics, it is important to establish orthologous relationships when comparing homologous sequences. Due to the slight sequence dissimilarity between orthologs and paralogs, it is prone to regarding paralogs as orthologs. For this reason, several methods based on evolutionary distance, phylogeny and BLAST have tried to detect orthologs with more precision. Depending on their algorithmic implementations, each of these methods sometimes has increased false negative or false positive rates. Here, we developed a novel algorithm for orthology detection that uses a distance method based on the phylogenetic criterion of minimum evolution. Our algorithm assumes that sets of sequences exhibiting orthologous relationships are evolutionarily less costly than sets that include one or more paralogous relationships. Calculation of evolutionary cost requires the reconstruction of a neighbor-joining (NJ) tree, but calculations are unaffected by the topology of any given NJ tree. Unlike tree reconciliation, our algorithm appears free from the problem of incorrect topologies of species and gene trees. The reliability of the algorithm was tested in a comparative analysis with two other orthology detection methods using 95 manually curated KOG datasets and 21 experimentally verified EXProt datasets. Sensitivity and specificity estimates indicate that the concept of minimum evolution could be valuable for the detection of orthologs.
INTRODUCTION
Since its introduction in 1843 by Owen (1), the concept of homology has been adopted as the basis of phylogenetics and comparative biology. Although there have been many arguments about its interpretation (2), homology can be defined as a similarity relationship between features that is due to shared ancestry. Homology can be categorized into orthology and paralogy when evolutionary relationships arise from gene duplication and speciation (3). Homologous sequences are orthologous when they diverge from a common ancestor and are separated by a speciation event. On the other hand, paralogous sequences arise as direct products of gene duplication within the lineage of a single species (3–6). Recent gene duplication without any further speciation produces co-orthologs, which are paralogous within the genome of a species, but can also be orthologous to genes in other species (6).
Gene duplication events are prevalent during evolution of life. For example, comparative genomic analysis of paralogous gene families in the genomes of Drosophila melanogaster, Caenorhabditis elegans and Saccharomyces cerevisiae showed that the proportion of genes with paralogous relationships ranged from 30% to 49% in their genomes (7). When gene duplication occurs, the duplicated genes can experience different evolutionary fates, leading to the pseudogenization of one of the two daughter genes (e.g. immunoglobulins), homogenization of duplicate genes with the same gene function (e.g. ribosomal RNAs), subfunctionalization of two descendant genes to which part of the function of their ancestral gene is assigned, and neofunctionalization producing a new functional gene (8).
Many methods have been developed to automatically identify orthologs and paralogs in complex datasets [for a summary, see (9)]. These can be categorized into BLASTp-based, phylogeny-based and evolutionary distance-based approaches. Because of computational complexity, most recently developed methods have depended on the BLASTp algorithm [e.g. Inparanoid (10); OrthoMCL (11); reciprocal best hit (RBH) and reciprocal smallest distance (RSD), (12)]. However, BLASTp-based methods do not take into consideration evolutionary ortholog divergence, although RSD uses an evolutionary distance for orthology detection that is estimated with a maximum likelihood method (12). This lack of evolutionary information can mistakenly detect homoplasious paralogs as orthologs (13). On the other hand, the phylogeny-based approaches are more time-consuming and computationally demanding than the BLASTp-based methods, and are prone to error due to topological variation among phylogenetic trees caused by uncertainties in phylogenetic parameters (14). However, phylogenies truly reflect the divergent histories of genes, and make it possible to detect orthologs among homologs under the original concept of orthology and paralogy (4). Moreover, recently developed phylogeny-based methods [e.g. Orthostrapper (14); and RIO (15)] have adopted bootstrap methods to overcome the topological instability problems of phylogenetic trees, enhancing the reliability of these approaches.
Despite recent advances, all existing ortholog detection methods still suffer from false negative or false positive rates. Any advance in our ability to reduce error rates in detecting orthologs is therefore desirable. Here, we describe the development and implementation of a novel evolutionary distance-based approach to extract orthologs from homologs. In theory, the daughter genes produced by gene duplication events have various evolutionary fates (8) and two or more sets of orthologous genes exhibit different functional constraints (6,8,16). We therefore postulate that sequences consisting only of orthologs require evolutionary cost less than those including one or more paralogous relationships. Under this assumption, our approach can detect orthologous sequences from a given homologous sequence dataset. Although several problems of orthology detection remain to be addressed, a comparative reliability test using experimentally verified and manually curated orthologous sequence datasets revealed that our approach under the concept of minimum evolution offers advantages in orthology detection.
METHODS
Algorithmic concept
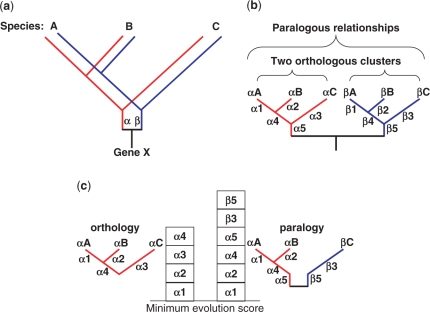
Among the many phylogenetic methods that are used to reconstruct evolutionary history, the maximum parsimony (MP) method selects phylogenetic trees with minimum character changes. The minimum evolution (ME) method, an analog of the MP method that is based on genetic distance, regards a tree with the smallest sum of branch lengths among all possible phylogenetic trees as the most reliable one (17). In this study, we developed an algorithm based on the ME method. The phylogenetic relationships that result from a gene duplication event are represented using a simple tree diagram (Figure 1a). In the tree, two descendants (α and β) have diverged from an ancestral gene along with speciation (Figure 1a), forming two orthologous clusters and some paralogous relationships (Figure 1b). If a subset of homologous sequences consists of orthologs (αA, αB and αC), the sum of branch lengths (SBL) is α1+α2+α3+α4, which is less than SBL (α1+α2+α4+α5+β3+β5) of αA, αB and βC with paralogous relationships between αC and βC (Figure 1c). Therefore, under the ME criterion, it can be postulated that the evolutionary cost of one cluster composed of purely orthologous sequences is less than that of clusters that include paralogous relationships. In this study, we adopted the neighbor-joining (NJ) method, in which SBL is referred to as ‘minimum evolution score’ (MES) and SBL was calculated using the MES of an NJ tree (18).
Figure 1.
A conceptual representation describing orthologous and paralogous relationships related to gene duplication. (a) A simple phyletic history of gene duplication. In the hypothetical phylogenetic tree, the primary ancestral gene X duplicated into two ancestral descendants α (red) and β (blue). It follows that they have diverged along with speciation into three species. (b) Unfolded phylogenetic trees showing orthologous and paralogous relationships. There are two orthologous clusters [(αA, αB), αC] and [(βA, βB), βC]. Within each orthologous cluster, the letters marked beside a branch indicates the length of a branch. (c) Comparison of minimum evolution scores between orthologous and paralogous relationships.
Algorithmic implementation
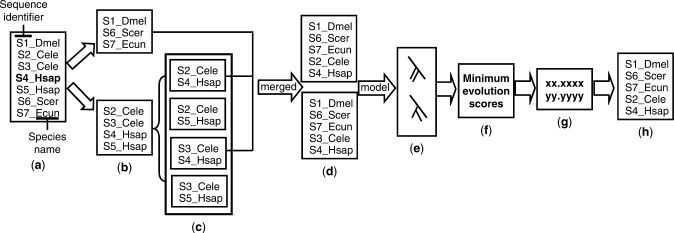
To implement the algorithm, we developed a novel program called Mestortho (minimum evolution score to orthology). For a given multiple sequence alignment, the program automatically considers more than one sequence per species as having a paralogous relationship. For each of all datasets which are generated by all possible combinations of candidate orthologs, the program generates an MES by reconstructing an NJ tree and then calculating SBL. Finally, the sequence set with the smallest MES is determined as a reliable orthologous cluster. The program requires a multiple sequence alignment in which the name of each sequence should consist of the sequence identifier and species information (Figure 2a). In general, paralogous relationships of homologous sequences occur when there are more than one orthologous cluster (Figure 1b). Thus, the program requires a user-defined reference sequence to determine which orthologous cluster should be detected (shown in bold in Figure 2a). Given an alignment, the sequences are classified into two groups (Figure 2b): group 1 consists of the sequences with one occurrence per species, and group 2 is composed of the sequences with more than one occurrence per species. For group 2, exhaustive combinatorial sets with one sequence per species are created (Figure 2c). If the reference sequence is included in group 2, only the datasets with a reference sequence are selected for further analyses. In addition, sequences separated by a genetic distance of zero are regarded as one sequence to reduce the number of combinations. Then, the group 1 sequences are merged with each dataset obtained from group 2 (Figure 2d). For each merged dataset, an NJ tree is reconstructed and its MES is calculated (Figure 2e–g). Finally, the merged dataset with the smallest MES is chosen as a set of orthologs (Figure 2h).
Figure 2.
Conceptual representation of the detection of orthologs in a given alignment. (a) In an input alignment, a sequence name consists of a sequence identifier and the species information. The letters before an underbar denote a sequence identifier, while those after the underbar indicate an abbreviation of the scientific name of a species. (b) An upper box includes sequences with one occurrence per species, while the lower box includes paralogous sequences with more than one occurrence per species. (c) All possible combinations of sequences in which one species is represented only once by one of its sequences in a combination. (d) Datasets in which (b) dataset was merged into one of (c) datasets. (e) Collection of phylogenetic trees reconstructed from the merged datasets. (f) Calculation of minimum evolution scores for obtained phylogenetic trees. (g) Selection of the smallest minimum evolution score. (h) Determination of orthologous sequences.
Once orthologous relationships have been detected, the program examines co-orthologies as follows: (i) on the topology of the NJ tree obtained from the complete original input multiple alignment, monophyletic groups limited to a single species are searched. (ii) If a group includes an orthologous sequence obtained from the step of Figure 2h, the group is regarded as a candidate co-ortholog group. As co-orthology indicates recent gene duplication without any further speciation, all paired branch length distances among sequences within a co-ortholog group should be less than any distance between a co-ortholog and a sequence of all other species. (iii) Under these conditions, the program calculates SBL between two sequences in the NJ for all possible cases, and identifies co-orthologs.
Software implementation
Mestortho is implemented as a Python program, and its web application is available at http://snugenome.snu.ac.kr/Mestortho. The program accepts sequence alignments in three formats (ClustalW, FASTA and Phylip). The modules ‘dnadist’, ‘protdist’ and ‘neighbor’ of the Phylip package ver. 3.66 (19) are used to generate distance matrices for DNA and protein datasets and reconstruct NJ trees. Mestortho analyzes an input alignment and provides the following results: (i) the smallest MES of the sequences inferred as an orthologous group; (ii) a list of orthologous sequences; (iii) data on sequences with co-orthology relationships and no genetic distance; (iv) the initial input alignment, together with the alignment of orthologs detected by the program and (v) clustering information in Newick and image formats for orthologous sequences. Executables can be downloaded from the Mestortho website at the URL given above.
Datasets for reliability tests
We compared the reliabilities of three programs in this study: Mestortho, RBH and Orthostrapper. To prepare datasets for testing, we retrieved a sequence dataset from the EXProt database (http://www.cmbi.kun.nl/EXProt), which consists of bacterial protein sequences with experimentally verified functions (20). Since two different proteins with the same enzyme commission (EC) number have a same enzyme reaction and are likely to express an orthologous relationship (11), we collected protein sequences with the same EC numbers from the database and considered them as ortholog candidates. However, a single gene can be associated with several EC numbers and can contain domains involved in multiple enzymatic reactions. Furthermore, a same EC number can be assigned to proteins that catalyze the same reaction but harbor functionally different subunits (21). For this reason, the transitive relationships between protein orthology and an identical EC number cannot be guaranteed. Since EC number is not a perfect indicator of sequence orthology, we used the recently developed KEGG ortholog (KO) system to define the orthology of proteins related to metabolic pathways (21). In the system, a KO identifier (KOI) reveals an orthologous group of proteins. Using the EC numbers of the putatively orthologous EXProt datasets, we checked whether each dataset had only one KOI in the KEGG database (http://www.genome.jp/kegg/). For datasets with more than one KOI, orthologous relationships were checked using the KEGG automatic annotation server (KAAS; http://www.genome.jp/kegg/kaas/) with the option of bidirectional best hits (22). For datasets still having more than one KOI, EXProt sequences whose KOIs were not identical to those of reference Escherichia coli EXProt sequences were regarded as False sequences. Detailed procedures and results for the curation of the 21 EXProt datasets are described in Supplementary Materials Part 1 and Supplementary Table 1.
To collect homologous sequences for each orthologous EXProt dataset, the protein sequence of E. coli was used as the query. For this reason, we selected datasets that included at least one E. coli sequence among the orthologous datasets. This resulted in 21 final orthologous datasets. Using the E. coli sequence of each dataset, a BLASTp ver. 2.2.10 search was conducted against the nonredundant protein database of the National Center for Biotechnology Information (NCBI), using the protein weight matrix of BLOSUM62 and an E-value cutoff of 0.001. Accession numbers of homologous sequences were collected from the BLAST result of each query and corresponding protein sequences were obtained from the local NCBI protein database using the module FASTACMD of the BLAST package. Among the homologous sequences, the sequences whose species were included in the species list of each of 21 EXProt orthologous datasets were chosen, and merged into their corresponding EXProt orthologous datasets. We then used CLUSTALW ver. 1.83 (23) to generate multiple alignments of the 21 merged datasets with the slow–accurate option, the BLOSUM30 matrix and default gap penalties (pairwise and multiple gap open penalties of 10 and 0.1; pairwise and multiple gap extension penalties of 10 and 0.2). Ambiguous and uninformative variable sites in the aligned datasets were excluded using the BioEdit program ver. 5.0.9 (24). Finally, 21 curated multiple sequence alignments were imported into three programs Mestortho, RBH and Orthostrapper.
One limitation of the EXProt database is that it provides experimentally verified protein sequences for bacterial species only. To test the reliability of methods examined in this study with eukaryotic homologous sequences, we retrieved manually curated clusters of orthologs for seven eukaryotic genomes from the Clusters of Eukaryotic Orthologous Groups (KOG) database [ftp://ftp.ncbi.nih.gov/pub/COG/KOG; (25)]. Using an in-house developed Python script, we extracted 95 KOG clusters, each consisting of one sequence per species. We followed the procedures outlined above to perform homology searches using a KOG sequence of Homo sapiens as the query, merge the homologous sequences into their corresponding KOG clusters, conduct multiple sequence alignment and edit the aligned sequences. Finally, the 95 multiple alignments were prepared for reliability testing of the three programs. When using Mestortho, the query sequences used in the homology searches of the EXProt and KOG datasets were also used as the reference sequences.
Reliability tests
Defining true orthologs and tests
We tested the reliability of Mestortho, RBH and Orthostrapper with KOG and EXProt datasets using measures of sensitivity and specificity, which are defined as probability of true positives among true orthologous sequences and probability of true negatives among nonorthologous sequences, respectively. First, we defined True orthologs in homologous sequences of each KOG dataset as follows: (i) seven KOG sequences from seven eukaryotic genomes were regarded as True sequences; and (ii) using the modules ‘protdist’ and ‘neighbor’ in the Phylip package, homologous sequences with no genetic distance from or co-orthologous relationships with the seven KOG sequences were determined and added to the True sequence set. With the information of True sequences in a KOG dataset, the remaining factors for the calculation of sensitivity and specificity were determined as follows: (i) False = sequences excluding True sequences in a given input alignment; (ii) True positive (TP) = among True, the number of sequences detected as orthologs by a program; (iii) False negative (FN) = True – TP; (iv) False positive (FP) = among False, the number of sequences detected as orthologs by a program; and (v) True negative (TN) = False – FP. The sensitivity and specificity of each dataset were calculated using the following equations: (i) sensitivity = TP/(TP + FN); and (ii) specificity = TN/(FP + TN). In general, sequences detected as orthologs by RBH include those with no genetic distance, but not those with co-orthologous relationships. To adjust the balance among the results of RBH and the other two methods, the sequences which are co-orthologous to the positive sequences of RBH were included in the final results of RBH.
Although the KOG database has been curated manually, its orthologous clusters were defined using RBH, suggesting that there may be paralogs in KOG clusters (25). Therefore, we also calculated the sensitivity and specificity for Mestortho, RBH and Orthostrapper using the 21 EXProt datasets under the same conditions as described above. However, the True sequence set of each EXProt dataset was determined according to the following conditions: (i) EXProt sequences with a KOI identical to that of the E. coli EXProt sequence; and (ii) the sequences with no genetic distance from or co-orthologous relationships with each of the EXProt sequences obtained from (i). The 21 EXProt datasets with the True sequences that were curated under the conditions described above are called EXProt-1 in this study. Here, the EXProt-1 datasets are not free from data attributes based on sequence similarity because True sequences in 6 out of the 21 datasets were determined using the RBH-based KAAS server (for details, see Supplementary Table 1). We therefore generated True sequence sets of the 21 EXProt datasets using a tree reconciliation approach (28,29). We first reconstructed rooted NJ trees of genes and species for each EXProt dataset, which includes homologous sequences. Subsequently, we obtained new True sequence sets by comparing the gene trees with the corresponding species trees (for detailed procedures, see Supplementary Materials Part 2). In this study, we call the 21 EXProt datasets with these True sequence sets EXProt-2. The positive sequences detected by Mestortho, RBH and Orthostrapper for each EXProt-2 dataset were compared to the corresponding True sequence set and the reliabilities of the three programs were established.
Unlike the large number of KOG datasets that were generated, we only produced 21 EXProt datasets. The small number of these datasets could bias the calculation of mean sensitivities and specificities of the three programs. To test whether the number of the EXProt datasets was sufficient for reliability evaluation, we conducted random sampling with a sample size of 21 from 95 KOG datasets. Because positive sequences detected by Mestortho and RBH for 95 KOG datasets were previously determined, we calculated the mean sensitivity and specificity of the two methods for each sample based on information of positive sequences (for details, see Supplementary Materials Part 4).
Running time
We assessed the running time of Mestortho for each of the total 116 datasets including 95 KOG and 21 EXProt alignments. The algorithmic complexity of each dataset was evaluated under the concept of big O with the following conditions: (i) P indicates the number of all paralogs, where the number of sequences with no genetic distance was treated as one sequence; (ii) S is the number of species with paralogs and (iii) N (calculated as P/S) indicates the average number of paralogs per species. Using a computer with a 2.66 GHz processor, we estimated the computational complexity of each dataset with regard to the number of all possible combinations of putative orthologs (NS).
Parameters
Sequence alignment parameters (e.g. weight matrices and gap penalties) can influence the outcome multiple sequence alignments, which can cause changes of evolutionary distance among the aligned sequences. Initially, we aligned the 21 EXProt datasets of the FASTA format using CLUSTALW with the BLOSUM30 weight matrix and the following gap penalties: (i) pairwise gap open penalty = 10; (ii) pairwise gap extension penalty = 0.10; (iii) multiple gap open penalty = 10 and (iv) multiple gap extension penalty = 0.20. We call the 21 EXProt alignments obtained from the parameters above ‘EXProt reference alignments’. We then tested the robustness of Mestortho results against changes of alignment parameters, setting the following values for each of five parameters: (i) weight matrices of BLOSUM30, GONNET250 and PAM350 in pairwise and multiple alignments; (ii) gap open penalties of 1, 5 and 20 in pairwise alignment; (iii) gap extension penalties of 0.01, 0.5 and 1 in pairwise alignment; (iv) gap open penalties of 1, 5 and 20 in multiple alignment and (v) gap extension penalties of 0.1, 0.5 and 1 in multiple alignment. Any combination of these five parameters produced gap penalties different from default parameters of the 21 EXProt reference alignments. For all possible combinations of values of five parameters, we generated multiple sequence alignments using CLUSTALW with the prealigned EXProt sequence datasets. Subsequently, the alignment score of each reference EXProt alignment was compared to scores of newly generated alignments. The datasets with the alignment scores different from that of their corresponding reference alignment were collected and imported into Mestortho. The list of orthologous sequences given by Mestortho for each of the modified alignments was compared to that for their corresponding reference alignment.
Horizontal gene transfer
To examine the existence of horizontal gene transfer (HGT) events in the 21 EXProt datasets, protein ID (PID) information of putative HGT genes for 409 prokaryotic genomes was obtained from http://cbcsrv.watson.ibm.com/HGT_SVM/ [hereafter, CBCSRV_DB; (26)]. DNA sequences corresponding to the PIDs were obtained using FASTACMD, and formatted as a local database using the module FORMATDB of the BLAST package. Almost all sequences in the 21 EXProt datasets were searched against the local CBCSRV_DB using BLASTn ver. 2.2.10 with an E-value cutoff of 0.0 000 001. Species with genomes that were not covered in the original CBCSRV_DB were excluded in the present analysis. The sequence of the EXProt datasets, whose species name was matched to that of the best hit sequence of the local database, was regarded as the candidate of HGT. The rate of HGT occurrence was defined as the number of the HGT candidates divided by the number of the EXProt sequences whose species have genomes in CBCSRV_DB, and given as a percentage.
Manual inspection
Manual inspection of phylogenetic trees describing the KOG and EXProt datasets ensures that we know how many orthologous groups exist in each of the datasets. In order to assess whether Mestortho can detect multiple orthologous groups, we analyzed the sequences of 14 globin-related genes in three different orthologous groups (myoglobin, and α- and β-hemoglobins). Following previously reported results (27), the following protein sequences were retrieved from the NCBI website (http://www.ncbi.nlm.nih.gov/), and aligned as described above: (i) myoglobin: CAI23587, NP_005359, NP_976311 and NP_976312 of human, P04247 of mouse and P02206 of Heterodontus portusjacksoni (the Port Jackson shark); (ii) α-hemoglobin: P69905 of human, P01942 of mouse and P02021 of shark; and (iii) β-hemoglobin: P68226, P68871 and P68872 of human, P02088 of mouse and P02143 of shark. We then used Mestortho with the Jones–Taylor–Thornton weight matrix to reconstruct an NJ tree.
Confidence interval for MESs of true orthologous sets
For each of the 116 datasets, True and False sequences were determined according to the conditions defined in section ‘Defining true orthologs and tests’ of ‘Reliability tests’. Using a sample size corresponding to the number of False sequences of each dataset, 100 subsets of True sequences were extracted randomly and their MESs were calculated using the modules ‘protdist’ and ‘neighbor’ of the Phylip package. With 100 MESs of each dataset, we determined the confidence interval of True sequences under a one-tailed 95% significance level. The definitions of True and False provided in the session were used below.
RESULTS
Algorithmic and software implementation of Mestortho
We developed a novel algorithm for detecting orthologs from homologous sequences using MES, a measure derived from the minimum evolution criterion. The algorithm was implemented in Mestortho, a program written in Python that analyzes DNA or protein sequence alignments in three different formats (ClustalW, FASTA and Phylip) and identifies orthologous sequences. A web application of Mestortho can process only one sequence alignment. In contrast, the local executables can analyze a series of multiple input alignments simultaneously.
Performance tests using the EXProt and KOG datasets
The reliability of the three programs Mestortho, RBH and Orthostrapper was tested using 95 KOG and 21 EXProt datasets using measures of sensitivity and specificities. For the KOG datasets, the mean sensitivity was 0.941 when using Mestortho, higher than the mean sensitivities obtained using RBH (0.838) and Orthostrapper (0.779–0.930) at any bootstrap support level (Table 1; Supplementary Table 2). In addition, Mestortho produced the mean sensitivity with SDs that were smaller (0.185) than RBH (0.257) and Orthostrapper (0.228–0.375). On the other hand, the mean specificity of Mestortho (0.894) was lower than the mean specificity of RBH (0.943), although it was ∼2.0–2.6 times larger than the mean specificities of Orthostrapper (0.353–0.432). Increasing bootstrap support levels in the analysis of Orthostrapper decreased mean sensitivities but increased mean specificities (Table 1; Supplementary Table 2).
Table 1.
Reliability estimates of Mestortho, RBH and Orthostarpper
| Dataseta | Program | Meanb sensitivity (SD)c | Mean specificity (SD) |
|---|---|---|---|
| KOG | Mestortho | 0.941 (0.185) | 0.894 (0.243) |
| RBH | 0.838 (0.257) | 0.943 (0.213) | |
| Orthostrapper (700)d | 0.930 (0.228) | 0.353 (0.422) | |
| Orthostrapper (1000) | 0.779 (0.375) | 0.432 (0.461) | |
| EXProt-1 | Mestortho | 0.970 (0.096) | 0.773 (0.292) |
| RBH | 0.670 (0.205) | 0.715 (0.374) | |
| Orthostrapper (700) | 0.980 (0.051) | 0.132 (0.307) | |
| Orthostrapper (1000) | 0.753 (0.334) | 0.464 (0.457) | |
| EXProt-2 | Mestortho | 0.763 (0.222) | 0.287 (0.402) |
| RBH | 0.569 (0.234) | 0.675 (0.418) | |
| Orthostrapper (700) | 0.901 (0.341) | 0.301 (0.416) | |
| Orthostrapper (1000) | 0.646 (0.098) | 0.365 (0.394) |
aThree programs (Mestortho, RBH and Orthostrapper) were tested using 95 KOG and 21 EXProt datasets. Among two versions of the 21 EXProt datasets, True sequences of EXProt-1 were curated using an EC number identity criterion with the aid of KEGG orthology system and KAAS server while those of EXProt-2 were determined using tree reconciliation.
bThe average value for sensitivities (specificities) of 21 EXProt or 95 KOG datasets.
cThe SD of the mean values.
dThe value in parentheses indicates the bootstrap support level in the analysis of Orthostrapper. We conducted Orthostrapper analyses with bootstrap support levels ranging from 700 to 1000, at intervals of 50. For details, see Supplementary Table 2.
True sequences of each of the EXProt datasets were defined: (i) using an EC number identity criterion and with the aid of the KO system and KAAS server (EXProt-1 datasets); and (ii) using the tree reconciliation criterion (EXProt-2 datasets). For the EXProt-1 datasets, the mean sensitivity and specificity of Mestortho (0.970 and 0.773, respectively) were higher than those of RBH (0.670 and 0.715) (Table 1). All mean sensitivities and specificities of Orthostrapper were lower than those of Mestortho except for a higher mean sensitivity (0.980) at 70% bootstrap support level (Table 1; Supplementary Table 2). The data curation of the EXProt datasets under tree reconciliation (EXProt-2) decreased the reliabilities of the three methods in comparison to the other datasets (KOG and EXProt-1) (Table 1). Reliability tests showed a higher mean sensitivity of Mestortho (0.763) in comparison to that of RBH (0.569), but lower mean sensitivities than Orthostrapper (0.901) at 70% bootstrap support level. On the other hand, the mean specificity of Mestortho (0.287) for the EXProt-2 was lower than the mean specificities of RBH and Orthostrapper (Table 1).
In order to test the validity of the EXProt dataset size, we calculated reliabilities for each of 100 random 21 dataset samples obtained from the 95 KOG datasets. The mean sensitivities of Mestortho were clearly higher than those of RBH, only with five exceptions among 100 samples. On the other hand, the mean specificities of Mestortho were higher than those of RBH only in 16 random samples (Supplementary Figure 2).
Influence of alignment parameters
To evaluate robustness of Mestortho results against changes of alignment parameters, we constructed 243 alignments for each of the 21 EXProt reference alignments under combinations of alignment parameters described in Methods section. When the alignment scores of the newly obtained alignments were compared to those of their corresponding reference alignments, the scores of 1548 out of 5103 alignments were different from those of their reference alignments. When the 1548 alignments were imported into Mestortho, 32 175 sequences were detected as orthologs. Among them, 29 379 sequences (91.3%) were matched to orthologs of the 21 EXProt reference alignments. Mestortho generated the same results of the reference alignments in 927 out of 1548 alignments (Supplementary Figure 3). In addition, we described how many orthologs detected by Mestortho for each of the 21 reference alignments are maintained in the Mestortho outputs of the 243 corresponding new alignments (for details, see the tables at http://agbiotech.snu.ac.kr/PARA/stability.php).
Influence of horizontal gene transfer
In terms of HGT, 32 species (504 sequences) among 51 species (537 sequences) in the 21 EXProt datasets were included in 409 genomes of CBCSRV_DB. When 504 sequences were searched against the local CBCSRV_DB using BLASTn, the names of five species following EXProt sequences were matched to those of the top hits: (i) EXP0100924 (Bacillus subtilis, EC1.15.1.1); (ii) EXP0100758 (B. licheniformis, EC1.15.1.1); (iii) EXP0102685 (Neisseria meningitidis, EC1.15.1.1); (iv) EXP000114 (Pseudomonas aeruginosa, EC4.1.3.7) and (v) EXP0000293 (P. aeruginosa, EC1.15.1.1). Since 19 species including 33 sequences in the 21 EXProt datasets were not covered to the CBCSRV_DB, we estimated an HGT rate to evaluate the possibility of HGT occurrence. The estimated rate was 0.992% (5/504 × 100).
Running time and algorithmic complexity
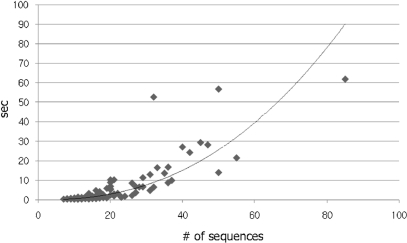
We assessed the running time of Mestortho in the analysis of the 116 total datasets examined in this study. The running time of each dataset was plotted in Figure 3. The nonlinear curve of y = 0.002x2.365, where y is running time and x is the number of sequences in a given dataset, fitted well to the running times of 116 datasets with the smallest R2-value of 0.798. Under the big O concept, most datasets had a computational complexity of the polynomial big O (N > 1 and S > 2), although 13 and 4 datasets showed constant (N = 1) and quadratic (N > 1 and S = 2) time complexity, respectively. On a computer with a 2.66 GHz processor, each dataset with less than 20 000 possible combinations of putative orthologs was completely analyzed by Mestortho within 60 s. Five KOG datasets with more than 20 000 possible combinations (e.g. 279 936 – 107) took 274, 539, 4540, 18 200 and 26 301 s to complete the Mestortho processes. As a guide for the approximate running time of Mestortho for any input dataset, we developed the module ‘time estimator’, which calculates the number of all possible combinations of putative orthologs and subsequently estimates the approximate running time (http://snugenome.snu.ac.kr/Mestortho/).
Figure 3.
Real running time of Mestortho according to the number of sequences for each dataset. To plot the graph, 21 EXProt and 90 KOG alignments were used. The remaining five KOG alignments with sequence sizes of 67, 76, 49, 60 and 71 had running times of 26301, 18200, 274, 539 and 4540 s, respectively. The equation of the fit curve is y = 0.002x2.365 (R2 = 0.798).
Orthologous relationships in globin-related genes
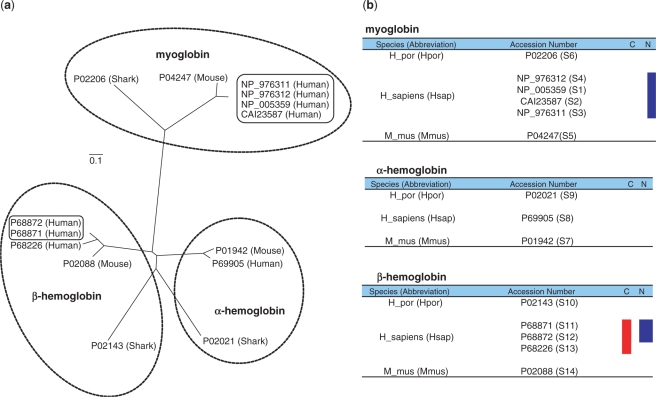
We collected protein sequences of 14 globin-related genes from the NCBI and reconstructed an NJ tree from an alignment with 160 amino acid sites. For human, mouse and shark, myoglobin genes were clustered monophyletically (Figure 4a). Four sequences of human in the cluster of myoglobin showed no genetic distance from each other (Figure 4b). The α- and β-hemoglobin clusters had paraphyletic relationships due to shark hemoglobin sequences (Figure 4a), but were identified as orthologous groups by Mestortho (Figure 4b). The human β-hemoglobin sequences P68871 and P68872, which showed no genetic distance to each other, were co-orthologs of P68226 (Figure 4a and b). Under the concept of tree reconciliation, two hemoglobin sequences of shark showed co-orthology (Figure 4a), but were identified as paralogs by Mestortho (Figure 4b).
Figure 4.
Tree reconciliation versus Mestortho. (a) Phylogenetic tree reconstructed from the sequences of myoglobin, and α- and β-hemoglobins. Boxed sequences have no genetic distance between them. The sequences within the three dotted ellipses have orthologous relationships according to previous reports. (b) Results of Mestortho for the aligned dataset. The letter C and N in the first row of each Table denotes ‘co-orthology’ and ‘no genetic distance’. The sequences having relationships of co-orthology and no genetic distance were marked by red and blue bars, respectively. The abbreviations Hpor, Hsap and Mmus indicate H. portusjacksoni (Port Jackson shark), H. sapiens (human) and Mus musculus (mouse).
Confidence interval for MESs of true orthologous sets
There were always more True sequences than False sequences in the 116 datasets analyzed. Since phylogenetic reconstruction requires more than two sequences, we examined the number of False sequences for each dataset. As a result, 33 out of the 116 datasets had more than two False sequences. For each of these 33 datasets, the confidence intervals of MES of True sequences were determined with 100 random subsets under a one-tailed 95% significance level, and were plotted together with the MES of false sequences (Figure 5). In every case, the MES of False sequences was significantly higher than those of True sequences.
Figure 5.
The confidence interval of True datasets at a one-tailed 95% significance level. Among 116 datasets (KOG+EXProt), 33 alignments were used to calculate the confidence interval. For each of 33 datasets, the black dot indicates the MES of False sequences, while the shaded rectangle indicates the MES range of the one-tailed 95% confidence interval of True sequences. A number in parentheses is the number of False sequences in each of the datasets.
DISCUSSION
Tree reconciliation is a standard approach for phylogeny-based orthology detection. This method identifies orthologous and paralogous relationships by comparing gene and species trees (28,29) and thus requires accurate trees of both types. While the divergence patterns among several representative species (e.g. human, mouse, rat and nematode) can be elucidated in detail, the phylogenetic relationships among numerous or unfamiliar species can lead to inaccurate species trees. Furthermore, uncertainties in phylogenetic parameters and lineage sorting can cause inconsistencies among gene trees obtained from a given sequence dataset (3,14), leading to inaccurate orthology detection. To obtain reliable phylogenetic trees for detecting orthology, recent phylogeny-based methods have used bootstrap analyses (14,15). The two representative programs, Orthostrapper and Rio, generate a set of bootstrapped trees that provide more confidence than a gene tree. However, the results are dependent on arbitrary levels of bootstrap supports (15). In theory, decrease in bootstrap support levels would increase the probability of detecting orthologs, but may also produce many false positives, thereby increasing sensitivity but reducing specificity. Alternatively, the increase of the level of the bootstrap support would lead to lower sensitivity and higher specificity. The weaknesses of bootstrap analyses have been demonstrated in this (Table 1; Supplementary Table 2) and previous studies (14,15).
To overcome the weaknesses of tree reconciliation and bootstrap analyses, we developed a novel algorithm for detecting orthologs based on the concept of minimum evolution and implemented Mestortho. We then compared the reliability of Mestortho to phylogeny-based (Orthostrapper) and BLASTp-based RBH methods using three kinds of datasets (KOG, EXProt-1 and EXProt-2). Since the KOG datasets were originally curated based on the BLASTp-based reciprocal best hit method, the reliability of RBH may be overestimated in the datasets. It is therefore noteworthy that the reliability of RBH with the KOG datasets was higher (mean sensitivity: 83.8%; mean specificity: 94.3%) than the reliabilities for the EXProt-1 (67%; 71.5%) and EXProt-2 (56.9%; 67.5%) datasets (Table 1), where their true orthologs were respectively defined using the KO system and BLAST-based KAAS server, and the tree reconciliation from the EXProt datasets curated using an EC number identity criterion. Since Mestortho and Orthostrapper did not show these patterns, the KOG datasets appear to be artificially better references for the RBH method. We then checked the EXProt-1 datasets. Because the 14 EXProt datasets with more than one KOI were involved in sequence similarity searches using the KAAS, it is evident that the EXProt-1 datasets are more or less driven by the attribute of sequence similarity. However, the decrease of the reliability of RBH for the EXProt-1 datasets (sensitivity: 67%; specificity: 71.5%) in comparison to that for the KOG datasets (83.8%; 94.3%) suggested that the degree of the sequence similarity attribute of the EXProt-1 was less than that of the KOG datasets. Finally, we used the EXProt-2 datasets to evaluate reliability. Unexpectedly, the reliabilities of Orthostrapper with the EXProt-2 datasets were not higher than those obtained with the other datasets (Table 1). This indicates that the EXProt-2 datasets curated by tree reconciliation are not biased, even if using a phylogeny-based method (Orthostrapper).
Regardless of dataset attribute considerations, the mean sensitivities of Mestortho (94.1, 97 and 76.3%) were consistently higher than those of RBH (83.8, 67 and 56.9%) for the KOG, EXProt-1 and EXProt-2 datasets (Table 1). Moreover, the higher mean sensitivity of Mestortho (76.3%) in comparison to that of RBH (56.9%) in the analysis of the EXProt-2 datasets indicates that Mestortho is more powerful than RBH in its ability to discriminate true positives from false negatives even for datasets that were not defined by sequence similarity (EXProt-2; Table 1). On the other hand, the RBH method had the maximum mean specificity for the KOG datasets and even for the EXProt-2 datasets, although the mean specificity of Mestortho (77.3%) was slightly higher than that of RBH (71.5%; Table 1). Because the RBH method involves a sequence similarity-based approach, this result suggests that the method could be more useful to detect true negatives in any given dataset, in comparison to the other methods. In contrast, the mean specificity of Mestortho for the EXProt-2 datasets (28.7%) was lower than any mean specificity obtained with Orthostrapper (Table 1; Supplementary Table 2), suggesting that Mestortho's weakness is its ability to discriminate false positives from true negatives. In a previous study using the KOG database of eukaryotic genomes (9), RSD showed reliability similar to that of RBH. Furthermore, a recent study of orthology datasets curated by the BLASTp method also showed that RSD has no advantage over RBH in ortholog detection (30). Although the reliability of Mestortho was not directly compared with that of RSD in this study, we expect Mestortho to be at least more sensitive than RSD.
We also argue that the reliability of Mestortho, RBH and Orthostrapper is not limited by the small number of EXProt datasets analyzed and is not biased by the occurrence of HGT. The sensitivities of Mestortho in the analysis of 100 random samples of size 21 obtained from the 95 KOG datasets were clearly higher than those of RBH (Supplementary Figure 2), with the exception of five random samples, despite the small difference (∼10%; Mestortho: 94.1%, RBH: 83.8%) between the mean sensitivities of the two methods for the KOG datasets (Table 1). These results indicate that the higher sensitivity of Mestortho over RBH appears displayed more reliably in the EXProt datasets than the KOG datasets, mainly due to the relatively large difference of the mean sensitivities (∼30% in the EXProt-1; ∼20% in the EXProt-2; Table 1). Although HGT events in any bacterial sequence set can generate incorrect orthologs, our estimate of HGT occurrence (0.992%) for the 21 EXProt datasets showed that such events were rare and had minimum impact on the reliabilities obtained in this study.
Sequence alignment can also affect our minimum evolution approach. A change in alignment parameters can affect the evolutionary distance between any two sequences in a given dataset, which can subsequently affect the MES of any putative ortholog set. Mestortho results showed they were indeed dependent on changes of alignment parameters (Supplemental Figure 3); only ∼60% out of newly generated alignments resulted in results being identical to those of the reference alignments. At the level of individual sequences, however, most sequences (91.3%) were detected as orthologs regardless of alignment details. Although our simulation did not show the precise degree of robustness against changes in input alignment, Mestortho appears to generate moderately consistent results, especially at the individual sequence level.
Mestortho calculates the MES of each of the NJ trees describing evolution of every putative ortholog subset within a set of homologous sequences. Because the MES depends only on the sum of branch lengths of a phylogenetic tree, our algorithm is independent of topological changes and instabilities of reconstructed trees, and the program has the potential to detect orthologs that are not orthologous on the topology of a phylogenetic tree. To verify this property, we performed orthology detection using sequences of globin-related genes. It is generally believed that an ancestral globin gene duplicated to generate globin and myoglobin about 800 million years ago, and that the families of α- and β-globins have arisen by the duplication of the recent globin gene ∼450–500 million years ago (3). If the occurrences of speciation and duplication events have been mixed in evolutionary time, each of related species would have different numbers of paralogous genes. However, a recent study on globin evolution showed that each of human, mouse and Port Jackson shark has three globin-related genes; myoglobin, α- and β-hemoglobins. This implies that two duplication events of globin were followed by speciation events of three species (27). In the phylogenetic tree shown in Figure 4a, the topology of the myoglobin group is completely congruent with the divergence of the three species. Therefore, the group can be easily detected as orthologous by tree reconciliation. However, the groups of α- and β-hemoglobins have incongruent phylogenetic topologies in comparison to the species divergence (Figure 4a). According to the standpoint of tree reconciliation, the hemoglobin sequences of shark should be excluded from each orthologous clusters of α- and β-hemoglobins. However, the previous study reported that the sequences of α- and β-hemoglobins for shark were orthologous in each gene family (27). Despite conflict between the phylogenetic tree and the prior evolutionary knowledge, Mestortho regarded α- and β-hemoglobin sequences of shark as orthologs to α- and β-hemoglobins of human and mouse, respectively (Figure 4b). There are two co-ortholog groups in the phylogenetic tree (Figure 4a). According to tree reconciliation, each of two groups had only co-orthologous sequences. However, the previous evolutionary study of globin-related genes showed that the α- and β-hemoglobin duplication of shark precedes the speciation of the species (27), revealing that the co-orthology of two sequences of shark on the phylogenetic tree is not correct. Unlike manual inspection of the phylogenetic tree (Figure 4a), Mestortho detected three human sequences of α- and β-hemoglobin as co-orthologs, but rejected shark-related co-orthology (Figure 4b), indicating that our approach for detecting orthology may be free from errors inherent to tree reconciliation.
Several methods are available for detecting orthologs among homologous sequences. Unfortunately, all of them, including Mestortho, produce different false positive and negative rates depending on the algorithm used (9). We assume that orthologs of a reference sequence have different functional constraints than other orthologous groups in a given homologous sequence dataset. If an orthologous group with the reference sequence has evolutionary constraints stronger than or similar to other orthologous groups in a given dataset, Mestortho will probably detect the sequence members of the orthologous group more accurately because the evolutionary cost of the ancestral divergence between the two groups (α5+β5 in Figure 1b) is sufficient to yield a difference in their MESs. In our simulation, the MES confidence interval of 33 orthologous groups showed that True orthologous groups including reference sequences evolved slower than False groups (Figure 5), indicating that our assumption for orthology detection is reliable. However, there are some exceptional cases that lead to incorrect orthology detection. First, if an orthologous group of a reference sequence includes a pseudogene-like sequence (αB; Figure 6a) that evolved faster than other sequences in a given dataset, Mestortho would detect the paralog of the pseudogene-like sequence as an ortholog (βB; Figure 6a). Similarly, if a gene was missed in a species (αB; Figure 6a), the false ortholog would be detected as an ortholog (βB; Figure 6a). Second, if an ortholog of the reference (αB; Figure 6b) is more closely clustered to the paralogs of the reference than the other true orthologs (αA; Figure 6b), our approach would detect a false positive as an orthologous sequence (Figure 6b). For example, in the EXProt dataset of superoxide dismutase (EC 1.15.1.1), the paralogous gene of P. aeruginosa, instead of the true ortholog of the species, was identified as being orthologous to the reference sequence, due to its closer clustering with the reference sequence of E. coli (Figure 6c).
Figure 6.
Two examples of errors when running Mestortho. (a and b) In the phylogenetic trees, A, B and C indicate species, and α and β denote two descendants after gene duplication. In each of the trees, red branches indicate the lineages detected as orthologous sequences by Mestortho. (c) The phylogenetic tree of (b) is obtained from an EXProt dataset (EC 1.15.1.1) corresponding to the model (b). The monophyletic sequence group is marked by a triangle. The branches of genes detected as orthologs by Mestortho are indicated in bold. Among taxa of the phylogenetic tree, True sequences are also marked in bold and red. The asterisk symbol indicates the reference sequence of the EXProt dataset. The dotted line shows the paralogous relationships between sequences.
CONCLUSIONS
In recent years, the rapid accumulation of gene and genome sequences has made it possible to predict the function and role of numerous genes. However, comparative analyses require reliable methods of orthology detection. In this study, we developed a novel orthology detection algorithm motivated by the criterion of minimum evolution. The algorithm was implemented with web and stand-alone versions. Advantages of our approach include high reliability, the potential to detect orthologs regardless of tree-topology, and the ability to analyze both DNA and protein data, a feature which is not possible using other evolutionary distance-based and phylogeny-based methods. Our approach therefore is sufficiently useful for orthology detection and provides a new tool with which to enhance the set of existing orthology detection methods. The validation and comparative analysis of Mestortho and other orthology detection programs revealed the importance of the reference datasets that were used to evaluate correctly the reliability of the methods. Further studies will be needed to develop better and more thorough standards of validation for future orthology detection methods.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
We are sincerely grateful to Mr Sunjin Moon for providing valuable comments. This work is supported by a grant from BioGreen 21 Program of the Korean Rural Development Administration and by the Brain Korea 21 Project of the Ministry of Education. Funding to pay the Open Access publication charges for this article was provided by the Brain Korea 21 Project.
Conflict of interest statement. None declared.
REFERENCES
- 1.Owen R. London: Longman, Brown, Green and Longmans; 1843. Lectures on the Comparative Anatomy and Physiology of the Invertebrate Animals. [Google Scholar]
- 2.Fitch WM. Homology a personal view on some of the problems. Trends Genet. 2000;16:227–231. doi: 10.1016/s0168-9525(00)02005-9. [DOI] [PubMed] [Google Scholar]
- 3.Graur D, Li W-H, editors. 2nd edn. Sunderland, MA: Sinauer Associates; 2000. Fundamentals of Molecular Evolution. [Google Scholar]
- 4.Fitch WM. Distinguishing homologous from analogous proteins. Syst. Zool. 1970;19:99–106. [PubMed] [Google Scholar]
- 5.Sonnhammer ELL, Koonin EV. Orthology, paralogy and proposed classification for paralog subtypes. Trends Genet. 2002;18:619–620. doi: 10.1016/s0168-9525(02)02793-2. [DOI] [PubMed] [Google Scholar]
- 6.Koonin EV. Orthologs, paralogs, and evolutionary genomics. Annu. Rev. Genet. 2005;39:309–338. doi: 10.1146/annurev.genet.39.073003.114725. [DOI] [PubMed] [Google Scholar]
- 7.Rubin GM, Yandell MD, Wortman JR, Miklos GLG, Nelson CR, Hariharan IK, Fortini ME, Li PW, Apweiler R, Fleischmann W, et al. Comparative genomics of the eukaryotes. Science. 2000;287:2204–2215. doi: 10.1126/science.287.5461.2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang J. Evolution by gene duplication: an update. Trends Ecol. Evol. 2003;18:292–298. [Google Scholar]
- 9.Chen F, Mackey AJ, Vermunt JK, Roos DS. Assessing performance of orthology detection strategies applied to eukaryotic genomes. PLoS One. 2007;2:e383. doi: 10.1371/journal.pone.0000383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.O’Brien KP, Remm M, Sonnhammer ELL. Inparanoid: a comprehensive database of eukaryotic orthologs. Nucleic Acids Res. 2005;33:D476–D480. doi: 10.1093/nar/gki107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li L, Christian J, Stoeckert J, Roos DS. OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res. 2003;13:2178–2189. doi: 10.1101/gr.1224503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wall DP, Fraser HB, Hirsh AE. Detecting putative orthologs. Bioinformatics. 2003;19:1710–1711. doi: 10.1093/bioinformatics/btg213. [DOI] [PubMed] [Google Scholar]
- 13.Koski LB, Golding GB. The closest BLAST hit is often not the nearest neighbor. J. Mol. Evol. 2001;52:540–542. doi: 10.1007/s002390010184. [DOI] [PubMed] [Google Scholar]
- 14.Storm CEV, Sonnhammer ELL. Automated ortholog inference from phylogenetic trees and calculation of orthology reliability. Bioinformatics. 2002;18:92–99. doi: 10.1093/bioinformatics/18.1.92. [DOI] [PubMed] [Google Scholar]
- 15.Zmasek CM, Eddy SR. RIO: analyzing proteomes by automated phylogenomics using resampled inference of orthologs. BMC Bioinform. 2002;3:14. doi: 10.1186/1471-2105-3-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ohno S. Evolution by Gene Duplication. New York: Springer; 1970. [Google Scholar]
- 17.Cavalli-Sforza LL, Edwards WF. Phylogenetic analysis: models and estimation procedures. Evolution. 1967;21:550–570. doi: 10.1111/j.1558-5646.1967.tb03411.x. [DOI] [PubMed] [Google Scholar]
- 18.Saitou N, Nei M. The neighbor-joining methods: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 19.Felsenstein J. PHYLIP – phylogeny inference package (version 3.2) Cladistics. 1989;5:164–166. [Google Scholar]
- 20.Ursing BM, van Enckevort FHJ, Leunissen JAM, Siezen RJ. EXProt: a database for proteins with an experimentally verified function. Nucleic Acids Res. 2002;30:50–51. doi: 10.1093/nar/30.1.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kanehisa M, Goto S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moriya Y, Itoh M, Okuda S, Yoshizawa AC, Kanehisa M. KAAS: an automatic genome annotation and pathway reconstruction server. Nucleic Acids Res. 2007;35:W182–W185. doi: 10.1093/nar/gkm321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999;41:95–98. [Google Scholar]
- 25.Tatusov RL, Fedorova ND, Jackson JD, Jacobs AR, Kiryutin B, Koonin EV, Krylov DM, Mazumder R, Mekhedov SL, Nikolskaya AN, et al. The COG database: an updated version includes eukaryotes. BMC Bioinform. 2003;4:41. doi: 10.1186/1471-2105-4-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tsirigos A, Rigoutsos I. A sensitive, support-vector-machine method for the detection of horizontal gene transfers in viral, archaeal and bacterial genomes. Nucleic Acids Res. 2005;33:3699–3707. doi: 10.1093/nar/gki660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Burmester T, Ebner B, Weich B, Hankeln T. Cytoglobin: a novel globin type ubiquitously expressed in vertebrate tissues. Mol. Biol. Evol. 2002;19:416–421. doi: 10.1093/oxfordjournals.molbev.a004096. [DOI] [PubMed] [Google Scholar]
- 28.Goodman M, Czelusniak K, Moore GW, Romero-Herrera AE, Matsuda G. Fitting the gene lineage into its species lineage: a parsimony strategy illustrated by cladograms constructed from globin sequences. Syst. Zool. 1979;28:132–169. [Google Scholar]
- 29.Page RDM. Maps between trees and cladistic analysis of historical associations among genes, organisms, and areas. Syst. Biol. 1994;43:58–77. [Google Scholar]
- 30.Moreno-Hagelsieb G, Latimer K. Choosing BLAST options for better detection of orthologs as reciprocal best hits. Bioinformatics. 2008;24:319–324. doi: 10.1093/bioinformatics/btm585. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.