Abstract
Bacterial and mammalian AlkB proteins are iron(II)- and 2-oxoglutarate-dependent dioxygenases that reverse methylation damage, such as 1-methyladenine and 3-methylcytosine, in RNA and DNA. An AlkB-domain is encoded by the genome of numerous single-stranded, plant-infecting RNA viruses, the majority of which belong to the Flexiviridae family. Our phylogenetic analysis of AlkB sequences suggests that a single plant virus might have acquired AlkB relatively recently, followed by horizontal dissemination among other viruses via recombination. Here, we describe the first functional characterization of AlkB proteins from three plant viruses. The viral AlkB proteins efficiently reactivated methylated bacteriophage genomes when expressed in Escherichia coli, and also displayed robust, iron(II)- and 2-oxoglutarate-dependent demethylase activity in vitro. Viral AlkB proteins preferred RNA over DNA substrates, and thus represent the first AlkBs with such substrate specificity. Our results suggest a role for viral AlkBs in maintaining the integrity of the viral RNA genome through repair of deleterious methylation damage, and support the notion that AlkB-mediated RNA repair is biologically relevant.
INTRODUCTION
Alkylating agents are found both in the environment and inside cells and can cause alkylation of the majority of N- and O-atoms in DNA and RNA bases (1). Some of these alkylations interfere with Watson-Crick base pairing, thereby blocking replication and transcription of DNA, as well as mRNA translation. Alkylations may also cause mispairing between bases, leading to mutations. Thus, alkylation damage poses a serious threat to the transmission and integrity of the genetic information stored in DNA and RNA, and all organisms are equipped with various repair systems to counteract the deleterious effects of alkylation damage (2).
The AlkB protein from Escherichia coli (EcAlkB) is expressed as part of an adaptive response when bacteria are exposed to alkylating agents, and repairs base lesions resulting from the methylation of an equivalent position in purines (N1) and pyrimidines (N3), i.e. 1-methyladenine (1-meA) and 3-methylcytosine (3-meC), as well as the less abundant 1-methylguanine and 3-methylthymine (3–5). Since these positions are involved in Watson–Crick base pairing, and therefore shielded within the structure of double-stranded (ds) nucleic acids, the corresponding lesions are preferentially introduced into single-stranded (ss) regions. EcAlkB belongs to the iron(II)- and 2-oxoglutarate-dependent [Fe(II)/2OG-dependent] dioxygenase superfamily (6), which comprises enzymes that require 2OG as cosubstrate and ferrous iron as a cofactor. EcAlkB utilizes molecular oxygen to oxidize its methylated substrate and the resulting unstable hydroxymethyl group is spontaneously released as formaldehyde (7,8). It has also been shown that bulkier lesions, such as ethyl and propyl groups, as well as exocyclic etheno and ethano groups, can be repaired by AlkB proteins, but usually with lower efficiencies than for methylated bases (9–12).
AlkB homologues (ABHs) are found in all multicellular organisms, as well as in many bacteria and fungi, and mammalian genomes encode eight different ABHs (13). Two of these, ABH2 and ABH3 (hABH2/hABH3 in humans; mABH2/mABH3 in mice), have been characterized biochemically, and were shown to have a repair activity similar to that of EcAlkB (9,14–16). Interestingly, ABH3 and EcAlkB can also remove lesions from RNA substrates, leading to functional recovery of damaged RNA (14,17). This suggests a possible role for AlkB-mediated demethylation in RNA repair, but the biological relevance is yet uncertain.
Conspicuously, an AlkB domain is present in the replicase polyprotein of a number of plant viruses, most of them belonging to the family Flexiviridae (6,18). The presence of an AlkB domain is remarkable, considering the limited coding capacity of these positive-stranded RNA viruses with genome sizes as small as 7 kb. In this study, we have performed a functional characterization of AlkB proteins from Grapevine virus A (GVA) (19), Blueberry scorch virus (BlScV) (20) and Blackberry virus Y (BVY) (21), representing diverse genera of Flexiviridae, and a yet unnamed new genus in the Potyviridae family, respectively. These proteins were all shown to remove methyl lesions from RNA and DNA, but with substantially higher activity on RNA substrates. The results support a role for viral AlkBs in maintaining the integrity of the viral RNA genome through removal of deleterious RNA damage.
MATERIALS AND METHODS
Protein sequence analysis
The set of viral, bacterial and eukaryotic AlkB proteins for phylogenetic analysis was identified using PSI-BLAST searches (22) against the nonredundant database of protein sequences (NCBI, NIH). Multiple alignments were constructed using the MUSCLE program (23). Maximum likelihood trees were generated using the ProtML program of the MOLPHY package (24) by optimizing the least-squares tree with local rearrangements [Jones–Taylor–Thornton evolutionary model (25) with adjustment for observed amino acid frequencies]. Reliability of the internal tree branches were estimated with the RELL bootstrap method (10 000 replications) using the ProtML program (26).
Plasmid construction
For phage reactivation assays, viral AlkB-coding sequences were amplified by polymerase chain reaction (PCR) on full-length or partial cDNAs derived from viral genomes, using primers containing NdeI (fwd) and BamHI (rev) restriction sites, and subsequently cloned into the same sites in the low-copy-number, toluic acid inducible vector pJB658 (27). For purification of N-terminally 6xHis-tagged recombinant protein expressed in E. coli, NdeI-BamHI fragments were subcloned from the pJB658-derived expression construct into the corresponding sites in pET28a (Novagen, Madison, WI, USA). For the expression of C-terminally 6xHis-tagged GVA-36, the GVA-36 coding sequence was obtained by PCR and the resulting fragment was inserted between the SacI and SalI sites in pET28a. The corresponding expression plasmids for EcAlkB, pJB658-AlkB and pET28-AlkB, have been described previously (14).
Protein purification
EcAlkB was expressed and purified as previously described (14). C-terminally (GVA-36) or N-terminally (all others) 6xHis-tagged viral AlkB proteins were expressed from the corresponding pET28a-derived constructs in the E. coli strain BL21-CodonPlus(DE3)-RIPL (Stratagene, La Jolla, CA, USA). Expression was performed overnight at 16°C in 1 l of LB medium containing 0.075 mM IPTG. Bacterial extracts were obtained by French press treatment, and proteins were subsequently purified from the extracts using TALON Metal Affinity Resin (Clontech, Mountain View, CA, USA) according to the manufacturers instructions. Protein purity and yield was assessed by 15% SDS–PAGE followed by coomassie brilliant blue-staining of the gel.
Phage reactivation assay
DNA bacteriophage M13mp18 and RNA bacteriophage MS2 were methyl methanesulphonate (MMS) treated to examine reactivation of methylated ss phage DNA and RNA, respectively, essentially as previously described (14). MMS-inactivated bacteriophages were mixed with E. coli alkB mutant cells (strain HK82/F′), expressing EcAlkB or viral AlkB from the pJB658 vector in the presence of 2 mM toluic acid. This mixture in soft agar medium was layered on top of an agar plate that was subsequently placed overnight at 37°C. Phage survival was scored by counting plaques.
Assay for AlkB-mediated decarboxylation of 2-oxoglutarate
The assay was performed essentially as described before (4), using the method developed by Kaule and Günzler (28). Reactions were performed for 30 min at 37°C in a 50 µl reaction mixture containing 50 mM HEPES–KOH pH 7.5, 4 mM ascorbic acid, 16 µM [5-14C]2-oxoglutarate (Moravek Biochemicals, Brea, CA, USA), 144 µM unlabelled 2-oxoglutarate and 100 pmol of EcAlkB or 500 pmol of viral AlkB. After incubation, remaining [5-14C]2-oxoglutarate was precipitated with 2,4-dinitrophenylhydrazine, and [1-14C]succinate in the supernatant was measured by scintillation counting. When indicated, 400 µM 1-methyladenosine was present in the reaction mixture. Fe2+ (80 µM FeSO4) was present as indicated, while 1 mM EDTA was added when Fe2+ was omitted.
Assay for oxidative demethylation of [3H]methylated DNA and RNA oligonucleotides
The preparation of [3H]methylated oligonucleotides (ssDNA: TAAAATAATAAATTAAA, ssRNA: CAUGAUAACCGCGACUACACUGAC, polyA and polyC) using N-[3H]methyl-N-nitrosourea was performed as described previously (15). [3H]methylated dsRNA substrates were generated by annealing a non-methylated complementary strand as described (15). [3H]methylated ss and ds substrates (typically 1000 d.p.m.) were incubated with AlkB proteins for 30 min at 37°C in a 50 µl reaction mixture containing 50 mM HEPES–KOH pH 7.5, 2 mM ascorbic acid, 100 µM 2-oxoglutarate, 50 µg/ml BSA, 40 µM FeSO4 and 20 U RNasin Plus RNase Inhibitor (Promega, Madison, WI, USA). The released ethanol-soluble radioactivity was measured by scintillation counting as described earlier (15).
RESULTS
Phylogenetic analysis of viral AlkB proteins
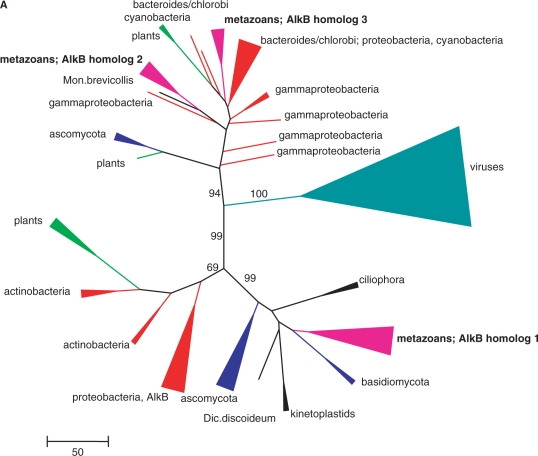
In an attempt to gain insight into the origin and evolution of the AlkB homologues encoded in genomes of plant viruses, a two-tier phylogenetic analysis of the AlkB family was performed. First, a large alignment was constructed that included representative sets of AlkB proteins from bacteria, eukaryotes and viruses (Figure S1), and a phylogenetic tree was built, aiming at deciphering the ultimate origin of the viral AlkBs. In the resulting tree, the viral sequences formed one of the three major, well-separated branches that is strongly supported by bootstrap analysis, whereas the remaining two branches both included assortments of bacterial and eukaryotic members of the family (Figures 1A and S2). Because no independent information was available to root this tree, the origin of viral AlkBs could formally not be inferred. However, viral AlkBs were significantly more similar to bacterial homologues than to eukaryotic ones. This was also reflected by the fact that the first iteration of BLASTp searches with viral AlkB sequences yielded statistically significant hits (E-values in the range of 10−5 to 10−4) to many bacterial AlkBs, especially those from cyanobacteria, but not to eukaryotic AlkBs. Notably, the viral AlkB clade in the family tree is well separated from the plant homologues (Figure 1A).
Figure 1.
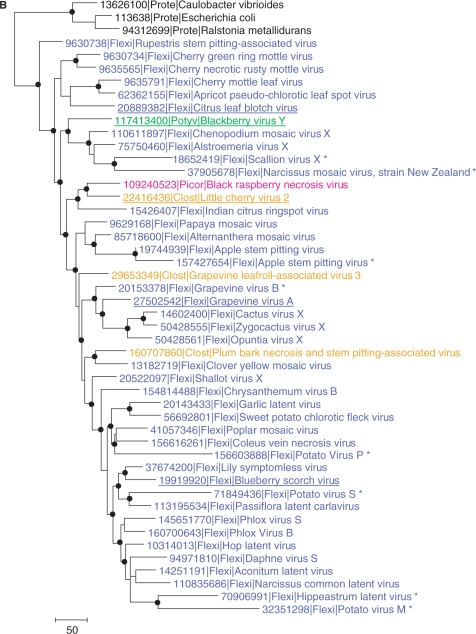
Phylogenetic analysis of viral AlkB proteins. (A) The overall tree of the AlkB family. For clarity, individual viruses are grouped and major groups of bacteria and eukaryotes are collapsed, colour-coded and labelled with the name of the group. The complete tree is given in Figure S2 and the complete alignment used for tree construction is given in Figure S1. Numbers indicate bootstrap support for central tree nodes. The scale bar represents the number of substitutions per 100 residues. (B) Phylogenetic tree of selected viral AlkB proteins rooted by proteobacteria (Prote) outgroup (E. coli, Ralstonia metallidurans and Caulobacter vibrioides). Dots indicate tree nodes with bootstrap support ≥70%. Members of different viral families are colour-coded: blue, Flexiviridae (Flexi); orange, Closteroviridae (Clost); green, Potyviridae (Potyv); magenta, Sadwavirus (Picornavirales, Picor). Species studied in this work are underlined and inactivated AlkBs are indicated with an asterisk. Numbers represent Genbank Identifier (gi) numbers.
Next, a more complete alignment of viral AlkBs was generated using proteobacteria as the outgroup to examine the evolutionary relationships of these proteins among AlkB-containing plant viruses (Figure S3). The resulting topology of the phylogenetic tree is compatible with considerable intervirus horizontal transfer of AlkB-coding sequences (Figure 1B). The single potyvirus sequence clusters with a distinct subset of flexiviruses in a strongly supported branch. The closterovirus sequences clearly do not form a clade and are instead scattered among the flexiviruses. However, there is a well-supported clade that unites a flexivirus (ICRV), a closterovirus (LChV-2) and a sadwavirus (BRNV) of the order Picornavirales, so far the only AlkB-carrying virus with an icosahedral capsid.
Notably, several viruses encode inactivated forms of AlkB as indicated by the replacement of one or more conserved amino acid residues implicated in enzymatic activity (Figure S3). In the phylogenetic tree, the inactivated AlkBs typically comprised long branches that were scattered among the active enzymes (Figure 1B).
Bioinformatics prediction of a repair function for viral AlkBs
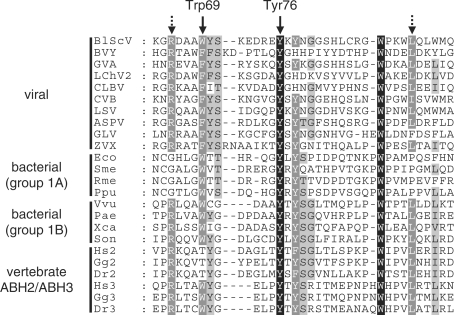
As discussed in the previous paragraph, viral AlkBs display a high degree of similarity to bacterial AlkB proteins, particularly those belonging to group 1B (29), as well as to ABH2 and ABH3, which represent vertebrate AlkB proteins already reported to be involved in nucleic acid repair (9,14). Interestingly, the viral, vertebrate and bacterial AlkBs shared several conserved amino acid residues in a region corresponding to the EcAlkB nucleotide recognition lid (Figure 2). Two of these residues, Trp69 and Tyr76, were found to be in close contact with the 1-meA substrate in the EcAlkB 3D-structure (30). Therefore, our initial bioinformatics analysis suggested that the viral AlkB proteins were likely to possess a repair activity similar to that previously reported for EcAlkB, ABH2 and ABH3, and experiments were designed to further explore this possibility.
Figure 2.
Sequence alignment of a part of the nucleotide recognition lid region from various AlkB proteins. The region shown corresponds to aa 63–98 of EcAlkB (gi|113638). Filled arrows indicate residues Trp69 and Tyr76 in EcAlkB, which have been shown to be involved in coordinating 1-meA (30). Dotted arrows indicate residues that are conserved in viral, vertebrate and bacterial AlkB proteins belonging to group 1B, but not to group 1A (29). BlScV, Blueberry scorch virus; BVY, Blackberry virus Y; GVA, Grapevine virus A; LChV-2, Little cherry virus 2; CLBV, Citric leave blotch virus, CVB, Chrysanthemum virus B; LSV, Lily symptomless virus; ASPV, Apple stem pitting virus; GLV, garlic latent virus; ZVX, Zygocactus virus X; Eco, E. coli; Sme, Sinorhizobium meliloti; Rme, Ralstonia metallidurans; Ppu, Pseudomonas putida; Vvu, Vibrio vulnificus; Pae, Pseudomonas aeruginosa; Xca, Xanthomonas campestris; Son, Shewanella oneidensis; Hs, Homo sapiens; Gg, Gallus gallus; Dr, Danio rerio.
Repair of methylation damage in bacteriophage genomes
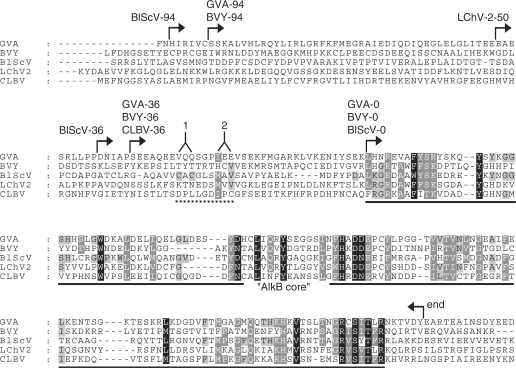
AlkB proteins from five plant viruses, i.e. GVA, BlScV, BVY, Citrus leaf blotch virus (CLBV) and Little cherry virus 2 (LChV-2) were selected for initial functional characterization. These viruses represent three divergent families, Flexiviridae (GVA, BlScV and CLBV), Closteroviridae (LChV-2) and Potyviridae (BVY). The viral AlkB domain is part of a large replicase polyprotein, where it is flanked by long stretches of low sequence conservation. It is therefore not trivial to define the N- and C-terminal borders of the functionally active viral AlkB proteins. However, based on sequence homology within the viral AlkB family, as well as sequence comparison with bacterial AlkBs, a viral AlkB core region could be defined (Figure 3). This core region of ∼132 aa is substantially smaller than a typical AlkB protein from bacteria (190–220 aa), suggesting that parts of the less-conserved regions flanking the viral AlkB core may be required to form a functional AlkB protein. Indeed, some sequence conservation within the viral AlkB group, not shared with other AlkB proteins, is observed in a region of 20–25 aa upstream of the viral AlkB core (Figure 3). Based on this observation, viral AlkB proteins with N-terminal extensions of different length were constructed (Figure 3). The border of these extensions was chosen rather arbitrarily, but without disrupting predicted secondary structure elements. This resulted in viral AlkB proteins with N-terminal extensions of 0, 36, 50 or 94 aa, designated accordingly (GVA-36, BlScV-94, etc.). The C-terminal border of the viral AlkB proteins was set at 6 aa downstream of the characteristic RX5R motif (Figures 3, S1 and S3), which is found close to the C-terminus of most AlkB proteins, and is usually followed by 5–20 non-conserved residues.
Figure 3.
Viral AlkB proteins used in the present study. A sequence alignment was generated from 23 viral AlkB protein sequences, and the five sequences shown were extracted from this alignment. The shading pattern of this 23 sequence alignment has been maintained (explaining why some residues are shaded without being conserved among the five sequences shown). Arrows indicate the borders of the AlkB-coding regions included in the various expression constructs. The solid line indicates the ‘AlkB core’, i.e. the region which displays homology to other members of the AlkB family, and extensive sequence homology within the subfamily of viral AlkB proteins. The dotted line indicates a region where sequence conservation is observed within the group of 23 viral AlkB sequences (but not shared by other AlkB family members): (1) CXC in 14 sequences; (2) I/L/V/M-X-I/L/V in 15 sequences; 12 sequences adhered to the consensus CXCX3-I/L/V/M-X-I/L/V.
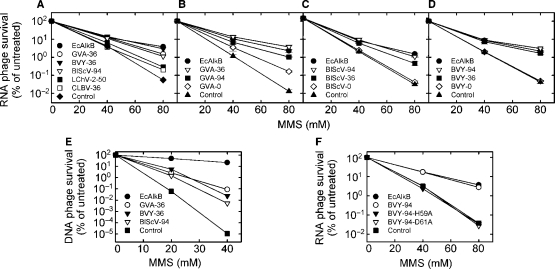
An AlkB protein active on RNA (e.g. EcAlkB or hABH3) is, when expressed in AlkB-deficient E. coli, able to increase the survival of RNA bacteriophage MS2 inactivated by treatment with a methylating compound (e.g. MMS), due to repair of deleterious methyl lesions in the viral RNA (14). In initial experiments, AlkB proteins derived from the five selected viruses were tested for such repair activity. Robust repair activity, comparable to that of EcAlkB, was observed for GVA-36, BVY-36 and BlScV-94, while only low activity was detected in the case of LChV-2-50 and CLBV-36 (Figure 4A). Based on these results, the viral AlkB proteins from GVA, BVY and BlScV were selected for further analysis.
Figure 4.
Reactivation of MMS-treated bacteriophages by expression of AlkB proteins in E. coli. (A) Reactivation of methylated ssRNA phage MS2 by AlkB proteins from GVA, BVY, BlScV, LChV-2 or CLBV. Reactivation of methylated MS2 by different variants of AlkB from (B) GVA, (C) BlScV and (D) BVY. (E) Reactivation of methylated ssDNA phage M13 by GVA-36, BVY-36 and BlScV-94. (F) Reactivation of methylated MS2 by BVY-94 and mutants BVY-94-H59A and BVY-94-D61A. Expression plasmid pJB658 without insert was used as control. Error bars in B–D represent the standard deviation of triplicate measurements.
Our initial experiments demonstrated repair activity for viral AlkBs containing N-terminal extensions beyond the AlkB core. To address whether these extensions are required for AlkB activity, each of the three selected viral AlkBs was provided with N-terminal extension of 0, 36 or 94 aa (Figure 3). The activity of the resulting proteins was again assessed by measuring their ability to reactivate methylated MS2 when expressed in E. coli. The results showed that the three shortest proteins were inactive (BVY-0 and BlScV-0) or showed low activity (GVA-0), whereas the longer forms displayed moderate to high activity (Figure 4B–D). GVA-36, BVY-36, BVY-94 and BlScV-94 were as active as EcAlkB, while the activity of GVA-94 and BlScV-36 was somewhat lower. In conclusion, sequences N-terminal to the AlkB core appeared to be required for viral AlkB activity.
The results presented above strongly suggest that viral AlkBs represent nucleic acid repair proteins. Since viral AlkBs originate from RNA viruses, RNA repair is the most obvious candidate function for these proteins. However, all AlkB proteins studied to date are at least as active on DNA as on RNA substrates (14,15). To assess the ability of the viral AlkBs to repair damaged DNA, an experiment identical to the phage reactivation experiment described above was performed, except that the RNA phage MS2 was replaced by the ssDNA phage M13. For this experiment, three proteins that displayed high activity on methylated RNA were selected, i.e. GVA-36, BVY-36 and BlScV-94. The expression of these proteins in E. coli increased the survival of methylated DNA phage to a similar extent, indicating that they also can repair DNA (Figure 4E). However, while expression of EcAlkB increased the survival of DNA phage M13 exposed to 40 mM MMS by 6–7 orders of magnitude, the observed increase was only 3–4 orders of magnitude for the viral AlkB proteins. Combined with the observation that these proteins were as effective as EcAlkB in reactivating methylated RNA phage (Figure 4A–D), these results may suggest that DNA is not the preferred substrate for the viral AlkB proteins.
All AlkB proteins contain a His-X-Asp-Xn-His motif, which is involved in iron(II) binding, and represents a hallmark of Fe(II)/2OG-dependent dioxygenases (30,31). It was shown for hABH3 that mutating either the first histidine or the aspartic acid residue completely abolished enzymatic activity (32). To test whether corresponding mutations in viral AlkB would affect activity, amino acid substitutions His59Ala and Asp61Ala were introduced in BVY-94 (amino acid numbering refers to the AlkB core; Figure 3). As expected, both mutants, BVY-94-H59A and BVY-94-D61A, were unable to rescue methylated RNA phage MS2, supporting that His59 and Asp61 are important residues for catalysis (Figure 4F).
In vitro characterization of recombinant viral AlkB proteins
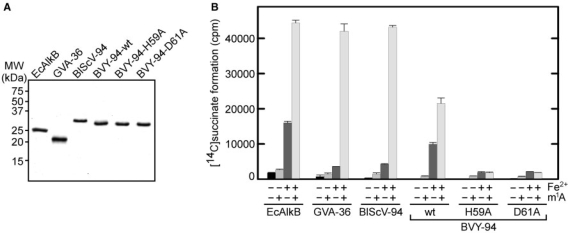
Many Fe(II)/2OG-dependent dioxygenases are able to catalyse the decarboxylation of the cosubstrate 2-oxoglutarate to succinate at a low rate even in the absence of their primary substrate (‘uncoupled reaction’), and this is also the case for EcAlkB (7,8). To confirm that viral AlkBs represent 2OG-dependent enzymes, recombinant GVA-36, BVY-94 and BlScV-94, were expressed in E. coli and purified as hexahistidine-tagged proteins (Figure 5A), and then tested for their ability to carry out the conversion of 2-oxoglutarate to succinate. All three proteins showed robust decarboxylation activity in the absence of any substrate, and the reaction could be stimulated by the substrate-mimic 1-methyladenosine (Figure 5B), which was previously shown to stimulate this reaction in the case of EcAlkB (33). Omitting Fe2+ from the reaction buffer reduced decarboxylation activity drastically (Figure 5B). The mutants BVY-94-H59A and BVY-94-D61A were both unable to convert 2-oxoglutarate to succinate (Figure 5B). Taken together, these results confirmed that the viral AlkBs are iron(II)- and 2-oxoglutarate-dependent repair enzymes, and indicated that these proteins are able to accommodate 1-meA in their active site.
Figure 5.
AlkB-mediated decarboxylation of 2-oxoglutarate. (A) Purified recombinant His-tagged AlkB proteins used in this study. Proteins were visualized by coomassie staining of a 15% SDS–PAGE gel. (B) Reaction mixtures containing [5-14C]2-oxoglutarate, 100 pmol of EcAlkB and 500 pmol of GVA-36, BlScV-94, BVY-94, BVY-94-H59A or BVY-94-D61A were incubated for 30 min at 37°C, and the released [1-14C]succinate was measured by scintillation counting. Cofactor Fe2+ and substrate-mimic 1-methyladenosine (m1A) were present as indicated. Error bars represent the standard deviation of duplicate measurements.
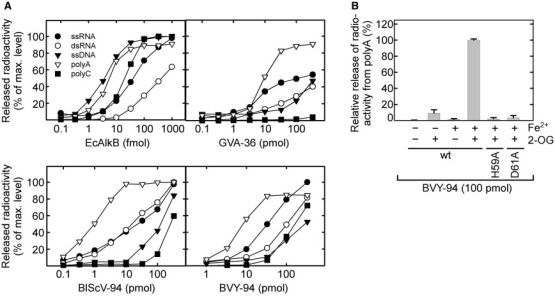
When ss nucleic acids are treated with methylating agents such as MMS or N-methyl-N-nitrosourea (MNU), substantial amounts of the AlkB substrates 1-meA and, to a lesser extent, 3-meC are introduced (1). To investigate the in vitro repair activity and the substrate specificity of viral AlkB, recombinant GVA-36, BVY-94 and BlScV-94 were incubated with RNA or DNA oligonucleotides that had been [3H]methylated through treatment with [3H]MNU. When comparing the activity on ssRNA relative to ssDNA, EcAlkB was approximately 10-fold more active on ssDNA (Figure 6A), confirming previous observations (14). In contrast, the activity of the viral AlkB proteins was generally higher (up to 10-fold) on ssRNA than on ssDNA (Figure 6A).
Figure 6.
Activity of AlkB proteins on various [3H]methylated RNA and DNA substrates. (A) [3H]methylated oligonucleotides were incubated with varying amounts of EcAlkB, GVA-36, BlScV-94 or BVY-94 and the ethanol soluble radioactivity released was measured by scintillation counting. (B) [3H]methylated poly(A) was incubated with 100 pmol of BVY94, BVY-94-H59A or BVY-94-D61A. Fe2+ and 2OG were present as indicated. Error bars represent the standard deviation of duplicate measurements.
Next, it was tested whether the viral AlkBs were also active on a dsRNA substrate, which was generated by annealing an unlabelled complementary RNA oligonucleotide to the [3H]methylated ssRNA molecule. In agreement with previous observations (15), EcAlkB displayed a somewhat lower activity on dsRNA substrates than on ssRNA substrates (Figure 6A). This was also the case for GVA-36 and BVY-94, while BlScV-94 was equally active on both substrates.
Treatment of nucleic acids with methylating agents generates a variety of methyl adducts. To study whether the viral AlkB proteins are able to specifically repair 1-meA and 3-meC, their demethylating activity was tested on the homopolymeric [3H]methylated substrates polyA and polyC, respectively. As previously found (14), EcAlkB was able to efficiently demethylate both substrates (Figure 6A). However, the viral AlkB proteins displayed a substantially lower activity on polyC than on polyA, suggesting that 1-meA rather than 3-meC is the preferred substrate for these enzymes. As expected, omitting the cofactor Fe2+ and/or the cosubstrate 2OG from the reaction, or introducing the inactivating mutations His59Ala or Asp61Ala drastically reduced the demethylase activity of BVY-94 on polyA (Figure 6B).
In summary, the data from our enzymatic assays indicate, as already suggested by the phage reactivation experiments (Figure 4), that the viral AlkB proteins are repair enzymes capable of reversing 1-meA, and somewhat less efficiently, 3-meC lesions in nucleic acids, with an appreciable preference for RNA over DNA.
DISCUSSION
This report describes the first functional characterization of AlkB proteins from plant viruses. A sequence comparison with other AlkB proteins suggested that the viral AlkB proteins are involved in repairing methylation damage, and this indeed turned out to be the case. The three proteins studied in detail represent two diverse viral superfamilies, Alphavirus- and Picornavirus-like (34), and they had very similar properties, indicating that they perform the same biological function in planta. They all displayed a repair activity similar to EcAlkB, but preferred RNA over DNA, and are the first AlkB proteins with such substrate specificity. Moreover, the viral AlkB proteins displayed robust demethylation activity both on ssRNA and dsRNA, presumably enabling them to repair alkylation damage both in the ssRNA viral genome and in the dsRNA replication intermediates generated in the host cell during infection. Thus, viral AlkB represents the first example of virally encoded alkylation repair proteins.
It has been demonstrated that AlkB-mediated repair can functionally reactivate methylated tRNA and mRNA, indicating that RNA repair may be biologically relevant (17). This notion was further supported by the observation that while mice lacking mABH2 displayed defective repair of 1-meA lesions in DNA, no such defect was observed in mice lacking the putative RNA repair enzyme mABH3 (35). However, since the AlkB proteins reported to repair RNA are equally (ABH3) or more (EcAlkB) active on DNA, it has been difficult to rule out the possibility that AlkB-mediated RNA demethylation is an irrelevant side activity. On the other hand, since the AlkB-containing viruses have a positive-strand RNA genome, replicate in cytosolic membrane-bound compartments (36), and the AlkB domain in many cases remains part of an unprocessed (cytosolic) replicase polyprotein, it is very difficult to imagine a DNA repair function for viral AlkB. Thus, the current finding that viral AlkB proteins are able to efficiently repair methyl lesions in RNA, and even prefer RNA over DNA, provides a very strong argument for the biological significance of AlkB-mediated RNA repair.
Only a few examples of viral RNA repair have so far been demonstrated. The 3'-end of viral RNAs is vulnerable to degradation by host RNases, and several mechanisms can mend the resulting truncated ends (37,38). Also, when bacteria are infected with bacteriophage T4, a host response is mounted involving the cleavage of the Lys-specific tRNA in the anticodon loop, thereby preventing the synthesis of proteins required for the phage infection to proceed. The T4 RNA ligase counteracts this response by repairing the cleaved tRNA (39,40). Furthermore, mechanisms for removing aberrant bases from viral DNA genomes have been reported. Some DNA viruses encode photolyases or glycosylases to remove UV-induced pyrimidine dimers from the genome (41,42), whereas others carry uracil-DNA glycosylases to remove uracils resulting from cytosine deamination or uracil misincorporation (43). Several of these DNA repair mechanisms depend on an undamaged template strand (present in dsDNA). The AlkB-mechanism, on the other hand, is template-independent, making it well suited for RNA repair.
In plants, small interfering RNAs (siRNAs) are mediators of a host RNA interference (RNAi) response causing degradation of viral RNAs. It was originally suggested that the role of viral AlkB may be to counteract a host response involving RNA methylation (6). Interestingly, it was subsequently shown that siRNAs are methylated at the 2′-OH of the 3′-terminal nucleotide by the methylase HEN1 (44). Several virus-encoded suppressors of silencing have been shown to interfere with HEN1-mediated methylation (45). Since the viral AlkBs displayed robust activity on 1-meA and contain key conserved residues involved in the coordination of this substrate, we find it less likely that the 2′-O-methylated ribose of siRNAs is a substrate for viral AlkB. In agreement with this, we were unable to detect RNAi suppression activity of viral AlkBs using conventional transient in planta assays or stable transformation of Arabidopsis thaliana by viral AlkBs (Peremyslov,V.V. and Dolja,V.V., unpublished data). However, although our presented results clearly indicate a role for viral AlkB in RNA repair, one cannot completely rule out the possibility that they are involved in counteracting a yet undiscovered host antiviral defence system, involving the introduction of AlkB substrates into host or viral RNA.
Alkylating agents are found both in the environment and intracellularly, and it is believed that both these sources can contribute significantly to the cellular load of alkylation damage. It has been suggested that plant viruses may have acquired AlkB recently in response to the use of methylating agents, e.g. methylbromide, as pesticides (46). We consider this unlikely, because many AlkB-containing viruses infect plant species not treated with such agents, and because the time elapsed since the relatively recent introduction of such pesticides is probably too short for the observed spread and phylogenetic divergence of viral AlkBs from their cellular ancestors. Viruses that have AlkB often infect woody or perennial plants, where they may survive for years in a hostile environment within the phloem sieve elements to ensure their eventual transmission to new plants. We favour the hypothesis that the viral genome may occasionally be subjected to considerable methylation damage in this environment, and that viral AlkBs substantially enhance virus propagation by repairing such damage upon virus entry into the host cell.
Viral AlkBs represent a remarkable example of viral proteins with readily identifiable homologues in cellular genomes. Based on the observation that viral AlkB proteins are more similar to AlkB homologues from bacteria than from plants, it appears most likely that the first viral AlkB was derived via horizontal gene transfer from a bacterial source, possibly, from a bacterial pathogen or symbiont of plants. The lack of clustering between viral AlkBs and any specific bacterial branch (Figure 1A) could be explained by the well-known fast evolution of viral RNA sequences and/or by the absence of close relatives of the actual source bacterium in the current databases. With the exception of Flexiviridae, AlkB-containing viruses represent a minor proportion of viruses within their corresponding taxa, with BVY being the only known AlkB-containing member of the vast family Potyviridae. This distribution pattern suggests that AlkB was acquired relatively recently, likely by a virus within the family Flexiviridae where AlkB-containing viruses are more common than in other viral families, and then disseminated horizontally, via recombination between viruses including very distantly related ones such as BRNV. The spread of AlkB proteins to new viruses might have occurred through gene transfer between flexiviruses and viruses from other plant RNA virus families, which frequently occurs in mixed infections of the same, often woody or perennial host. Also, given the rapid evolution of RNA viruses, the very existence of a compact viral clade in the AlkB family tree (Figure 1A) is best compatible with a horizontal mode of their spread among viruses.
It is an intriguing question why only a subset of the plant viruses within a given family possesses an AlkB homologue. Possibly, having an AlkB protein gives the virus a selective advantage under conditions of high methylation load, and viruses that do not experience such conditions may have disposed of, or never acquired AlkBs. The notion that viral AlkB proteins may be disposed of when no longer needed, e.g. when a virus evolves to infect a different host, is supported by the observation that a handful of plant viruses have AlkB domains where several of the residues thought to be important for enzymatic activity are mutated. The scattered presence of inactivated AlkBs among active ones in the phylogenetic tree is best compatible with multiple, independent inactivations of AlkB that potentially might have been linked to a switch to environments with lower alkylation pressure. Interestingly, three of the viruses that apparently have inactivated AlkBs, infect potato; a non-woody and non-perennial plant, where the evolutionary pressure for maintaining the AlkB function might be absent (Figures 1B and S3).
To further understand the role of AlkB in viral infection, it will be of interest to study the infectivity in planta of viruses in which the AlkB region has been inactivated by mutation, as well as to study the possible accumulation of methylation damage in wild-type and AlkB-mutated viral genomes during infection.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
FRIBIOMOL and FUGE programs in the Research Council of Norway (P.Ø.F.); DHHS (National Library of Medicine) intramural funds (M.V.O. and E.V.K.); National Institutes of Health award (GM053190 to V.V.D.) and BARD award (IS-3784-05 to V.V.D.). Funding for open access charges: NIH intramural funds.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The [3H]methylated ssDNA AT-oligonucleotide was a kind gift from Hanne Korvald and Ingrun Alseth. The plasmids pJB658-AlkB and pET28-AlkB were kindly provided by Hans Krokan. The authors are thankful to M. Mawassi, V. Peremyslov, R. Martin, I. Tzanetakis, D. Rochon, J. Guerri and B. Hillman for providing viral cDNAs. We thank Stefan Kernstock for critical reading of the manuscript.
REFERENCES
- 1.Singer B, Grunberger D. Molecular Biology of Mutagens and Carcinogens. New York: Plenum Press; 1983. [Google Scholar]
- 2.Sedgwick B, Bates PA, Paik J, Jacobs SC, Lindahl T. Repair of alkylated DNA: recent advances. DNA Repair (Amst) 2007;6:429–442. doi: 10.1016/j.dnarep.2006.10.005. [DOI] [PubMed] [Google Scholar]
- 3.Delaney JC, Essigmann JM. Mutagenesis, genotoxicity, and repair of 1-methyladenine, 3-alkylcytosines, 1-methylguanine, and 3-methylthymine in alkB Escherichia coli. Proc. Natl Acad. Sci. USA. 2004;101:14051–14056. doi: 10.1073/pnas.0403489101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Falnes PO. Repair of 3-methylthymine and 1-methylguanine lesions by bacterial and human AlkB proteins. Nucleic Acids Res. 2004;32:6260–6267. doi: 10.1093/nar/gkh964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Koivisto P, Robins P, Lindahl T, Sedgwick B. Demethylation of 3-methylthymine in DNA by bacterial and human DNA dioxygenases. J. Biol. Chem. 2004;279:40470–40474. doi: 10.1074/jbc.M407960200. [DOI] [PubMed] [Google Scholar]
- 6.Aravind L, Koonin EV. The DNA-repair protein AlkB, EGL-9, and leprecan define new families of 2-oxoglutarate- and iron-dependent dioxygenases. Genome Biol. 2001;2 doi: 10.1186/gb-2001-2-3-research0007. research0007.1–0007.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Falnes PO, Johansen RF, Seeberg E. AlkB-mediated oxidative demethylation reverses DNA damage in Escherichia coli. Nature. 2002;419:178–182. doi: 10.1038/nature01048. [DOI] [PubMed] [Google Scholar]
- 8.Trewick SC, Henshaw TF, Hausinger RP, Lindahl T, Sedgwick B. Oxidative demethylation by Escherichia coli AlkB directly reverts DNA base damage. Nature. 2002;419:174–178. doi: 10.1038/nature00908. [DOI] [PubMed] [Google Scholar]
- 9.Duncan T, Trewick SC, Koivisto P, Bates PA, Lindahl T, Sedgwick B. Reversal of DNA alkylation damage by two human dioxygenases. Proc. Natl Acad. Sci. USA. 2002;99:16660–16665. doi: 10.1073/pnas.262589799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Delaney JC, Smeester L, Wong C, Frick LE, Taghizadeh K, Wishnok JS, Drennan CL, Samson LD, Essigmann JM. AlkB reverses etheno DNA lesions caused by lipid oxidation in vitro and in vivo. Nat. Struct. Mol. Biol. 2005;12:855–860. doi: 10.1038/nsmb996. [DOI] [PubMed] [Google Scholar]
- 11.Frick LE, Delaney JC, Wong C, Drennan CL, Essigmann JM. Alleviation of 1,N6-ethanoadenine genotoxicity by the Escherichia coli adaptive response protein AlkB. Proc. Natl Acad. Sci. USA. 2007;104:755–760. doi: 10.1073/pnas.0607377104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mishina Y, Yang CG, He C. Direct repair of the exocyclic DNA adduct 1,N6-ethenoadenine by the DNA repair AlkB proteins. J. Am. Chem. Soc. 2005;127:14594–14595. doi: 10.1021/ja055957m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kurowski MA, Bhagwat AS, Papaj G, Bujnicki JM. Phylogenomic identification of five new human homologs of the DNA repair enzyme AlkB. BMC Genomics. 2003;4:48. doi: 10.1186/1471-2164-4-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Aas PA, Otterlei M, Falnes PO, Vagbo CB, Skorpen F, Akbari M, Sundheim O, Bjoras M, Slupphaug G, Seeberg E, et al. Human and bacterial oxidative demethylases repair alkylation damage in both RNA and DNA. Nature. 2003;421:859–863. doi: 10.1038/nature01363. [DOI] [PubMed] [Google Scholar]
- 15.Falnes PO, Bjoras M, Aas PA, Sundheim O, Seeberg E. Substrate specificities of bacterial and human AlkB proteins. Nucleic Acids Res. 2004;32:3456–3461. doi: 10.1093/nar/gkh655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee DH, Jin SG, Cai S, Chen Y, Pfeifer GP, O’Connor TR. Repair of methylation damage in DNA and RNA by mammalian AlkB homologues. J. Biol. Chem. 2005;280:39448–39459. doi: 10.1074/jbc.M509881200. [DOI] [PubMed] [Google Scholar]
- 17.Ougland R, Zhang CM, Liiv A, Johansen RF, Seeberg E, Hou YM, Remme J, Falnes PO. AlkB restores the biological function of mRNA and tRNA inactivated by chemical methylation. Mol. Cell. 2004;16:107–116. doi: 10.1016/j.molcel.2004.09.002. [DOI] [PubMed] [Google Scholar]
- 18.Martelli GP, Adams MJ, Kreuze JF, Dolja VV. Family Flexiviridae: a case study in virion and genome plasticity. Annu. Rev. Phytopathol. 2007;45:73–100. doi: 10.1146/annurev.phyto.45.062806.094401. [DOI] [PubMed] [Google Scholar]
- 19.Minafra A, Saldarelli P, Martelli GP. Grapevine virus A: nucleotide sequence, genome organization, and relationship in the Trichovirus genus. Arch. Virol. 1997;142:417–423. doi: 10.1007/s007050050088. [DOI] [PubMed] [Google Scholar]
- 20.Cavileer TD, Halpern BT, Lawrence DM, Podleckis EV, Martin RR, Hillman BI. Nucleotide sequence of the carlavirus associated with blueberry scorch and similar diseases. J. Gen. Virol. 1994;75:711–720. doi: 10.1099/0022-1317-75-4-711. [DOI] [PubMed] [Google Scholar]
- 21.Susaimuthu J, Tzanetakis IE, Gergerich RC, Martin RR. A member of a new genus in the Potyviridae infects Rubus. Virus Res. 2008;131:145–151. doi: 10.1016/j.virusres.2007.09.001. [DOI] [PubMed] [Google Scholar]
- 22.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hasegawa M, Kishino H, Saitou N. On the maximum likelihood method in molecular phylogenetics. J. Mol. Evol. 1991;32:443–445. doi: 10.1007/BF02101285. [DOI] [PubMed] [Google Scholar]
- 25.Jones DT, Taylor WR, Thornton JM. The rapid generation of mutation data matrices from protein sequences. Comput. Appl. Biosci. 1992;8:275–282. doi: 10.1093/bioinformatics/8.3.275. [DOI] [PubMed] [Google Scholar]
- 26.Waddell PJ, Kishino H, Ota R. Very fast algorithms for evaluating the stability of ML and Bayesian phylogenetic trees from sequence data. Genome Inform. 2002;13:82–92. [PubMed] [Google Scholar]
- 27.Blatny JM, Brautaset T, Winther-Larsen HC, Karunakaran P, Valla S. Improved broad-host-range RK2 vectors useful for high and low regulated gene expression levels in gram-negative bacteria. Plasmid. 1997;38:35–51. doi: 10.1006/plas.1997.1294. [DOI] [PubMed] [Google Scholar]
- 28.Kaule G, Günzler V. Assay for 2-oxoglutarate decarboxylating enzymes based on the determination of [1-14C]succinate: application to prolyl 4-hydroxylase. Anal. Biochem. 1990;184:291–297. doi: 10.1016/0003-2697(90)90683-z. [DOI] [PubMed] [Google Scholar]
- 29.Falnes PO, Rognes T. DNA repair by bacterial AlkB proteins. Res. Microbiol. 2003;154:531–538. doi: 10.1016/S0923-2508(03)00150-5. [DOI] [PubMed] [Google Scholar]
- 30.Yu B, Edstrom WC, Benach J, Hamuro Y, Weber PC, Gibney BR, Hunt JF. Crystal structures of catalytic complexes of the oxidative DNA/RNA repair enzyme AlkB. Nature. 2006;439:879–884. doi: 10.1038/nature04561. [DOI] [PubMed] [Google Scholar]
- 31.Schofield CJ, Zhang Z. Structural and mechanistic studies on 2-oxoglutarate-dependent oxygenases and related enzymes. Curr. Opin. Struct. Biol. 1999;9:722–731. doi: 10.1016/s0959-440x(99)00036-6. [DOI] [PubMed] [Google Scholar]
- 32.Sundheim O, Vagbo CB, Bjoras M, Sousa MM, Talstad V, Aas PA, Drablos F, Krokan HE, Tainer JA, Slupphaug G. Human ABH3 structure and key residues for oxidative demethylation to reverse DNA/RNA damage. EMBO J. 2006;25:3389–3397. doi: 10.1038/sj.emboj.7601219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Welford RW, Schlemminger I, McNeill LA, Hewitson KS, Schofield CJ. The selectivity and inhibition of AlkB. J. Biol. Chem. 2003;278:10157–10161. doi: 10.1074/jbc.M211058200. [DOI] [PubMed] [Google Scholar]
- 34.Koonin EV, Dolja VV. Evolution and taxonomy of positive-strand RNA viruses: implications of comparative analysis of amino acid sequences. Crit. Rev. Biochem. Mol. Biol. 1993;28:375–430. doi: 10.3109/10409239309078440. [DOI] [PubMed] [Google Scholar]
- 35.Ringvoll J, Nordstrand LM, Vagbo CB, Talstad V, Reite K, Aas PA, Lauritzen KH, Liabakk NB, Bjork A, Doughty RW, et al. Repair deficient mice reveal mABH2 as the primary oxidative demethylase for repairing 1meA and 3meC lesions in DNA. EMBO J. 2006;25:2189–2198. doi: 10.1038/sj.emboj.7601109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ahlquist P. Parallels among positive-strand RNA viruses, reverse-transcribing viruses and double-stranded RNA viruses. Nat. Rev. Microbiol. 2006;4:371–382. doi: 10.1038/nrmicro1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hema M, Gopinath K, Kao C. Repair of the tRNA-like CCA sequence in a multipartite positive-strand RNA virus. J. Virol. 2005;79:1417–1427. doi: 10.1128/JVI.79.3.1417-1427.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nagy PD, Carpenter CD, Simon AE. A novel 3-end repair mechanism in an RNA virus. Proc. Natl Acad. Sci. USA. 1997;94:1113–1118. doi: 10.1073/pnas.94.4.1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Amitsur M, Levitz R, Kaufmann G. Bacteriophage T4 anticodon nuclease, polynucleotide kinase and RNA ligase reprocess the host lysine tRNA. EMBO J. 1987;6:2499–2503. doi: 10.1002/j.1460-2075.1987.tb02532.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang LK, Shuman S. Mutational analysis defines the 5'-kinase and 3'-phosphatase active sites of T4 polynucleotide kinase. Nucleic Acids Res. 2002;30:1073–1080. doi: 10.1093/nar/30.4.1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Garvish JF, Lloyd RS. The catalytic mechanism of a pyrimidine dimer-specific glycosylase (pdg)/abasic lyase, Chlorella virus-pdg. J. Biol. Chem. 1999;274:9786–9794. doi: 10.1074/jbc.274.14.9786. [DOI] [PubMed] [Google Scholar]
- 42.Srinivasan V, Schnitzlein WM, Tripathy DN. Fowlpox virus encodes a novel DNA repair enzyme, CPD-photolyase, that restores infectivity of UV light-damaged virus. J. Virol. 2001;75:1681–1688. doi: 10.1128/JVI.75.4.1681-1688.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen R, Wang H, Mansky LM. Roles of uracil-DNA glycosylase and dUTPase in virus replication. J. Gen. Virol. 2002;83:2339–2345. doi: 10.1099/0022-1317-83-10-2339. [DOI] [PubMed] [Google Scholar]
- 44.Yu B, Yang Z, Li J, Minakhina S, Yang M, Padgett RW, Steward R, Chen X. Methylation as a crucial step in plant microRNA biogenesis. Science. 2005;307:932–935. doi: 10.1126/science.1107130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yu B, Chapman EJ, Yang Z, Carrington JC, Chen X. Transgenically expressed viral RNA silencing suppressors interfere with microRNA methylation in Arabidopsis. FEBS Lett. 2006;580:3117–3120. doi: 10.1016/j.febslet.2006.04.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bratlie MS, Drablos F. Bioinformatic mapping of AlkB homology domains in viruses. BMC Genomics. 2005;6:1. doi: 10.1186/1471-2164-6-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.