Abstract
It is unclear whether Mediator complex in yeast is necessary for all RNA polymerase II (Pol II) transcription or if it is limited to genes activated by environmental stress. In mammals, amino acid limitation induces SNAT2 transcription through ATF4 binding at an amino acid response element. ATF4 is the functional counterpart to the yeast amino acid-dependent regulator GCN4 and GCN4 recruits Mediator during transcriptional activation. Consistent with enhanced SNAT2 transcription activity, the present data demonstrate that amino acid limitation increased SNAT2 promoter association of the general transcription factors that make up the preinitiation complex, including Pol II, but there was no increase in Mediator recruitment. Furthermore, siRNA knockdown of eight Mediator subunits caused no significant decrease in SNAT2 transcription. The estrogen-dependent pS2 gene was used as a positive control for both the ChIP and the siRNA approaches and the data demonstrated the requirement for Mediator recruitment. These results document that activation of the SNAT2 gene by the mammalian amino acid response pathway occurs independently of enhanced Mediator recruitment.
INTRODUCTION
Mediator, consisting of about 30 protein subunits (1), has been proposed to function as a general transcription factor (GTF) and is therefore necessary for most, if not all, RNA polymerase II (Pol II)-mediated transcription (2). However, Fan et al. (3) recently showed that there is not always a correlation between recruitment of Pol II and Mediator on many highly active genes in yeast, such as these for ribosomal proteins or glycolytic enzymes. Those authors concluded that thus far, the in vivo data suggest that Mediator is ‘recruited to enhancers in an activator-specific manner, and it does not seem to be a stoichiometric component of the basic Pol II machinery’. Fan et al. also suggested that Mediator might be selectively recruited to genes that are transcriptionally activated by environmental stress or sub-optimal growth conditions. In a commentary on the Fan et al. (3) report, Lewis and Reinberg (4) suggested that in metazoans some promoters may use TFIID, instead of Mediator, as a link between enhancer-binding proteins and the preinitiation complex.
To test the hypothesis that Mediator is required for stress-responsive genes in mammalian cells, the present studies focused on the transcriptional control of an amino acid-regulated gene, the sodium-dependent neutral amino acid transporter 2 (SNAT2). In yeast, general control nonderepressible-4 (GCN4) is the transcription factor that activates genes in response to amino acid deprivation6. GCN4 binding results in recruitment of enhanced levels of the Mediator complex to amino acid responsive genes (5,6). Activating transcription factor 4 (ATF4) is the functional mammalian homologue to yeast GCN4 (7). Like GCN4, increased de novo ATF4 synthesis (8,9) and enhanced transcription of ATF4 target genes is observed after activation of the amino acid response (AAR) pathway by protein deprivation (in vivo) or amino acid limitation (in vivo or in vitro) (10,11). The AAR pathway results in activated transcription from genes containing an amino acid response element (AARE) that functions as a positive enhancer element, either proximal or distal to the promoter (12). Activation of these AARE-containing genes is associated with increased ATF4 binding and subsequent enhanced recruitment of the preinitiation complex to the promoter (11). The SNAT2 gene contains an AARE within the first intron, downstream of the promoter by ∼700 bp (13,14). It was hypothesized that even if Mediator is not required for all Pol II-driven transcription in mammalian cells, it may be required for SNAT2 activation by this distal, stress-responsive enhancer element.
In the present report, results obtained by chromatin immunoprecipitation (ChIP) show that as a consequence of AARE-enhanced transcription of the SNAT2 gene in human HepG2 hepatoma and MCF-7 breast cancer cells, there is an enhanced association of the GTFs that make up the preinitiation complex, but no increase in recruitment of the Mediator complex. Accordingly, there is no correlation between Mediator binding and the binding of the GTFs, including Pol II, at the SNAT2 promoter. Furthermore, siRNA knockdown of eight individual Mediator subunits did not block induction of AARE-driven transcription. As a positive control for both the ChIP and the siRNA approaches, the observations by others were confirmed showing that the estrogen-dependent activation of the pS2 gene required Mediator recruitment to the pS2 promoter (15,16).
MATERIALS AND METHODS
Cell culture
HepG2 human hepatoma cells were cultured in modified Eagle's MEM and MCF-7 human breast carcinoma cells were cultured in Dulbecco's Modified Eagle's MEM (DMEM) (Mediatech, Herndon, VA, USA). Both media contained 10% (v/v) fetal bovine serum (FBS) and supplements as described (11). For amino acid deprivation, cells were incubated in either complete medium or medium lacking histidine, both containing 10% dialyzed FBS. For β-estradiol (E2) treatment of MCF-7 cells, the DMEM medium was replaced with DMEM containing no phenol red and 10% charcoal-stripped FBS for 48 h and then the cells were treated with either ethanol (control) or 100 nM E2 for the time indicated.
Immunoblotting and antibodies
Nuclear protein extracts were prepared (NE-PER, Pierce Chemicals, Rockford, IL, USA) at the time points indicated and subjected to immunoblotting (11). The following antibodies were purchased from Santa Cruz Biotechnology (Santa Cruz, CA, USA): TAF1 (TAF250), sc-17134; TAF9 (TAF32), sc-1248; TFIIA, sc-5316; TFIIB, sc-274; TFIIE, sc-237; TRAP220 (Med1), sc-5334; CRSP130 (Med23), sc-12454; MED15, sc-101185; CDK8, sc-5612; RNA Polymerase II, sc-899; rabbit IgG, sc-2027. Antibody against MED15 (anti-PCQAP, positive cofactor 2 glutamine/Q rich-associated protein) was also purchased from Sigma Chemical Company, St. Louis, MO (# HPA003179). Antibody for acetylated histone H3, (#06–599, specific for acetylated Lys-9 and Lys-14) was purchased from Upstate Biotechnology (Charlottesville, VA, USA) and antibody for total H3 protein was from Abcam, Cambridge, MA (#1791).
Transcription activity and mRNA determination
Total RNA was isolated from MCF-7 cells using the Qiagen RNeasy Kit (Qiagen, Valencia, CA, USA), including a DNase I treatment before final elution to eliminate DNA contamination. Steady state mRNA levels for the Mediator subunits were assayed by RT–qPCR, as described previously (11), using the primers in Supplementary Table I. To measure the transcription activity, oligonucleotide primers (Supplementary Table I) were chosen that amplify a region across an intron–exon boundary to measure the unspliced heteronuclear RNA (hnRNA), as described by Lipson and Baserga (17), except that the hnRNA levels in the present studies were analyzed by RT–qPCR using the DNA Engine Opticon 2 system (MJ Research, Reno, NV, USA) and SYBR Green chemistry. Reactions without reverse transcriptase were performed as a negative control to rule out amplification from any residual genomic DNA, and these tests were always negative. After PCR, melting curves were acquired by stepwise increase of the temperature from 55°C to 95°C to ensure that a single product was amplified in the reaction. The qPCR analysis for each sample was done in duplicate with samples from at least three independent experiments and the means ± the SEM calculated.
siRNA treatment
MCF-7 cells were transfected with either a siControl nontargeting siRNA pool or SMARTpool siRNA against the indicated Mediator subunit using 100 nM siRNA and Dharmafect-4 reagent (Dharmacon, Lafayette, CO, USA). After 24 h, the cells were transferred to fresh DMEM medium containing 10% FBS and maintained for another 24 h, and then subjected to treatment as indicated in each figure.
Chromatin immunoprecipitation
ChIP analysis of HepG2 and MCF-7 cells was performed as described previously (11). Quantitative PCR was performed with the DNA Engine Opticon 2 system and SYBR Green chemistry using the primers listed in Supplementary Table 2. The qPCR analysis for each sample was done in duplicate with samples from at least three independent experiments and the means ± the SEM calculated.
RESULTS
Histone modification and preinitiation complex assembly on the SNAT2 gene
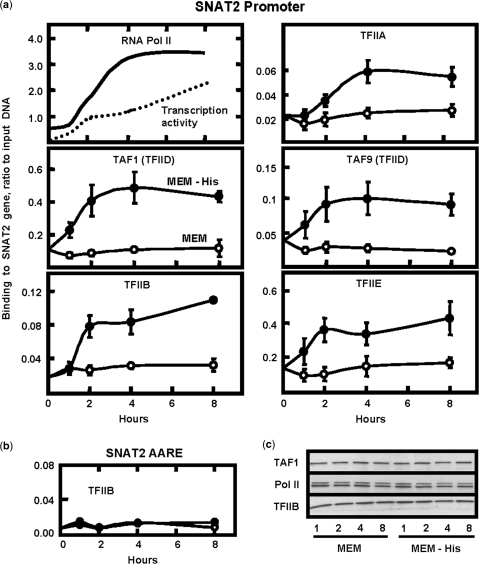
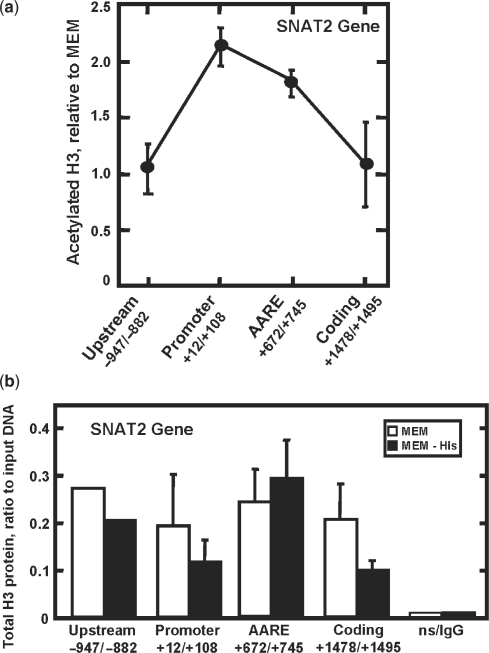
The SNAT2 gene has an AARE in intron 1 (nucleotides +709 to +717) that functions as an ATF4-responsive enhancer to activate transcription within 1 h after amino acid limitation and the transcription remains elevated for about 10–12 h (13,14). The time course of SNAT2 transcription activity and recruitment of RNA Pol II have been published previously (14), but for the purpose of comparison a graphical representation is shown in the first panel of Figure 1a. It has been suggested that in the absence of Mediator, the TFIID complex may provide the link between enhancer elements and RNA Pol II (4). For SNAT2, the activator ATF4 binds to the intronic AARE, but does not appear to interact directly with the promoter region (12). To provide mechanistic insight into assembly of the preinitiation complex following amino acid limitation, binding of the TFIIA, TFIIB, TFIID (TAF1 and TAF9) and TFIIE was measured by ChIP analysis (Figure 1a). Following amino acid removal, all of the GTFs exhibited increased binding to the SNAT2 promoter in a manner that qualitatively paralleled the enhancement in transcription activity. In contrast, as shown by the example of TFIIB (Figure 1b), none of the GTFs showed significant binding to the intronic region containing the SNAT2 AARE (TFIIA, TFIID and TFIIE, not shown). The stress-induced recruitment of the GTFs was not a consequence of a change in their absolute concentration, as illustrated by immunoblotting of nuclear extracts (Figure 1c). Increased acetylation of histone H3 (AcH3) is a general marker for induced gene expression. To monitor the localization of amino acid-dependent chromatin modification for the SNAT2 gene, ChIP analysis for AcH3 was performed at specific regions across the SNAT2 gene 8 h after amino acid limitation (Figure 2a). The SNAT2 promoter exhibited the largest increase in AcH3, but acetylation was also elevated at the AARE. In contrast, at regions only about 1 kb upstream or downstream within the gene, there was little or no increase in histone modification relative to the MEM values. Except for an area within the SNAT2 coding region, total H3 protein was largely unchanged by amino acid limitation (Figure 2b). These results demonstrate that there is region-specific chromatin remodeling and an active recruitment of the preinitiation complex to the SNAT2 promoter during amino acid stress.
Figure 1.
Amino acid limitation induces region-specific changes in histone acetylation, SNAT2 transcription activity and GTF binding at the SNAT2 gene. (a) The SNAT2 transcription activity and the binding of RNA Pol II, both published elsewhere (14), are provided as a graphical representation to provide a comparison with the ChIP data for the remaining GTFs. To obtain the data of (a), HepG2 cells were incubated in MEM (open circle) or MEM lacking histidine (closed circle) for 1 h, 2 h, 4 h or 8 h and protein association with the SNAT2 promoter was monitored by ChIP assay. For each of the GTF proteins shown in (a) binding to the SNAT2 AARE was also tested, but was not significantly different from the background value obtained with a nonspecific antibody. An example is illustrated by the data for TFIIB (b). Nuclear extracts were prepared and subjected to immunoblot analysis for several GTFs, those for TAFI, Pol II and TFIIB are shown as examples (c).
Figure 2.
Increased histone acetylation occurs in response to amino acid limitation. HepG2 cells incubated for 8 h in MEM or MEM lacking histidine were subjected to ChIP analysis with an antibody specific for either acetylated histone H3 (a) or total histone H3 protein (b). Primer sets (see Supplementary Table 1) spanning the SNAT2 gene were used for qPCR analysis. The data shown in (a) represent three independent experiments. For (b), the ‘upstream’ values are the average of two experiments, whereas all of the remaining values are from three or more experiments. The qPCR was run in duplicate for each sample.
Increased recruitment of the Mediator complex does not occur during AARE-dependent activation of transcription
Mediator complex is thought to link enhancer-associated activator proteins with the promoter-bound general transcription machinery (2,18) and in yeast it has been proposed to be particularly relevant for stress-induced genes (3). Consequently, it was plausible that enhanced Mediator recruitment is required for AARE-containing genes that respond to amino acid stress in mammalian cells. To investigate Mediator association with the SNAT2 gene in HepG2 hepatoma cells, antibodies specific for the Mediator subunits MED1 or MED23 were used for ChIP analysis in cells incubated for 0–8 h in complete MEM or MEM lacking histidine (Figure 3a). At either the promoter or the AARE region of the gene, the association for either Mediator subunit was 0.01 or less (ratio to input DNA), regardless of whether the cells were amino acid deprived. This level of detection is about the same as the background value obtained by incubation with a nonspecific antibody. These results suggest that there may be little if any association of Mediator to the SNAT2 promoter or AARE enhancer regions in the basal or ‘fed’ state, and more importantly, there is no additional recruitment following amino acid stress.
Figure 3.
Activation of SNAT2 transcription by the AAR does not require enhanced recruitment of Mediator. HepG2 cells were incubated in MEM or MEM lacking histidine for 1 h, 2 h, 4 h or 8 h and then MED1 or MED23 association with the SNAT2 promoter or AARE region was monitored by ChIP analysis (a). Immunoprecipitation with a nonspecific IgG (n/s IgG) served as a negative control. MCF-7 cells were incubated in DMEM or DMEM plus 100 nM β-estradiol (E2) for 1 h and MED1, MED23 or CDK8 association with the pS2 promoter was monitored by ChIP analysis (b). On the left side, the data are presented as the ratio of protein bound to total input DNA. On the right side, the same data are shown as the fold change induced by E2 treatment. MCF-7 cells were incubated in DMEM or DMEM lacking histidine for 2 h and MED1, MED23 or CDK8 association with the SNAT2 gene was monitored by ChIP (c).
To provide a positive control, ChIP analysis of the estrogen-responsive pS2 gene was performed in MCF-7 cells treated with either vehicle (ethanol) or β-estradiol (E2) (Figure 3b). Increased Mediator binding to the pS2 promoter in response to E2 treatment has been documented by others (15,16,19). In those previous studies, an E2-dependent enrichment of MED1, MED23 and CDK8 was documented. CDK8 is a kinase subunit which may function as a component of either a repressive or an activating Mediator complex (20,21). Binding of RNA Pol II and all three Mediator subunits, was detectable in the basal state, although the degree of MED1 and MED23 association was not much greater than that for the nonspecific IgG (Figure 3b). Following E2 treatment, the recruitment of all three Mediator subunits was substantially increased, although the overall involvement of CDK8 was less than that for MED1 and MED23.
To ensure that the lack of Mediator binding to SNAT2 in HepG2 cells was not a cell-specific result, the presence of Mediator on the SNAT2 gene in MCF-7 cells was measured by ChIP analysis (Figure 3c). Whereas RNA Pol II recruitment to the SNAT2 promoter was strongly induced by amino acid deprivation, once again, the association of Mediator did not change. Although the values for all three Mediator subunits were close to that for the nonspecific IgG at the 5′-distal, the 3′-distal and the AARE sites, a basal amount of Mediator binding to the SNAT2 promoter cannot be completely ruled out because those values were somewhat larger than the nonspecific IgG and the other three sites tested. Regardless, the data of Figure 3 demonstrate that increased recruitment of Mediator to the SNAT2 gene does not occur during the nutrient stress of amino acid limitation, nor does the level of Mediator association correlate with the increased recruitment for RNA Pol II and the remainder of the preinitiation complex.
Knockdown of Mediator does not block transcriptional activation of SNAT2
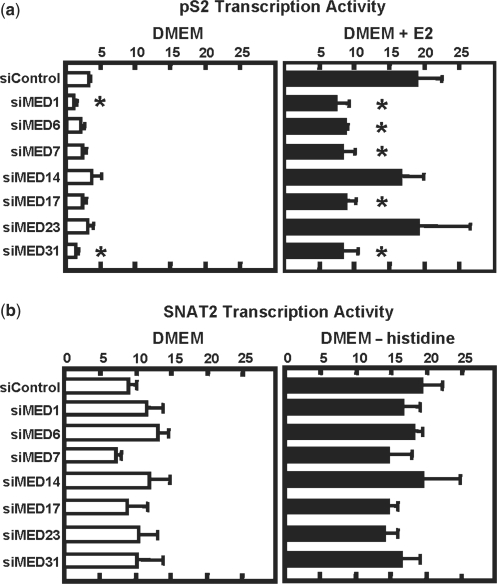
Given the limited number of Mediator antibodies that function for ChIP analysis and to obtain data from an independent approach, MCF-7 cells were treated with an siRNA specific for one of seven different Mediator subunits, MED1, MED6, MED7, MED14, MED17, MED23 or MED31. To monitor the effectiveness of the siRNA action, 48 h after transfection, cellular RNA was isolated and subjected to qRT–PCR to monitor steady state mRNA levels (Figure 4). The results demonstrate that each of the siRNA treatments was effective in reducing the corresponding Mediator subunit expression by 50–90%, relative to the siControl. For MED1 and MED6, the mRNA results were confirmed by immunoblotting for protein (data not shown). To monitor the effect of Mediator knockdown on transcription activity, cells were treated with either E2 to activate transcription from the pS2 gene (Figure 5a) or medium lacking histidine to induce SNAT2 transcription (Figure 5b). Using the pS2 gene as a positive control, the E2 transcriptional activation was blocked by >50% when MED1, MED6, MED7, MED17 and MED31 were reduced in their expression. For unknown reasons, knockdown of MED14 and MED23 had no effect on the E2 induction. In contrast, for each of the seven Mediator subunits there was no effect on the transcription activity from the SNAT2 gene in the basal or ‘fed’ state nor was the activation of the SNAT2 gene by amino acid limitation reduced (Figure 5b).
Figure 4.
Knockdown of Mediator subunits by siRNA. MCF-7 cells were treated for 24 h with either Dharmacon's ‘control’ siRNA (siControl) or siRNA (siMED subunit) specific for seven individual Mediator subunits, incubated in fresh DMEM for another 24 h, and then incubated in either DMEM, DMEM lacking histidine or DMEM plus 100 nM E2 for 4 h. To determine the decline in expression for individual Mediator subunits, mRNA was analyzed by RT–qPCR. The siMED data are plotted as the fraction of the mRNA levels in the siControl condition, which was set to 1.
Figure 5.
Reduction in Mediator expression does not block induction of SNAT2 transcription. MCF-7 cells were treated with siRNA and incubated in DMEM, DMEM plus E2 or DMEM lacking histidine as described in Figure 4 and then pS2 (a) or SNAT2 (b) transcription activity was analyzed by RT–qPCR. Asterisks denote transcription activity in the siMed condition that is significantly different (P < 0.05) from the siControl condition.
Zhang et al. (22) have shown that when Sin4p, a protein that links the ‘tail’ module to the body module in yeast, is deleted from the genome, a triad of proteins that make up the remainder of the tail (gal11/Med2/Pgd1), can be recruited to and activate transcription from GCN4-induced genes independently of the rest of the Mediator complex. Although mammalian cells may not have paralogs to Med2 and Pgd1 (1,18,23), to determine if MED15, the human counterpart to yeast gal11, was recruited to SNAT2 independently of the remainder of Mediator, siRNA knockdown and ChIP analysis were employed for this subunit as well. The data show that despite a 50–80% reduction of the MED15 expression (Figure 6b), the activated transcription from the pS2 gene by E2 and transcription from the SNAT2 gene was unaffected (Figure 6a). ChIP assays for MED15 (antibody from Santa Cruz Biotechnology) association with the SNAT2 promoter or AARE region revealed a relatively low level of binding (Figure 6c), yielding values that were comparable to those for a nonspecific IgG (Figure 3), and there was no additional recruitment of MED15 following amino acid limitation. When ChIP analysis was performed on the pS2 promoter to determine if MED15 recruitment was enhanced after E2 treatment, in a manner similar to other Mediator subunits shown in Figure 3, no association of MED15 with the pS2 gene was observed (Figure 6c). To extend this result, a second MED15 antibody was tested (Sigma Chemical Company), but the results were the same (data not shown).
Figure 6.
MED15 is not required for induction of SNAT2 transcription by amino acid limitation. MCF-7 cells were treated for 24 h with either ‘control’ siRNA (siControl) or siRNA for MED15, incubated in fresh DMEM for another 24 h, and then incubated in either DMEM, DMEM lacking histidine or DMEM plus 100 nM E2 for 4 h. Transcription activity for the pS2 and SNAT2 genes was analyzed as described in the Materials and methods section (a). To determine the decline in expression for MED15, mRNA was analyzed by RT–qPCR (b). MCF-7 cells were incubated in medium with or without histidine for 8 h, as well as in control medium or medium containing 100 nM E2 for 1 h and then ChIP analysis was performed for the SNAT2 promoter, SNAT2 AARE and the pS2 promoter to determine the degree of MED15 association (c).
DISCUSSION
Mammalian genes, such as SNAT2, for which transcription is regulated by amino acid availability contain an AARE enhancer that is recognized by ATF4 (14,24,25). The present study illustrates amino acid-dependent changes in histone modification at the SNAT2 promoter and the intronic AARE enhancer, but not at other regions within the gene. The ChIP analysis also demonstrates that subsequent to ATF4 binding there is recruitment of the GTFs to the SNAT2 promoter in conjunction with transcriptional activation. Despite the fact that the SNAT2 AARE is only 700-bp downstream of the transcription start site, immunoprecipitation with AARE binding proteins such as ATF4 does not precipitate SNAT2 promoter fragments (14), and vice versa, GTFs are not associated with the AARE (Figure 1). Thus, ChIP analysis provides no evidence for a looping model or other direct interactions between the SNAT2 promoter and the AARE enhancer. These results lead us to test the hypothesis that Mediator could be important for SNAT2 amino acid-regulated gene activation by bridging the promoter and enhancer binding proteins. In yeast, Fan et al. (3) observed that Mediator was recruited by activators associated with genes induced by ‘environmental stress or nonoptimal growth conditions’. These Mediator-associated activators include GCN4, the yeast counterpart to ATF4 (5,6). Surprisingly, in the human cells, none of the several Mediator subunits tested showed a high degree of association with the SNAT2 gene in the basal or fed state, nor was there increased Mediator recruitment to the SNAT2 promoter or enhancer during amino acid deprivation.
The number of studies investigating Mediator interaction with mammalian nutrient responsive genes is limited. Toth et al. (26) showed that the cholesterol and fatty-acid sensing factor sterol regulatory element binding protein-1a interacts with DRIP150 (i.e. MED14) and that DRIP150 exhibits enhanced association with responsive genes when cells are deprived of sterol. Pavri et al. (27) reported that retinoic acid treatment results in a poly ADP ribose polymerase-1-dependent shift from a repressive Mediator complex containing CDK8 to an activating complex lacking this subunit. Mo et al. (28) showed that Ras-induced phosphorylation of C/EBPβ in chicken fibroblasts also leads to a switch from a C/EBPβ-Mediator complex containing CDK8 to one lacking this subunit, although MED23 (CRSP70/Sur2) was present in both the repressive and active C/EBPβ-interacting complexes. C/EBPβ is constitutively bound to the AARE sites of six different genes and after amino acid deprivation its binding is increased during a time of transcriptional decline (25). Therefore, along with MED1, which is thought to be a component of the ‘body’ or ‘middle’ of the complex (19,23), MED23 and CDK8 were the initial subunits tested for recruitment to the SNAT2 gene. ChIP analysis revealed little or no binding of these factors regardless of nutritional status or the cell type (Figure 3a and c). In contrast, all three were enhanced in their association with the pS2 gene after estrogen treatment. The pS2 gene was chosen as a positive control to demonstrate that the ChIP analyses were valid as it has been documented by other groups that Mediator is a component of estrogen-dependent pS2 activation (15,16). To provide independent support for the ChIP analysis, cells were transfected with siRNA against MED1 and MED23, as well as five additional subunits, so that components of the ‘head, body/middle and tail’ were included. The results illustrate that despite data from the pS2 gene showing that five of the seven subunits had an adverse effect on transcription, none of the subunits appeared to be required for either basal or activated transcription of the SNAT2 gene.
In both Saccharomyces cerevisiae (29) and Schizosaccharomyces pombe (30), ChIP on chip studies have shown that Mediator recruitment does not always correlate with transcription activity or the recruitment of the GTF complex, and that Mediator associates with coding regions as well as upstream activating sequences. In S. cerevisiae, the three proteins Med2/Pgd1/Gal11, along with Sin4, form what is referred to as the ‘tail’ of the Mediator complex (23). This tail module is thought to be linked to the middle or body module as a result of the binding of Sin4 (human MED16) in the tail portion to Rgr1 (human MED14) in the body (31). When the Sin4 gene was deleted, the Med2/Pgd1/Gal11 triad was recruited to GCN4-activated genes independently of the remainder of the Mediator complex and activated transcription (22). However, it has not been demonstrated that the yeast tail triad is recruited to genes as a functionally independent unit in wild type yeast cells, that is, when Sin4 is expressed. Interestingly, in both biochemical studies and computer analysis designed to identify the protein composition of the Mediator complex in mammalian cells, no evidence was obtained for human paralogs to the yeast tail proteins Med2 (putative mammalian MED2) and Pgd1 (putative mammalian MED3) (1,18,23). Given those results, it is possible that the tail module composition and/or structure for the human Mediator complex may be different than that for yeast. Consequently, the function of the remaining member of the human tail triad complex, MED15 (yeast gal11), may also be different. Nonetheless, the yeast data raise the question of whether or not MED15 recruitment occurs during activation of ATF4 responsive genes. The data obtained from the present studies indicate that MED15 siRNA knockdown shows no effect on pS2 activation by E2 or induction of the SNAT2 gene by amino acid limitation. Furthermore, using two different antibodies, no evidence could be obtained in vivo for the increased recruitment of MED15 to the promoter of either of these two genes. In addition, siRNA knockdown of MED14 (Rgr1) did not suppress either the pS2 gene activation by estradiol or activation of the SNAT2 gene by amino acid limitation. This lack of an effect of MED14 suppression in the human cells is consistent with the observation in yeast that deletion of Rgr1, such that the tail module is not bound to the head/body portion of the Mediator complex, does not block Mediator binding and induction of GCN4-activated genes (22). Thus, the apparent absence of what should be MED2 or MED3 in the human complex and the lack of evidence for a role for MED14 and MED15 in the ATF4-dependent regulation of the SNAT2 gene suggest that there is no recruitment or activation by a human tail module independent of the rest of the Mediator complex. This conclusion is consistent with the remainder of the data indicating that recruitment of Mediator is not required for ATF4 action in the context of amino acid-regulated gene expression in mammalian cells.
It is plausible that an AARE-specific coactivator complex other than Mediator is necessary. Given that ATF4 is expressed during many different stress conditions (amino acid limitation, UPR, hypoxia and viral infection) in mammalian cells and that it contributes to functions such as osteoblast differentiation (32), the use of different co-activators may be a mechanism to selectively regulate these independent cellular responses. In fact, Yu et al. (33) have documented that TFIIA-γ binds to ATF4 and promotes its activation of the osteoblast associated gene osteocalcin. Lewis and Reinberg (4) suggested that in metazoans perhaps a subset of genes use TFIID instead of Mediator. For the SNAT2 promoter, the data presented here show that recruitment of TFIID parallels Pol II binding during activation of the gene by amino acid limitation. In addition to TFIIA-γ (33), ATF4 has been shown to interact directly with several other GTFs including TBP, TFIIB and TFIIF (34) and the RPB3 subunit of RNA polymerase II (35). Indeed, coexpression of RPB3 with ATF4 enhances its transcriptional activation capabilities (35). The latter observation is interesting in that yeast Mediator may bind RNA Pol II via the Rpb3/Rpb11 heterodimer (36) and mutations in Rpb3 have been observed to alter activated transcription, but not basal activity (37).
Collectively, the data presented argue against the hypothesis that mammalian Mediator is required for all Pol II-dependent transcription. The results also provide a striking contrast to the amino acid sensitive GCN4-dependent transcription in yeast by documenting that enhanced recruitment of the mammalian Mediator complex is not a mandatory event for ATF4-activated transcription from an amino acid-regulated gene.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health (DK-52064, DK70647 to M.S.K.). Funding for open access charge: NIH grant DK-52064.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors wish to thank other members of the laboratory for technical advice, reagents and helpful discussion.
REFERENCES
- 1.Sato S, Tomomori-Sato C, Parmely TJ, Florens L, Zybailov B, Swanson SK, Banks CA, Jin J, Cai Y, Washburn MP, et al. A set of consensus mammalian mediator subunits identified by multidimensional protein identification technology. Mol. Cell. 2004;14:685–691. doi: 10.1016/j.molcel.2004.05.006. [DOI] [PubMed] [Google Scholar]
- 2.Takagi Y, Kornberg RD. Mediator as a general transcription factor. J. Biol. Chem. 2006;281:80–89. doi: 10.1074/jbc.M508253200. [DOI] [PubMed] [Google Scholar]
- 3.Fan X, Chou DM, Struhl K. Activator-specific recruitment of Mediator in vivo. Nat. Struct. Mol. Biol. 2006;13:117–120. doi: 10.1038/nsmb1049. [DOI] [PubMed] [Google Scholar]
- 4.Lewis BA, Reinberg D. Promoter activation when the ChIPs are down. Nat. Struct. Mol. Biol. 2006;13:96–97. doi: 10.1038/nsmb0206-96. [DOI] [PubMed] [Google Scholar]
- 5.Swanson MJ, Qiu H, Sumibcay L, Krueger A, Kim SJ, Natarajan K, Yoon S, Hinnebusch AG. A multiplicity of coactivators is required by Gcn4p at individual promoters in vivo. Mol. Cell Biol. 2003;23:2800–2820. doi: 10.1128/MCB.23.8.2800-2820.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Qiu H, Hu C, Zhang F, Hwang GJ, Swanson MJ, Boonchird C, Hinnebusch AG. Interdependent recruitment of SAGA and Srb mediator by transcriptional activator Gcn4p. Mol. Cell Biol. 2005;25:3461–3474. doi: 10.1128/MCB.25.9.3461-3474.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Harding HP, Novoa I, Zhang Y, Zeng H, Wek R, Schapira M, Ron D. Regulated translation initiation controls stress-induced gene expression in mammalian cells. Mol. Cell. 2000;6:1099–1108. doi: 10.1016/s1097-2765(00)00108-8. [DOI] [PubMed] [Google Scholar]
- 8.Vattem KM, Wek RC. Reinitiation involving upstream ORFs regulates ATF4 mRNA translation in mammalian cells. Proc. Natl Acad. Sci. USA. 2004;101:11269–11274. doi: 10.1073/pnas.0400541101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lu PD, Harding HP, Ron D. Translation reinitiation at alternative open reading frames regulates gene expression in an integrated stress response. J. Cell Biol. 2004;167:27–33. doi: 10.1083/jcb.200408003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Siu F, Bain PJ, LeBlanc-Chaffin R, Chen H, Kilberg MS. ATF4 is a mediator of the nutrient-sensing response pathway that activates the human asparagine synthetase gene. J. Biol. Chem. 2002;277:24120–24127. doi: 10.1074/jbc.M201959200. [DOI] [PubMed] [Google Scholar]
- 11.Chen H, Pan YX, Dudenhausen EE, Kilberg MS. Amino acid deprivation induces the transcription rate of the human asparagine synthetase gene through a timed program of expression and promoter binding of nutrient-responsive bZIP transcription factors as well as localized histone acetylation. J. Biol. Chem. 2004;279:50829–50839. doi: 10.1074/jbc.M409173200. [DOI] [PubMed] [Google Scholar]
- 12.Kilberg MS, Pan YX, Chen H, Leung-Pineda V. Nutritional control of gene expression: how mammalian cells respond to amino acid limitation. Annu. Rev. Nutr. 2005;25:59–85. doi: 10.1146/annurev.nutr.24.012003.132145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Palii SS, Chen H, Kilberg MS. Transcriptional control of the human sodium-coupled neutral amino acid transporter system A gene by amino acid availability is mediated by an intronic element. J. Biol. Chem. 2004;279:3463–3471. doi: 10.1074/jbc.M310483200. [DOI] [PubMed] [Google Scholar]
- 14.Palii SS, Thiaville MM, Pan YX, Zhong C, Kilberg MS. Characterization of the amino acid response element within the human SNAT2 system A transporter gene. Biochem. J. 2006;395:517–527. doi: 10.1042/BJ20051867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Burakov D, Crofts LA, Chang CP, Freedman LP. Reciprocal recruitment of DRIP/mediator and p160 coactivator complexes in vivo by estrogen receptor. J. Biol. Chem. 2002;277:14359–14362. doi: 10.1074/jbc.C200099200. [DOI] [PubMed] [Google Scholar]
- 16.Metivier R, Penot G, Hubner MR, Reid G, Brand H, Kos M, Gannon F. Estrogen receptor-alpha directs ordered, cyclical, and combinatorial recruitment of cofactors on a natural target promoter. Cell. 2003;115:751–763. doi: 10.1016/s0092-8674(03)00934-6. [DOI] [PubMed] [Google Scholar]
- 17.Lipson KE, Baserga R. Transcriptional activity of the human thymidine kinase gene determined by a method using the polymerase chain reaction and an intron-specific probe. Proc. Natl Acad. Sci. USA. 1989;86:9774–9777. doi: 10.1073/pnas.86.24.9774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Conaway RC, Sato S, Tomomori-Sato C, Yao T, Conaway JW. The mammalian Mediator complex and its role in transcriptional regulation. Trends Biochem. Sci. 2005;30:250–255. doi: 10.1016/j.tibs.2005.03.002. [DOI] [PubMed] [Google Scholar]
- 19.Malik S, Roeder RG. Dynamic regulation of pol II transcription by the mammalian Mediator complex. Trends Biochem. Sci. 2005;30:256–263. doi: 10.1016/j.tibs.2005.03.009. [DOI] [PubMed] [Google Scholar]
- 20.Donner AJ, Szostek S, Hoover JM, Espinosa JM. CDK8 is a stimulus-specific positive coregulator of p53 target genes. Mol. Cell. 2007;27:121–133. doi: 10.1016/j.molcel.2007.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Furumoto T, Tanaka A, Ito M, Malik S, Hirose Y, Hanaoka F, Ohkuma Y. A kinase subunit of the human mediator complex, CDK8, positively regulates transcriptional activation. Genes Cells. 2007;12:119–132. doi: 10.1111/j.1365-2443.2007.01036.x. [DOI] [PubMed] [Google Scholar]
- 22.Zhang F, Sumibcay L, Hinnebusch AG, Swanson MJ. A triad of subunits from the Gal11/tail domain of Srb mediator is an in vivo target of transcriptional activator Gcn4p. Mol. Cell Biol. 2004;24:6871–6886. doi: 10.1128/MCB.24.15.6871-6886.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Boube M, Joulia L, Cribbs DL, Bourbon HM. Evidence for a mediator of RNA polymerase II transcriptional regulation conserved from yeast to man. Cell. 2002;110:143–151. doi: 10.1016/s0092-8674(02)00830-9. [DOI] [PubMed] [Google Scholar]
- 24.Siu F, Bain PJ, LeBlanc-Chaffin R, Chen H, Kilberg MS. ATF4 is a mediator of the nutrient-sensing response pathway that activates the human asparagine synthetase gene. J. Biol. Chem. 2002;277:24120–24127. doi: 10.1074/jbc.M201959200. [DOI] [PubMed] [Google Scholar]
- 25.Pan YX, Chen H, Thiaville MM, Kilberg MS. Activation of the ATF3 gene through a co-ordinated amino acid-sensing response programme that controls transcriptional regulation of responsive genes following amino acid limitation. Biochem. J. 2007;401:299–307. doi: 10.1042/BJ20061261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Toth JI, Datta S, Athanikar JN, Freedman LP, Osborne TF. Selective coactivator interactions in gene activation by SREBP-1a and -1c. Mol. Cell Biol. 2004;24:8288–8300. doi: 10.1128/MCB.24.18.8288-8300.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pavri R, Lewis B, Kim TK, Dilworth FJ, Erdjument-Bromage H, Tempst P, de MG, Evans R, Chambon P, Reinberg D. PARP-1 determines specificity in a retinoid signaling pathway via direct modulation of mediator. Mol. Cell. 2005;18:83–96. doi: 10.1016/j.molcel.2005.02.034. [DOI] [PubMed] [Google Scholar]
- 28.Mo X, Kowenz-Leutz E, Xu H, Leutz A. Ras induces mediator complex exchange on C/EBP beta. Mol. Cell. 2004;13:241–250. doi: 10.1016/s1097-2765(03)00521-5. [DOI] [PubMed] [Google Scholar]
- 29.Andrau JC, van de PL, Lijnzaad P, Bijma T, Koerkamp MG, van de PJ, Werner M, Holstege FC. Genome-wide location of the coactivator mediator: binding without activation and transient Cdk8 interaction on DNA. Mol. Cell. 2006;22:179–192. doi: 10.1016/j.molcel.2006.03.023. [DOI] [PubMed] [Google Scholar]
- 30.Zhu X, Wiren M, Sinha I, Rasmussen NN, Linder T, Holmberg S, Ekwall K, Gustafsson CM. Genome-wide occupancy profile of mediator and the Srb8-11 module reveals interactions with coding regions. Mol. Cell. 2006;22:169–178. doi: 10.1016/j.molcel.2006.03.032. [DOI] [PubMed] [Google Scholar]
- 31.Dotson MR, Yuan CX, Roeder RG, Myers LC, Gustafsson CM, Jiang YW, Li Y, Kornberg RD, Asturias FJ. Structural organization of yeast and mammalian mediator complexes. Proc. Natl Acad. Sci. USA. 2000;97:14307–14310. doi: 10.1073/pnas.260489497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ameri K, Harris AL. Activating transcription factor 4. Int. J. Biochem. Cell Biol. 2008;40:14–21. doi: 10.1016/j.biocel.2007.01.020. [DOI] [PubMed] [Google Scholar]
- 33.Yu S, Jiang Y, Galson DL, Luo M, Lai Y, Lu Y, Ouyang HJ, Zhang J, Xiao G. General transcription factor IIA-gamma increases osteoblast-specific osteocalcin gene expression via activating transcription factor 4 and runt-related transcription factor 2. J. Biol. Chem. 2008;283:5542–5553. doi: 10.1074/jbc.M705653200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liang G, Hai T. Characterization of human activating transcription factor 4, a transcriptional activator that interacts with multiple domains of cAMP-responsive element-binding protein (CREB)-binding protein. J. Biol. Chem. 1997;272:24088–24095. doi: 10.1074/jbc.272.38.24088. [DOI] [PubMed] [Google Scholar]
- 35.De Angelis R, Lezzi S, Bruno T, Corbi N, Di Padova M, Floridi A, Fanciulli M, Passananti C. Functional interaction of the subunit 3 of RNA polymerase II (RPB3) with transcription factor-4 (ATF4) FEBS Lett. 2003;547:15–19. doi: 10.1016/s0014-5793(03)00659-8. [DOI] [PubMed] [Google Scholar]
- 36.Davis JA, Takagi Y, Kornberg RD, Asturias FA. Structure of the yeast RNA polymerase II holoenzyme: mediator conformation and polymerase interaction. Mol. Cell. 2002;10:409–415. doi: 10.1016/s1097-2765(02)00598-1. [DOI] [PubMed] [Google Scholar]
- 37.Tan Q, Linask KL, Ebright RH, Woychik NA. Activation mutants in yeast RNA polymerase II subunit RPB3 provide evidence for a structurally conserved surface required for activation in eukaryotes and bacteria. Genes Dev. 2000;14:339–348. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.