Abstract
The smallest catalytic RNA identified to date is a manganese-dependent ribozyme that requires only a complex between GAAA and UUU to effect site-specific cleavage. We show here that this ribozyme occurs naturally in the 3′-UTR of Vg1 and β-actin mRNAs. In accord with earlier studies with model RNAs, cleavage occurs only in the presence of manganese or cadmium ions and proceeds optimally near 30°C and physiological pH. The time course of cleavage in Vg1 mRNA best fits a two-step process in which both steps are first-order. In Vg1 mRNA, the ribozyme is positioned adjacent to a polyadenylation signal, but has no influence on translation of the mRNA in Xenopus oocytes. Putative GAAA ribozyme structures are also near polyadenylation sites in yeast and rat actin mRNAs. Analysis of sequences in the PolyA Cleavage Site and 3′-UTR Database (PACdb) revealed no particular bias in the frequency or distribution of the GAAA motif that would suggest that this ribozyme is currently or was recently used for cleavage to generate processed transcripts. Nonetheless, we speculate that the complementary strands that comprise the ribozyme may account for the origin of sequence elements that direct present-day 3′-end processing of eukaryotic mRNAs.
INTRODUCTION
RNA-based catalysis is an integral part of key steps in gene expression that include RNA replication, RNA splicing, and peptide bond formation (1). With the exception of the latter, most ribozymes catalyze phosphoryl transfer reactions that require a metal cofactor and result in strand cleavage. Many of the present-day ribozymes, such as the hammerhead, hepatitis delta virus (HDV) and hairpin ribozymes, are limited to the processing of multimeric viral RNA replication products into individual monomeric units. However, there is recent evidence that the activity of a self-cleaving element in the 3′ flanking region of human β-globin transcripts is required for efficient termination of transcription (2). Thus, ribozyme activity, at least in some cases, may be important for the 3′ processing of mRNA and termination of transcription, significantly expanding the scope of ribozyme activity. It also appears that ribozyme activity continues to evolve in modern organisms. An HDV-like ribozyme has been identified in the second intron of human cytoplasmic polyadenylation element-binding protein 3 (3). Orthologs of the ribozyme are highly conserved, but are limited to mammals, leading to speculation that this ribozyme not only arose recently, but also within a protein milieu.
The localization of Vg1 mRNA to the vegetal cortex of Xenopus oocytes is mediated by a 340-nucleotide (nt) region in the 3′-UTR designated the Vg1 localization element (VLE) (4). A 250-nt sequence, positioned 118 nt downstream of the VLE, controls translation of Vg1 mRNA, and is referred to as the Vg1 translational element (VTE) (5,6). We have provided evidence that a site between the VLE and VTE is used for cytoplasmic polyadenylation during the late stages of oogenesis when translation of Vg1 mRNA is initiated (7). During experiments to delineate the cytoplasmic processing of Vg1 mRNA, we detected cleavage of the RNA in the absence of protein and have determined that this activity can be attributed to a previously described manganese-dependent ribozyme (8,9). The structural motif that defines this ribozyme also occurs in the 3′-UTR of another localized mRNA, chicken β-actin mRNA, which we find undergoes metal-dependent self-cleavage as well. The position of the ribozyme in Vg1 mRNA and in several actin mRNAs is close to a polyadenylation site, which raises the question whether they represent remnants of an ancient mechanism to produce transcripts of defined sizes and, in light of the ribozyme activity in β-globin mRNA (2), to terminate transcription.
MATERIALS AND METHODS
Nucleic acids
RNA representing residues 210–350 of the Vg1 localization element was prepared by run-off transcription of the plasmid p1650Ase linearized with AseI using T7 RNA polymerase. The β-actin zipcode RNA (residues 4–42) was similarly transcribed from a synthetic oligonucleotide containing the T7 promoter. Internally radiolabeled RNAs were synthesized in the presence of [α-32P]CTP and 3′ end-labeled RNAs were prepared using cytidine 3′,5′-[5′-32P]bisphosphate and T4 RNA ligase.
Cleavage reactions
The cleavage reactions contained the indicated RNA in buffer containing 50 mM Tris–HCl, pH 7.5, 200 mM NaCl, 50 mM KCl and 10 mM MnCl2. Reactions were kept at ambient temperature for the indicated time and stopped by addition of 50 mM EDTA. The reactions with Vg1 mRNA contained 0.4 mM m7GpppA. Samples were diluted with two volumes of loading buffer containing 8 M urea, and kept at 60°C for 5 min before loading on 8% polyacrylamide gels containing 7 M urea. The pH dependence of cleavage was measured using 50 mM sodium acetate (pH 4.5 and 5.0), 2-(N-morpholino)ethanesulfonate (pH 5.5 and 6.0), HEPES (pH 7.0) or Tris (pH 7.5, 8.0, 8.5 and 9.0). Metal dependence was tested using the indicated chloride salt at 10 mM. The amount of cleavage was measured by scanning autoradiographs with a laser densitometer and these data fit to curves using TableCurve 2D software.
Characterization of cleavage products
The ability to phosphorylate the 3′ cleavage fragment with polynucleotide kinase was tested. After self-cleavage of Vg1 RNA labeled at the 3′-end, a sample of gel-purified 3′ product was incubated with 0.5 mM ATP and 5 U of T4 polynucleotide kinase for 20 min at 37°C. The mobility of this sample was then analyzed on a denaturing gel alongside a control sample incubated in the same conditions, but without enzyme, and an alkaline hydrolysate. The polyadenylation of the Vg1 substrate and cleavage product were tested in a reaction containing internally radiolabeled RNA and 1.5 U of Escherichia coli poly(A) polymerase (TaKaRa). The reactions were run in 10 mM Tris–HCl, pH 8.0, 100 mM KCl, 5 mM MgCl2, 0.1 mM CaCl2, 4 mM ATP, 50 mM sucrose, 0.4 mM EGTA, 20 μM vanadylribonucleoside complex, 30 mM creatine phosphate, 10 μg/ml creatine kinase and 0.5 U/μl RNasin for 1 h at ambient temperature. The reactions were stopped by the addition of three volumes of a solution containing 134 mM Tris–HCl, pH 8.0, 200 mM NaCl, 17 mM EDTA, 1.3% SDS, 0.4 mg/ml tRNA and 0.67 mg/ml proteinase K, and then kept at 37°C for 30 min. The RNA was precipitated with ethanol and analyzed by electrophoresis on a denaturing urea gel.
In vivo expression of Vg1 mRNA
The GAAA ribozyme sequence (nucleotide positions 1771–1774) was changed to CCCA using the QuikChange mutagenesis kit (Stratagene). Capped wild-type and mutant Vg1 mRNAs were synthesized by run off transcription using SP6 RNA polymerase. Mixed staged oocytes were incubated with collagenase (2 mg/ml) at room temperature for 45 min in OR2(–) solution. After multiple washes with OR2, stage VI oocytes were microinjected in the vegetal pole with 27.6 nl of 0.36 μM (8 ng) wild-type or mutant mRNA, producing an 10 nM final concentration in each oocyte. Control samples were prepared by injecting oocytes with water. Microinjected oocytes were placed at 18°C for 2 h before adding 250 μCi [35S] methionine/cysteine translabel. After overnight incubation in OR2 solution, 20 oocytes from each sample were homogenized in 300 μl of harvest buffer (0.1 M Tris–HCl, pH 8.9 and 1% SDS). A second set of oocytes was incubated in the presence of 25 mM manganese. The samples were boiled for 5 min and centrifuged for 1 min at 16 000g and 100 μl of NET-2 buffer (50 mM Tris–HCl, pH 7.4, 150 mM NaCl and 0.05% NP-40) was added to the supernatant, which was then incubated with protein A-Sepharose beads (Sigma) adsorbed with Vg1 polyclonal antibody. The precipitation reactions were incubated for 2 h at 4°C with end-to-end rotation. Beads were washed three times with NET-2 buffer and then boiled for 5 min in 20 μl of SDS loading buffer. Samples were analyzed by SDS–PAGE followed by autoradiography to visualize in vivo synthesized Vg1 protein.
Sequence analyses
The PolyA Cleavage Site and 3′-UTR Database (PACdb) is a catalog of putative 3′-processing sites (10). This database was used to compare the frequency of tetranucleotide sequences within 200 nt of polyadenylation signals and to compare the frequency of the GAAA ribozyme sequence upstream and downstream of polyadenylation signals. The analyses were restricted to only polyadenylation signals that are external to the open reading frame and that occur on the (+) strand.
A database containing 3′-UTR sequences (available at: http://www.ba.itb.cnr.it/UTR/) was used to search 1000 sequences each from Caenorhabditis elegans and Drosophila melanogaster mRNAs for the presence of three tetranucleotide sequences (GAAA, CAAA and UAUG) within 15 nt upstream or downstream of AAUAAA polyadenylation signals. The control sequence, UAUG, was generated using a random number generator. The number of 3′-UTRs containing the indicated tetranucleotide was plotted versus their distance from the AAUAAA hexanucleotide.
RESULTS AND DISCUSSION
A manganese-dependent ribozyme in the 3′-UTR of Vg1 mRNA
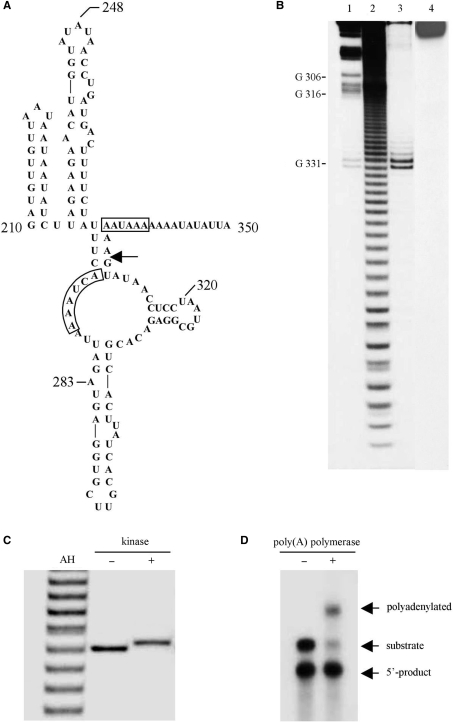
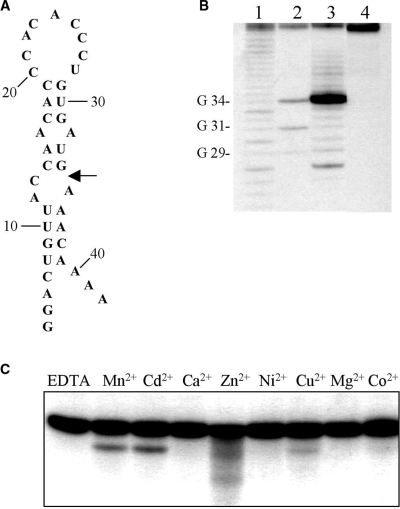
The predicted secondary structure at the 3′-end of the localization element in Vg1 mRNA includes a short helix having UUU paired with GAAA (Figure 1A), which is the minimum structural motif required for activity in the manganese-dependent ribozyme (9). We tested an RNA fragment, encompassing nucleotides 210–350 of the VLE, for cleavage in the presence of 10 mM manganese. Cleavage occurs specifically between G331 and A332 (Figure 1B), which is the expected site based upon previous studies of this ribozyme (8,9). In initial assays for cleavage activity, we detected an additional product that exhibited considerably lower mobility relative to the starting substrate on denaturing polyacrylamide gels (data not shown). The appearance of this species was eliminated by the inclusion of cap analog (m7GpppA) in the reaction mixture. This alternative reaction and the identity of the secondary product are being investigated.
Figure 1.
A manganese-dependent ribozyme in the 3′-UTR of Vg1 mRNA. (A) The secondary structure of the region encompassing residues 210–350 of the Vg1 localization element (VLE). An arrow indicates the site of cleavage and boxes enclose a consensus polyadenylation signal at position 334 and a nonconsensus signal at 272. The structure was generated using mfold version 3.1 (51). (B) Vg1 RNA (radiolabeled at the 3′-end) was incubated overnight in cleavage buffer in the presence (lane 3) or absence (lane 4) of 10 mM manganese and then analyzed on a denaturing polyacrylamide gel alongside ribonuclease T1 and alkaline hydrolysates (lanes 1 and 2, respectively). The doublet in lane 3 is also apparent in the T1 digest (lane 1) and is due to length heterogeneity in the substrate RNA. (C) A 5′ hydroxyl group at the cleavage site. The 3′ cleavage product was incubated with polynucleotide kinase and ATP and compared to untreated fragment and an alkaline hydrolysate. (D) A 3′ phosphate at the cleavage site. A portion of a cleavage reaction containing an equal mixture of substrate and product RNA was incubated with poly(A) polymerase and compared to an untreated sample. Only the substrate RNA is polyadenylated.
Like the majority of self-cleaving ribozymes, the manganese ribozyme yields products with a 2′,3′-cyclic phosphate and a free 5′-hydroxyl at the site of cleavage (8). The cleavage product of the Vg1 RNA substrate, radiolabeled at the 3′ terminus, comigrates with a fragment generated by limited alkaline hydrolysis, which also yields products with a 5′-hydroxyl (Figure 1B and C). Incubation of the 3′ cleavage product with polynucleotide kinase and ATP shifts the mobility of the fragment to an intermediary position relative to the products from alkaline digestion. Together, these results establish that a free 5′-hydroxyl is formed at the site of cleavage in Vg1 mRNA (Figure 1C). This conclusion is supported by a second experiment in which self-cleavage of internally radiolabeled Vg1 RNA was terminated after the reaction had proceeded approximately half way to completion in order to generate a mixture of substrate and product RNAs. A portion of this sample was then incubated with E. coli poly(A) polymerase (PAP), which requires a 3′-hydroxyl for addition of a poly(A) tail (Figure 1D). Although the full-length VLE RNA is a substrate for the polymerase, none of the 5′ cleavage product was polyadenylated. This result supports the other evidence that the autocatalytic cleavage of Vg1 RNA produces products with a 3′-phosphate and a free 5′-hydroxyl.
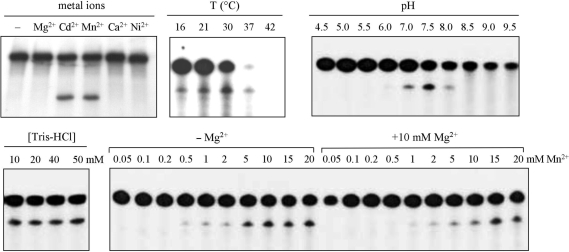
Cleavage of the Vg1 3′-UTR was characterized further by determining the metal, temperature and pH dependence of the reaction (Figure 2). In addition to manganese, the only other divalent metal that supports this activity is cadmium. The amount of cleavage increases with temperature between 16°C and 30°C, above which there is an appreciable degradation of the RNA. The pH optimum of the reaction is centered near 7.5. The concentration of Tris buffer has no effect on the extent of the reaction, eliminating the possibility that it is participating in the reaction as a general base. The inclusion of magnesium has little effect on the activity of the ribozyme, indicating that formation of the catalytically competent structure in the RNA does not depend on this divalent cation, nor is there any apparent competitive binding of magnesium to the manganese site. The cleavage activity in Vg1 mRNA is strikingly similar to that first described for the manganese ribozyme in the Tetrahymena group I intron (8,9).
Figure 2.
Characterization of the self-cleavage reaction. All reactions were carried out in standard conditions, excepting the indicated variable. The metal dependence of cleavage was tested with the indicated divalent cation at a concentration of 10 mM.
Kinetic properties of the Vg1 ribozyme
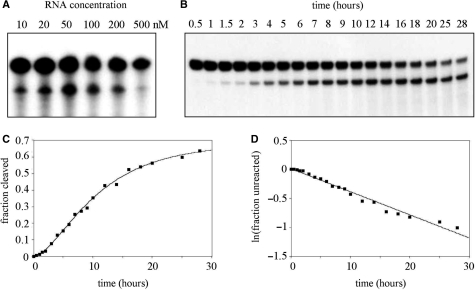
The extent of Vg1 mRNA cleavage exhibits little dependence on RNA concentration (Figure 3A), indicating that the reaction is first order. Decreased activity at high concentrations of the RNA actually suggests that intermolecular interactions may block or hinder formation of the active conformation in the RNA needed for catalysis. The time course of the cleavage reaction not only provides further evidence for a first-order reaction, but also that it is a two-step kinetic process (Figure 3B–D). Reaction products were separated on a denaturing polyacrylamide gel and the resulting autoradiograph was scanned with a laser densitometer in order to quantitate the intensity of the individual bands. The progress curve (Figure 3C) is best fit to a rate expression that represents a two-step mechanism in which both steps are first order. This two-step mechanism suggests that a conformational change in the RNA to a catalytically competent structure may be partially rate limiting. Biphasic kinetic behavior, also ascribed to an equilibrium between conformational states, has been reported for group I (11) and group II (12,13) self-splicing introns. The plot of ln(fraction unreacted) versus time (Figure 3D) yielded an observed first-order rate constant of 6.6 × 10–4 min−1 which is similar to rate constants measured for naturally occurring and model manganese-dependent GAAA ribozymes (14,15). This slow reaction rate indicates that the catalytically competent conformation of the ribozyme may not be highly populated in the free RNA.
Figure 3.
Kinetics of the cleavage reaction. (A) A trace amount of internally labeled Vg1 RNA was added to the indicated concentration of unlabeled Vg1 RNA. The reactions proceeded in standard conditions overnight. (B) Vg1 RNA (100 nM) was incubated for the indicated amount of time in standard conditions. Reactions were stopped with EDTA and analyzed on a denaturing polyacrylamide gel. (C) The autoradiograph was scanned with a laser densitometer to generate the individual data points. The best-fit curve (solid line) is defined by a kinetic equation that represents a two-step process with both steps being first order. (D) The plot of ln(fraction unreacted) versus time yields an observed first-order rate constant of 6.6 × 10−4 min−1.
In vivo activity of the Vg1 ribozyme
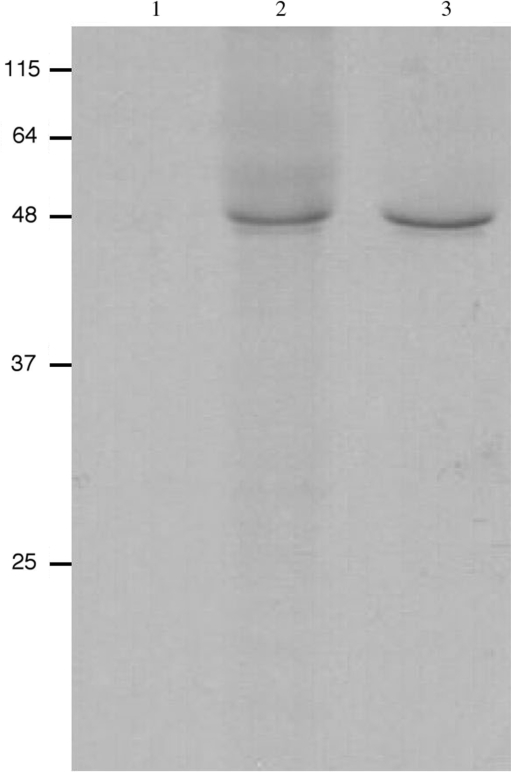
In order to test whether the manganese ribozyme has any effect on the expression of Vg1 mRNA in vivo, we mutated the GAAA sequence to CCCA and then compared translation of this and wild-type mRNAs in Xenopus oocytes. Capped transcripts were injected into stage VI oocytes, which were kept overnight in OR2 solution containing [35S]-labeled amino acids. The amount of injected RNA (8 ng) represents an 80-fold excess compared to the amount of endogenous Vg1 mRNA (16). Equal numbers of oocytes were disrupted and Vg1 protein was retrieved by immunoprecipitation and analyzed by SDS–PAGE/autoradiography (Figure 4). The two mRNAs exhibit comparable levels of translation, indicating that the ribozyme sequence has no physiological activity. Inclusion of manganese in the culture medium had no effect on the relative levels of translation of wild-type and mutant Vg1 mRNAs (data not shown). This situation with Vg1 mRNA appears similar to the processing of Xenopus U16 snoRNA. This small RNA, which is located within the third intron of mRNA encoding ribosomal protein L1, is generated by endonucleolytic cleavage of the mRNA as opposed to splicing-dependent excision. Two functional manganese-dependent ribozymes are found close to the actual in vivo processing sites that are, nonetheless, cleaved by an endoribonuclease which, remarkably, requires manganese as a cofactor (17,18).
Figure 4.
In vivo translation of wild-type and mutant Vg1 mRNA lacking the GAAA ribozyme sequence. Stage VI oocytes were injected with capped mRNA and incubated overnight in OR2 buffer containing [35S]methionine/cysteine. Vg1 protein was immunoprecipitated from whole-cell extract prepared from 20 oocytes and analyzed by SDS–PAGE followed by autoradiography. Lane 1, oocytes injected with water only; lane 2, oocytes injected with wild-type mRNA; lane 3, oocytes injected with mRNA in which the GAAA ribozyme sequence was changed to CCCA.
A manganese ribozyme in the 3′-UTR of actin mRNA
β-Actin mRNA is localized to the leading edge of fibroblasts or the growth cones of developing neurites (19). A 54-nt element in the 3′-UTR, called the zipcode, mediates localization (Figure 5A). We have detected manganese-dependent cleavage of an oligoribonucleotide that contains the zipcode hairpin (Figure 5B). As with all other GAAA ribozymes, cleavage occurs specifically between the G and A residues. Similarly, cadmium is the only other metal that supports appreciable site-specific cleavage of this RNA. There is one notable difference between the zipcode RNA and the canonical manganese-dependent ribozyme. In the former, the consensus A:U base pair immediately flanking the cleavage site is apparently an A:C or possibly an A:A pair. Other examples of manganese-dependent ribozymes in which the GAAA is not paired with UUU have been detected within the group I intron of Chlamydomonas reinhardti chloroplast 23S rRNA (15). The structure of a manganese-dependent ribozyme has not been determined; however, in a theoretical model, the uracil residue in question is not involved in either the chemistry of the cleavage reaction or as a ligand to the metal (9). The structure of the ribozyme in the zipcode RNA supports an earlier proposal that the UUU segment serves only as a guide sequence to orient the metal-binding adenosine residues (9).
Figure 5.
A manganese-dependent ribozyme in the zipcode of β-actin mRNA. (A) The predicted secondary structure of the zipcode element in the 3′-UTR of β-actin mRNA (52). An arrow marks the site of manganese-dependent cleavage. (B) Zipcode RNA (radiolabeled at the 3′-end) was incubated overnight in cleavage buffer in the presence (lane 3) or absence (lane 4) of 10 mM manganese and then analyzed on a denaturing polyacrylamide gel alongside alkaline and ribonuclease T1 hydrolysates (lanes 1 and 2, respectively). (C) The metal dependence of self-cleavage in zipcode RNA. RNA was incubated overnight in the presence of the indicated metal. Weak cleavage with Cu2+ has been reported previously for the manganese ribozyme (53); whereas, Zn2+ triggers nonspecific degradation.
The occurrence of GAAA relative to polyadenylation sites
The manganese ribozymes in Vg1 mRNA and β-actin mRNA occur in close proximity to polyadenylation sites; thus, we questioned whether these ribozymes might be molecular fossils that were active before the advent of the proteins that now mediate the cleavage reaction associated with this process. Seemingly, mechanisms to generate RNA products of defined length must have existed in an RNA world. We examined the occurrence of GAAA in the PACdb to determine whether there is any bias in the frequency or position of this sequence (10). The frequency of GAAA was compared to CAAA, AAAA, TAAA and a randomly generated sequence, UAUG (Table 1). The occurrence of these tetranucleotide sequences was limited to the 200 nt immediately preceding polyadenylation signals in the organisms listed. Since the number of entries for each species examined is different, the occurrence of each sequence is expressed as the percent of the total number of the five tetranucleotide sequences. GAAA does not occur significantly more or less frequently than the other sequences containing a triplet of adenosines, indicating no selective bias for or against the ribozyme sequence in the 3′-UTR. The sequences AAAA and UAAA are consistently present at a somewhat higher frequency, which likely reflects the A/U-rich nature of 3′-UTRs in general. Similarly, the randomly generated sequence occurs less frequently than the A-rich sequences.
Table 1.
Frequency of tetranucleotide sequences within 200 nt upstream of polyadenylation sites
| M. musculus | D. rerio | D. melanogaster | C. elegans | S. cerevisiae | S. pombe | A. thaliana | |
|---|---|---|---|---|---|---|---|
| GAAA | 18.7 | 17.0 | 17.2 | 21.6 | 20.0 | 17.7 | 19.6 |
| CAAA | 15.9 | 16.6 | 19.3 | 22.2 | 15.2 | 16.4 | 18.1 |
| AAAA | 24.1 | 23.2 | 24.9 | 25.0 | 27.1 | 25.6 | 22.1 |
| UAAA | 29.8 | 29.9 | 28.4 | 20.1 | 26.1 | 28.2 | 22.2 |
| UAUG | 11.5 | 13.3 | 10.2 | 10.2 | 11.6 | 12.1 | 18.1 |
The frequency of each sequence is expressed as the percentage of the total of all five sequences in order to normalize the different number of entries for each species.
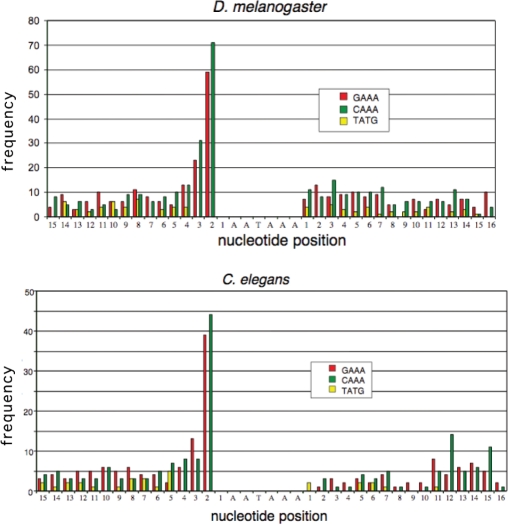
We also analyzed the sequences flanking polyadenylation signals to determine whether there is any apparent bias in the location of GAAA sequences. We determined the number of times the ribozyme sequence occurs within 50 or 100 nt upstream or downstream of polyadenylation signals (Table 2). This analysis did not reveal any notable positional effects. In addition, we examined 1000 sequences each from C. elegans and D. melanogaster, scanning the sequence immediately surrounding the polyadenylation signal in 1-nt increments using a 4-nt window (Figure 6). There is a marked increase in the frequency of the GAAA sequence immediately before and including the first 2 nt of the polyadenylation signal. However, the occurrence of the ribozyme sequence is no greater than CAAA. This analysis simply demonstrates the greater likelihood of a tetranucleotide sequence with three consecutive adenosines due to the polyadenylation signal.
Table 2.
Frequency of GAAA within various distances of polyadenylation sites
| Distancea (nt) | M. musculus | D. rerio | D. melanogaster | C. elegans | S. cerevisiae | S. pombe | A. thaliana |
|---|---|---|---|---|---|---|---|
| − 50 | 91 889 | 10 457 | 6 666 | 3 589 | 210 | 397 | 19 463 |
| +50 | 105 725 | 9 553 | 5 918 | 5 732 | 239 | 416 | 19 963 |
| − 100 | 162 272 | 19 425 | 12 296 | 7 840 | 452 | 768 | 36 348 |
| +100 | 181 328 | 18 637 | 11 633 | 13 632 | 541 | 770 | 42 992 |
a(−) and (+) designate upstream and downstream, respectively.
Figure 6.
The frequency of GAAA immediately upstream and downstream of the polyadenylation signal. One thousand mRNA sequences from C. elegans and D. melanogaster were scanned in 1-nt increments using a 4-nt window. The frequency of the sequences GAAA, CAAA and TATG are presented as histograms relative to the position of the AATAAA hexanucleotide sequence.
We considered the question whether the ribozyme sequence is correlated with mRNA stability. Genome-wide microarray analyses of mRNA decay have been reported (20–23). In addition to AU-rich elements (ARE's) that are found in many mRNAs with short half-lives, other sequence motifs have been identified that seem to be correlated with RNA decay rates (20,21). However, GAAA has not emerged as a conserved sequence in any of these elements that appear to influence stability. We compared the frequency of GAAA in a group of short-lived transcripts in human T cells (24) relative to a group of constitutive transcripts (list available from authors) and found no statistical difference (P = 0.482). A purine-rich instability element in the open reading frame of c-fos (25), likewise, does not have an overrepresentation of GAAA. Thus, there is no evidence to connect the ribozyme sequence to mRNA stability.
Origins of 3′ processing signals
The simplicity of the manganese-dependent ribozyme means its inclusion would put remarkably little structural constraint on an RNA molecule. Our analysis indicates that there has been no obvious evolutionary pressure to retain or restrict the GAAA ribozyme sequence in the 3′ terminal regions of RNAs once cleavage reactions became protein-based. Yet, if these simple ribozymes were at one time important for processing at the 3′-ends of nascent transcripts, then one may speculate how the transition from RNA- to protein-catalyzed cleavage occurred. One model that we posit is that the two sequences that comprise the manganese ribozyme, GAAA and UUU, gave rise to the two signal sequences, AAUAAA and U-rich element, that define the site for the cleavage reaction associated with polyadenylation. This scenario is appealing because these sequences are used solely as identity elements with cleavage occurring at a somewhat flexible distance between the two. The role of early protein factors could reasonably have been to promote a catalytically competent conformation in the ribozyme RNA, just as is seen in present-day RNP complexes that promote RNA-based cleavage reactions, such as the protein subunit of RNase P (26,27) and tyrosyl-tRNA synthetase enhancement of group I intron excision (28).
In the case of the cleavage reaction associated with polyadenylation, two factors, CPSF-160 (29,30) and CstF-64 (31), specifically bind to the AAUAAA and downstream U-rich elements, respectively, thereby determining the site of cleavage by CPSF-73 (32). Thus, one possibility is that an RNP complex that formed on the ribozyme initially to facilitate or modulate self-cleavage underpinned the transition from an RNA-based to a protein-based cleavage reaction by direct recruitment of an endonuclease. This may have a bearing on the nature of CPSF-73 itself, a member of the metallo-β-lactamase superfamily that has a wide variety of substrates ranging far beyond nucleic acids (33). Consistent with this model, CPSF-73 on its own has no sequence specificity and, thus, requires the complexes that form on the AAUAAA and U-rich elements to determine the site of cleavage (32).
This model is equally applicable to yeast and plant polyadenylation signals that, likewise, contain an AAUAAA positioning element and a U-rich sequence. The only difference is that the latter element occurs upstream rather than downstream of the AAUAAA motif in both (34).
Recent evidence demonstrates that CPSF-73 or a paralog also participates in the 3′-end processing of poly(A)– histone mRNA (35,36) and U-type snRNA (37). Remarkably, all five subunits of CPSF and two subunits of CstF, most notably CstF-64, are found together in a complex that supports the 3′-end processing of histone mRNA (35). An RNA interference screen has also provided evidence that some factors of the cleavage/polyadenylation complex are required for 3′-end processing of histone mRNAs (38).
The processing signal for histone mRNA also consists of two elements flanking the cleavage site: a conserved upstream hairpin and a purine-rich histone downstream element (HDE) (39). The 3′-end cleavage of histone mRNA requires base-pairing between the purine-rich HDE and the 5′-end of U7 snRNA (40). Dávila López and Samuelsson (41) have identified candidate U7 snRNAs genes in a greatly expanded number of organisms. In each instance, and others that we have investigated, an A/G-rich sequence in the HDE is paired with a U-rich sequence in U7 snRNA that, together, closely approximates the structure of the manganese ribozyme (Figure 7). Thus, it is plausible that this structure likewise evolved from a functional manganese ribozyme, thereby accounting for the observation that some factors are common to the 3′-end processing of both poly(A)+ and poly(A)– mRNAs.
Figure 7.
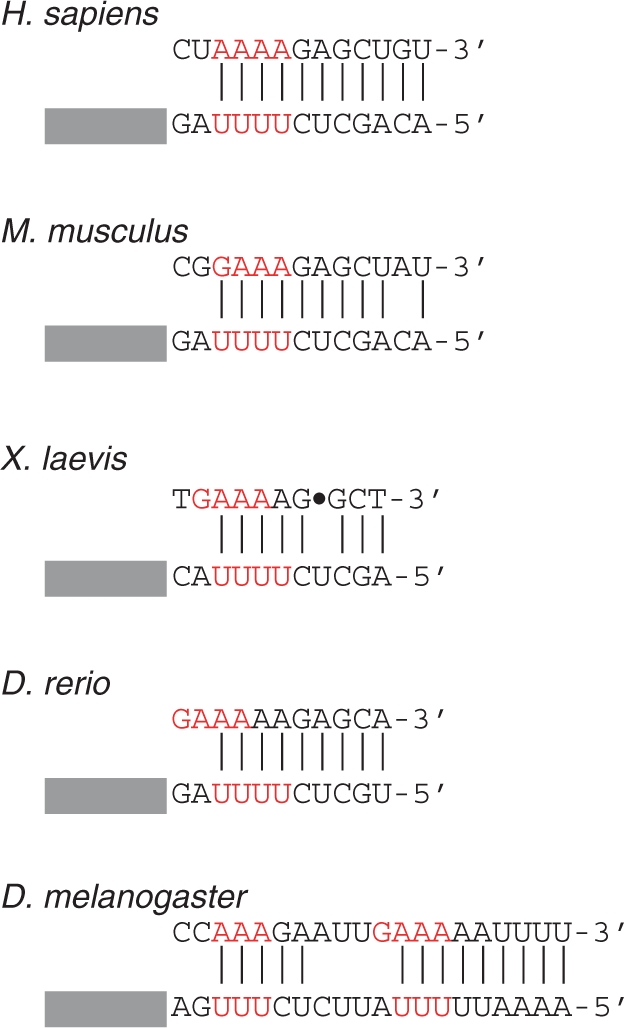
Predicted base-pairing interactions between the HDE of histone mRNAs and U7 snRNAs. Selected histone mRNAs (top strand) and U7 snRNAs (bottom strand) are presented; only the region immediately flanking the Sm site (designated by a rectangle) of U7 is shown in each case. Adapted from ref. 41.
The 3′ processing sites in the U1, U2, U4 and U5 snRNAs also have a bipartite structure, consisting of a poorly defined sequence upstream of the cleavage site and a downstream ‘3′ box’ that resembles the HDE in histone mRNA (42–44). A large multicomponent complex, Intergrator, which mediates the 3′ processing of these U-type RNAs, contains homologs of CPSF-73 and CPSF-100, but apparently, none of the other proteins found in either CPSF or CstF (37). Interestingly, maximum 3′ processing activity of these snRNAs in S100 extract occurs in the presence of 3 mM manganese (42). The 3′ processing signals for RNA polymerase II (pol II) transcripts all appear to have a bipartite structure comprised of purine- and uridine-rich sequences that determine the site of endonucleolytic cleavage. We speculate that these elements may have evolved from the simple manganese ribozyme that was used for 3′-end processing of early RNAs.
The termination of transcription by pol II is inextricably tied to 3′-end cleavage of the transcript (2,45–48). In the case of human β-globin genes, the co-transcriptional cleavage (CoTC) site positioned downstream of the polyadenylation site is required for proper termination of transcription (49). There is evidence that the CoTC element has self-cleaving activity (2) that generates a point of entry for the exonuclease Xrn2, which degrades the downstream product, leading to the dissociation of the polymerase from the DNA template (48). Whether self-cleaving elements are required for efficient termination of transcription in other extant genes remains to be determined (50). Nonetheless, this ‘torpedo’ mechanism for termination of transcription by pol II possibly reflects another function for ribozymes in the 3′-ends of early RNAs. Even after RNA synthesis became protein based, autocatalytic cleavage of the transcript may have been the means to terminate transcription by early polymerases. If so, then features of this mechanism are still utilized for termination by pol II.
We have detected the occurrence of manganese-dependent ribozymes in the 3′ UTR of two mRNAs. The catalytic activity of these ribozymes is the same as that reported for the founding member of this class identified in the Tetrahymena group I intron (8,9). As is the case with the manganese-dependent ribozymes that occur in U16 snoRNA, the ribozyme in Vg1 mRNA is not functional in vivo, but may represent a molecular fossil. We propose that these ribozymes may have been important in the processing of prebiotic transcripts and possibly account for the purine- and uridine-rich identity elements that comprise the signals used for 3′-end processing of present-day pol II transcripts.
FUNDING
American Heart Association (0151445Z); Office of Research, University of Notre Dame. Funding for open access charge: College of Science, University of Notre Dame.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We thank Joseph Piccirilli and Ira Wool for comments on the manuscript, and especially Qingshun Quinn Li for several helpful discussions concerning sequence analysis of UTRs.
REFERENCES
- 1.Fedor MJ, Williamson JR. The catalytic diversity of RNAs. Nat. Rev. Mol. Cell Biol. 2005;6:399–412. doi: 10.1038/nrm1647. [DOI] [PubMed] [Google Scholar]
- 2.Teixeira A, Tahiri-Alaoui A, West S, Thomas B, Ramadass A, Martianov I, Dye M, James W, Proudfoot NJ, Akoulitchev A. Autocatalytic RNA cleavage in the human β-globin pre-mRNA promotes transcription termination. Nature. 2004;432:526–530. doi: 10.1038/nature03032. [DOI] [PubMed] [Google Scholar]
- 3.Salehi-Ashtiani K, Luptak A, Litovchick A, Szostak JW. A genomewide search for ribozymes reveals an HDV-like sequence in the human CPEB3 gene. Science. 2006;313:1788–1792. doi: 10.1126/science.1129308. [DOI] [PubMed] [Google Scholar]
- 4.Mowry KL, Melton DA. Vegetal messenger RNA localization directed by a 340-nt RNA sequence element in Xenopus oocytes. Science. 1992;255:991–994. doi: 10.1126/science.1546297. [DOI] [PubMed] [Google Scholar]
- 5.Wilhelm JE, Vale RD, Hegde RS. Coordinate control of translation and localization of Vg1 mRNA in Xenopus oocytes. Proc. Natl Acad. Sci. USA. 2000;97:13132–13137. doi: 10.1073/pnas.97.24.13132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Otero LJ, Devaux A, Standart N. A 250-nucleotide UA-rich element in the 3′ untranslated region of Xenopus laevis Vg1 mRNA represses translation both in vivo and in vitro. RNA. 2001;7:1753–1767. [PMC free article] [PubMed] [Google Scholar]
- 7.Kolev NG, Huber PW. VgRBP71 stimulates cleavage at a polyadenylation signal in Vg1 mRNA, resulting in the removal of a cis-acting element that represses translation. Mol. Cell. 2003;11:745–755. doi: 10.1016/s1097-2765(03)00071-6. [DOI] [PubMed] [Google Scholar]
- 8.Dange V, Van Atta RB, Hecht SM. A Mn2+-dependent ribozyme. Science. 1990;248:585–588. doi: 10.1126/science.2185542. [DOI] [PubMed] [Google Scholar]
- 9.Kazakov S, Altman S. A trinucleotide can promote metal ion-dependent specific cleavage of RNA. Proc. Natl Acad. Sci. USA. 1992;89:7939–7943. doi: 10.1073/pnas.89.17.7939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brockman JM, Singh P, Liu D, Quinlan S, Salisbury J, Graber JH. PACdb: PolyA Cleavage Site and 3′-UTR Database. Bioinformatics. 2005;21:3691–3693. doi: 10.1093/bioinformatics/bti589. [DOI] [PubMed] [Google Scholar]
- 11.Herschlag D, Khosla M. Comparison of pH dependencies of the Tetrahymena ribozyme reactions with RNA 2′-substituted and phosphorothioate substrates reveals a rate-limiting conformational step. Biochemistry. 1994;33:5291–5297. doi: 10.1021/bi00183a036. [DOI] [PubMed] [Google Scholar]
- 12.Chin K, Pyle AM. Branch-point attack in group II introns is a highly reversible transesterification, providing a potential proofreading mechanism for 5′-splice site selection. RNA. 1995;1:391–406. [PMC free article] [PubMed] [Google Scholar]
- 13.Podar M, Perlman PS, Padgett RA. The two steps of group II intron self-splicing are mechanistically distinguishable. RNA. 1998;4:890–900. doi: 10.1017/s1355838298971643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bombard S, Kozelka J, Favre A, Chottard JC. Probing the mechanism of an Mn2+-dependent ribozyme by means of platinum complexes. Eur. J. Biochem. 1998;252:25–35. doi: 10.1046/j.1432-1327.1998.2520025.x. [DOI] [PubMed] [Google Scholar]
- 15.Kuo TC, Herrin DL. Quantitative studies of Mn2+-promoted specific and non-specific cleavages of a large RNA: Mn2+-GAAA ribozymes and the evolution of small ribozymes. Nucleic Acids Res. 2000;28:4197–4206. doi: 10.1093/nar/28.21.4197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rebagliati MR, Weeks DL, Harvey RP, Melton DA. Identification and cloning of localized maternal RNAs from Xenopus eggs. Cell. 1985;42:769–777. doi: 10.1016/0092-8674(85)90273-9. [DOI] [PubMed] [Google Scholar]
- 17.Prislei S, Fatica A, De Gregorio E, Arese M, Fragapane P, Caffarelli E, Presutti C, Bozzoni I. Self-cleaving motifs are found in close proximity to the sites utilized for U16 snoRNA processing. Gene. 1995;163:221–226. doi: 10.1016/0378-1119(95)00344-6. [DOI] [PubMed] [Google Scholar]
- 18.Laneve P, Altieri F, Fiori ME, Scaloni A, Bozzoni I, Caffarelli E. Purification, cloning, and characterization of XendoU, a novel endoribonuclease involved in processing of intron-encoded small nucleolar RNAs in Xenopus laevis. J. Biol. Chem. 2003;278:13026–13032. doi: 10.1074/jbc.M211937200. [DOI] [PubMed] [Google Scholar]
- 19.Kislauskis EH, Zhu X, Singer RH. Sequences responsible for intracellular localization of β-actin messenger RNA also affect cell phenotype. J. Cell Biol. 1994;127:441–451. doi: 10.1083/jcb.127.2.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang E, van Nimwegen E, Zavolan M, Rajewsky N, Schroeder M, Magnasco M, Darnell J.E., Jr Decay rates of human mRNAs: correlation with functional characteristics and sequence attributes. Genome Res. 2003;13:1863–1872. doi: 10.1101/gr.1272403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Foat BC, Houshmandi SS, Olivas WM, Bussemaker HJ. Profiling condition-specific, genome-wide regulation of mRNA stability in yeast. Proc. Natl Acad. Sci. USA. 2005;102:17675–17680. doi: 10.1073/pnas.0503803102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Grigull J, Mnaimneh S, Pootoolal J, Robinson MD, Hughes TR. Genome-wide analysis of mRNA stability using transcription inhibitors and microarrays reveals posttranscriptional control of ribosome biogenesis factors. Mol. Cell Biol. 2004;24:5534–5547. doi: 10.1128/MCB.24.12.5534-5547.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang Y, Liu CL, Storey JD, Tibshirani RJ, Herschlag D, Brown PO. Precision and functional specificity in mRNA decay. Proc. Natl Acad. Sci. USA. 2002;99:5860–5865. doi: 10.1073/pnas.092538799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Raghavan A, Bohjanen PR. Microarray-based analyses of mRNA decay in the regulation of mammalian gene expression. Brief Funct. Genomic Proteomic. 2004;3:112–124. doi: 10.1093/bfgp/3.2.112. [DOI] [PubMed] [Google Scholar]
- 25.Chen CY, You Y, Shyu AB. Two cellular proteins bind specifically to a purine-rich sequence necessary for the destabilization function of a c-fos protein-coding region determinant of mRNA instability. Mol. Cell Biol. 1992;12:5748–5757. doi: 10.1128/mcb.12.12.5748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Buck AH, Dalby AB, Poole AW, Kazantsev AV, Pace NR. Protein activation of a ribozyme: the role of bacterial RNase P protein. EMBO J. 2005;24:3360–3368. doi: 10.1038/sj.emboj.7600805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Buck AH, Kazantsev AV, Dalby AB, Pace NR. Structural perspective on the activation of RNAse P RNA by protein. Nat. Struct. Mol. Biol. 2005;12:958–964. doi: 10.1038/nsmb1004. [DOI] [PubMed] [Google Scholar]
- 28.Mohr G, Rennard R, Cherniack AD, Stryker J, Lambowitz AM. Function of the Neurospora crassa mitochondrial tyrosyl-tRNA synthetase in RNA splicing. Role of the idiosyncratic N-terminal extension and different modes of interaction with different group I introns. J. Mol. Biol. 2001;307:75–92. doi: 10.1006/jmbi.2000.4460. [DOI] [PubMed] [Google Scholar]
- 29.Murthy KG, Manley JL. The 160-kD subunit of human cleavage-polyadenylation specificity factor coordinates pre-mRNA 3′-end formation. Genes Dev. 1995;9:2672–2683. doi: 10.1101/gad.9.21.2672. [DOI] [PubMed] [Google Scholar]
- 30.Moore CL, Chen J, Whoriskey J. Two proteins crosslinked to RNA containing the adenovirus L3 poly(A) site require the AAUAAA sequence for binding. EMBO J. 1988;7:3159–3169. doi: 10.1002/j.1460-2075.1988.tb03183.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.MacDonald CC, Wilusz J, Shenk T. The 64-kilodalton subunit of the CstF polyadenylation factor binds to pre-mRNAs downstream of the cleavage site and influences cleavage site location. Mol. Cell Biol. 1994;14:6647–6654. doi: 10.1128/mcb.14.10.6647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mandel CR, Kaneko S, Zhang H, Gebauer D, Vethantham V, Manley JL, Tong L. Polyadenylation factor CPSF-73 is the pre-mRNA 3′-end-processing endonuclease. Nature. 2006;444:953–956. doi: 10.1038/nature05363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Callebaut I, Moshous D, Mornon JP, de Villartay JP. Metallo-β-lactamase fold within nucleic acids processing enzymes: the β-CASP family. Nucleic Acids Res. 2002;30:3592–3601. doi: 10.1093/nar/gkf470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhao J, Hyman L, Moore C. Formation of mRNA 3′ ends in eukaryotes: mechanism, regulation, and interrelationships with other steps in mRNA synthesis. Microbiol. Mol. Biol. Rev. 1999;63:405–445. doi: 10.1128/mmbr.63.2.405-445.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kolev NG, Steitz JA. Symplekin and multiple other polyadenylation factors participate in 3′-end maturation of histone mRNAs. Genes Dev. 2005;19:2583–2592. doi: 10.1101/gad.1371105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dominski Z, Yang XC, Marzluff WF. The polyadenylation factor CPSF-73 is involved in histone-pre-mRNA processing. Cell. 2005;123:37–48. doi: 10.1016/j.cell.2005.08.002. [DOI] [PubMed] [Google Scholar]
- 37.Baillat D, Hakimi MA, Naar AM, Shilatifard A, Cooch N, Shiekhattar R. Integrator, a multiprotein mediator of small nuclear RNA processing, associates with the C-terminal repeat of RNA polymerase II. Cell. 2005;123:265–276. doi: 10.1016/j.cell.2005.08.019. [DOI] [PubMed] [Google Scholar]
- 38.Wagner EJ, Burch BD, Godfrey AC, Salzler HR, Duronio RJ, Marzluff WF. A genome-wide RNA interference screen reveals that variant histones are necessary for replication-dependent histone pre-mRNA processing. Mol. Cell. 2007;28:692–699. doi: 10.1016/j.molcel.2007.10.009. [DOI] [PubMed] [Google Scholar]
- 39.Dominski Z, Marzluff WF. Formation of the 3′ end of histone mRNA: getting closer to the end. Gene. 2007;396:373–390. doi: 10.1016/j.gene.2007.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bond UM, Yario TA, Steitz JA. Multiple processing-defective mutations in a mammalian histone pre-mRNA are suppressed by compensatory changes in U7 RNA both in vivo and in vitro. Genes Dev. 1991;5:1709–1722. doi: 10.1101/gad.5.9.1709. [DOI] [PubMed] [Google Scholar]
- 41.Davila Lopez M, Samuelsson T. Early evolution of histone mRNA 3′ end processing. RNA. 2008;14:1–10. doi: 10.1261/rna.782308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Uguen P, Murphy S. The 3′ ends of human pre-snRNAs are produced by RNA polymerase II CTD-dependent RNA processing. EMBO J. 2003;22:4544–4554. doi: 10.1093/emboj/cdg430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yuo CY, Ares M., Jr, Weiner AM. Sequences required for 3′ end formation of human U2 small nuclear RNA. Cell. 1985;42:193–202. doi: 10.1016/s0092-8674(85)80115-x. [DOI] [PubMed] [Google Scholar]
- 44.Hernandez N. Formation of the 3′ end of U1 snRNA is directed by a conserved sequence located downstream of the coding region. EMBO J. 1985;4:1827–1837. doi: 10.1002/j.1460-2075.1985.tb03857.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Buratowski S. Connections between mRNA 3′ end processing and transcription termination. Curr. Opin. Cell Biol. 2005;17:257–261. doi: 10.1016/j.ceb.2005.04.003. [DOI] [PubMed] [Google Scholar]
- 46.Chodchoy N, Pandey NB, Marzluff WF. An intact histone 3′-processing site is required for transcription termination in a mouse histone H2a gene. Mol. Cell Biol. 1991;11:497–509. doi: 10.1128/mcb.11.1.497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kim M, Krogan NJ, Vasiljeva L, Rando OJ, Nedea E, Greenblatt JF, Buratowski S. The yeast Rat1 exonuclease promotes transcription termination by RNA polymerase II. Nature. 2004;432:517–522. doi: 10.1038/nature03041. [DOI] [PubMed] [Google Scholar]
- 48.West S, Gromak N, Proudfoot NJ. Human 5′→3′ exonuclese Xrn2 promotes transcription terminatin at co-transcriptional cleavage sites. Nature. 2004;432:522–525. doi: 10.1038/nature03035. [DOI] [PubMed] [Google Scholar]
- 49.Dye MJ, Proudfoot NJ. Multiple transcript cleavage precedes polymerase release in termination by RNA polymerase II. Cell. 2001;105:669–681. doi: 10.1016/s0092-8674(01)00372-5. [DOI] [PubMed] [Google Scholar]
- 50.West S, Zaret K, Proudfoot NJ. Transcriptional termination sequences in the mouse serum albumin gene. RNA. 2006;12:655–665. doi: 10.1261/rna.2232406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zuker M, Mathews DH, Turner DH. Algorithms and Thermodynamics for RNA Secondary Structure Prediction: A Practical Guide. In: Barciszewski J, Clark BFC, editors. RNA Biochemistry and Biotechnology. Dordrecht, NL: Kluwer Academic Publishers; 1999. pp. 11–43. [Google Scholar]
- 52.Ross AF, Oleynikov Y, Kislauskis EH, Taneja KL, Singer RH. Characterization of a β-actin mRNA zipcode-binding protein. Mol. Cell Biol. 1997;17:2158–2165. doi: 10.1128/mcb.17.4.2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Van Atta RB, Hecht SM. A ribozyme model: site-specific cleavage of an RNA substrate by Mn2+ Adv. Inorg. Biochem. 1994;9:1–40. [PubMed] [Google Scholar]