Abstract
The throughput of DNA reading (sequencing) has dramatically increased recently due to the incorporation of in vitro clonal amplification. The throughput of DNA writing (synthesis) is trailing behind, with cloning and sequencing constituting the main bottleneck. To overcome this bottleneck, an in vitro alternative for in vivo DNA cloning must be integrated into DNA synthesis methods. Here we show how a new single molecule PCR (smPCR)-based procedure can be employed as a general substitute to in vivo cloning thereby allowing for the first time in vitro DNA synthesis. We integrated this rapid and high fidelity in vitro procedure into our earlier recursive DNA synthesis and error correction procedure and used it to efficiently construct and error-correct a 1.8-kb DNA molecule from synthetic unpurified oligos completely in vitro. Although we demonstrate incorporating smPCR in a particular method, the approach is general and can be used in principle in conjunction with other DNA synthesis methods as well.
INTRODUCTION
The broad availability of synthetic DNA oligonucleotides enabled the development of many powerful applications in biotechnology. Longer synthetic DNA molecules and libraries (made by the assembly of these oligonucleotides) in the 0.5–5 kb range are now becoming increasingly available thanks to newly developed synthesis and error correction methods (1–7). Broad availability of such molecules, much needed since the advent of synthetic biology and modern genetic engineering, is expected to enable routine creation of new genetic material as well as offer an alternative to obtaining DNA from natural sources.
Unfortunately, the synthetic DNA oligonucleotides used as building blocks for making the longer constructs are error prone. Such errors accumulate linearly with the length of the constructed molecule and result in an exponential decrease in the fraction of error-free molecules (Supplementary Figure 1). Hence an exponentially increasing number of molecules have to be screened, i.e. cloned into a host organism and sequenced, in order to obtain ever longer error-free molecules (Supplementary Figure 2, blue and green plots). In order to mitigate this effect a two-step assembly process (4,7) is often used, in which fragments in the 500–1000 bp range are first screened via cloning and sequencing and then synthesis proceeds from the error-free clones (Supplementary Figure 2, red plot).
In vivo cloning (1–7) is time consuming, manual-labor intensive, difficult to scale up and automate. This combined with the sheer number of clones that need to be screened to obtain long error-free synthetic DNA (Supplementary Figure 2) makes the cloning phase a bottleneck in de novo DNA synthesis and prevents synthetic DNA from being routinely produced in a fast, cheap and high-throughput manner. Reducing the number of clones required to obtain an error-free molecule is the subject of intensive ongoing research (1,2,4,6), also recently addressed by us (5) with a method that relieves much of this burden (Supplementary Figure 2, cyan plot).
In this report we address the second major issue, namely replacing the time consuming and labor intensive in vivo cloning procedure associated with synthetic DNA synthesis with a faster and less laborious in vitro cloning procedure.
Since its introduction, PCR (8) has been implemented in a myriad of variations, one of which is PCR on a single DNA template molecule (9), which essentially creates a PCR ‘clone’. Single molecule PCR (smPCR) is a faster, cheaper, scalable and automatable alternative to traditional in vivo cloning. Its standard application in molecular biology has been nonsystematic, most commonly for the amplification of single molecules for sequencing, genotyping or downstream translation purposes (8–12). Recently, it has been systematically integrated into high-throughput DNA reading (sequencing) (13,14). High-throughput DNA writing (synthesis) technology can also benefit from smPCR, as demonstrated by our work reported here. The use of smPCR is described in the context of our recently introduced DNA synthesis procedure (5), which combines recursive synthesis and error-correction, and operates as follows. Divide and Conquer (D&C), the quintessential recursive problem solving technique, is used in silico to divide the target DNA sequence to be constructed into fragments short enough to be synthesized by conventional oligo synthesis, albeit with errors (15); these oligos are synthesized and are recursively combined in vitro, forming target DNA molecules with roughly the same error rate as the source oligos; error-free parts of these molecules identified by cloning and sequencing are extracted and used as new, typically longer and more accurate inputs to another iteration of the recursive synthesis procedure. Typically, an error-free clone is obtained after one iteration of this procedure.
In this article we show that in vitro cloning based on smPCR can be used as a practical alternative to conventional in vivo cloning in our DNA synthesis protocol. In particular, we successfully constructed a 1.8-kb long DNA molecule from synthetic unpurified oligos using our recursive synthesis and error correction procedure with smPCR, and as a control also constructed the same molecule using conventional in vivo cloning. The results are compared below.
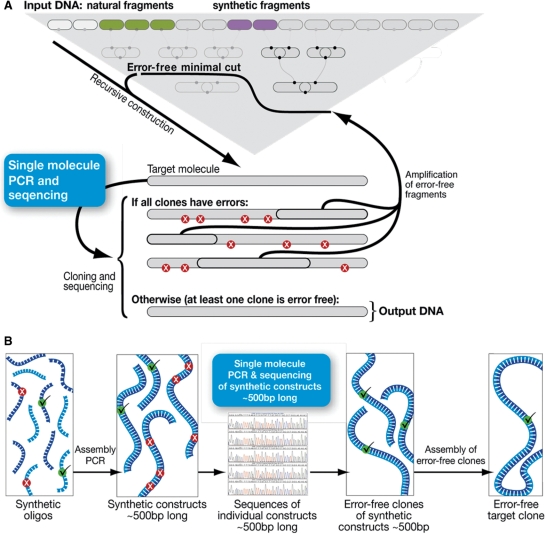
We expect that our methods may be used to incorporate smPCR also in other DNA synthesis procedures, for example in conjunction with the widely used two step assembly PCR method (7) (Figure 1b).
Figure 1.
Overview—Although de novo DNA synthesis is traditionally performed with in vivo cloning, which is time consuming and labor intensive, it can, in principle, be performed instead in vitro using a modified smPCR protocol. (a) Work reported here: Target synthetic molecules are recursively constructed (5) from oligos and then error-corrected using the new smPCR procedure instead of in vivo cloning. In brief, Preparation of the target DNA molecules for smPCR amplification is carried out by a PCR that introduces sites for the smPCR primer (see text). This PCR is stopped at the exponential phase of amplification so that heterodimers are not formed (see text). The PCR products are then diluted according to calculations and experimental results (see text) and used as template for smPCR with a special primer (C–A primer) that doesn't produce nonspecific amplification products (see text). The DNA ‘clones’ amplified using smPCR are then sequenced and an error-correction process (5) (Also see Supplementary Data Methods section for error-correction description) is carried out using the smPCR amplified molecules as starting material until an error free molecule is obtained. (b) Conceptual illustration of how the smPCR procedure could also be used in principle, with a two-step assembly PCR. From left to right, oligos are assembled in groups and amplified to yield fragments 400–500 bp long. These could be cloned using exactly the same smPCR procedure described in this work and sequenced. The error-free clones are then selected for further assembly of the target sequence using various methodologies.
MATERIALS AND METHODS
Cloning
Fragments were cloned into the pGEM T easy Vector System1 from PROMEGA. Vectors containing cloned fragments were transformed into JM109 competent cells from PROMEGA1 and sequenced.
smPCR
smPCR was performed with hot-start Accusure (BioLINE, Taunton, MA, USA) for the longer Mitochondrial and with Taq Polymerase (ABgene, Epsom, United Kingdom) for the GFP fragment. Template concentration is according to calculations described in the paper and dissolved in 5 μl DDW; 10 pmol of the CA primer dissolved in 10 µl DDW. Reaction contained 25 mM TAPS pH 9.3 at 25°C, 2 mM MgCl2, 50 mM KCl, 1 mM β-mercaptoethanol, 200 μM each of dNTP, 1.9 U AccuSure DNA Polymerase (BioLINE).
Real-time PCR (RT-PCR) Thermal Cycler program: Enzyme activation at 95°C for 10 min, denaturation 95°C 30 s, annealing at Tm of primers 30 s, extension 72°C 1.5 min/kb, 50 cycles. It is important that the PCR is prepared in a sterile environment using sterile equipment and uncontaminated reagents.
Methods for recursive construction and error correction
The core recursive construction and reconstruction (error-correction) step requires four basic enzymatic reactions: phosphorylation, elongation, PCR and Lambda exonucleation. They are described in the order of execution by our protocol.
Phosphorylation
Phosphorylation of all PCR primers used by the recursive construction protocol is performed beforehand simultaneously, according to the following protocol: A total of 300 pmol of 5′ DNA termini in a 50 µl reaction containing 70 mM Tris–HCl, 10 mM MgCl2, 7 mM dithiothreitol, pH 7.6 at 37°C, 1 mM ATP, 10 U T4 polynucleotide kinase (NEB, Ipswich, MA, USA). Incubation is at 37°C for 30 min and inactivation at 65°C for 20 min.
Overlap extension elongation between two ssDNA fragments
One to five Picomoles of 5′ DNA termini of each progenitor in a reaction containing 25 mM TAPS pH 9.3 at 25°C, 2 mM MgCl2, 50 mM KCl, 1 mM β-mercaptoethanol 200 μM each of dNTP, 4 U Thermo-Start DNA polymerase (ABgene). Thermal cycling program is as follows: enzyme activation at 95°C for 15 min, slow annealing 0.1°C/s from 95°C to 62°C and elongation at 72°C for 10 min.
PCR amplification of the above elongation product with two primers, one of which is phosphorylated
A total of 1–0.1 fmol template, 10 pmol of each primer in a 25 µl reaction containing 25 mM TAPS pH 9.3 at 25°C, 2 mM MgCl2, 50 mM KCl, 1 mM β-mercaptoethanol 200 μM each of dNTP, 1.9 U AccuSure DNA Polymerase (BioLINE). Thermal cycling program is: enzyme activation at 95°C for 10 min, denaturation 95°C, annealing at Tm of primers, and extension 72°C for 1.5 min/kb to be amplified 20 cycles.
Lambda exonuclease digestion of the above PCR product to re-generate ssDNA
One to five Picomoles of 5′ phosphorylated DNA termini in a reaction containing 25 mM TAPS pH 9.3 at 25°C, 2 mM MgCl2, 50 mM KCl, 1 mM β-mercaptoethanol 5 mM 1,4-Dithiothreitol, 5 U Lambda Exonuclease (Epicentre). Thermal cycling program is: enzyme activation at 37°C for 15 min, 42°C for 2 min and enzyme inactivation at 70°C 10 min.
RESULTS
We have constructed an error-free 1.8 kb molecule from synthetic unpurified oligos using recursive synthesis and error correction with in vitro cloning based on smPCR. At the same time we followed the exact same procedure but with traditional in vivo cloning as a control. Our results show that the smPCR-based procedure is comparable to traditional cloning in terms of the fidelity of the clones. Although the accuracy of in vivo cloning is higher than smPCR, this has a minor effect on the number of clones required to obtain an error-free clone for molecules in several kilo base range. The relatively small difference in fidelity is greatly outweighed by the improved time, cost and throughput offered by the in vitro procedure. We had to integrate several modifications into smPCR methodology in order for it to be suitable for de novo DNA synthesis, as discussed in the results section below. These included improved primer selection, computational optimization and experimental calibration of template concentration, real-time diagnosis of faulty reactions, avoiding the cloning of heteroduplexes, bar-coding molecules and creating a process with adequate fidelity.
Careful selection of adequate primers is needed to enable single molecule amplification
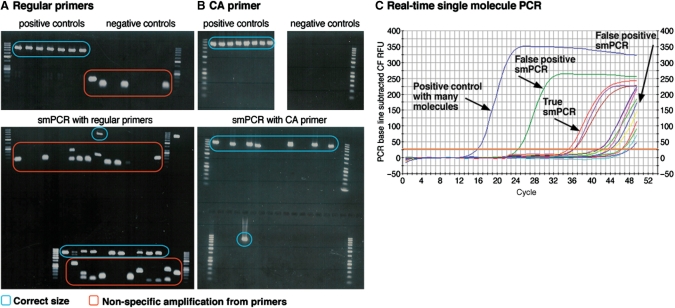
smPCR amplification requires extensive cycling (9–12). This often leads to the amplification of nonspecific products originating from interaction between the PCR primers (Figure 2a). This often inhibits the amplification of the single molecule template, typically resulting in either no amplification of the target molecule due to dimer formation or in amplification of the primer dimer on top of the correct PCR product (Figure 2a). Consequently, a large fraction of the smPCRs performed cannot be used for synthesis since they did not amplify or have nonspecific amplification products. This has to be compensated for by performing more smPCRs than are actually needed for synthesis. To solve this problem we designed a special primer for smPCR consisting of a single sequence (complementary to both ends of the single molecule template), which contains a sequence of cytosine and adenine DNA bases only (see Supplementary Data for sequence). We reasoned that this should reduce the formation of PCR products that originate from primer-primer interactions due to the noncomplementary nature of the cytosine and adenine bases. This successfully eliminated nonspecific amplification resulting from interaction between primers and its inhibiting effect on single molecule amplification (Figure 2b), which in turn significantly decreased the total number of PCRs needed to obtain the minimal number of smPCR clones required for synthesis of error-free DNA. The sites for the C–A primer (as well as the random bar coding bases to be discussed later on) at the termini of the target molecules are incorporated by either an a priori PCR (16) or during the synthesis of the molecule as part of the target sequence.
Figure 2.
Primer, dimers and anticipation. Adequate selection of primers leads to improved specificity in smPCR; RT-PCR can distinguish true smPCRs from false positives. (a) smPCRs with regular primers show many nonspecific amplification products. Top gel: Lanes 1–7: positive control (many template molecules) PCRs show bands at the correct size. Lanes 8–15: no-template control PCRs have nonspecific amplification from primers. Bottom gel: smPCR experiments—a large fraction of reactions show nonspecific amplification from primers which inhibit smPCR and hinder its use. (b) smPCRs with the C–A primer shows specific amplification. Top left gel: positive control (multiple template molecules) PCRs show bands at the correct size. Top right gel: no-template control PCRs do not have nonspecific amplification. Bottom gel: smPCR experiments bands at the correct size and frequency with no nonspecific amplification C. RT-PCR helps determining whether PCRs are true smPCRs or false positives due to nonspecific amplification from primers or contamination.
Computational optimization and experimental calibration of template DNA concentration
smPCR reactions are generally similar to regular PCR reactions in their basic biochemistry, the difference is that while PCR typically start the amplification with multiple copies of the template molecule, the goal in smPCR is to amplify a single template molecule. This is achieved by diluting a solution with template molecules in a known concentration so that the template aliquot is expected to have about one molecule. As the dilution is a stochastic process, at any such dilution some aliquots would have no template molecule and some would have multiple template molecules. As these two cases cannot be avoided, smPCR is done as a batch of multiple parallel reactions, with the hope that at least some would be true smPCRs, namely successful PCR reactions that amplify single template molecules. ‘False positive’ smPCRs, which amplify multiple template molecules, are identified using sequencing (Figure 3 and Supplementary Figure 6). The cost of sequencing is a major component of synthetic DNA synthesis, and the sequencing of false positives can render smPCR unpractical if their fraction in the total number of reactions is too high. Standard gel/capillary electrophoreses (CE)/RT-PCR analyses can be used to differentiate no template (negative) reactions from (positive) PCRs with template, however, they cannot be used to differentiate a true smPCR from false positive reactions. Diluting the template to one molecule per well on average maximizes the fraction of true smPCRs out of all the reactions in the batch (Supplementary Figure 3a, blue plot). However, it does not maximize the ratio of true smPCRs to false positives (Supplementary Figure 3a, green plot) which is important for avoiding futile sequencing. For example, aiming for one molecule per well on average leads to >50% futile sequencing of false positives (Supplementary Figure 3a, green plot). Further reducing template concentration reduces the extent of futile sequencing of PCRs with multiple template molecules, however, it increases the extent of futile PCRs due to no template reactions. Determining the template concentration that would result in an optimal ratio between true smPCRs, false positives and no template reactions can only be determined by associating a cost to performing sequencing and smPCR reactions. We calculated the optimal concentration to be ∼0.6 template molecules per smPCR well if an equal cost is associated with smPCR and sequencing (Supplementary Figure 3b), and ∼0.2 molecules per well if sequencing is assigned the more realistic cost of eight times that of smPCR (Supplementary Figure 3c). Performing smPCRs at the optimal template concentration reduces the overall cost of obtaining each sequenced true smPCR and the overall cost of using smPCR with de novo DNA synthesis since it reduces futile sequencing from 50% (with 1 molecule/well) to 10% (with ∼0.2 molecules/well) (Supplementary Figure 3a). A standard 260 nm OD measurement can be used to determine the optimal concentration.
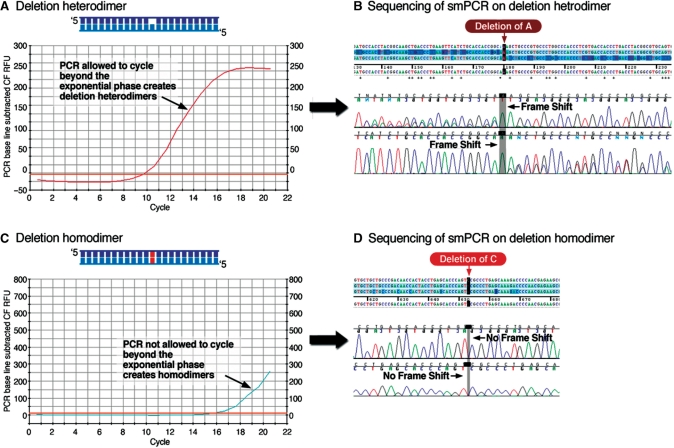
Figure 3.
Heterodimers hinder smPCR. The template for smPCR is produced with an ordinary PCR reaction. If this PCR is not not terminated at the exponential phase of amplification it produces heterodimers, which hinder smPCR. (a) Overcycling of the PCR past the exponential phase of amplification leads to the formation of hetero-dimers by re-annealing of different elongated strands. (b) The sequencing chromatograms of both sense and antisense strands of a PCR amplified heterodimer are frame-shifted and unreadable from the site of the (insertion or deletion) mutation and on. (c) A PCR terminated before the end of the exponential amplification generates homodimers, not heterodimers. (d) The sequencing chromatogram of a PCR amplified homodimer is readable and not frame-shifted even if a mutations (with respect to the target sequence) are present.
Even though most of the smPCRs performed using 0.2 molecules/well (i.e. 80% of reactions having no template), these no-template PCRs are easily identified and distinguished from ‘true’ smPCRs, and their sequencing is avoided. Additionally, the cost of no template PCRs is further diminished by performing the reactions in very low volume (down to 2 μl in standard liquid handling robots). We also found that RT-PCR can be used to accurately determine the dilution required to dilute the template to the calculated optimal concentration (0.2 molecules/well). A one-time calibration (see Supplementary Data Methods for description) allows the routine use of RT-PCR to determine the dilution required before each smPCR experiment. This strategy proved as accurate and as robust as performing the dilution according to a 260 nm OD measurement and was used throughout the work presented in this paper.
RT-PCR facilitates the diagnosis of faulty reactions
We used RT-PCR to confirm that the efficiency at which our C–A primer amplifies DNA is close to 100%. Given this efficiency, we predict the number of PCR cycles required to reach PCR amplification saturation from the initial and typical final template concentrations (Supplementary Figure 4, green plot). Our RT-smPCR results confirm that this prediction is accurate all the way down to single molecule amplification, which displays an amplification curve that is detectable from approximately cycle 32 and saturates after ∼42 cycles (Figure 2c and Supplementary Figure 4, blue plot). This prediction allows real-time determination of whether PCRs are true smPCRs or false positives (e.g. contaminated, actually had many template molecules or primer dimers) since they do not exhibit a typical amplification curve which indicates single molecule amplification (Figure 2c), eschewing their further analysis.
Heteroduplexes prevent in vitro cloning of synthetic DNA
Initially, the sequencing of all our true smPCR experiments resulted in shifted sequencing chromatograms which could not be read properly, despite the fact that in vivo clones from the same DNA sequenced fine. The cause of this turned out to be that de novo constructed DNA is double stranded (1–4,6,7), with each strand having different errors originating from different synthetic oligo species. Performing smPCR on such a heteroduplex creates two distinct populations of amplified molecules, one from each strand. The abundance of deletions and insertions in synthetic oligos (4,15) causes the sequencing chromatograms of these dual population PCRs to be frame shifted and their sequence cannot be determined (Figure 3b).
These smPCR cloning results were reinforced by calculations that show that, according to the error-rate of oligos (4,15), heteroduplexes are much more abundant than homoduplexes at the typical cloning length (Supplementary Figure 5). In practice almost all synthetic clones were heteroduplexes (due to insertions or deletions) which could not be sequenced properly. Rare exceptions were clones that were heteroduplexes only due to substitutions in one or both strands (which do not result in frame-shifts) (Supplementary Figure 6) and were therefore sequenced properly.
The reason that heteroduplexes were not reported to be a problem so far in de novo synthesis (1–4,6,7) is probably the ubiquitous use of in vivo cloning, which converts the erroneous mismatched DNA into perfectly matched DNA, albeit erroneous compared to the target sequence. A true smPCR should therefore be performed on either one ssDNA molecule or on two perfectly complemented molecules, i.e. one homoduplex dsDNA. Initially we treated synthetic dsDNA constructs labeled with a 5′ phosphate at one end with Lambda exonuclease to convert them into ssDNA. smPCR on ssDNA templates generated by this enzymatic treatment indeed resulted in a larger fraction of smPCRs which can be sequenced. However, a complete and simpler solution to this problem was achieved not by generating ssDNA but by generating homoduplex dsDNA. Homoduplex dsDNA was generated by terminating the PCR amplification of synthetic DNA prematurely, not allowing it past the exponential phase of amplification, as monitored by RT-PCR (Figure 3c). Terminating the PCR at the exponential phase of amplification assures that each dsDNA molecule is formed by primer-directed polymerization which forms homoduplexes (Figure 3d and Supplementary Figure 5, primer directed polymerization plot), and not by the annealing of previously elongated strands which forms heteroduplexes (Figure 3b and Supplementary Figure 5, annealing plot). A comparison between smPCRs executed using templates generated by primer-directed polymerization and by annealing of previously elongated strands are shown in Figure 3c and d, and Figure 3a and b, respectively.
Single-molecule verification with random oligos
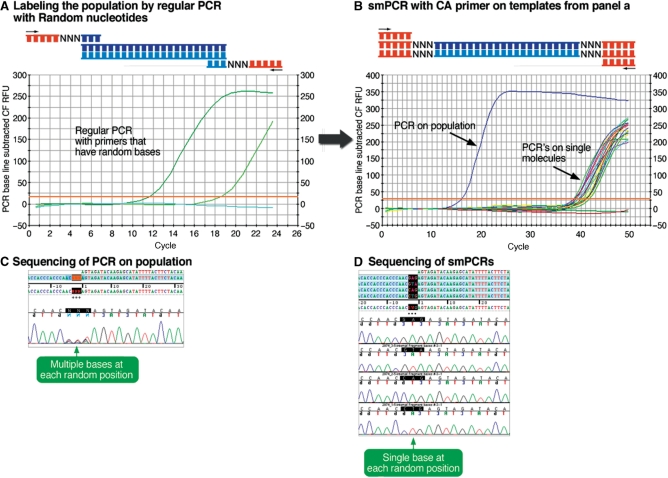
To facilitate the simple identification of rare smPCRs that despite the measures reported above were still not performed on single molecules, we integrated another feature in our procedure, previously proposed for other smPCR applications (16). We incorporated oligos with three random bases at both ends of the synthetic DNA constructs that are to be cloned, effectively bar-coding the molecules with a four-letter code at six positions (46 = 4096 tags) (Figure 4a). Sequencing these molecules show that the sequence at the location of the random bases is always singular in the sequencing of a true smPCR (Figure 4d) and multiple in PCRs performed on >1 template molecules (Figure 4c).
Figure 4.
Randomized primers. (a) Primers with random bases are inserted into the termini of the molecules by PCR and the reaction is terminated at the exponential phase to avoid hetero-dimers. (b) DNA molecules from the light green PCR shown in panel A are diluted and used as template for smPCR with the C–A primer (PCRs on single molecules). As control a ‘false positive’ smPCR with the same DNA but with many template molecules was also performed. (c) On the left: the sequencing chromatogram of the ‘false positive’ smPCR from panel b shows al 4 bases at the 3 random positions, indicating that the reaction was not a true smPCR. On the right: the sequencing chromatograms of four different smPCRs from panel B show only one base call at each of the three random positions, indicating they were true smPCRs.
Fidelity of single molecule amplification
Errors produced by smPCR pose a minor problem in sequencing and genotyping applications since they can only produce artifacts if inserted during the first few rounds of amplification (11). Errors inserted after the first few cycles (i.e. the remaining ∼36–37 cycles) are represented in a low fraction of the population (Supplementary Figure 7) and are not detectable by sequencing. Nevertheless, errors are inserted during all cycles of smPCR at a fixed rate (Supplementary Figure 8). Although this hardly affects DNA reading applications (11) (Supplementary Figure 7) it dramatically affects DNA writing using smPCR since the smPCR amplified molecules are used as building blocks for further synthesis. Using a standard Taq polymerase with an error-rate of 1/8000 (17) to amplify single error-free DNA molecules results in amplified copies that have an average error rate of 1/200 compared to the original sequence after the 40 PCR cycles required for single molecule amplification (Supplementary Figure 8). This linear increase of error-rate with polymerase cycling results in an exponential increase in the number of clones that have to be sequenced in order to obtain an exact copy of a template molecule 1 kb long (Supplementary Figure 9).
We initially recursively constructed and error corrected (Figure 1a for overview of procedure) the 800-bp long DNA coding for the GFP from synthetic unpurified oligos using our smPCR-based procedure with a Taq DNA polymerase. The clones produced from the uncorrected GFP constructs were sequenced and had an error rate of 1/129 (Supplementary Table 1). Only error-free fragments from them were used for the reconstruction of the full-length molecule. The error rate of full-length error corrected GFP molecules (after reconstruction) with the smPCR procedure was determined by traditional cloning of the error corrected molecules into Escherichia coli and sequencing. The results were poor, as expected, reflecting an error-rate of 1/215 (Supplementary Table 2) and no error-free GFP molecules were found among the 12 clones, reinforcing our calculations (Supplementary Figures 8 and 9, respectively). The error-corrected clones turned out to be error-prone (Supplementary Table 2) even though the segments used for their reconstruction were error-free. These segments seemed error free in the sequencing of smPCR clones since most of the errors inserted during smPCR amplification (i.e. approximately during the last 37 of the 40 cycles required) are invisible in the sequencing chromatogram (Supplementary Figure 7). To make sure the errors originated from smPCR and not from the oligos we repeated the exact same error-correction procedure using traditional in vivo cloning of the GFP fragments into E. coli instead of smPCR. As with the smPCR procedure, error-free segments were chosen and used for reconstruction of the target GFP molecule. This control procedure yielded error-free GFP molecules out of almost every clone (Supplementary Table 2).
Therefore, the entire procedure using Taq is noneffective for de novo DNA synthesis since the error-rate resulting from smPCR amplification is roughly the error-rate of the synthetic molecules before any error-correction. Moreover, error-correction using smPCR with Taq may even increase the number of clones needed compared to construction with no error-correction, depending on the error-rate of the oligos used (Figure 5c, dark blue and green plots).
Figure 5.
Error-free molecules are readily cloned using smPCR. smPCR provides an alternative to in vivo cloning in de novo DNA synthesis up to the 2 Kb range at least. (a) For a 1-kb molecule and (b) for a 2-kb molecule show the probability that at least one of the molecules after error correction is error-free as a function of the number of molecules screened: blue plot—no error-correction or error-correction with smPCR using Taq (error-rate 1/200); green plot—error-correction with smPCR using a proofreading polymerase; red plot—error-correction with in vivo cloning. (c) The total (including clones of construction) number of clones needed for the construction of at least one error-free molecule with 90% probability as a function of the length of the molecule.
Nevertheless, technically the procedure was successful (i.e. there were no frame-shifting heteroduplexes, properly calculated limiting dilution, no primer–dimer problems, etc.), indicating that the remaining difficulty is indeed the error rate of the polymerase.
De novo synthesis of a 1.8-kb mitochondrial DNA using the smPCR procedure
We set out to test the procedure using Accusure, a more accurate (proof-reading) DNA polymerase (Materials and Methods). This time we also attempted to construct a longer synthetic construct 1.8 kb long, since a fragment of this length would demonstrate that the procedure can be used for the complete in vitro synthesis and error correction of most synthetic genes. Its synthesis and error correction was conducted as a comparative analysis between our in vitro smPCR-based procedure and an in vivo cloning-based procedure.
We constructed the molecule from unpurified oligos up to the cloning phase (Supplementary Figure 10) and then split the error-correction process into two separate and parallel courses executed side-by-side using the same starting material, one with smPCR and the other with in vivo cloning. Clones generated by both methods before error-correction were sequenced and their error-rate was the same (Supplementary Tables 3 and 4), as expected, reflecting the error-rate of the synthetic oligos used in synthesis (4,15). We identified the same set of error-free of segments (i.e. the minimal cut, see Supplementary Data Methods section for definition) in both sets of clones and used them to reconstruct the target 1.8-kb molecule twice, once from each set of clones (Supplementary Figures 11 and 12) and using the exact same protocol for reconstruction. Once reconstructed from error-free segments, the two 1.8-kb synthetic constructs were cloned into E. coli and sequenced in order to evaluate their error-rate. Target constructs from the smPCR procedure had an error-rate of 1/1128 (Supplementary Table 6) (we have no reference to compare this with as the Accusure error-rate is not known), giving a ∼6-fold improvement compared to the same procedure using Taq polymerase (see GFP results) and to the error-rate of initial uncorrected synthetic DNA. Error-free synthetic 1.8-kb target molecules were easily obtained from a small number of clones with this improved error-rate (Figure 5, red plot). The control in vivo cloning procedure also yielded error-free clones at an error-rate of 1/2193 (Supplementary Table 5).
The 1/1128 error-rate obtained using a proof-reading enzyme for the smPCR-procedure is sufficient for the synthesis of most genes with a reasonable number of clones (see Figure 5c, green plot). This error-rate is a result of two factors, namely the errors inserted during smPCR amplification and errors inserted during the PCR amplifications required for the reconstruction process. The 1/2193 error rate obtained from error correction using traditional cloning is most probably largely due to the errors inserted during the PCR amplifications required for reconstruction since in vivo amplification of DNA is very accurate. Although the overall error rate of the procedure using in vivo cloning is better than with the in vitro cloning presented here, this ∼2-fold difference in error rates only slightly affects the number of clones required for obtaining error-free synthetic molecules of most genes (Figure 5c, light blue and red plots). In general, the probability that a given synthesis process yields error-free molecules largely depends on the number of clones that are sequenced. For example, even synthesis without error correction can, in principle, produce error-free clones with high probability if a very large number of clones are screened (Supplementary Figure 2, blue and green plots). Conversely, the same process is unlikely to produce error-free molecules if a small number of clones are screened (Supplementary Figure 2, blue and green plots). Therefore, it is useful to describe for different synthesis methods how the number of sequenced clones influences the probability of obtaining error-free clones and, more practically, vice versa, how the required probability of success of obtaining error-free clones determines the number of clones that one should sequence (Figure 5a and b). We aimed at designing a process that yields error-free clones with high probability. Our results show that even with high success requirements (90% probability) the difference between our smPCR procedure and traditional cloning is negligible up to the 2-kb range at least (Figure 5a and b). For example, finding error-free fragments after error correction 1 kb and 2 kb long with probability of at least 90% requires only 4 and 8 clones respectively after using our smPCR method compared to 2 and 3 clones after using in vivo cloning (Figure 5a and b).
DISCUSSION
Our results show that, even though smPCR has typically been used in DNA reading applications to date (11–14), by following the procedures outlined in this work it can also be used for the typically cloning intensive de novo DNA writing (3–9). We demonstrated for the first time a general method for the synthesis of long synthetic fragments from unpurified oligos completely in vitro. The entire method as reported here is highly accessible to every lab since it is performed using off-the-shelf reagents, standard lab equipment and requires no special expertise.
In this report we show that the total construction and error correction of synthetic error free fragments of at least ∼2 kb can be made from a small number of clones using our in vitro method and that these results are comparable to construction using traditional in vivo cloning (Figure 5c, red and light blue plots). The use of other thermostable enzymes with improved fidelity (18) is expected to enable synthesis of even larger synthetic DNA molecules using the same procedure and is a subject of current work. Alternatives to high fidelity DNA amplification with thermostable polymerases, for example mesophilic amplification based on the isothermal strand displacement polymerization activity of the phi29 polymerase may also be considered in the future. The phi29 polymerase, already shown to be useful in the amplification of single DNA molecules (19) is comparable in accuracy to high fidelity thermostable polymerases (20), however its integration into a DNA synthesis scheme is not straightforward.
Although, in this report we have demonstrated the integration of in vitro cloning based on smPCR with a specific DNA synthesis method (5), it is conceivable that it can be used as an alternative to the cloning phase of other DNA synthesis methods as well (Figure 1b) and for the cloning of synthetic DNA in general. Cloning of synthetic DNA molecules using smPCR is more rapid (∼3 h), it is amenable to automation (using standard liquid handling robots) and scalable (using 96- or 384-well PCR plates), whereas traditional cloning is time consuming (∼1–2 days), manual labor intensive and difficult to automate.
A major requirement for automated DNA synthesis is robustness and reproducibility. Our experience with performing PCR directly on colonies is that it is not as robust and reproducible as traditional production and purification of plasmids. Additionally, although automated colony picking does exist it requires relatively expensive specialty equipment, while the process reported in this manuscript only requires standard lab equipment and turned out to be a highly robust and reproducible process.
Furthermore, automation of traditional cloning doesn't sum up to only automated colony picking. It also requires inoculation of bacteria in sterile conditions into a Petri dish and overnight growing of colonies. These are difficult to automate and time consuming, respectively. It should be noted that automated colony picking may be substituted by in vivo cloning-by-dilution, but this may hold difficulties of its own such as the absence of selection for blue/white colonies which helps avoid futile sequencing.
In any case, all this is preceded by the process of inserting DNA into cells (the transformation itself) which may be performed in 96-well electroporation devices or by heat shock but usually requires some manual labor and is not easily performed in an automated robotic setup. The new procedure described here does not require the use of cells of any kind and therefore reduces potential biohazards associated with replicating specific DNA fragments in vivo, with overusing antibiotic resistance for cloning and allows processing of fragments that are difficult to replicate in vivo.
Although this is a proof of concept paper, intended only to demonstrate the feasibility of using smPCR for de novo DNA synthesis, the amenability of the method for scaling-up is apparent. Therefore, we reason that its simplicity, rapidness and amenability to automation make it a possible alternative to traditional cloning practice in DNA synthesis.
Combining the in vitro synthetic DNA cloning and sequencing methodology reported here with our previously published automated construction and error correction method (5) has been straightforward. In the future we hope to mange to apply these methodologies to construct DNA originating from high-throughput synthesis on chips (4). Such capabilities should, in turn, facilitate ever more ambitious synthetic biology efforts involving the synthesis of synthetic DNA.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENT
This Research was supported by the Yeshaya Horowitz association through the center for Complexity Science, the Research Grant from Dr. Mordecai Roshwald, the Grant from Kenneth and Sally Leafman Appelbaum Discovery Fund, the Estate of Karl Felix Jakubskind, the Estate of Funnie Sherr, the Clore Center for Biological Physics and the Louis Chor Memorial Trust. Funding to pay the Open Access publication charges for this article was provided by these grants.
Conflict of interest statement. E. S. is the Incumbent of The Harry Weinrebe Professorial Chair of Computer Science and Biology and of The France Telecom–Orange Excellence Chair for Interdisciplinary Studies of the Paris “Centre de Recherche Interdisciplinaire” (FTO/CRI). All the other authors have declared no conflicts of interest.
REFERENCES
- 1.Bang D, Church GM. Gene synthesis by circular assembly amplification. Nat. Methods. 2008;5:37–39. doi: 10.1038/nmeth1136. [DOI] [PubMed] [Google Scholar]
- 2.Carr PA, Park JS, Lee YJ, Yu T, Zhang S, Jacobson JM. Protein-mediated error correction for de novo DNA synthesis. Nucleic Acids Res. 2004;32:e162. doi: 10.1093/nar/gnh160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kodumal SJ, Patel KG, Reid R, Menzella HG, Welch M, Santi DV. Total synthesis of long DNA sequences: synthesis of a contiguous 32 kb polyketide synthase gene cluster. Proc. Natl Acad. Sci. USA. 2004;101:15573–15578. doi: 10.1073/pnas.0406911101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tian J, Gong H, Sheng N, Zhou X, Gulari E, Gao X, Church G. Accurate multiplex gene synthesis from programmable DNA microchips. Nature. 2004;432:1050–1054. doi: 10.1038/nature03151. [DOI] [PubMed] [Google Scholar]
- 5.Linshiz G, Yehezkel TB, Kaplan S, Gronau I, Ravid S, Adar R, Shapiro E. Recursive construction of perfect DNA molecules from imperfect oligonucleotides. Mol. Syst. Biol. 2008;4:191. doi: 10.1038/msb.2008.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xiong AS, Yao QH, Peng RH, Duan H, Li X, Fan HQ, Cheng ZM, Li Y. PCR-based accurate synthesis of long DNA sequences. Nat. Protoc. 2006;1:791–797. doi: 10.1038/nprot.2006.103. [DOI] [PubMed] [Google Scholar]
- 7.Xiong AS, Yao QH, Peng RH, Li X, Fan HQ, Cheng ZM, Li Y. A simple, rapid, high-fidelity and cost-effective PCR-based two-step DNA synthesis method for long gene sequences. Nucleic Acids Res. 2004;32:e98. doi: 10.1093/nar/gnh094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Saiki RK, Gelfand DH, Stoffel S, Scharf SJ, Higuchi R, Horn GT, Mullis KB, Erlich HA. Primer-directed enzymatic amplification of DNA with a thermostable DNA polymerase. Science. 1988;239:487–491. doi: 10.1126/science.2448875. [DOI] [PubMed] [Google Scholar]
- 9.Ohuchi S, Nakano H, Yamane T. In vitro method for the generation of protein libraries using PCR amplification of a single DNA molecule and coupled transcription/translation. Nucleic Acids Res. 1998;26:4339–4346. doi: 10.1093/nar/26.19.4339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nakano M, Komatsu J, Kurita H, Yasuda H, Katsura S, Mizuno A. Adaptor polymerase chain reaction for single molecule amplification. J. Biosci. Bioeng. 2005;100:216–218. doi: 10.1263/jbb.100.216. [DOI] [PubMed] [Google Scholar]
- 11.Kraytsberg Y, Khrapko K. Single-molecule PCR: an artifact-free PCR approach for the analysis of somatic mutations. Expert Rev. Mol. Diagn. 2005;5:809–815. doi: 10.1586/14737159.5.5.809. [DOI] [PubMed] [Google Scholar]
- 12.Lukyanov KA, Matz MV, Bogdanova EA, Gurskaya NG, Lukyanov SA. Molecule by molecule PCR amplification of complex DNA mixtures for direct sequencing: an approach to in vitro cloning. Nucleic Acids Res. 1996;24:2194–2195. doi: 10.1093/nar/24.11.2194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Margulies M, Egholm M, Altman WE, Attiya S, Bader JS, Bemben LA, Berka J, Braverman MS, Chen YJ, Chen Z, et al. Genome sequencing in microfabricated high-density picolitre reactors. Nature. 2005;437:376–380. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shendure J, Porreca GJ, Reppas NB, Lin X, McCutcheon JP, Rosenbaum AM, Wang MD, Zhang K, Mitra RD, Church GM. Accurate multiplex polony sequencing of an evolved bacterial genome. Science. 2005;309:1728–1732. doi: 10.1126/science.1117389. [DOI] [PubMed] [Google Scholar]
- 15.Hecker KH, Rill RL. Error analysis of chemically synthesized polynucleotides. Biotechniques. 1998;24:256–260. doi: 10.2144/98242st01. [DOI] [PubMed] [Google Scholar]
- 16.Nakano H, Kobayashi K, Ohuchi S, Sekiguchi S, Yamane T. Single-step single-molecule PCR of DNA with a homo-priming sequence using a single primer and hot-startable DNA polymerase. J. Biosci. Bioeng. 2000;90:456–458. [PubMed] [Google Scholar]
- 17.Tindall KR, Kunkel TA. Fidelity of DNA synthesis by the Thermus aquaticus DNA polymerase. Biochemistry. 1988;27:6008–6013. doi: 10.1021/bi00416a027. [DOI] [PubMed] [Google Scholar]
- 18.Cline J, Braman JC, Hogrefe HH. PCR fidelity of pfu DNA polymerase and other thermostable DNA polymerases. Nucleic Acids Res. 1996;24:3546–3551. doi: 10.1093/nar/24.18.3546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hutchison CA, III, Smith HO, Pfannkoch C, Venter JC. Cell-free cloning using phi29 DNA polymerase. Proc. Natl Acad. Sci. USA. 2005;102:17332–17336. doi: 10.1073/pnas.0508809102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Esteban JA, Salas M, Blanco L. Fidelity of phi 29 DNA polymerase. Comparison between protein-primed initiation and DNA polymerization. J. Biol. Chem. 1993;268:2719–2726. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.