Abstract
Over recent years small submicroscopic DNA copy-number variants (CNVs) have been highlighted as an important source of variation in the human genome, human phenotypic diversity and disease susceptibility. Consequently, there is a pressing need for the development of methods that allow the efficient, accurate and cheap measurement of genomic copy number polymorphisms in clinical cohorts. We have developed a simple competitive PCR based method to determine DNA copy number which uses the entire genome of a single chimpanzee as a competitor thus eliminating the requirement for competitive sequences to be synthesized for each assay. This results in the requirement for only a single reference sample for all assays and dramatically increases the potential for large numbers of loci to be analysed in multiplex. In this study we establish proof of concept by accurately detecting previously characterized mutations at the PARK2 locus and then demonstrating the potential of quantitative interspecies competitive PCR (qicPCR) to accurately genotype CNVs in association studies by analysing chromosome 22q11 deletions in a sample of previously characterized patients and normal controls.
INTRODUCTION
It is well recognized that rare large microscopic genomic abnormalities are associated with disease (1,2). Over recent years a number of landmark studies have shown that smaller submicroscopic DNA copy-number variants (CNVs) (typically segments >1 kb that are deleted, duplicated or inserted) are an important source of variation in the human genome (3). That CNVs are a common source of genetic variation in healthy individuals (4–7) implies that some result in no obvious phenotypic changes. However, as CNVs can disrupt entire genes and regulatory regions (4,6,8,9) an increasing number have been shown to make an important contribution to human phenotypic diversity and play a role in disease susceptibility (10–13). Consequently, the identification of disease-related CNVs is important both clinically and for studies aiming to identify disease-related aetiological pathways. A key step in the investigation of CNVs will involve the analysis of specific loci in disease association studies and to achieve this, CNVs will have to be accurately typed in large clinical cohorts.
Array-based methodologies (14) have allowed large numbers of CNVs to be detected and characterized. Such methods have however been developed mainly for studies aimed at accurately detecting CNVs and in order to maintain a low false positive detection rate typically set stringent inclusion criteria which can often result in an inflated false negative rate (15). Therefore, it is generally accepted that a confirmatory analysis using an independent method is required to accurately determine the frequency CNVs identified by array-based methods.
Association studies require an extremely accurate determination of the genotype at each locus for large numbers of samples. To perform association analysis on CNVs it is essential that all levels of copy number are accurately determined in each sample (0, 1, 2, 3, etc.). Estimations of DNA copy number from data generated by array-based studies often does not fall into discrete categories and instead can typically form a continuous distribution. In fact, a recent assessment of 1500 CNVs identified by array-based discovery studies revealed that the call data of only 70 were of the standard expected of genotyping assays (15). This problem is likely to be an even greater issue for smaller CNVs, which contain lower numbers of probes. It is well recognized that even relatively small error rates can dramatically reduce the power of an association study (16), therefore, inaccurate estimates of copy number are unacceptable and influence both type I and type II error. To address this, some have attempted to fit such data into the appropriate discrete categories (7,17), however, such approaches have major limitations (18) and are likely to be more amenable to some CNVs than others. Given this, a more satisfactory approach would be to develop assays that estimate DNA copy number with sufficiently high quality that artefacts introduced by experimental noise are suitably reduced. Such approaches would also facilitate the economic association analysis of CNVs in large replication samples. As a result of these issues, the development of methods that allow efficient, accurate and cheap measurement of genomic copy number polymorphisms in clinical cohorts has been recently recognized as one of the most pressing needs in CNV research (15,19).
A number of methods exist that allow accurate quantitation of DNA copy number (20–25), however, none are particularly well suited to large-scale association analysis, either because the assay requires intensive optimization or because the analysis platform on which they have been developed is not best applied to high throughput studies. Competitive PCR is an established technique that involves amplifying a test DNA sequence in the presence of a competitor sequence, which is identical apart from a single nucleotide, which allows both sequences to be distinguished. Given a known concentration of the competitor, the test sequence can be accurately quantified. Typically, the source of the competitor is a synthetic DNA sequence, however, the requirement for a competitor to be synthesized to match each test locus is often impractical for studies analysing large numbers of loci. To avoid this, others have shown that very accurate estimates of DNA copy number can be obtained by amplifying the test locus in the presence of a paralogous locus of known copy number (25). However, not all test loci will have a non-deleted/duplicated paralogous sequence, limiting the number of assays that can be analysed by this approach.
We have applied competitive PCR to determine DNA copy number by exploiting the high degree of conservation between orthologous human and chimpanzee genomic sequence. To achieve this we have co-amplified orthologous loci and targeted non-conserved nucleotides by primer extension to distinguish between template from the two species (Figure 1). Quantitative interspecies qicPCR uses the entire genome of a single chimpanzee as a competitor. This results in the requirement for only a single competitor sample for all assays and dramatically increasing the potential of large numbers of loci to be analysed in multiplex. As the rate of fixed (non-polymorphic) interspecies sequence divergence is ∼1% (26) then >3 × 107 single nucleotides could potentially be targeted. Therefore, qicPCR can potentially be used to analyse the majority of CNVs. These factors, together with accuracy, low cost and potential application to array platforms make it potentially amenable to studies performing multiplex high throughput association analysis of CNVs. Here, we establish proof of concept by first accurately detecting previously characterized mutations at the PARK2 locus and then demonstrating the potential of qicPCR to analyse CNVs in association studies by genotyping chromosome 22q11 deletions in a sample of previously characterized patients and normal controls.
Figure 1.
Schematic representation of the principle of quantitative interspecies competitive PCR. Human and P. troglodytes specific alleles are represented by alleles ‘C’ and ‘G’, respectively.
METHODS
Samples
All subjects were unrelated and provided written informed consent to participate in genetic studies.
Parkinsons disease (PD) samples
Our initial analysis was based on two patients diagnosed with PD who had been previously characterized as carrying PARK2 mutations; patient PD-patient1 was hemizygously deleted at exon 3 while patient PD-patient2 carried a heterozygous duplication of exon 6. All mutations were molecularly confirmed by MLPA.
22q11DS samples
We analysed 20 unrelated individuals who carried hemizygous deletions at 22q11 as determined by fluorescence in situ hybridization using the N25 probe (Oncor Inc., Gaithersburg, MD, USA).
Control samples
All 733 unrelated individuals had been collected for use as controls, details of which have been previously described (27). The 10 samples used as controls against the PD patients had previously not been screened for the PARK2 mutations, however, given the rarity of such pathogenic mutations it is unlikely that they were carriers. All samples had however been previously excluded for the presence of typical 22q11 deletions region (28).
Chimpanzee
Pan troglodytes DNA was obtained from the cell line EB176 (JC) deposited at ECACC (http://www.ecacc.org.uk).
Molecular assays
MLPA assay
Samples were analysed using the SALSA P051/P052 Parkinson MLPA probe kit (MRC Holland, Amsterdam, The Netherlands) following manufacturer's instruction. Each kit contains a probe for exons 1 to 12 of the PARK2 gene. All steps were performed on the same MJ thermocycler. Briefly, 100 ng of DNA was denatured at 98°C and hybridized to the probes by incubation (16–17 h, 60°C) with 1.5 μl SALSA probemix and 1.5 μl of MLPA buffer. The ligation reaction was carried out by incubating the 8 μl of hybridized product and 32 μl of Ligase-65 mix for 15 min at 54°C, followed by 5 min at 98°C. The PCR was performed in a 50 μl reaction using 10 μl of ligation product, 4 μl of 10 × SALSA PCR buffer and 10 μl of Polymerase mix. The PCR cycling conditions were 60°C hotstart, followed by 35 cycles of 95°C for 30 s, 60°C for 30 s and 72°C for 60 s, followed by 72°C for 20 min. The PCR product was analysed on an ABI 3100 Sequencer (Applied Biosystems, Foster City, CA, USA) with a GeneScan500 Rox internal size standard.
qicPCR assays
PCR
PCR was performed on MJ thermocyclers in a 5 μl reaction using approximately 12 ng of dried-down genomic human DNA (hsDNA), 12 ng of P. troglodytes DNA (ptDNA), 0.5 pmol of each primer in multiplex, 250 µM dNTPs, 0.325 μl of 10 × 20 mM MgCl PCR Buffer and 0.1 units of Hotstar Taq polymerase (Qiagen, Valencia, CA, USA). The PCR cycling parameters were 95°C for 15 min, followed by 45 cycles of 94°C for 20 s, 56°C for 30 s and 72°C for 1 min, followed by 72°C for 3 min.
Primer extension
Primer extension was performed on MJ thermocyclers in 9 μl reactions using the 7 μl SAP-treated PCR product, 6.6–13.3 pmol of each primer in multiplex, 0.25 μl of iplex buffer, 0.25 μl of iplex termination mix and 0.05 μl of iplex enzyme (Sequenom, San Diego, CA, USA). The cycling parameters were 94°C for 30 s, followed by 40 cycles of 94°C for 5 s and a nested 5 cycles of 52°C for 5 s and 80°C for 5 s, followed by 72°C for 3 min.
qicPCR assay selection
Interspecies sequence differences were identified by aligning the respective genomic DNA sequences of human (homo sapiens) and chimpanzee (P. troglodytes) using UCSC Blat (http://genome.ucsc.edu/). Nucleotides that differed between human and chimp were identified and with reference to the UCSC genome browser human March 2006 build and the chimpanzee genome database at the Broad Institute (http://www.broad.mit.edu/ftp/pub/assemblies/mammals/chimp_SNPs/), known human and chimpanzee SNPs were excluded. Single nucleotide extension assays were then designed to target the non-conserved nucleotides using Sequenom Assay Design v3.1 software.
To study the mutations in the PD patients the human reference sequence at the PARK2 locus (NCBI build 36.1, chr6:161,689,662-162,784,495) with its orthologous sequence in P. troglodytes (genome build 2 version 1, chr6:164,335,097-165,462,200). To study the deletion at 22q11 we aligned the human genome NCBI build 36.1, chr22:18131905-18,222,087 with its orthologous region in P. troglodytes (genome build 2 version 1, chr22:18221742-18316567). Primer extension assays (Sequenom iplex™, San Diego, CA, USA) were then designed to target nucleotides that were not conserved between the two species at the PARK2 locus (20 assays) and at 22q11 (10 assays) (Supplementary Table 1). Internal control assays were designed by identifying nucleotides that were not conserved between human and chimpanzee at a number of autosomal regions that had showed no previous evidence for harbouring common CNVs (Supplementary Table 2). qicPCR assays targeting the PARK2 locus were designed as two independent multiplex reactions each containing 9 and 11 independent test assays as well as a single reference assay. qicPCR assays targeting 22q11 were designed as two independent multiplex reactions each containing five 22q11 test assays and five reference assays.
Standard curve
The genomic DNA from a single human reference sample was quantified to 32 ng/ul by pico green analysis and then serially diluted to 16, 8, 4, 2 and 1 ng/ul. For each titrate, PCR followed by primer extension was then performed using 3 ul of hsDNA and 3 ul of ptDNA (4 ng/ul) as per the manufacturer's instructions with the iplex chemistry. The peak areas of the extended primers specific to the respective human and chimpanzee alleles were determined following analysis on a Sequenom MassARRAY™ system. The amount of hsDNA relative to that of ptDNA at each locus (hsDNArelative) was then estimated by
A reference standard curve was generated by plotting hsDNArelative (x-axis) against the log of known hsDNA concentration (y-axis) and from this the slope and intercept was estimated from the equation of the straight line, y = mx + c. This simple and rapid procedure was performed for each primer extension assay.
qicPCR analysis
For each test sample, the assay was performed using equal concentrations of genomic hsDNA and ptDNA (3 ul of each at ∼4 ng/ul) as described earlier. The area under peaks corresponding to extended primer peaks of the respective human and chimpanzee nucleotides (Figure 1) was then determined and the data were used to estimate hsDNArelative for each primer extension assay. With reference to the respective standard curve, the quantity of hsDNA present in each primer extension assay (hsDNAquant) was then estimated by
where m and c were derived from the reference standard curve.
The estimated hsDNA copy number at the test locus was determined as the ratio of hsDNAquant determined at the test and internal control loci, respectively: a ratio of 1:1 being expected in the absence of a CNV, 0.5 for a heterozygous deletion, 1.5 for a heterozygous duplication. Dividing each ratio by 0.5 provided an estimate of the human DNA copy number at the test locus (hsCN). When multiple test assays from the same locus were used we first determined the average hsDNAquant at the test locus and then estimated the average hsDNA copy number by calculating the ratio to the hsDNAquant determined at each independent reference assay.
Silhouette scores
Silhouette scores are a graphical aid for interpretation and validation of data clusters that provides a measure of how well a data point was classified when it was assigned to a cluster (29). In brief, they are determined by comparing the distance between each data point within a cluster to the distance between each data point in any other cluster. The overall average silhouette score was calculated for each qicPCR assay using the software ClusterA (29).
RESULTS AND DISCUSSION
Evaluation of qicPCR
To initially evaluate qicPCR we analysed a total of 11 probes designed to assay exons at the PARK2 locus and compared the results to data generated by the established technique of MPLA (23). The standard curves of each qicPCR assay being presented in Supplementary Table 3. All probes were analysed in 10 healthy controls as well as two PD individuals who had previously been characterized to carry PARK2 mutations. Analysis of the human DNA copy number (hsCN) determined for the assays targeting PARK2 exons 3 and 6 established that qicPCR was capable of detecting both the heterozygous deletion at exon 3 (mean hsCN = 1.08) and the heterozygous duplication at exon 6 (mean hsCN = 2.88) in the respective PD samples (Table 1). Neither assay detected any evidence for a change in copy number in any of the 10 control samples [exon 3a: mean hsCN = 1.92 (1.8–2.0); exon 3b: mean hsCN = 1.76 (1.66–1.88); exon 6: mean hsCN = 2.16 (2.04–2.26)], Table 1. Direct comparison of the hsCN estimates of qicPCR to those generated by MPLA showed that the results of both methods were highly correlated, r = 0.82, P = 0.005 (Table 1 and Figure 2). No evidence for change in copy number was observed in either the PD patients or the 10 control samples at the qicPCR assays that were not targeting PARK2 exons 3 or 6 (mean hsCN = 1.98, 95% CI 1.9–2.06), Table 1.
Table 1.
Copy number estimates at the PARK2 locus
| ID | Test | PARK2 assays |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Exon 3a | Exon 3b | Exon 4b | Exon 5a | Exon 5b | Exon 6a | Exon 8a | Exon 8b | Exon 9b | Exon 10a | Exon 11a | ||
| 05-180 | MPLA | 1.06 | 1.06 | 2.59 | 2.17 | 2.17 | 1.88 | 2.14 | 2.14 | 2.12 | 2.15 | 2.27 |
| qicPCR | 1.10 | 1.07 | 2.07 | 1.94 | 2.06 | 1.91 | 2 13 | 2.02 | 1.93 | 2.08 | 1.96 | |
| 04-115 | MPLA | 1.96 | 1.96 | 1.95 | 1.71 | 1.71 | 2.80 | 1.86 | 1.86 | 1.68 | 1.86 | 1.43 |
| qicPCR | 2.32 | 1.84 | 1.99 | 1.91 | 1.96 | 2.89 | 2.14 | 1.79 | 2.01 | 2.04 | 1.88 | |
| Control 1 | qicPCR | 2.05 | 1.96 | 1.77 | 1.85 | 2 24 | 2.19 | 1.92 | 1.94 | 1.93 | 2.07 | 1.98 |
| Control 2 | qicPCR | 1.94 | 1.83 | 2.00 | 2.01 | 2.24 | 2.31 | 2.00 | 2.03 | 2.01 | 2.23 | 2.05 |
| Control 3 | qicPCR | 1.63 | 1.60 | 2.28 | 1.65 | 1.99 | 2 27 | 1.97 | 2.08 | 1.95 | 1.76 | 1.93 |
| Control 4 | qicPCR | 1.91 | 1.63 | 2.14 | 1.88 | 2.00 | 2.01 | 1.90 | 1.82 | 1.63 | 2.05 | 1.84 |
| Control 5 | qicPCR | 1.94 | 1.87 | 2 14 | 2.08 | 1.94 | 2 22 | 1.81 | 1.74 | 2.16 | 1.85 | 2.06 |
| Control 6 | qicPCR | 2.10 | 2.01 | 2.17 | 1.89 | 1.91 | 2.35 | 2.16 | 2.12 | 2.04 | 2.02 | 2.07 |
| Control 7 | qicPCR | 1.71 | 1.66 | 2.05 | 1.63 | 2.15 | 1.94 | 1.68 | 2.02 | 1.87 | 1.70 | 2.09 |
| Control 8 | qicPCR | 1.91 | 1.62 | 1.90 | 1.93 | 2.06 | 1.98 | 1.91 | 2.17 | 1.94 | 1.98 | 2.11 |
| Control 9 | qicPCR | 1.99 | 1.73 | 2.11 | 1.94 | 1.96 | 2.19 | 1.88 | 2.19 | 1.78 | 2.08 | 1.77 |
| Control 10 | qicPCR | 1.94 | 1.66 | 2.09 | 1.79 | 2 12 | 2.04 | 1.87 | 2.01 | 1.87 | 1.88 | 2.15 |
| All controls | Mean | 1.91 | 1.76 | 2.06 | 1.86 | 2.06 | 2.15 | 1.91 | 2.01 | 1.92 | 1.96 | 2.01 |
| SD | 0.14 | 0.15 | 0.14 | 0.14 | 0.12 | 0.15 | 0.12 | 0.14 | 0.15 | 0.16 | 0.12 | |
| CV | 0.07 | 0.09 | 0.08 | 0.08 | 0.06 | 0.07 | 0.06 | 0.07 | 0.08 | 0.08 | 0.06 | |
Copy number at PARK2 exons was determined by MLPA and qicPCR in two patients containing previously characterized mutations (highlighted in grey) and 10 healthy controls. Mean, SD and coefficient of variance were calculated for each qicPCR assay in the 10 control samples only.
Figure 2.
Comparison of the copy number at the PARK2 locus as determined by MPLA and qicPCR. Copy number estimates of PARK2 exons 3, 4, 5, 6, 8, 9, 10 and 11 as determined by qicPCR and MLPA in two patients with previously characterized PARK2 mutations. The assays of exons 3 (A and B) and exon 6, which detect the deletion and duplication in our test samples PD-patient1 and PD-patient2, respectively, are highlighted.
A total of four replicate experiments were performed for each sample. Comparison of the average hsCN determined at each technical replicate indicated that the results of qicPCR were reproducible (coefficient of variance for samples with a gain in copy, a loss in copy and also those showing no evidence of variation being 0.035, 0.09 and 0.01, respectively).
Three PARK2 exons were each assayed using two independent probes. Comparison of the hsCN determined for the 10 control samples revealed that the results for each pair of probes were consistent: exon 3 [exon 3a: mean hsCN = 0.96 (95% CI 0.90–1.01); exon 3b: mean hsCN = 0.88 (95% CI 0.83–0.94); CV = 0.06], exon 5 [exon 5a: mean hsCN = 0.93 (95% CI 0.88–0.98); exon 5b: mean hsCN = 1.03 (95% CI 0.99–1.07); CV = 0.07] and exon 8 [exon 8a: mean hsCN = 0.96 (95% CI 0.91–1.0); exon 8b: mean hsCN = 1.01 (95% CI 0.95–1.06); CV = 0.04], Table 1.
Evaluation of qicPCR in large samples
To be applicable to large-scale association analysis of a given CNV, qicPCR should allow hsCN at the locus to be automatically called in >95% of samples with an error rate <0.1%, which are the typical standards of current high throughput SNP genotyping platforms (30). It is therefore essential that the hsCN estimates at each locus are sufficiently consistent over large numbers of samples to allow the clear distinction of those with different levels of copy number.
We used two criteria to assess the quality of the data generated by qicPCR. First, analysis of the coefficient of variance of the hsCN estimated for all samples within each category of copy number known to be present in our dataset (normal, single copy loss) allowed a specific assessment of the concordance of the data within each group. Second, we used silhouette scores (29) to assess the quality with which the hsDNA estimated by qicPCR formed distinct clusters. This is effectively a measure of the accuracy with which data points can be blindly assigned to different groups based on the relative location of all other data points. Silhouette scores range from −1 to + 1 with assays >0.65 generally being considered high quality, having minimal distance between the data points within each group and large distances between groups. We empirically determined the CV expected of clusters of data points in a high quality genotyping assay by calculating the CV for the data generated by 6 independent SNP genotyping assays (iplex, Sequenom) that we had previously scored as being high quality (>95% of samples called with an error rate <0.1%). All had silhouette scores >0.65 and the CV for all clusters of data points were <0.1 [mean = 0.05 (0.04min−0.07max)]. Based on this we set our criteria for genotyping quality to be silhouette score >0.65 and CV <0.1.
To assess whether qicPCR was sufficiently robust to meet the criteria expected of a genotyping assay we analysed two independent multiplex panels each consisting of five independent test assays designed to detect copy number variation at chromosome 22q11 (qicPCR22q11a and qicPCR22q11b). Both multiplex assays were initially analysed in 10 individuals previously diagnosed with 22q11DS and 10 healthy controls.
In comparison to the preliminary data generated at the PARK2 locus, analysing a larger number of samples revealed that while determining the hsDNA copy number of each 22q11 test probe with comparison to a single reference probe allowed the 22q11DS samples [mean hsCN = 1.10 (0.58–1.84)] to be generally differentiated from the non-deleted control subjects [mean hsCN = 2.14 (1.16–3.62)], the quality of the data fell short of that expected for a genotyping assay [mean silhouette score = 0.16 (−0.03 to –0.32); CV of hsCN for 22q11DS samples and non-deleted controls = 0.25 and 0.28, respectively].
We reasoned that the measurement error in the hsCN determined by qicPCR could be reduced by; (1) averaging multiple independent unlinked reference loci and/or (2) averaging multiple independent test probes targeting the same test locus. We therefore set out to systematically assess the variability associated with these parameters. Estimating hsCN using a single test probe with reference to an increasing number of independent control assays (1 to 5) resulted in tighter confidence intervals for the average estimated hsCN for both 22q11DS and non-deleted samples (Figure 3A). However, the modest reduction in the average CV (22q11DS 0.25–0.19; controls 0.28–0.21) and a minor increase in the silhouette scores [0.16 (−0.03min to 0.32max) to 0.32 (0.26min–0.36max)] (Figure 3A), did not achieve the required performance.
Figure 3.
(A) Effect of increasing the number of reference samples to estimate copy number by qicPCR. (B) Effect of increasing the number of test assays to estimate copy number by qicPCR. Blue vertical bars represent the range of hsCN determined by qicPCR for 22q11DS patients and controls while histograms represent the CV. The ranges of silhouette scores determined at each parameter are displayed as black vertical bars. For each measure the mean is indicated as a small horizontal bar.
To assess the impact of increasing the number of test assays we considered all 22q11 test probes to be independent assays targeting a single CNV (the 22q11 locus). hsDNAabsolute values were then calculated for each sample using a single 22q11 test probe and also from the average of 2, 3, 4 and 5 independent 22q11 test probes. Estimates of hsCN were then determined with reference to a single control assay amplified within the same multiplex panel. This again revealed that increasing the number of independent test assays from one to five resulted in tighter confidence intervals for the hscopy number for both 22q11DS and non-deleted samples (Figure 3B), but that the minor reduction in the CV (22q11DS 0.25–0.19; controls 0.28–0.21) and minor improvement in the silhouette scores [0.16 (−0.03min to 0.32max) to 0.40 (−0.02min to 0.65max)] did not the meet the criteria expected for a genotyping assay (Figure 3B).
We next assessed the effect of increasing both the number of independent test probes and also the number of independent control probes within the same multiplex reaction. Calculating the hsDNAquant for the locus as an average of at least 4 independent test assays and then determining the mean hsCN for the locus by comparing to at least 4 reference assays resulted in a considerable improvement in data quality (Figure 4) where the low CV <0.09 and high silhouette scores (>0.77) met our predefined criteria for a genotyping assay. We performed an identical assessment of qicPCR to detect changes in copy number at 22q11 using an entirely independent multiplex panel of 22q11 test assays (qicPCR22q11b). Analysis in the same set of samples revealed that the estimates of hsCN at 22q11 by each multiplex panel of markers were similar (Figure 5) and that both CV and silhouette scores met our predefined criteria when at least 4 independent test and reference assays were used (data not presented).
Figure 4.
Effect of increasing both the number of reference and test assays to estimate copy number by qicPCR. hsCN was determined at 22q11 for all combinations of 1 to 5 test and reference assays in 10 patients with 22q11DS and 10 non-deleted controls. For simplicity the CV of only the 22q11DS samples is presented, however, an analogous pattern was seen in the data for the non-deleted controls.
Figure 5.
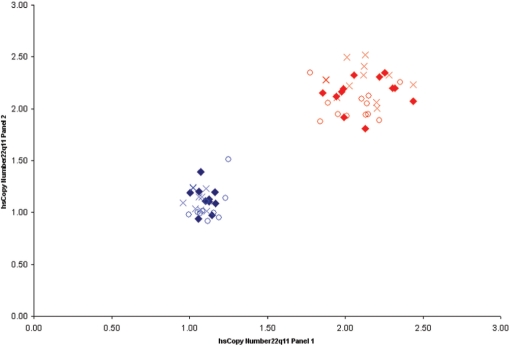
Reproducibility of multiplex qicPCR assays. hsCN was determined at 22q11 using two independent multiplex qicPCR panels in ten 22q11DS patients (blue) and 10 non-deleted controls (red). Each panel was analysed in triplicate with replicates 1, 2 and 3 for each qicPCR panel being represented as closed diamonds, crosses and circles, respectively.
Given our experience that the performance of some assays can be less robust when scaled up, we analysed a larger series of 753 samples, composed of 733 non-deleted controls and 20 new 22q11DS samples independent of those in which the assay was optimized, dispersed among the control samples. For each sample the hsCN at the 22q11 locus was determined as an average of five locus-specific test probes and five reference probes all located within the same multiplex reaction. All samples were analysed in duplicate and similar results were obtained from both experimental replicates (Figure 6). All 22q11DS samples could be clearly distinguished from the 20 control samples (silhouette score = 0.81) (Figure 6). The mean hsCN estimate of the 22q11DS samples was 0.96 (0.46min−1.48max), mean CV = 0.20 (0.14min−0.23max), while that of the non-deleted controls was 2.07 (1.68min−2.82max), mean CV = 0.06 (0.05min−0.07max). High-throughput genotyping assays are expected to be sufficiently robust to analyse large numbers of samples (drop-out rate <5%) while maintaining very low genotype error rates (<0.1%). Blind genotyping of our samples using a simple Excel (Microsoft) spreadsheet, which adhered to a simple semi-automated protocol whereby samples were called as carrying either 1 or 2 copy numbers if the hsCN determined by qicPCR fell within ± 40% of the expected integer (1 copy = 0.6–1.4, 2 copies = 1.6–2.4). Estimates of hsCN that fell outside these ranges were classed as genotype failures. Analysis of each single pass experiment resulted in >97% of samples being genotyped with 100% accuracy (a total of 46/1506 samples failed to genotype). Moreover, determining the hsCN as the average of the two experimental replicates of each multiplex panel further improved the genotyping quality, with >99.4% of samples genotyped with 100% accuracy (a total of 5/753 samples analysed in duplicate failed to be genotyped), suggesting that further improvements in the quality of quantitative genotyping of CNVs by qicPCR are possible simply by performing one replicate experiment.
Figure 6.
Estimated copy number at 22q11 in 753 samples. Replicate experiments of a single multiplex qicPCR panel to determine hsCN at 22q11 in twenty 22q11DS patients (blue) and 733 controls (red).
The accuracy of qicPCR is highly dependent on the reference locus itself not varying in copy number. While it is clearly impossible to absolutely eliminate rare CNVs at any given reference locus, the risk could clearly be reduced by selecting reference loci from genomic regions that have previously shown no evidence of common CNVs. Moreover, this possibility could be further reduced by designing each multiplex reaction to include multiple reference loci selected from different genomic regions, allowing rare CNVs present at just one control region to be detected and excluded from the analysis.
Another potential problem is the presence of SNPs at the primer-binding sites. Most SNPs are specific to either human or chimpanzee (31). Human-specific SNPs (hsSNPs) located at the primer-binding sites of either the test or reference locus would result in polymorphic mismatches which could lead to variability in the estimates of copy number by qicPCR due to allele specific ‘drop out’. Consequently, hsSNPs located under the primers of the test and reference assays would lead to an apparent reduction and increase of the estimated copy number at the test locus, respectively. Both test and reference assays should be excluded for the presence of hsSNPs, the unintentional selection, which at either the target or reference locus would likely result in a highly unstable assay. Given that the qicPCR assay estimates the hscopy number with reference to only a single chimpanzee genome the presence of chimpanzee-specific SNPs (ptSNPs) are likely to have minimal effect on the accuracy of a qicPCR assay, either when they have been inadvertently selected as assay targets or as when they result in mismatches under the primer sequences. However, qicPCR assays that inadvertently target ptSNPs will fail if the genotype of the reference chimpanzee DNA sample is homozygous for the same nucleotide conserved in human. Ideally ptSNPs should therefore be avoided, however as the reference chimpanzee genome sequence was primarily derived from a single donor (Clint) then this task could be aided by utilizing the DNA from ‘Clint’ in qicPCR assays.
Single nucleotide substitutions have been estimated to occur at an average rate of 1.23% between the human and chimpanzee genome, with ∼1.06% corresponding to fixed divergence between the two species (26). Non-conserved nucleotides are, however, not consistently distributed throughout the human genome with CpG islands, and distal regions showing increased rates of divergence (26). Given this, analyses of the human genome dissected into 1-Mb segments indicate that the rate of divergence with chimpanzee varies from 0.006 to 0.022 (26), implying that even in the most conserved regions of the human genome we could expect non-conserved nucleotides to occur on average once every ∼200 bp. Therefore, as the most conservative estimate suggests that non-conserved nucleotides are at least as common as SNPs it is likely that qicPCR assays could be designed to target a large majority of the human genome. Clearly, it would be impossible to analyse human sequences that do not have a chimpanzee homologue by qicPCR, but, comparative analysis of the human and chimpanzee genomic sequence indicate that this number is very small, with only 53 genes (∼0.2%) being identified as being deleted in either species (26).
The results of large-scale studies aimed at genotyping CNVs by array methods can be ambiguous, and typically require validation by an independent assay. As qicPCR assays target diverged nucleotides then they can act as an independent assay to validate CNVs detected by more conventional methods (SNPs, CGH). Moreover, given the high frequency of fixed non-conserved nucleotides it will be potentially feasible to design qicPCR assays to analyse small CNVs (<2 kb) which are becoming increasingly the target of CNV studies.
In this study we have shown that qicPCR can accurately distinguish normal copy number (2 copies) from a single copy deletion (1 copy) and a single copy gain (3 copies). A number of reports of disease association have involved more complex CNVs, typically having ≥4 DNA copies (32,12). While we have not analysed assays of this nature in this study, qicPCR using the paralogue ratio test (25) has been previously demonstrated to have sufficient sensitivity to genotype complex CNVs (25,32). Given this and the sensitivity of the data reported in this study, it is possible that qicPCR will be sufficiently sensitive to allow the genotyping of more complex copy number polymorphisms, however further analysis will be required to determine this. All assays performed in this study were performed using the Sequenom Massarray genotyping platform which is best suited to analysing multiplex panels of 30–40 assays and consequently limited to analysing ∼6 CNVs simultaneously. Given that qicPCR can potentially be performed using any SNP genotyping assay that generates a quantitative readout of the signal intensity for each allele, it can be applied to currently available array-based platforms and therefore offers a potential that could be used in very large-scale CNV genotyping studies.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENT
Collection of the controls, 22q11DS and PD samples was funded by the MRC, Wellcome Trust and Parkinson's Disease Society UK, respectively. Funding to pay the Open Access publication charges for this article was provided by the Department of Psychological Medicine, Cardiff University.
Conflict of interest statement. None declared.
REFERENCES
- 1.Chance PF, Alderson MK, Leppig KA, Lensch MW, Matsunami N, Smith B, Swanson PD, Odelberg SJ, Disteche CM, Bird TD. DNA deletion associated with hereditary neuropathy with liability to pressure palsies. Cell. 1993;72:143–151. doi: 10.1016/0092-8674(93)90058-x. [DOI] [PubMed] [Google Scholar]
- 2.Driscoll DA, Spinner NB, Budarf ML, McDonald-McGinn DM, Zackai EH, Goldberg RB, Shprintzen RJ, Saal HM, Zonana J, Jones MC, et al. Deletions and microdeletions of 22q11.2 in velo-cardio-facial syndrome. Am. J. Med. Genet. 1992;44:261–268. doi: 10.1002/ajmg.1320440237. [DOI] [PubMed] [Google Scholar]
- 3.Feuk L, Carson AR, Scherer SW. Structural variation in the human genome. Nat. Rev. Genet. 2006;7:85–97. doi: 10.1038/nrg1767. [DOI] [PubMed] [Google Scholar]
- 4.McCarroll SA, Hadnott TN, Perry GH, Sabeti PC, Zody MC, Barrett JC, Dallaire S, Gabriel SB, Lee C, Daly MJ, et al. Common deletion polymorphisms in the human genome. Nat. Genet. 2006;38:86–92. doi: 10.1038/ng1696. [DOI] [PubMed] [Google Scholar]
- 5.Conrad DF, Andrews TD, Carter NP, Hurles ME, Pritchard JK. A high-resolution survey of deletion polymorphism in the human genome. Nat. Genet. 2006;38:75–81. doi: 10.1038/ng1697. [DOI] [PubMed] [Google Scholar]
- 6.Hinds DA, Kloek AP, Jen M, Chen X, Frazer KA. Common deletions and SNPs are in linkage disequilibrium in the human genome. Nat. Genet. 2006;38:82–85. doi: 10.1038/ng1695. [DOI] [PubMed] [Google Scholar]
- 7.Redon R, Ishikawa S, Fitch KR, Feuk L, Perry GH, Andrews TD, Fiegler H, Shapero MH, Carson AR, Chen W, et al. Global variation in copy number in the human genome. Nature. 2006;444:444–454. doi: 10.1038/nature05329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sebat J, Lakshmi B, Troge J, Alexander J, Young J, Lundin P, Maner S, Massa H, Walker M, Chi M, et al. Large-scale copy number polymorphism in the human genome. Science. 2004;305:525–528. doi: 10.1126/science.1098918. [DOI] [PubMed] [Google Scholar]
- 9.Sharp AJ, Locke DP, McGrath SD, Cheng Z, Bailey JA, Vallente RU, Pertz LM, Clark RA, Schwartz S, Segraves R, et al. Segmental duplications and copy-number variation in the human genome. Am. J. Hum. Genet. 2005;77:78–88. doi: 10.1086/431652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sebat J, Lakshmi B, Malhotra D, Troge J, Lese-Martin C, Walsh T, Yamrom B, Yoon S, Krasnitz A, Kendall J, et al. Strong association of de novo copy number mutations with autism. Science. 2007;316:445–449. doi: 10.1126/science.1138659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lucito R, Suresh S, Walter K, Pandey A, Lakshmi B, Krasnitz A, Sebat J, Wigler M, Klein AP, Brune K, et al. Copy-number variants in patients with a strong family history of pancreatic cancer. Cancer Biol. Ther. 2007;6:1592–1599. doi: 10.4161/cbt.6.10.4725. [DOI] [PubMed] [Google Scholar]
- 12.Aitman TJ, Dong R, Vyse TJ, Norsworthy PJ, Johnson MD, Smith J, Mangion J, Roberton-Lowe C, Marshall AJ, Petretto E, et al. Copy number polymorphism in Fcgr3 predisposes to glomerulonephritis in rats and humans. Nature. 2006;439:851–855. doi: 10.1038/nature04489. [DOI] [PubMed] [Google Scholar]
- 13.Gonzalez E, Kulkarni H, Bolivar H, Mangano A, Sanchez R, Catano G, Nibbs RJ, Freedman BI, Quinones MP, Bamshad MJ, et al. The influence of CCL3L1 gene-containing segmental duplications on HIV-1/AIDS susceptibility. Science. 2005;307:1434–1440. doi: 10.1126/science.1101160. [DOI] [PubMed] [Google Scholar]
- 14.Carter NP. Methods and strategies for analyzing copy number variation using DNA microarrays. Nat. Genet. 2007;39:S16–21. doi: 10.1038/ng2028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McCarroll SA, Altshuler DM. Copy-number variation and association studies of human disease. Nat. Genet. 2007;39:S37–42. doi: 10.1038/ng2080. [DOI] [PubMed] [Google Scholar]
- 16.Moskvina V, Craddock N, Holmans P, Owen MJ, O'Donovan MC. Effects of differential genotyping error rate on the type I error probability of case-control studies. Hum. Hered. 2006;61:55–64. doi: 10.1159/000092553. [DOI] [PubMed] [Google Scholar]
- 17.Stranger BE, Forrest MS, Dunning M, Ingle CE, Beazley C, Thorne N, Redon R, Bird CP, de Grassi A, Lee C, et al. Relative impact of nucleotide and copy number variation on gene expression phenotypes. Science. 2007;315:848–853. doi: 10.1126/science.1136678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McCarroll SA. Copy-number analysis goes more than skin deep. Nat. Genet. 2008;40:5–6. doi: 10.1038/ng0108-5. [DOI] [PubMed] [Google Scholar]
- 19.Todd JA. Statistical false positive or true disease pathway? Nat. Genet. 2006;38:731–733. doi: 10.1038/ng0706-731. [DOI] [PubMed] [Google Scholar]
- 20.Heid CA, Stevens J, Livak KJ, Williams PM. Real time quantitative PCR. Genome Res. 1996;6:986–994. doi: 10.1101/gr.6.10.986. [DOI] [PubMed] [Google Scholar]
- 21.Charbonnier F, Raux G, Wang Q, Drouot N, Cordier F, Limacher JM, Saurin JC, Puisieux A, Olschwang S, Frebourg T. Detection of exon deletions and duplications of the mismatch repair genes in hereditary nonpolyposis colorectal cancer families using multiplex polymerase chain reaction of short fluorescent fragments. Cancer Res. 2000;60:2760–2763. [PubMed] [Google Scholar]
- 22.Armour JA, Sismani C, Patsalis PC, Cross G. Measurement of locus copy number by hybridisation with amplifiable probes. Nucleic Acids Res. 2000;28:605–609. doi: 10.1093/nar/28.2.605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schouten JP, McElgunn CJ, Waaijer R, Zwijnenburg D, Diepvens F, Pals G. Relative quantification of 40 nucleic acid sequences by multiplex ligation-dependent probe amplification. Nucleic Acids Res. 2002;30:e57. doi: 10.1093/nar/gnf056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Suls A, Claeys KG, Goossens D, Harding B, Van Luijk R, Scheers S, Deprez L, Audenaert D, Van Dyck T, Beeckmans S, et al. Microdeletions involving the SCN1A gene may be common in SCN1A-mutation-negative SMEI patients. Hum. Mutat. 2006;27:914–920. doi: 10.1002/humu.20350. [DOI] [PubMed] [Google Scholar]
- 25.Armour JA, Palla R, Zeeuwen PL, den Heijer M, Schalkwijk J, Hollox EJ. Accurate, high-throughput typing of copy number variation using paralogue ratios from dispersed repeats. Nucleic Acids Res. 2007;35:e19. doi: 10.1093/nar/gkl1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Consortium TCSaA. Initial sequence of the chimpanzee genome and comparison with the human genome. Nature. 2005;437:69–87. doi: 10.1038/nature04072. [DOI] [PubMed] [Google Scholar]
- 27.Williams NM, Preece A, Morris DW, Spurlock G, Bray NJ, Stephens M, Norton N, Williams H, Clement M, Dwyer S, et al. Identification in 2 independent samples of a novel schizophrenia risk haplotype of the dystrobrevin binding protein gene (DTNBP1) Arch. Gen. Psychiatry. 2004;61:336–344. doi: 10.1001/archpsyc.61.4.336. [DOI] [PubMed] [Google Scholar]
- 28.Ivanov D, Kirov G, Norton N, Williams HJ, Williams NM, Nikolov I, Tzwetkova R, Stambolova SM, Murphy KC, Toncheva D, et al. Chromosome 22q11 deletions, velo-cardio-facial syndrome and early-onset psychosis. Molecular genetic study. Br. J. Psychiatry. 2003;183:409–413. doi: 10.1192/bjp.183.5.409. [DOI] [PubMed] [Google Scholar]
- 29.Lovmar L, Ahlford A, Jonsson M, Syvanen AC. Silhouette scores for assessment of SNP genotype clusters. BMC Genomics. 2005;6:35. doi: 10.1186/1471-2164-6-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sawcer SJ, Maranian M, Singlehurst S, Yeo T, Compston A, Daly MJ, De Jager PL, Gabriel S, Hafler DA, Ivinson AJ, et al. Enhancing linkage analysis of complex disorders: an evaluation of high-density genotyping. Hum. Mol. Genet. 2004;13:1943–1949. doi: 10.1093/hmg/ddh202. [DOI] [PubMed] [Google Scholar]
- 31.Kehrer-Sawatzki H, Cooper DN. Understanding the recent evolution of the human genome: insights from human-chimpanzee genome comparisons. Hum. Mutat. 2007;28:99–130. doi: 10.1002/humu.20420. [DOI] [PubMed] [Google Scholar]
- 32.Hollox EJ, Huffmeier U, Zeeuwen PL, Palla R, Lascorz J, Rodijk-Olthuis D, van de Kerkhof PC, Traupe H, de Jongh G, den Heijer M, et al. Psoriasis is associated with increased beta-defensin genomic copy number. Nat. Genet. 2008;40:23–25. doi: 10.1038/ng.2007.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.