Abstract
Several observations indicate that compatible ends of separate, yet closely linked, transposable elements (TEs) can interact in alternative transposition reactions. First, pairs of TEs cause chromosome breaks with frequencies inversely related to the intertransposon distance. Second, some combinations of two TEs produce complex rearrangements that often include DNA adjacent to one or both elements. In pairs of TEs in direct orientation, alternative reactions involving the external ends of the two TEs should lead to the transposition of a macrotransposon consisting of both elements plus the intervening chromosomal segment. Such macrotransposons have been hypothesized previously based on deletions, but no macrotransposon insertions have been recovered. To detect macrotransposition, we have analyzed heritable chromosomal rearrangements produced by a chromosome-breaking pair of Ac and Ds elements situated 6.5 kb apart in direct orientation in a part of the maize (Zea mays) genome dispensable for viability. Here, we show that the postulated macrotransposon can excise and reinsert elsewhere in the genome. In addition, this transposon pair produces other complex rearrangements, including deletions, inversions, and reshuffling of the intertransposon segment. Thus, closely linked TE pairs, a common transposition outcome in some superfamilies, are adept at restructuring chromosomes and may have been instrumental in reshaping plant genomes.
INTRODUCTION
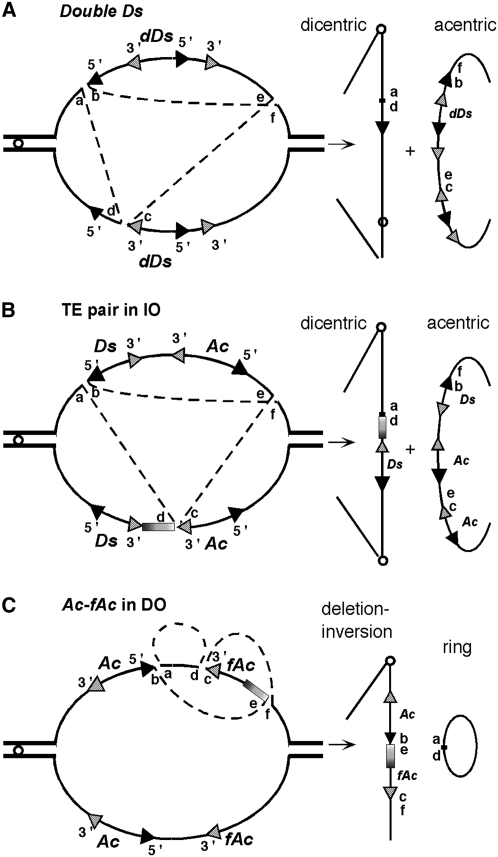
The first property of transposable elements (TEs) to be recognized was their ability to break chromosomes. McClintock (1947) identified a locus on the short arm of maize (Zea mays) chromosome 9 (9S) where breaks occurred regularly and named that locus Ds for Dissociation or breakage of chromosomes. Only later was the dissociation property (i.e., Ds) shown to be mobile (McClintock, 1949). The chromosome-breaking Ds element described by McClintock has a complex structure consisting of two identical 2-kb Ds elements inserted one inside the other in inverse orientation (IO) (Doring et al., 1984). This structure, known as DoubleDs, causes chromosome breaks because dicentric chromosomes are formed from a transposition reaction involving TE ends in sister chromatids (Figure 1A) (English et al., 1993, 1995; Kunze and Weil, 2002). Transpositions where compatible ends from separate TEs, rather than from a single TE, are mobilized together have been referred to as alternative events (Gray, 2000).
Figure 1.
Alternative Transposition Reactions in Compound Ac-Ds Structures.
(A) DoubleDs.
(B) Two TEs in IO.
(C) Ac and fAc in DO.
The diagrams illustrate DNA replication bubbles during chromosome replication and alternative transposition reactions that would lead to the formation of dicentrics plus acentrics and chromosome breakage ([A] and [B]) or a monocentric rearranged chromosome and an acentric ring (C). The TE 5′ ends are represented as solid arrowheads and the 3′ ends as hatched arrowheads. In each transposition reaction, the Ac transposase makes three cuts, one at each TE end to be mobilized and one at the receptor site, and generates six cut ends (a to f), identified as follows: a, host DNA adjacent to TE 5′ end; b, TE 5′ end; c: TE 3′ end; d: host DNA adjacent to TE 3′ end; e and f, host target site. Ligation of the cut ends leads to: a-d, excision or empty site; and either b-f and c-e or b-e and c-f reinsertion sites.
Pairs of separate, yet closely linked, transposons can also cause chromosome breaks, the frequency of breakage being an inverse function of the intertransposon genetic distance (Ralston et al., 1989; Dooner and Belachew, 1991). In pairs of TEs in IO, either two 5′ ends or two 3′ ends of different transposons face each other and are capable of undergoing alternative transposition reactions similar to those of DoubleDs (Figure 1B), which lead to dicentric formation and, therefore, chromosome breakage (Weil and Wessler, 1993). Transposon pairs in direct orientation (DO) do not have such a structure, and a model involving a macrotransposon has been proposed to explain their chromosome-breaking properties (Ralston et al., 1989). The postulated macrotransposon (MTn) extends from the 5′ end of one transposon to the 3′ end of the other and includes the chromosomal segment between them, but no evidence for its existence was presented at the time.
Combinations of a complete transposon and a closely linked partial copy in either IO or DO are proficient at rearranging chromosomes. Zhang and Peterson (1999, 2004, 2005) have analyzed chromosomal rearrangements generated in the vicinity of the maize P locus by a combination of an intact Ac element and a terminally deleted fAc (for fractured Ac) element in either IO or DO. The former twosome was shown to generate chromosomes containing either large inverted duplications or deletions with end points adjacent to one of the elements. However, the combination of directly oriented Ac and fAc elements in the P1-rr11 allele was capable of generating even more exotic rearrangements. In that allele, the 5′ and 3′ ends of nearby elements face each other and are separated by ∼13 kb of host DNA. Alternative transposition reactions involving these ends (Figure 1C) generated deletions, inversions, and a reshuffling of the segment in between. The rearrangements invariably included DNA adjacent to one or both transposons, and their size depended on the location of the reinsertion site. Recently, this same transposon configuration was shown to create novel chimeric genes by alternative transposition reactions that joined the coding and regulatory sequences of two linked, paralogous genes (Zhang et al., 2006).
Single transposons can also cause chromosomal rearrangements, such as adjacent deletions and inversions. These rearrangements, with one end point at a transposon terminus, were first detected next to insertion sequence elements in bacteria (Reif and Saedler, 1975) and have been reported for several plant transposon systems (Dooner, 1985; Taylor and Walbot, 1985; Martin et al., 1988). They are readily explained by abortive transposition reactions involving only one element end. In Drosophila, P and hobo elements often induce adjacent deletions during transposition, and this property has been exploited to create deletion mutations of nearby genes (Preston et al., 1996; Huet et al., 2002). The origin of these deletions has been explained by alternative transposition reactions that form hybrid elements containing complementary transposon ends from sister chromatids or homologous chromosomes (Gray et al., 1996; Gray, 2000). In maize, by contrast, single Ac elements rarely produce adjacent deletions (Dooner and Belachew, 1991). Nevertheless, single Ds elements in Arabidopsis were recently shown to create flanking deletions of ∼100 kb at a trackable frequency. These deletions differed in that both transposon junctions, rather than just one, had undergone sequence changes and their origin was proposed to involve an intermediate product with two closely linked Ds elements (Page et al., 2004).
Pairs of directly oriented TEs with intact ends have the potential to mobilize stretches of host sequences from one location of the genome to another. Recognition by the transposase of the external 5′ and 3′ ends from the two separate TEs should lead to the transposition of a MTn consisting of both TEs plus the intertransposon segment (ITS). Such MTns have been hypothesized on the basis of deletion products, but no MTn reinsertions have been recovered (Ralston et al., 1989). Here, we present evidence that such macrotranspositions do occur among an assortment of other rearrangements. In order to detect macrotranspositions, we analyzed the types of heritable chromosomal rearrangements produced by the combination of an Ac and a Ds element situated 6.5 kb apart in DO in a part of the maize genome known to be dispensable for viability. We show that transposon pairs in DO can produce multiple types of rearrangements. Transposition reactions involving the two external 5′ and 3′ ends lead to excision of the postulated MTn with and without its reinsertion in the genome. Reactions involving the two internal ends can lead to (1) deletion of the ITS and inversion of a transposon-adjacent segment and (2) split of the ITS and reshuffling of the split pieces in two possible ways, as described by Zhang and Peterson (2004), and, in addition, (3) the formation of a transposon with an extra end, if the reinsertion site is in one of the transposons, or (4) a chromatid breakage-fusion-bridge (BFB) cycle (McClintock, 1939), if the reinsertion site is in the sister chromatid. Recombination between the two transposons leads to deletion of the ITS and one of the two transposons, depending on the location of the exchange.
An important question regarding the ability of transposon pairs to rearrange chromosomes is, what is the maximum distance between transposons that can lead to chromosome breakage and to macrotransposition of a chromosome fragment? Dooner and Belachew (1991) reported that pairs of transposons even >1 centimorgan apart were efficient chromosome breakers. In order to determine the physical distance between transposons in chromosome-breaking pairs and their relative orientations, we have isolated the insertion sites of several transposed Ac (trAc) elements from bz-m2(Ac) (Cowperthwaite et al., 2002) and placed them on the physical map of the Bz-McC haplotype from which they were derived (Fu and Dooner, 2002; Fu et al., 2002). We found that Ac elements separated from a Ds element at bz by as little as 2.3 kb or as much as 100 kb on either the proximal or distal side of bz are highly efficient chromosome breakers and, therefore, potential genome restructurers. The proximal and distal 100-kb segments include both hypomethylated genes and methylated retrotransposons clusters, Helitrons, and various small insertions, suggesting that the Ac transposase can recognize transposon ends that are separated from each other by DNA of heterogeneous makeup and a distance >25 times the length of the transposon in which they normally reside. Transposon pairs located 6 kb apart in either IO or DO appear to break chromosomes with similar frequencies, suggesting that the length of the ITS, rather than element orientation, is the condition that determines whether closely linked transposon pairs display chromosome-breaking properties.
RESULTS
Selection and Analysis of bz-s Derivatives from bz-m2(D2) Ac6067
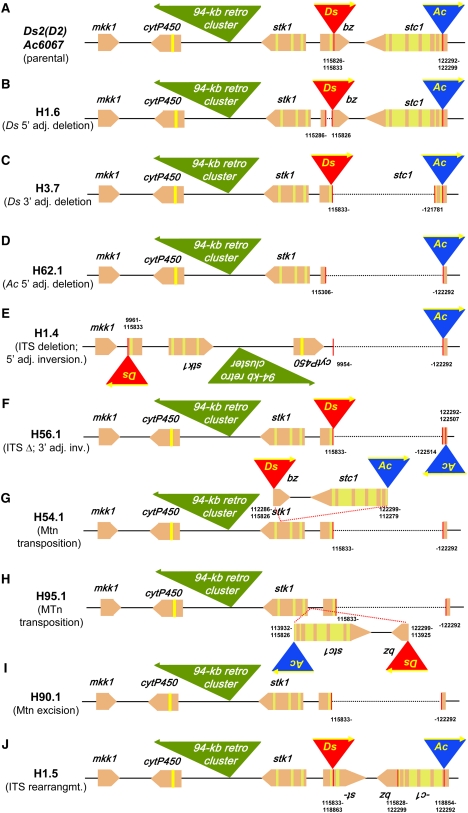
The structure of the parental chromosome used in our analysis is outlined in Figure 2A. The bz-m2(D2) Ac6067 chromosome carries a 3.7-kb Ds2(D2) element in bz and a 4.6-kb intact Ac element in the distal stc1 gene (Shen et al., 2000). The elements are in DO and separated by a 6.5-kb ITS consisting of part of the bz second exon, a 1.5-kb intergenic region, and most of the stc1 gene. Genetic tests have shown that the combination of these two elements causes breaks in 9S and sets up chromatid BFB cycles in the endosperm (Dooner and Belachew, 1991). Excision of a MTn stretching from Ds to Ac would result in a stable bronze derivative, easily distinguishable from the parental spotted (bz-m) phenotype. Thus, in order to detect transpositions of the MTn, we selected stable bronze (bz-s) derivatives among the majority of spotted seeds produced in crosses of bz-m2(D2) Ac6067 females and a sh-bz-X2 deletion line. Crossing to the deletion line allowed recovery of the exceptions in hemizygous condition, which greatly facilitated their subsequent molecular analysis. Two experiments were performed. The first experiment was designed to attempt to genetically identify transpositions of a MTn by selecting for the loss of bz mutability and retention of both Ac and chromosome-breaking (BFB) activity. The bz-s selections were crossed to a bz-m2(D1) tester to determine if Ac was still present and to a c Bz tester to determine if 9S breaks occurred (see Methods) and then analyzed molecularly. In the second experiment, the order of analyses was reversed. Several hundred bz-s derivatives were screened by PCR in order to identify heritable rearrangements produced by the Ds2(D2) Ac6067 transposon pair, and the selections of interest were then subjected to genetic tests.
Figure 2.
Structure of the Ds2(D2) Ac6067 Parental Chromosome and Some of Its bz-s Derivatives.
Ds2(D2) is shown as a red triangle and Ac6067 as a blue triangle. Genes are represented as pentagons pointing in the direction of transcription and are drawn approximately to scale: exons are in peach and introns are in yellow. The proximal 94-kb retrotransposon cluster is drawn as a solid green triangle in a smaller scale than the rest of the interval. The numbers identify the nucleotide positions of the new junctions relative to the sequence of the 226-kb McC bz haplotype contig (GenBank accession number AF391808).
(A) Ds2(D2) Ac6067 parent.
(B) bz-s derivative H1.6, a 546-bp deletion adjacent to the Ds 5′ end.
(C) bz-s derivative H3.7, a 5.9-kb Ds 3′ adjacent deletion.
(D) bz-s derivative H62.1, a 7.0-kb Ac 5′ adjacent deletion.
(E) bz-s derivative H1.4, an ITS deletion accompanied by inversion of Ds plus a 106-kb fragment adjacent to the Ds 5′ end, including the entire retrotransposon cluster.
(F) bz-s derivative H56.1, an ITS deletion accompanied by inversion of Ac plus a 215-bp fragment adjacent to the Ac 3′ end.
(G) bz-s derivative H54.1, transposition of the 14.8-kb MTn to the 3rd exon of the adjacent stk1 gene.
(H) bz-s derivative H95.1, transposition of the 14.8-kb MTn to the 5′ UTR of stk1.
(I) bz-s derivative H90.1, excision of the 14.8-kb MTn without reinsertion.
(J) bz-s derivative H1.5, a double-inversion ITS rearrangement.
In the first experiment, 60 putative bz-s derivatives were selected from a total of 1875 spotted kernels and tested for heritability and retention of both Ac and BFB activity, as indicated above. Eight selections (H1.1 to H1.8) passed this screen and were characterized further by PCR analysis and sequencing of the new DNA junctions. All retained Ds2(D2), as ascertained by PCR with a pair of Ds internal primers, one of which was specific for the unique deletion junction of Ds2(D2) (see Methods). Four of the eight derivatives were simple Ds transpositions from bz, probably to a nearby site because the new TE pair retained BFB activity. A fifth one was a Ds 5′ adjacent deletion of 546 bp from the bz first exon (H1.6; Figure 2B). The numbers in the figure refer to the nucleotide positions of the new junctions relative to the sequence of the 226-kb McC bz haplotype contig (GenBank accession number AF391808). The other three were unusual chromosomal rearrangements, reminiscent of those recovered by Zhang and Peterson (2004) at the P locus. Derivatives H1.1 and H1.4 carry a deletion of the ITS and an inversion of a chromosomal fragment that comprises the Ds element plus a Ds-5′–adjacent segment of variable length. To identify the proximal end of that segment, the sequence adjacent to the inverted Ds element was amplified by inverse (IPCR) with Ds2(D2) internal primers. In H1.1, the inverted segment is 4.8 kb and includes the entire stk1 gene located proximal (5′) to bz. In H1.4 (Figure 2E), the inverted segment is 106 kb and includes the stk1 gene, the 94-kb proximal retrotransposon cluster, and the cytP450 gene of the next proximal gene island (Fu et al., 2002). These two derivatives have two novel junctions: one adjacent to the Ds 3′ end and another one adjacent to the Ac 5′ end. The 8-bp 9S host sequences immediately adjacent to those transposon ends are identical in each case, as would be expected of Ac-Ds target site duplications (see Supplemental Table 1 online). Derivative H1.5 carries a rearrangement in which the ITS has been split in two at exon 6 of stc1 and the two segments have been inverted, creating two new bz-stc1 chimeric genes (Figure 2J). There are three novel junctions in this derivative: one adjacent to the Ds 3′ end (115,833 to 118,863), one adjacent to the Ac 5′ end (118,854 to 122,292), and one that joins the bz and stc1 sequences that were adjacent to those TE ends in the progenitor chromosome (115,828 to 122,299). The sequence at these junctions (see Supplemental Table 1 online) contains typical Ac-Ds transposition footprints, except that the stc1 sequence duplicated immediately next to the Ds 3′ and Ac 5′ ends is 10 bp long, whereas the standard target site duplication generated by Ac/Ds and other hAT elements is 8 bp long (Kunze and Weil, 2002). The structure of these and other bz-s derivatives was confirmed by DNA gel blots (Figure 3): either one or two new bands of the expected size were present in each derivative. The formation of the last three derivatives (H1.1, H1.4, and H1.5) can be explained by the transposition model of Zhang and Peterson (2004) for TEs in DO (Figure 1C). The Ac transposase would cleave the 5′ and 3′ internal ends of the Ac-Ds transposon pair and reinsert them at locations 4.8 kb proximal to bz in H1.1, 106 kb proximal to bz in H1.4, and in the stc1 sixth exon in H1.5, respectively. In the last of these derivatives, the ITS is rearranged by the transposition reaction, while in the first two, it is lost as an acentric ring.
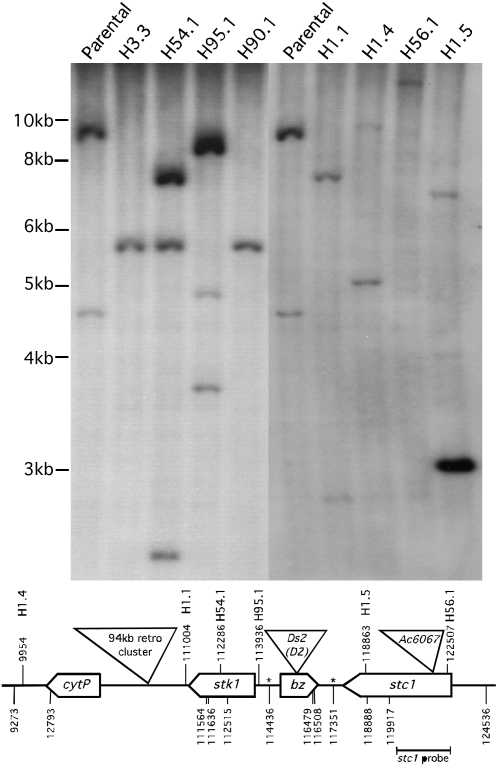
Figure 3.
DNA Gel Blot Analysis of bz-s Derivatives from bz-m2(D2) Ac6067.
PstI digests of genomic DNAs from bz-m2(D2) Ac6067 and bz-s derivatives H1.1, H1.4, H1.5, H3.3, H54.1, H56.1, H90.1, and H95.1 were blotted onto a membrane and hybridized to an stc1 probe (top) and a bz probe (see Supplemental Figure 1 online). The positions of the stc1 and bz fragments used as probes and the location of the PstI sites in the 226-kb McC bz haplotype contig (GenBank accession number AF391808) are shown below the map; the asterisks represent PstI sites known to be methylated in genomic DNA. The location of the inferred e-f transposition target site in each bz-s derivative is shown above the map. The following bands are detected by the stc1 probe: parental, 9.2 and 4.6 kb, Ac occupied and somatic excision sites, respectively; H3.3, a MTn transposition to an unmapped 9S site: 5.6 kb, MTn excision site (the transposed MTn was detected in a SacI digest: see Supplemental Figure 2 online); H54.1, a MTn transposition to stk1 exon 3: 7.2 and 2.6 kb, transposed MTn with Ac (germinal) and without Ac (somatic excision), respectively, and 5.6 kb, MTn excision site; H95.1, a MTn transposition in reverse orientation to the stk1 5′ UTR: 8.6 and 4.8 kb, MTn excision site plus transposed MTn with Ds (germinal) and without Ds (somatic excision), respectively, and 8.4 and 3.8 kb, transposed MTn with Ac (germinal) and without Ac (somatic excision), respectively; H90.1, a MTn excision: 5.6 kb, MTn excision site; H1.1, a 4.8-kb adjacent inversion: 7.4 and 2.8 kb, inversion with Ac (germinal) and without Ac (somatic excision), respectively; H1.4, a 106-kb adjacent inversion: 9.6 and 5.1 kb, inversion with Ac (germinal) and without Ac (somatic excision), respectively; H56.2, a 0.2-kb adjacent inversion on the Ac side: 12.7 kb, inversion with Ac and Ds; H1.5, an ITS rearrangement: 6.8 kb, rearrangement with Ac (the 2.3-kb Ac somatic excision product would have run out of the gel), and 3.1 kb, rearranged stc1 fragment from the middle of ITS.
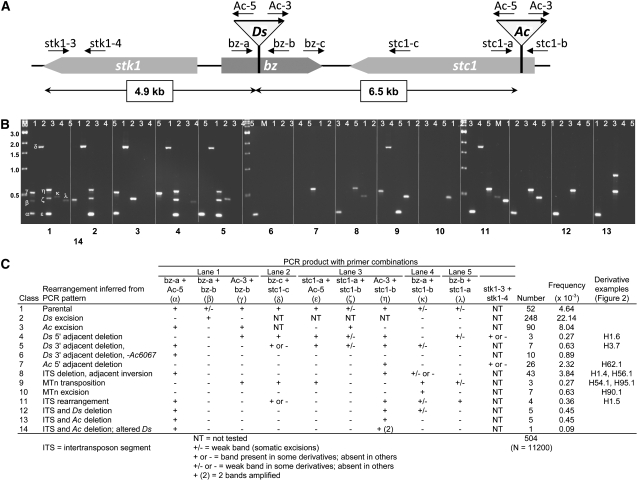
A second set of bz-s derivatives was directly screened by PCR tests using various combinations of primers adjacent to the TE insertion sites that permit a high-confidence assignment of each derivative to a particular rearrangement class (Figures 4A and 4B). For several complex rearrangements, the initial assignment was verified by sequencing of the new junctions. From an effective population of 11,200 kernels, 504 stable bronze seed selections were successfully analyzed by PCR (Figure 4C). Of them, 52 gave the same pattern as the bz-m2(D2) Ac6067 parental chromosome. They could have carried, conceivably, an extra copy of Ac that reduced or eliminated spotting (Brink and Nilan, 1952; McClintock, 1952), but they were not studied further. As expected, the majority of bronze selections represented simple excisions of Ds or Ac. There were 248 Ds excisions (2.2%) and 90 Ac excisions (0.8%). Because of the phenotype selected, all Ds excisions that do not restore Bz function (∼80%; Dooner and Belachew, 1989) are included here, whether or not Ds is retained in the genome. On the other hand, only Ac excisions that are lost from the genome are included among the bz-s selections. This bias would contribute to the difference in the recovery of simple Ds and Ac excisions, regardless of potential differences in the actual frequency of excision of Ds2(D2) from bz and Ac6067 from stc1.
Figure 4.
PCR Characterization of bz-s Derivatives from bz-m2(D2) Ac6067.
(A) Structure of the parental chromosome, showing the three adjacent genes covered by the analysis—stk1, bz, and stc1—and the locations and orientations of Ds2(D2) in bz and Ac6067 in stc1. Also shown are the locations of the various primers used in the PCR analysis of the derivatives.
(B) PCR patterns produced by the 14 classes of bz-s derivatives. Each of the 14 panels of five lanes corresponds to a representative of one of the 14 bz-s classes identified in (C). Lanes were loaded with PCRs using the following primers: lane 1, primers bz-a, Ac-5, Ac-3, and bz-b, which amplify the Ds-bz 5′ and 3′ junctions plus the Ds excision site in bz (PCR products α, γ, and β); lane 2, primers bz-c and stc1-c, which amplify the parental ITS (PCR product δ); lane 3, primers stc1-a, Ac-5, Ac-3, and stc1-b, which amplify the Ac-stc1 5′ and 3′ junctions plus the Ac excision site in stc1 (PCR products ɛ, η, and ζ); lane 4, primers bz-a and stc1-b, which amplify the MTn excision site (PCR product κ); lane 5, primers bz-b and stc1-a, which amplify the new a-d junction (Figure 1C) in an ITS ring chromosome (PCR product λ).
(C) Summary of the PCR amplification patterns given by the various derivatives. The derivatives fell into 14 classes, identified by number and inferred type of rearrangement in the two left columns. The PCR outcomes with specific primer combinations are shown in the next 10 columns. The number, overall frequency, and examples of derivatives in each class depicted in Figure 2 are given in the three right columns.
The remaining 114 derivatives gave PCR amplification patterns indicating that transposon-adjacent sequences had been either deleted or rearranged. Depending on the PCR pattern, they were assigned to one of several rearrangement classes and the novel DNA junctions in a representative sample of each class were sequenced to confirm these assignments (see Supplemental Table 1 online). In order of their presentation in Figure 4, the classes of chromosomal rearrangements recovered and the number of instances in each class were as follows: 3 Ds 5′ adjacent deletions (class 4), similar to derivative H1.6 (Figure 2B); 7 Ds 3′ adjacent deletions that did not extend distally beyond Ac6067 (class 5), as, for example, derivative H3.7 (Figure 2C), and 10 that did (class 6); 26 Ac 5′ adjacent deletions (class 7), as derivative H62.1 (Figure 2D), of which all extended proximally beyond Ds2(D2) and 4 extended beyond the stk1 gene; 43 ITS deletions and inversions of an adjacent fragment (class 8), either 5′ of Ds, as in H1.4 (Figure 2E), or 3′ of Ac, as derivative H56.1 (Figure 2F); 3 transpositions of the postulated MTn (class 9), as, for example, derivatives H54.1 and H95.1 (Figures 2G and 2H); 7 MTn excisions without reinsertion (class 10), as H90.1 (Figure 2I); 4 ITS rearrangements (class 11), similar to H1.5 (Figure 2J); 10 deletions of the ITS and one of the two transposons, 5 of Ds (class 12), 5 of Ac (class 13); and 1 deletion of the ITS and Ac, accompanied by the formation of a Ds element with a 524-bp duplication of the 3′ end (class 14). The most likely origin of each of these classes will be considered next, starting with the most novel.
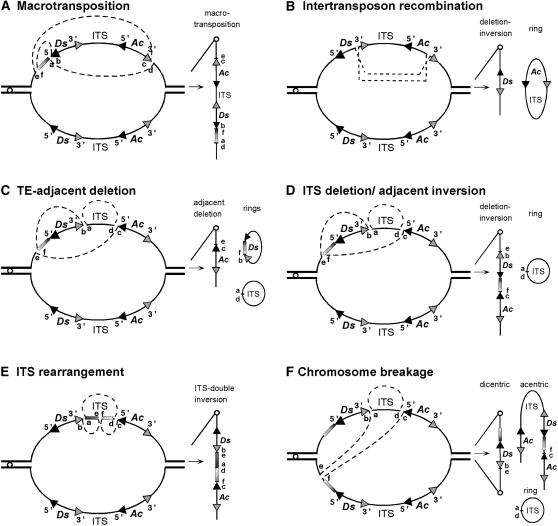
Excisions of a MTn extending from Ds2(D2) to Ac6067 and including the entire ITS (classes 9 and 10) occurred at a combined frequency of 0.9 × 10−3. In each of these 10 bz-s derivatives, the MTn empty site, which links bz and stc1 sequences adjacent to Ds and Ac, respectively, has a typical transposon excision footprint (see Supplemental Table 1 online). In 3 of the 10 derivatives (class 9), a reinserted MTn was recovered in the genome. Two of the MTn reinsertion sites isolated by IPCR lie in the proximal stk1 gene: within the third exon in derivative H54.1 (Figure 2G) and within the 5′ untranslated region (UTR) in derivative H95.1 (Figure 2H). In H54.1, the MTn has reinserted in the same orientation it had in the parental chromosome, whereas in H95.1, it has reinserted in the opposite orientation. The MTn reinsertion site of the third derivative, H3.3 (see Supplemental Table 1 online), corresponds to a low-copy-number sequence that has yet to be mapped in the maize genome. However, it must be linked to the donor site because all three bz-s derivatives retained 9S breakage properties (see below). The MTn is presumed to be intact on the basis of the sequence of its termini at the new insertion site (see Supplemental Table 1 online), the amplification of PCR bands of the expected size with primers from the ITS and the host target sequence (Figures 4A and 4B), and the recovery of bands of the expected size on DNA gel blots hybridized to stc1 and bz probes (Figure 3; see Supplemental Figures 1 and 2 online). As illustrated in Figure 5A, macrotranspositions arise by alternative reactions involving the two external TE ends of a transposon pair in DO and can lead to changes in the position of a chromosomal segment in the genome. In the present case, the ITS measures 6.5 kb, a length more than sufficient to contain an entire gene.
Figure 5.
Models for the origin of the various classes of bz-s derivatives and for chromosome breakage in bz-m2(D2) Ac6067.
(A) Macrotransposition: classes 9 and 10.
(B) Intertransposon recombination: classes 12 and 13.
(C) TE-adjacent deletion: classes 6 and 7.
(D) ITS deletion/adjacent inversion: class 8.
(E) ITS rearrangement: class 11.
(F) Chromosome breakage.
The Ac transposase can carry out two types of alternative transposition reactions, recognizing either the two external ends (A) or internal ends ([C] to [F]) of the MTn. The symbols follow Figure 1, except that when the two internal ends are cut ([C] to [F]), the b and c designations are reversed relative to Figure 1 and refer to the Ds 3′ end and the Ac 5′ end, respectively.
Ten derivatives carried precise deletions of the ITS and one or the other transposon (classes 12 and 13). Their simplest mode of origin is intrachromosomal recombination between the two elements, leading to deletions of the ITS, as diagrammed in Figure 5B. Whether the deletion derivative retains Ds or Ac depends on the location of the exchange relative to the internal deletion junction in Ds2(D2) (Yan et al., 1999): those to the left of (proximal to) the junction would generate an Ac-adjacent deletion; those to the right of (distal to) the junction would generate a Ds-adjacent deletion. Five derivatives of each kind were recovered, in general agreement with the 0.59:0.41 split expected based on the location of the deletion in the Ds2(D2) element.
One derivative carried a deletion of the ITS and a transposon with an extra 3′ end: H81.1 had a Ds element with a 524-bp duplication of the 3′ end (class 14). It probably arose by an alternative transposition of the internal ends of the transposon pair into the Ac element. Had the receptor site for the alternative transposition been inside of Ds, rather than Ac, an Ac element with two 5′ ends would have been generated.
Adjacent deletions, a common feature of prokaryotic and eukaryotic transposons, have been recovered at low frequencies from maize mutable alleles containing a single Ac or Ds element (Dooner et al., 1988). For example, Dooner and Belachew (1989) reported two adjacent deletion derivatives from the bz-m2(Ac) allele in a population of 5650 gametes. Yet, in our study, adjacent deletions (classes 4 to 7) were obtained at a 10-fold higher frequency (0.41%). The design of the experiment allows us to recover any deletion that produces a bronze phenotype: included here would be all deletions adjacent to either end of Ds and those deletions adjacent to the 5′ end of Ac that extend into bz sequences. Interestingly, only 10 of the 46 adjacent deletions retain both transposons: the 3 deletions adjacent to the Ds 5′ end (class 4) and 7 of the 17 deletions adjacent to the Ds 3′ end (class 5; e.g., derivative H3.7 in Figure 2C). These 10 deletions can be explained by single-ended abortive transposition reactions of Ds that involve, respectively, either the 5′ or the 3′ end. The great majority (36 of 46) of adjacent deletions—the remaining 10 deletions adjacent to the Ds 3′ end (class 6) and all 26 deletions adjacent to the 5′ end of Ac (class 7)—extend beyond the other transposon. The high frequency with which adjacent deletions are recovered from paired, as opposed to single, transposons and the frequent codeletion of one transposon suggests that the paired transposon structure may be involved in their origin. Most likely, these deletions originate from transposition reactions that involve the two internal ends of the transposons pair, as suggested by Zhang and Peterson (2004). As illustrated in Figure 5C, if the receptor site (e-f) is proximal to Ds, an Ac 5′ adjacent deletion will result, which will include the ITS, Ds, and sequences proximal to Ds. One such deletion derivative is H62.1, illustrated in Figure 2D. Of the 26 Ac 5′ adjacent deletions identified, only 4 extended beyond the 3′ end of the stk1 gene, located 4.9 kb proximal to Ds (Figures 4A and 4C). Thus, most of these alternative transpositions are to closely linked sites in the same gene island, sites that may be even closer than receptor sites for conventional Ac or Ds transpositions. If, on the other hand, the receptor site (e-f) is distal to Ac, a Ds 3′ adjacent deletion will result, which will include the ITS, Ac, and sequences distal to Ac. Ten such deletions were identified (class 6). Two were sized and found to extend only 19 and 69 bp beyond Ac6067, supporting the notion of very closely linked alternative transpositions.
Equally numerous to the sum of Ds 3′ and Ac 5′ adjacent deletions were deletions of the ITS accompanied by an inversion of a TE-adjacent fragment (class 8). These 43 derivatives are similar in structure to derivatives H1.4, and H56.1 (Figures 2E and 2F), and their origin can be explained by the same transposition reaction that gives rise to the adjacent deletions described above. The reaction, illustrated in Figure 5D, resembles that in Figure 5C, except that the internal TE ends (b and c) ligate with the opposite cut ends at the e-f receptor site, creating e-b and f-c, rather than e-c and f-b, junctions. It appears, from the similar frequencies of adjacent deletions and deletion-inversions, that the two outcomes depicted in Figures 5B and 5C are equally likely. It should be noted that if the e-f receptor site is located on a different chromosome, a dicentric chromosome or a reciprocal translocation would be produced, depending on how the b-c TE ends ligate with the e-f receptor site cut ends. The former probably would not be transmitted; the latter would give a class 8 PCR pattern, like the ITS deletion, adjacent inversion. However, heterozygous reciprocal translocations give semisterile ears in maize, and none were found in this class. We conclude that reciprocal translocations are not common products of alternative transpositions of two TEs in DO.
There were four ITS rearrangements (class 11), which most likely originate by alternative transposition of the two internal TE ends into the ITS itself (Figure 5E; see Supplemental Table 1 online). As a consequence, the 6.5-kb ITS is split into two fragments at locations 0.5, 0.6, 1.4, and 6.3 kb from the Ds 3′ end, respectively, in derivatives H57.3, H87.1, H72.3, and H75.1. In three of the ITS rearrangements (H57.3, H72.3, and H75.1), the two fragments were inverted, as in derivative H1.5 (Figures 2J and 5E). In the fourth (H87.1), they exchanged places. Again, these two outcomes are best explained as the products of alternative fusions of the b-c TE ends to the e-f reinsertion site in the ITS.
Chromosome-Breaking Properties of Derivatives
The bz-s derivatives in classes 4 to 14 (Figure 4C) were tested for 9S breakage activity by crossing them as pollen parents to a c Bz tester line (Figure 6). BFB activity was measured as the percentage of kernels showing pigmentation patterns clearly indicating chromosome breakage (more than five colorless sectors) in the test cross (BFB score; see Supplemental Table 2 online). Most derivatives that retained both elements, in either DO or IO, also retained BFB activity, whereas none that lost at least one element did. These data support the concept that transposon pairs in close proximity to each other promote chromosome breaks (Dooner and Belachew, 1991). A likely mechanism by which the Ds2(D2) Ac6067 transposon pair causes chromosome breaks is illustrated in Figure 5F. The two internal ends of the transposon pair undergo an alternative transposition reaction to the sister chromatid, generating a dicentric chromosome that initiates a BFB cycle at the next anaphase. Markers located distally on the same chromosome arm, such as C, will be lost at either this or later divisions and produce somatic sectors of various sizes.
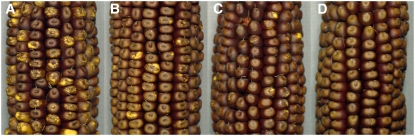
Figure 6.
Ears Illustrating the Genetic Assay for 9S Chromosome Breakage Activity.
bz-s derivatives (C bz-s/C sh-bz-X2) were crossed as pollen parents to c testers (c Bz). Breaks occurring during endosperm development are visualized as colorless (acyanic) sectors arising from the loss of the dominant C marker located distal to Bz in 9S. The phenotype of kernels receiving the sh-bz-X2 chromosome (∼45% due to its reduced male transmission) is solid color; that of kernels receiving the bz-s derivative chromosome is either mosaic or solid. Breakage activity is expressed as percentage of mosaic (BFB) kernels among those receiving a bz-s chromosome.
(A) bz-s derivative H74.2 (class 8, ITS deletion, adjacent inversion; 95.3% BFB).
(B) bz-s derivative H95.1 (class 9, MTn transposition; 82.4% BFB).
(C) bz-s derivative H3.12 (class 8, ITS deletion, adjacent inversion; 26.5% BFB).
(D) bz-s derivative H90.1 (class 10, MTn excision; 0% BFB).
Unlike the previous models of chromosome breakage, in which dicentric chromatid formation is the result of a predictable excision event (Figures 1A and 1B, a-d junction), the dicentric chromatid in the model of Figure 5F results from transposition to a variable site in the sister chromatid. Therefore, to demonstrate the existence of bridges in somatic tissues of the parental bz-m2(D2) Ac6067 stock, PCRs were set up with combinations of fixed primers from bz or the Ds 3′ end and variable primers in the same orientation from sites in stc1, bz, stk1, or mkk1. Four PCR products with the expected sequence of a Ds-containing bridge and its corresponding Ds excision footprint were detected (Figure 7), confirming the formation of the products predicted from alternative transpositions to a sister chromatid.
Figure 7.
Sequences of Four New Ds 3′ Junctions at Dicentric Chromatid Bridges and of Their Corresponding Excision Products.
Sixteen separate PCR amplification reactions on Ds2(D2) Ac6067 embryo DNA were performed with combinations of two primers oriented away from the centromere (i.e., in the same orientation) and covering the bz genomic region from mkk1 to stc1. Nested PCR was then performed with either a bz or a Ds 3′ end primer and a second set of 16 primers, each of which was downstream of the corresponding primer in the first set. The latter primer combination would amplify the b-e junction of the bridge in Figure 5F. Nested PCR products were pooled in groups of eight, cloned into pGEM-T Easy, and sequenced.
The parental Ds2(D2) Ac6067 chromosome had a BFB score of 89.3%, and the three chromosomes with transposed MTns (class 9) had scores between 78.5 and 89.6%, suggesting that, as with regular transposition, the frequency of alternative transposition reactions may vary with genomic location. The greatest variation in BFB activity was seen within class 8 derivatives. These derivatives retain both elements but have undergone a deletion of the original 6.5-kb ITS and an inversion of a chromosomal segment adjacent to, and including, one of the transposons. As a consequence, the two transposons are now in IO and separated by a chromosomal segment stretching from the inverted transposon to the receptor site in the alternative transposition event (Figure 5D). BFB activity in this class ranged from 0 to 100%, probably as a function of the distance separating the two TEs in the particular bz-s derivative. The highest BFB score (100%) was given by the adjacent deletion-inversion derivative H56.1, in which Ds and Ac are separated by only 215 bp (Figure 2F), the shortest intertransposon distance of any TE pair. However, the adjacent deletion-inversion derivative H1.4, in which Ds and Ac are separated by 106 kb of DNA (Figure 2E), including the large 94-kb retrotransposon cluster immediately proximal to bz, is also a strong breaker. This raises the question, what is the maximum distance that can separate a pair of transposons displaying chromosome-breaking activity?
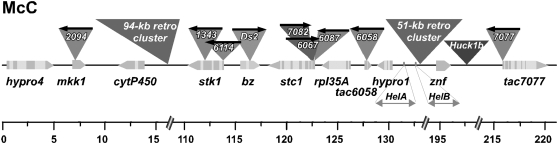
Dooner and Belachew (1991) reported that pairs of TEs as much as 1 centimorgan apart in the bz region were highly efficient chromosome breakers. In order to determine the physical distance separating transposons in those chromosome-breaking pairs, we have isolated tac sites (trAc adjacent sites) for several Ac elements characterized as efficient breakers in cis combinations with the Ds2(D1) element at bz (Dooner and Belachew, 1991; H.K. Dooner, unpublished data). Five of these tac sites have been reported previously (Cowperthwaite et al., 2002; Fu et al., 2002); three additional sites were isolated during the course of this investigation. The locations and orientations of the eight Ac elements relative to Ds2(D1) are shown in Figure 8. As seen, six of those Ac elements are located in the bz gene island, at distances from Ds2(D1) that range from 2.5 to 12 kb, and in either the same (two) or opposite (four) orientation relative to Ds. However, two are located more than 100 kb away, on either side of bz and in IO relative to Ds2(D1). The nature of the 9S DNA separating the elements is heterogeneous in both cases. The ITS separating the proximal Ac2094 from Ds2(D1) contains a mixture of genes, retrotransposons, and miniature inverted repeat TEs. The ITS separating the distal Ac7077 from Ds2(D1) contains, in addition, two Helitrons carrying fragments from several genes (Lai et al., 2005).
Figure 8.
Location of trAcs within the 226-kb McC bz Contig (GenBank Accession Number AF391808) That Form Chromosome-Breaking Pairs with Either Ds2(D1) or Ds2(D2).
The arrow above the Ac and Ds transposons points in a 5′ to 3′ direction. Ds is shown as a triangle on the bz second exon. Ac7082 and Ac6067 are in DO relative to Ds, while the other Acs are in IO relative to Ds. Genes are represented as pentagons pointing in the direction of transcription and are drawn approximately to scale: exons are dark and introns are light. The long terminal repeat retrotransposons in the region are shown above the line as clusters of 94 and 51 kb, respectively, and as a single Huck1b. The two helitrons in the interval, HelA and HelB, which contain gene fragments, are shown below the line. None of the recovered trAcs were in retrotransposons or helitrons.
DISCUSSION
A frequent outcome of hAT and CACTA element transposition is the generation of two identical TEs close to each other (Kunze and Weil, 2002). Such closely linked pairs of transposons efficiently break the chromosome arm in which they are inserted (Dooner and Belachew, 1991) and are, therefore, potential chromosomal engineers. Pairs of related, although nonidentical, transposons of the same family are also efficient breakers, as long as their ends are sufficiently similar to be recognized by that family's transposase. For example, in the Ac-Ds family of hAT transposons, combinations of Ac and either an internally deleted Ds derivative (Dooner and Belachew, 1991) or a terminally deleted fAc derivative (Ralston et al., 1989) are efficient chromosome breakers. Zhang and Peterson (1999, 2004, 2005) have reported that configurations of an intact and a fractured element in either IO or DO can give rise to a number of chromosomal rearrangements: deletions adjacent to one or the other transposon, rearrangements of the ITS, and deletions of the ITS plus inversions of one of the transposons and its adjacent sequence. Here, we describe the rearrangements produced by two intact-ended elements in DO that are separated by 6.5 kb of host DNA. The elements are a 3.7-kb internally deleted Ds element in bz and a 4.6-kb intact Ac element in the adjacent stc1 gene (Figure 2A).
In this combination, the percentage of germinally transmissible derivatives carrying chromosomal rearrangements other than single transposon excisions (classes 4 to 14) is high: over 1% (Figure 4C). Through alternative transpositions involving the external ends of the transposons, TE pairs can mobilize a MTn that extends between those ends and includes both TEs and the ITS. Homologous recombination between the closely linked transposons in DO results in loss of the ITS and one or the other transposon. Similar recombination events have been proposed for the I-R elements of the R-st complex, which also belong to the hAT transposon superfamily (Kermicle, 1984; Eggleston et al., 1995). Finally, through alternative transposition reactions involving the internal ends of the two transposons, TE pairs can give rise to the same types of chromosomal rearrangements described in the preceding paragraph, as well as to three- or four-ended transposons with different structures from those of SesquiDs (Martinez-Ferez and Dooner, 1997) or DoubleDs (Doring et al., 1984).
As is evident from the data presented in Figure 4C, TE pairs in DO undergo conventional or simple transpositions more frequently than alternative transpositions, but how much more so? We can determine the relative frequencies of the two types of transposition by making certain assumptions. As discussed earlier, classes 2 to 5 arise from transpositions of single elements in the transposon pair. bz-s derivatives are produced by ∼80% of Ds excisions (the other 20% produce Bz′ revertants) and 50% of Ac excisions (the other half is recovered as trAcs elsewhere in the genome and produce a bz-m parental phenotype) (Dooner and Belachew, 1989). Therefore, we can adjust the frequencies of classes 2 and 3 upward by multiplying them by the appropriate coefficients. Adding classes 4 and 5 to the total, we obtain an adjusted single element transposition frequency of 4.5%. Classes 6 to 11 most likely arise from alternative transpositions of either the external or internal ends of the paired transposons. Their overall frequency is 0.83%. Thus, conventional transpositions are ∼5.4 times more frequent in the TE configuration that we have studied, yet the frequency of alternative transpositions is remarkably high.
What are some of the potential evolutionary implications of the chromosomal restructuring described here? Macrotranspositions can move long stretches of host sequences from one location of the genome to another. Subsequent excision of the TE at either end of the MTn would stabilize the translocated chromosome segment at the new location (Dooner and Weil, 2007). The MTn described here translocates a 6.5-kb DNA fragment, which is long enough to contain one to two average-sized plant genes. Such macrotransposition could have contributed to the interrupted synteny, frequently involving single genes, detected in comparisons of cereal genomes (Lai et al., 2004). Because macrotranspositions are often to linked sites, they could also have been involved in the formation of closely linked paralogs found in many sequenced animal and plant genomes (Moore and Purugganan, 2005), including maize (Emrich et al., 2007). Closely linked TE pairs are particularly efficient at generating deletions, some of which can extend beyond 60 kb (Zhang and Peterson, 2005), and inversions (0.7% of gametes: classes 6 to 8). Thus, by alternative transposition reactions, TE pairs could have played a role in the rapid loss of duplicate genes that followed the whole genome duplication event in the maize ancestry (Ilic et al., 2003; Langham et al., 2004; Messing et al., 2004), in the small inversions in gene order that have occurred during grass genome evolution (Bennetzen and Ramakrishna, 2002), and in the origin of microRNA loci through inverted duplication events from protein-coding gene sequences (Fahlgren et al., 2007). Deletions or rearrangements of the ITS can join the coding and regulatory sequences of two nearby genes, generating new chimeric genes that express novel phenotypes, as shown by Zhang et al. (2006) at the P locus of maize. In bz-s derivative H1.5 (Figure 2J), a rearrangement of the ITS resulted in the production of two new chimeric genes that joined parts of bz and stc1. Although these genes are most likely nonfunctional, their origin illustrates how TE pairs in DO can shuffle exons between genes. Derivative H57.3, for example, carries a new chimeric gene (data not shown) made up of three gene fragments that specify a continuous open reading frame: the bz gene to the site of Ds insertion, an inverted 474 bp of bz exon 2, and the stc1 gene from the site of Ac insertion to the 3′ end. Surprisingly, the open reading frame created by the inverted bz gene fragment shares 38% identity over 104 amino acids with a rice (Oryza sativa) open reading frame of unknown function. Finally, recombination between two transposons in the same orientation also could have played a role in the deletion and duplication of genes. In humans, TE-induced genome instabilities appear to result principally from recombination between AluI retroposon repeats (Hedges and Deininger, 2007), yet recombination events in the hAT element system described here (classes 12 and 13) are less frequent than alternative transposition events (classes 6 to 11).
The breaker property of the Ds2(D2) Ac6067 chromosome can be readily explained by an alternative transposition reaction of the internal TE ends to a recipient site in the sister chromatid (Figure 5F). In this chromosome, the ITS is 6.5 kb long. In the fAc Ac2094 chromosome, in which breaks by separate TEs were first reported (Ralston et al., 1989), the ITS is 108 kb (Fu et al., 2002). To explain breaks in that chromosome, a model was proposed that invoked the transposition of a partially replicated MTn whose directly oriented TEs were at a great enough distance to be part of different replicons. At the time, the relative orientation of Ac2094 and fAc was not known, but it was presumed to be direct on the basis of the recovery of the bz-s3130 deletion, which contained the empty site predicted from the excision of a MTn extending from the fAc element in bz to the Ac2094 element in the proximal mkk1 gene (Ralston et al., 1989). However, isolation and sequencing of BACs from the Bz-McC genomic region established that the mkk1 gene was in the opposite orientation to that expected (Fu et al., 2002), and further analysis of the bz-s3130 derivative established that the ITS had not been excised, as predicted from the MTn excision, but had undergone an inversion and adjacent deletion (Shen, 2001). Therefore, Ac2094 and fAc are not in DO, but in IO, relative to each other. Whether two elements in DO and separated by 100 kb (or more) can break chromosomes and/or macrotranspose the intervening segment cannot be ascertained at this time. As shown in Figure 8, the two chromosome-breaking transposon pairs with ITSs that large, Ac2094 bz-m2(D1) and Ac7077 bz-m2(D1), are in IO. The TEs in those pairs are separated by >100 kb of mostly heterochromatic retrotransposon DNA, yet the Ac transposase is perfectly capable of recognizing their ends and carrying out alternative transposition reactions that lead to dicentric formation and BFB cycles in the endosperm (Figure 1B). It would be interesting to know the maximum distance that can separate TE pairs that display BFB activity. That question could be answered by isolating tac sites corresponding to trAc elements showing intermediate to low BFB activity, such as Ac2116, Ac6083, and Ac2106 (Dooner and Belachew, 1991), and placing them in the physical map. However, this would require us to expand the McC physical map beyond the 226 kb currently available in GenBank (accession number AF391808), because all of these trAcs were isolated in a McC haplotype and the extensive haplotype polymorphisms of the bz region (Fu and Dooner, 2002; Wang and Dooner, 2006) preclude using the B73 physical map generated by the Maize Sequencing Project (http://www.maizesequence.org/index.html) to answer the question.
The orientation of the two elements in a chromosome-breaking TE pair does not appear to affect BFB activity. Ac6087 and Ac6067 are inserted only 230 bp from each other in the first exon of the stc1 gene (Shen et al., 2000). Ac6087 is in the opposite orientation relative to the Ds element in bz, whereas Ac6067 is in the same orientation (Figure 8), yet both combinations of TEs have similarly high BFB activity (Dooner and Belachew, 1991). This is rather surprising, because the primary dicentric bridge in the BFB cycle is postulated to originate from two very different transposition reactions. In the case of two elements in IO, it arises from the excision product (empty site) of two transposon ends located in sister chromatids (Figure 1B), whereas in the case of two elements in DO, it arises from the transposition to a sister chromatid of two transposon ends located in the same strand (Figure 5F). Apparently, these distinct alternative transposition events do not differ greatly in frequency, at least not enough to be detected by our chromosome breakage assay. If this relationship holds at longer intertransposon distances, we may be able to estimate the upper size limit of a MTn based on the maximum distance that can separate pairs of TEs that display BFB activity, regardless of their relative orientation to each other.
To sum up, DNA transposons have been particularly adept at shaping present-day plant genomes because they associate preferentially with euchromatin, where they often escape silencing by the host (Dooner and Weil, 2007). This article documents the versatility of closely linked transposon pairs in DO—a common product of hAT and CACTA element transposition—in restructuring chromosomes.
METHODS
Genetic Stocks
All of the maize (Zea mays) stocks used in this study shared the common genetic background of the inbred W22. The bronze alleles and the aleurone phenotypes of the various stocks are described below.
Bz-McC (purple): the normal progenitor allele of the bz-m2(Ac) mutation.
bz-m2(Ac) (purple spots on a bronze background): an allele that arose from the insertion of the 4.6-kb Ac element at positions 755 to 762 in the second exon of Bz-McC (McClintock, 1955; Ralston et al., 1988). Most transposon excisions from this site fail to restore gene function (Dooner and Belachew, 1989).
bz-m2(D1) (bronze in the absence of Ac; spotted in its presence) is the first derivative from bz-m2(Ac) isolated by McClintock; it harbors a 3.3-kb internally deleted Ds element at the same position as Ac in bz-m2(Ac) (McClintock, 1962; Dooner et al., 1986).
bz-m2(D2) (bronze in the absence of Ac; spotted in its presence) is the second derivative from bz-m2(Ac) isolated by McClintock; it harbors a 3.7-kb internally deleted Ds element at the same position as Ac in bz-m2(Ac) (McClintock, 1962; Yan et al., 1999).
bz-m2(D2) Ac6067 (spotted): a bz-m2(D2) allele harboring a distal Ac element in DO. Ac6067 transposed from bz-m2(Ac) to positions 332 to 339 in the first exon of the adjacent stc1 gene (Dooner and Belachew, 1989; Shen et al., 2000).
bz-m2(D1) Ac6087 (spotted): a bz-m2(D1) allele harboring a distal Ac element in IO. Ac6087 transposed from bz-m2(Ac) to positions 95 to 102 in the first exon of the adjacent stc1 gene (Dooner and Belachew, 1989; Shen et al., 2000).
bz-m2(D1) Ac7077 (spotted): a bz-m2(D1) allele harboring a trAc element in the tac7077 gene located 101 kb distally (Dooner and Belachew, 1989; Cowperthwaite et al., 2002).
bz-s2094(fAc) Ac2094 (bronze): a derivative of bz-m2(Ac) harboring a 2-kb fractured Ac element at bz and a trAc element in the mkk1 gene located 108 kb proximally (Dooner and Belachew, 1989; Ralston et al., 1989; Fu et al., 2002).
bz-s3130: a deletion derivative from bz-s2094(fAc) Ac2094 lacking fAc and Ac and carrying a fusion of the insertion site of fAc in bz with tac2094 (Ralston et al., 1989).
sh-bz-X2 (shrunken, bronze): an x-ray–induced deletion of a large chromosomal fragment that includes sh, bz, and other loci between sh and bz, such as stc1 (Mottinger, 1973; Fu et al., 2001).
Selection and Analysis of bz-s Derivatives
Two experiments were performed. In the first experiment, stable bz derivatives (bz-s) were isolated from crosses between Sh Ac6067 bz-m2(D2) Wx and sh-bz-X2 wx. In those crosses, the majority of kernels are spotted. Exceptional unspotted kernels were selected and subjected to genetic and molecular analyses as described below. This experiment was designed to preselect for transpositions of a MTn based on the loss of bz mutability and the retention of both Ac and chromosome-breaking activity. The bz-s selections were crossed as pollen parents to bz-m2(D1) to determine if Ac was present and to c Bz to determine if 9S breaks occurred. In the second experiment, bz-s derivatives were selected as plump, bronze seeds from test crosses of Sh Ac6067 bz-m2(D2) Wx/sh-bz-X2 wx females to sh-bz-X2 wx males. The bz-s derivatives were first screened by PCR in order to identify different types of rearrangements, and a selected group was then subjected to genetic tests for Ac and chromosome-breaking activity.
Genetic Test for Chromosome Breaks in 9S
Chromosome breaks were assayed genetically by monitoring the loss of the dominant marker C located distal to bz on the short arm of chromosome 9 (9S). Stocks to be tested were used as pollen parents on c Bz females. Kernels where no breaks occur during the development of the endosperm will be purple; those where breaks occur in 9S will display colorless sectors on a purple background. In control crosses with bz-m2(Ac), a line with a single Ac element in 9S, kernels with more than five colorless sectors are rare, so that number was used as a cutoff. Breakage frequency is expressed as the percentage of kernels with more than five colorless sectors in crosses to a c Bz parent (Dooner and Belachew, 1991).
DNA Extraction, Blotting, and Hybridization
Leaf DNA for DNA gel blot hybridization was isolated by a urea extraction procedure (Greene et al., 1994). A modified cetyl-trimethyl-ammonium bromide extraction method (Svab and Maliga, 1993) was found to be more suitable for the large number of DNA preparations required to screen by PCR the several hundred bz-s derivatives from the second experiment. A 1.5-cm seedling leaf blade tip was placed in a 200-μL microcentrifuge tube. The leaf sample was frozen in liquid nitrogen and crushed with a toothpick. Cetyl-trimethyl-ammonium bromide buffer (200 μL) was added and the original method was followed thereafter, except that chemicals were added in proportion to the initial volume of extraction buffer. Restriction-digested DNA (10 μg) was resolved on 0.8% agarose gels and transferred to Hybond XL nylon membranes (Amersham Biosciences). 32P-labeled probes were generated with Ready-To-Go DNA labeling beads (Amersham Biosciences). The 1.1-kb stc1 probe was amplified from a cDNA clone (GenBank accession number AF296122) using primers in exon 4 (5′-CTCCCACGATCCAGAAGTAGCACT-3′) and exon 1 (5′-ATGACAGGCAGCAGCTACG-3′). The 0.9-kb bz probe was amplified from the 5′ end of a bz cDNA clone (Ralston et al. 1988) with primers in exon 1 (5′-CACGCTCTCGTTCCTCTCCA-3′) and exon 2 (5′-ATCCGCTGGTCGCCGAAGAA-3′).
PCR, IPCR, and Sequencing
PCR was performed according to the protocol of QiaTaq (Qiagen). Long-strand DNA fragment amplification was performed according to the protocol of Roche Expand Long Range (Hoffmann-La Roche). PCR products were cloned into pGEM-T Easy vector (Promega) and transformed into XL-Blue competent cells. Plasmids were purified with a Qiagen spin miniprep kit. DNA sequencing of plasmids or PCR products was performed in an ABI 3700 sequencer (Perkin-Elmer) following the manufacturer's instructions. Ac insertion sites tac6114 and tac7082 were isolated by IPCR, as described previously (Cowperthwaite et al., 2002). Transposon reinsertion sites in bz-s derivatives H1.1, H1.4, H54.1, and H95.1 were isolated by IPCR, using specific primers Ds-jct-A or Ds-jct-F, based on the unique deletion junction of Ds2(D2) (Conrad et al., 2007).
Primer sequences were as follows: bz-a, 5′-ATTGCGCGCGGGTTTGATGA-3′; bz-b, 5′-ACCGTGCCGAAGCTGACGTA-3′; bz-c, 5′-TGCAGGAAGAACTTCGACA-3′; stc-a, 5′-AGCGCAAGATTAAACGAGTCCAGA-3′; stc-b, 5′-ATGACAGGCAGCAGCTACG-3′; stc-c, 5′-AACGAGGAGTGGCTGAACAT-3′; Ac-5, 5′-ACCTCGGGTTCGAAATCGATCG-3′; Ac-3,5′-GTGTGCTCCAGATTTATATGGA-3′; Ds-jct-A, 5′-GCCTTGTCTTTGGACAACTACTA-3′; Ds-jct-F, 5′-CCATACTATCATTAGTAGTTGTCCAAAGA-3′.
PCR amplification of new DNA junctions in dicentric bridges was achieved with combinations of two primers oriented away from the centromere: bz-y3 and 16 primers that covered the region mkk1, stk1, bz, and stc1 (see Supplemental Table 3 online). Nested PCR was then performed with either bz-a (a downstream bz primer) or Ac-3 (a Ds 3′ end primer) and a second set of 16 primers, each of which is downstream of those in the first 16-primer set. Nested PCR products were pooled in groups of eight and ligated to pGEM-T Easy. The ligation product was transformed and plated; 48 clones were sequenced per ligation.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under the following accession numbers: McC bz haplotype contig (AF391808) and stc1-McC mRNA (AF296122).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. DNA Gel Blot Analysis of bz-s Derivatives (PstI Digest; bz Probe).
Supplemental Figure 2. DNA Gel Blot Analysis of bz-s Derivatives (SacI Digest; stc1 Probe).
Supplemental Table 1. Sequence of Excision Footprints and New TE Junctions in bz-s Derivatives.
Supplemental Table 2. BFB Activity of bz-s Derivatives from bz-m2(D2) Ac6067.
Supplemental Table 3. Primers Used in Dicentric Bridge PCR Amplification.
Supplementary Material
Acknowledgments
We thank members of the Dooner laboratory for comments on the manuscript. This research was supported by a Charles and Johanna Busch Predoctoral Fellowship to J.T.H. and by National Science Foundation Grant DBI-03-21494 to H.K.D.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Hugo K. Dooner (dooner@waksman.rutgers.edu).
Online version contains Web-only data.
References
- Bennetzen, J.L., and Ramakrishna, W. (2002). Numerous small rearrangements of gene content, order and orientation differentiate grass genomes. Plant Mol. Biol. 48 821–827. [DOI] [PubMed] [Google Scholar]
- Brink, R.A., and Nilan, R.A. (1952). The relation between light variegated and medium variegated pericarp in maize. Genetics 37 519–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrad, L.J., Bai, L., Ahern, K., Dusinberre, K., Kane, D.P., and Brutnell, T.P. (2007). State II Dissociation element formation following Activator excision in maize. Genetics 177 737–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowperthwaite, M., Park, W., Xu, Z., Yan, X., Maurais, S.C., and Dooner, H.K. (2002). Use of the transposon Ac as a gene-searching engine in the maize genome. Plant Cell 14 713–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dooner, H.K. (1985). A deletion adjacent to the Ac insertion site in a stable bz derivative from bz-m2(Ac). In Plant Genetics, M. Freeling, ed (New York: Alan R. Liss), pp. 561–573.
- Dooner, H.K., and Belachew, A. (1989). Transposition pattern of the maize element Ac from the bz-m2(Ac) allele. Genetics 122 447–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dooner, H.K., and Belachew, A. (1991). Chromosome breakage by pairs of closely linked transposable elements of the Ac-Ds family in maize. Genetics 129 855–862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dooner, H.K., English, J., and Ralston, E. (1988). The frequency of transposition of the maize element Ac is not affected by an adjacent deletion. Mol. Gen. Genet. 211 485–491. [DOI] [PubMed] [Google Scholar]
- Dooner, H.K., English, J., Ralston, E., and Weck, E. (1986). A single genetic unit specifies two transposition functions in the maize element Activator. Science 234 210–211. [DOI] [PubMed] [Google Scholar]
- Dooner, H.K., and Weil, C.F. (2007). Give-and-take: Interactions between DNA transposons and their host plant genomes. Curr. Opin. Genet. Dev. 17 486–492. [DOI] [PubMed] [Google Scholar]
- Doring, H.P., Tillmann, E., and Starlinger, P. (1984). DNA sequence of the maize transposable element Dissociation. Nature 307 127–130. [DOI] [PubMed] [Google Scholar]
- Eggleston, W.B., Alleman, M., and Kermicle, J.L. (1995). Molecular organization and germinal instability of R-stippled maize. Genetics 141 347–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emrich, S.J., Li, L., Wen, T.J., Yandeau-Nelson, M.D., Fu, Y., Guo, L., Chou, H.H., Aluru, S., Ashlock, D.A., and Schnable, P.S. (2007). Nearly identical paralogs: implications for maize (Zea mays L.) genome evolution. Genetics 175 429–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- English, J., Harrison, K., and Jones, J.D.G. (1993). A genetic analysis of DNA sequence requirements for Dissociation state I activity in tobacco. Plant Cell 5 501–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- English, J.J., Harrison, K., and Jones, J. (1995). Aberrant transpositions of maize Double Ds-like elements usually involve Ds ends on sister chromatids. Plant Cell 7 1235–1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahlgren, N., Howell, M.D., Kasschau, K.D., Chapman, E.J., Sullivan, C., Cumbie, J.S., Givan, S.A., Law, T.F., Grant, S.R., Dangl, J.L., and Carrington, J.C. (2007). High-throughput sequencing of Arabidopsis microRNAs: Evidence for frequent birth and death of miRNA genes. PLoS ONE 2 e219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu, H., and Dooner, H.K. (2002). Intraspecific violation of genetic colinearity and its implications in maize. Proc. Natl. Acad. Sci. USA 99 9573–9578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu, H., Park, W., Yan, X., Zheng, Z., Shen, B., and Dooner, H.K. (2001). The highly recombinogenic bz locus lies in an unusually gene-rich region of the maize genome. Proc. Natl. Acad. Sci. USA 98 8903–8908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu, H., Zheng, Z., and Dooner, H.K. (2002). Recombination rates between adjacent genic and retrotransposon regions differ by two orders of magnitude. Proc. Natl. Acad. Sci. USA 99 1082–1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray, Y.H. (2000). It takes two transposons to tango: Transposable-element-mediated chromosomal rearrangements. Trends Genet. 16 461–468. [DOI] [PubMed] [Google Scholar]
- Gray, Y.H., Tanaka, M.M., and Sved, J.A. (1996). P-element-induced recombination in Drosophila melanogaster: Hybrid element insertion. Genetics 144 1601–1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greene, B., Walko, R., and Hake, S. (1994). Mutator insertions in an intron of the maize knotted1 gene result in dominant suppressible mutations. Genetics 138 1275–1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedges, D.J., and Deininger, P.L. (2007). Inviting instability: Transposable elements, double-strand breaks, and the maintenance of genome integrity. Mutat. Res. 616 46–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huet, F., Lu, J.T., Myrick, K.V., Baugh, L.R., Crosby, M.A., and Gelbart, W.M. (2002). A deletion-generator compound element allows deletion saturation analysis for genomewide phenotypic annotation. Proc. Natl. Acad. Sci. USA 99 9948–9953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilic, K., SanMiguel, P.J., and Bennetzen, J.L. (2003). A complex history of rearrangement in an orthologous region of the maize, sorghum, and rice genomes. Proc. Natl. Acad. Sci. USA 100 12265–12270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kermicle, J.L. (1984). Recombination between components of a mutable gene system in maize. Genetics 107 489–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunze, R., and Weil, C.F. (2002). The hAT and CACTA superfamilies of plant transposons. In Mobile DNA II, N.L. Craig, R. Craigie, M. Gellert, and A.M. Lambowitz, eds (Washington, D.C.: ASM Press), pp. 565–610.
- Lai, J., Li, Y., Messing, J., and Dooner, H.K. (2005). Gene movement by Helitron transposons contributes to the haplotype variability of maize. Proc. Natl. Acad. Sci. USA 102 9068–9073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai, J., Ma, J., Swigonova, Z., Ramakrishna, W., Linton, E., Llaca, V., Tanyolac, B., Park, Y.J., Jeong, O.Y., Bennetzen, J.L., and Messing, J. (2004). Gene loss and movement in the maize genome. Genome Res. 14 1924–1931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langham, R.J., Walsh, J., Dunn, M., Ko, C., Goff, S.A., and Freeling, M. (2004). Genomic duplication, fractionation and the origin of regulatory novelty. Genetics 166 935–945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin, C., MacKay, S., and Carpenter, R. (1988). Large scale chromosome restructuring is induced by the transposable element Tam3 at the nivea locus of Antirrhinum majus. Genetics 119 171–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Ferez, I.M., and Dooner, H.K. (1997). Sesqui-Ds, the chromosome-breaking insertion at bz-m1, links double Ds to the original Ds element. Mol. Gen. Genet. 255 580–586. [DOI] [PubMed] [Google Scholar]
- McClintock, B. (1939). The behavior in successive nuclear divisions of a chromosome broken at meiosis. Proc. Natl. Acad. Sci. USA 25 405–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClintock, B. (1947). Cytogenetic studies of maize and Neurospora. Carnegie Inst. Washington Year Book 46 146–152. [Google Scholar]
- McClintock, B. (1949). Mutable loci in maize. Carnegie Inst. Washington Year Book 48 142–154. [PubMed] [Google Scholar]
- McClintock, B. (1952). Chromosome organization and gene expression. Cold Spring Harbor Symp. Quant. Biol. 16 13–47. [DOI] [PubMed] [Google Scholar]
- McClintock, B. (1955). Controlled mutation in maize. Carnegie Inst. Washington Year Book 54 245–255. [PubMed] [Google Scholar]
- McClintock, B. (1962). Topographical relations between elements of control systems in maize. Carnegie Inst. Washington Year Book 61 448–461. [Google Scholar]
- Messing, J., Bharti, A.K., Karlowski, W.M., Gundlach, H., Kim, H.R., Yu, Y., Wei, F., Fuks, G., Soderlund, C.A., Mayer, K.F., and Wing, R.A. (2004). Sequence composition and genome organization of maize. Proc. Natl. Acad. Sci. USA 101 14349–14354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore, R.C., and Purugganan, M.D. (2005). The evolutionary dynamics of plant duplicate genes. Curr. Opin. Plant Biol. 8 122–128. [DOI] [PubMed] [Google Scholar]
- Mottinger, J. (1973). Unstable mutants of bronze induced by premeiotic x-ray treatment in maize. Theor. Appl. Genet. 43 190–195. [DOI] [PubMed] [Google Scholar]
- Page, D.R., Kohler, C., Da Costa-Nunes, J.A., Baroux, C., Moore, J.M., and Grossniklaus, U. (2004). Intrachromosomal excision of a hybrid Ds element induces large genomic deletions in Arabidopsis. Proc. Natl. Acad. Sci. USA 101 2969–2974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preston, C.R., Sved, J.A., and Engels, W.R. (1996). Flanking duplications and deletions associated with P-induced male recombination in Drosophila. Genetics 144 1623–1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ralston, E.J., English, J., and Dooner, H.K. (1988). Sequence of three bronze alleles of maize and correlation with the genetic fine structure. Genetics 119 185–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ralston, E.J., English, J., and Dooner, H.K. (1989). Chromosome-breaking structure in maize involving a fractured Ac element. Proc. Natl. Acad. Sci. USA 86 9451–9455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reif, H.J., and Saedler, H. (1975). IS1 is involved in deletion formation in the gal region of E. coli K12. Mol. Gen. Genet. 137 17–28. [DOI] [PubMed] [Google Scholar]
- Shen, B. (2001). Transposon Ac-Facilitated Gene Search, Mutagenesis and Functional Characterization in Maize. PhD dissertation (Piscataway, NJ: Rutgers University).
- Shen, B., Zheng, Z., and Dooner, H.K. (2000). A maize sesquiterpene cyclase gene induced by insect herbivory and volicitin: Characterization of wild-type and mutant alleles. Proc. Natl. Acad. Sci. USA 97 14807–14812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svab, Z., and Maliga, P. (1993). High-frequency plastid transformation in tobacco by selection for a chimeric aadA gene. Proc. Natl. Acad. Sci. USA 90 913–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor, L.P., and Walbot, V. (1985). A deletion adjacent to the maize transposable element Mu1 accompanies loss of Adh1 expression. EMBO J. 4 869–876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Q., and Dooner, H.K. (2006). Remarkable variation in maize genome structure inferred from haplotype diversity at the bz locus. Proc. Natl. Acad. Sci. USA 103 17644–17649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weil, C.F., and Wessler, S.R. (1993). Molecular evidence that chromosome breakage by Ds elements is caused by aberrant transposition. Plant Cell 5 515–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan, X., Martinez-Ferez, I.M., Kavchok, S., and Dooner, H.K. (1999). Origination of Ds elements from Ac elements in maize: Evidence for rare repair synthesis at the site of Ac excision. Genetics 152 1733–1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, J., and Peterson, T. (1999). Genome rearrangements by nonlinear transposons in maize. Genetics 153 1403–1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, J., and Peterson, T. (2004). Transposition of reversed Ac element ends generates chromosome rearrangements in maize. Genetics 167 1929–1937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, J., and Peterson, T. (2005). A segmental deletion series generated by sister-chromatid transposition of Ac transposable elements in maize. Genetics 171 333–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, J., Zhang, F., and Peterson, T. (2006). Transposition of reversed Ac element ends generates novel chimeric genes in maize. PLoS Genet 2 e164. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.