Abstract
Plants have evolved robust mechanisms to respond and adapt to unfavorable environmental conditions, such as low temperature. The C-repeat/drought-responsive element binding factor CBF1/DREB1b gene encodes a transcriptional activator transiently induced by cold that controls the expression of a set of genes responding to low temperature (the CBF regulon). Constitutive expression of CBF1 confers freezing tolerance but also slows growth. Here, we propose that low temperature–induced CBF1 expression restrains growth at least in part by allowing the accumulation of DELLAs, a family of nuclear growth-repressing proteins, the degradation of which is stimulated by gibberellin (GA). We show that cold/CBF1 enhances the accumulation of a green fluorescent protein (GFP)–tagged DELLA protein (GFP-RGA) by reducing GA content through stimulating expression of GA-inactivating GA 2-oxidase genes. Accordingly, transgenic plants that constitutively express CBF1 accumulate less bioactive GA and as a consequence exhibit dwarfism and late flowering. Both phenotypes are suppressed when CBF1 is expressed in a line lacking two DELLA proteins, GA-INSENSITIVE and REPRESSOR OF GA1-3. In addition, we show that DELLAs contribute significantly to CBF1-induced cold acclimation and freezing tolerance by a mechanism that is distinct from the CBF regulon. We conclude that DELLAs are components of the CBF1-mediated cold stress response.
INTRODUCTION
Plants are sessile organisms and hence cannot escape unfavorable environmental conditions, such as cold. To overcome these limitations, plants have the potential to acclimate to low temperature by triggering a cascade of events leading to changes in gene expression and subsequently to biochemical and physiological modifications that enhance their freezing tolerance.
Transcript profiling experiments have shown that multiple regulatory pathways are activated during cold acclimation, and one such important pathway involves the CBF/DREB1 regulon (Thomashow, 1999; Shinozaki and Yamaguchi-Shinozaki, 2000; Fowler and Thomashow, 2002; Rabbani et al., 2003; Lee et al., 2005). The CBF/DREB1 proteins (CRT binding factor or DRE binding protein) are transcriptional regulators that bind specifically to the cold and dehydration-responsive cis-element, designated the CRT (C-repeat)/DRE (dehydration response element), present in the promoter of COR (cold-regulated) and a multitude of other stress-responsive genes known as the CBF regulon (Stockinger et al., 1997; Gilmour et al., 1998; Liu et al., 1998). CBF/DREB1 factors are members of the AP2/EREBP (Apetala2/ethylene-responsive element binding protein) family of transcription factors (Riechmann and Meyerowitz, 1998). In Arabidopsis thaliana, there are three genes encoding CBF/DREB1 proteins, known as CBF1, CBF2, and CBF3 (also referred to as DREB1b, DREB1c, and DREB1a, respectively) (Gilmour et al., 2004). Following cold exposure, these genes are rapidly and transiently induced via an abscisic acid–independent pathway, and their products activate the CBF regulon to enhance freezing tolerance (Agarwal et al., 2006; Nakashima and Yamaguchi-Shinozaki, 2006). Multiple mechanisms appear to contribute to this increase in freezing tolerance, including the synthesis of cryoprotective polypeptides (such as COR15a) and the accumulation of solutes, such as soluble sugars and Pro (Thomashow, 1998; Gilmour et al., 2004).
Expression of any one of the CBF proteins from a cDNA under control of the 35S cauliflower mosaic virus (CaMV) promoter in transgenic plants gives rise to strong constitutive expression of the CBF regulon and hence increased freezing tolerance (Jaglo-Ottosen et al., 1998; Kasuga et al., 1999; Gilmour et al., 2000, 2004). Arabidopsis CBF proteins are also effective in several other plant species; thus, a large number of transgenic plants overexpressing CBF proteins have been engineered to enhance freezing tolerance (Agarwal et al., 2006; Nakashima and Yamaguchi-Shinozaki, 2006). However, overexpression of these genes also resulted in severe growth retardation under normal growth condition (Jaglo-Ottosen et al., 1998; Kasuga et al., 1999; Gilmour et al., 2004).
The phytohormone gibberellin (GA) promotes important processes of plant growth and development, such as seed germination, growth through elongation, and floral transition (Richards et al., 2001). Key components of the GA-signaling pathway are the nuclear-localized growth repressing DELLA proteins (DELLAs) (Peng et al., 1997; Silverstone et al., 1998; Dill and Sun, 2001; King et al., 2001), a subset of the GRAS family of transcriptional regulators (Bolle, 2004; Zentella et al., 2007; De Lucas et al., 2008; Feng et al., 2008). The Arabidopsis genome encodes five DELLAs (GA-INSENSITIVE [GAI], REPRESSOR OF GA1-3 [RGA], RGA-LIKE1 [RGL1], RGL2, and RGL3) that all share the DELLA motif in their N-terminal domain (Peng et al., 1997; Dill and Sun, 2001; Silverstone et al., 2001; Lee et al., 2002; Wen and Chang, 2002). Recent genetic analyses have revealed both distinct but also overlapping functions for individual DELLAs in the regulation of plant development (Lee et al., 2002; Cheng et al., 2004; Tyler et al., 2004; Achard et al., 2006). According to the relief of restraint model, DELLAs restrain plant growth, whereas GA promotes growth by overcoming DELLA-mediated growth restraint (Harberd et al., 1998; Dill and Sun, 2001; King et al., 2001; Silverstone et al., 2001; Harberd, 2003). The GA signal is perceived by a GA receptor with homology to human hormone-sensitive lipase, GA-INSENSITIVE DWARF1 (GID1) (Ueguchi-Tanaka et al., 2005; Nakajima et al., 2006). The binding of bioactive GAs to GID1 promotes an interaction between GID1 and the DELLA domain of DELLAs (Griffiths et al., 2006; Ueguchi-Tanaka et al., 2007; Willige et al., 2007). Subsequently, this interaction enhances the affinity between DELLAs and a specific SCF E3 ubiquitin–ligase complex (involving the F-box protein called SLY1 [in Arabidopsis] or GID2 in rice [Oryza sativa]) (Griffiths et al., 2006; Willige et al., 2007), thus promoting the eventual destruction of DELLAs by the 26S proteasome (McGinnis et al., 2003; Sasaki et al., 2003; Dill et al., 2004; Fu et al., 2004). Thus, according to the current model, DELLAs restrain plant growth, whereas GA promotes growth by targeting the DELLAs for destruction.
Recent advances have revealed the role of DELLAs in many aspects of plant growth, including those that are influenced by environmental conditions (Lee et al., 2002; Cheng et al., 2004; Cao et al., 2005; Achard et al., 2006, 2007a, 2007b, 2008; Penfield et al., 2006). Interestingly, the growth of mutant plants lacking four of the five DELLAs (GAI, RGA, RGL1, and RGL2) is less inhibited by salt stress than is the growth of wild-type plants. By contrast, growth restraint conferred by DELLAs promotes survival (Achard et al., 2006, 2008). Thus, DELLA-dependent growth restraint is advantageous to plants and permits flexible and appropriate modulation of growth in response to changes in natural environments.
Although it is clear that both the cold-induced CBF and GA-signaling pathways regulate plant growth and stress tolerance, it is unclear whether and how these pathways interact. Recent experiments have shown that overexpression of DWARF AND DELAYED-FLOWERING1 (DDF1), an AP2 transcription factor closely related to the CBF/DREB1 family, conferred dwarfism, late flowering, and increased salt stress tolerance in Arabidopsis (Magome et al., 2004, 2008). Interestingly, GA treatment restored the wild-type phenotype. Similarly, constitutive expression of heterologous Arabidopsis CBF1 in tomato (Solanum lycopersicum) also conferred dwarfism, a phenotype that was prevented by application of GA (Hsieh et al., 2002). However, GA-treated transgenic tomato plants still exhibited a greater degree of dehydration and chilling tolerance compared with wild-type plants, which is not seen in Arabidopsis expressing DDF1 constitutively (Magome et al., 2004). Furthermore, the dwarf phenotype of transgenic Arabidopsis and tobacco (Nicotiana tabacum) plants overexpressing Arabidopsis CBF3 cannot be reversed by GA treatment (Agarwal et al., 2006). Thus, at present, it is unclear whether the CBF/DREB1 response pathway interacts with the GA pathway. Here, we undertook a molecular and genetic approach to evaluate the role of GA metabolism and signaling in the CBF1-dependent cold acclimation process in Arabidopsis. Essentially, we show that seedling roots of DELLA-deficient mutants have a reduced capacity for cold-induced root growth inhibition. We also show that cold-induced or constitutive expression of CBF1 promotes the accumulation of the green fluorescent protein (GFP)–tagged DELLA RGA and reduces plant growth. Accordingly, GA treatment or lack of functional GAI and RGA genes suppresses the growth defect of CBF1-ox plants. Thus, cold-induced CBF1 restrains growth at least in part via a DELLA-dependent mechanism. Moreover, we show that CBF1 reduces bioactive GA levels by increasing the abundance of transcripts encoding GA 2-oxidases, which may explain the CBF1-induced DELLA accumulation. Finally, we show that DELLA accumulation contributes significantly to the CBF1-induced cold acclimation and freezing tolerance by a mechanism that is independent of the well-known CBF regulon. We conclude that DELLA restraint is a component of the CBF1-mediated cold stress response.
RESULTS
Cold-Mediated Root Growth Inhibition Is Associated with DELLA Accumulation
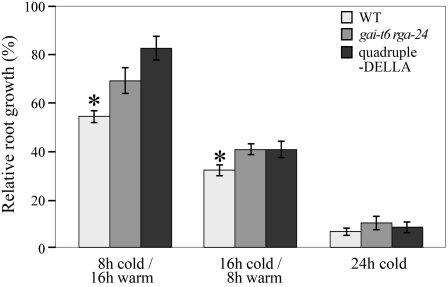
Although well characterized as growth inhibitors during adverse conditions, it is unknown whether the GA-signaling nuclear DELLA proteins inhibit growth during cold stress. We therefore investigated the involvement of DELLAs in the inhibition of primary root growth in response to cold using double-DELLA (gai-t6 rga-24) and quadruple-DELLA (gai-t6 rga-t2 rgl1-1 rgl2-1) mutant Arabidopsis lines that lack the corresponding DELLAs (King et al., 2001; Dill and Sun, 2001; Achard et al., 2006). In nature, plants are exposed to alternating periods of high and low temperatures that often correspond to day and night cycles (Guy, 1990). We found that DELLAs inhibit primary root growth to a degree proportionate to cold period length. Whereas the growth of gai-t6 rga-24 and quadruple-DELLA mutant roots was less affected by a daily 8-h cold exposure (4°C) than wild-type roots, the differences in root growth among these three lines decreased progressively with increase in cold period length (Figure 1). Thus, DELLA function restrains primary root growth during cold exposure but becomes less critical as the cold period length increases. Therefore, it is likely that additional (DELLA-independent) factors also restrict growth at low temperatures, particularly under long periods of exposure to cold.
Figure 1.
Low Temperature Slows Root Growth via a DELLA-Dependent Mechanism.
Mean (±se) relative growth of primary roots of 8-day-old wild-type (white), gai-t6 rga-24 (light gray), and the quadruple-DELLA mutant (gai-t6 rga-t2 rgl1-1 rgl2-1; dark gray) seedlings were grown for 5 d in alternating cold (4°C)/warm (22°C) temperatures as indicated (expressed as percentage of non-cold-treated control for each genotype). An asterisk indicates a significant difference between the wild type and gai-t6 rga24/quadruple-DELLA mutant (P < 0.05).
Next, we investigated whether cold-induced inhibition of root growth is associated with DELLA protein accumulation. For this purpose, we used pRGA:GFP-RGA transgenic seedlings expressing a GFP-RGA fusion protein that is detectable by protein gel blotting using an anti-GFP antibody (see Supplemental Figure 1 online; Silverstone et al., 2001). We found a twofold increase in GFP-RGA protein level within 4 h of transfer of pRGA:GFP-RGA seedlings from warm (22°C) to low temperature (4°C) (Figure 2A). This increase in GFP-RGA levels could be the consequence of an increase in GFP-RGA transcript levels or to a posttranscriptional effect, as both mechanisms are known to regulate DELLA protein accumulation (McGinnis et al., 2003; Sasaki et al., 2003; Dill et al., 2004; Fu et al., 2004; Oh et al., 2007). To discriminate between these possibilities, we performed an RNA gel blot experiment to compare GFP-RGA and RGA transcript levels in cold-treated and control seedlings. As shown in Figure 2B, exposure to cold had no detectable effect on either GFP-RGA or RGA transcript levels. Thus, cold enhances GFP-RGA accumulation (and probably that of other DELLAs) through a posttranslational mechanism, thereby inhibiting root growth.
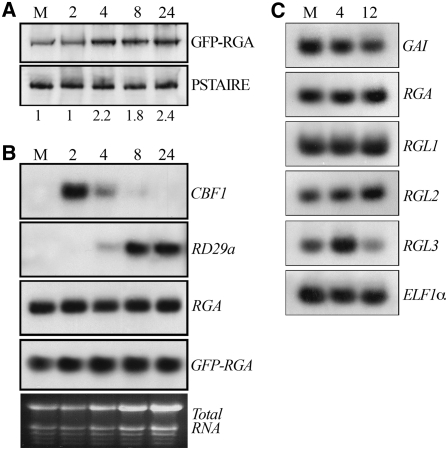
Figure 2.
Cold Enhances CBF1 Transcript and GFP-RGA Protein Levels.
(A) Immunodetection of GFP-RGA (with an antibody to GFP) in 7-d-old pRGA:GFP-RGA seedlings that had been subjected to cold (4°C) for the time (2, 4, 8, and 24 h) indicated (and controls, M). PSTAIRE serves as sample loading control. Numbers below the gels represent the fold increase in GFP-RGA protein levels relative to PSTAIRE levels.
(B) Expression of CBF1, RD29a, and RGA genes and the GFP-RGA transgene upon cold for the time as in (A) determined by RNA gel blot analysis. Each lane was loaded with 15 μg of total RNA prepared from 7-d-old pRGA:GFP-RGA seedlings.
(C) Levels of GAI, RGA, RGL1, RGL2, and RGL3 gene transcripts determined by a quantitative RT-PCR experiment in 7-d-old wild-type seedlings that had been subjected to cold for 4 and 12 h as indicated (and controls, M). ELF1α transcripts provide loading control.
To investigate whether cold affects the expression of other DELLA genes, we performed quantitative RT-PCR experiments for all five DELLAs. Interestingly, we found that RGL3 transcript levels were transiently increased by cold (Figure 2C), indicating that cold also specifically enhances RGL3 accumulation via its effect on gene expression.
CBF1 Restrains Plant Growth via the DELLA-Dependent Signaling Pathway
At present, the best-understood genetic system with a role in cold acclimation is the CBF cold response pathway. Upon exposure to low temperature, the CBF genes are rapidly induced and their products activate the expression of downstream CBF-regulated genes (Figure 2B; Jaglo-Ottosen et al., 1998; Gilmour et al., 2004). We found that GFP-RGA levels increase after 4 h of cold treatment, thus following CBF1 induction (Figure 2B). Furthermore, it was previously reported that transgenic Arabidopsis plants overexpressing CBF1 were dwarfed and exhibited a late-flowering phenotype compared with wild-type plants (Gilmour et al., 2004), a phenotype that is usually observed with GA-deficient plants (Richards et al., 2001). Altogether, these observations strongly suggest that low temperature enhances DELLA accumulation via a CBF-dependent activation.
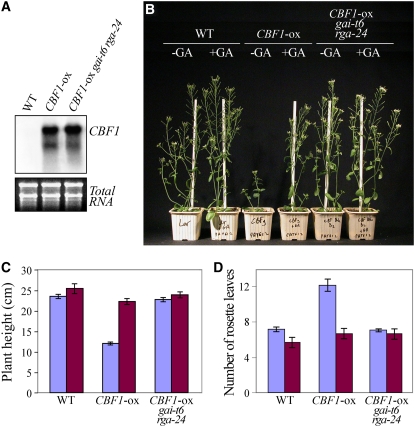
To examine the involvement of the GA/DELLA components in the CBF cold response pathway, we created transgenic Arabidopsis plants that constitutively express CBF1 under the control of the 35S CaMV promoter. Several T3 lines were identified, and a transgenic line that moderately expresses the corresponding CBF1 transgene (CBF1-ox) was selected for further studies (see Supplemental Figure 2 online). RNA gel blot analysis confirmed that CBF1-ox plants accumulated CBF1 transcripts in the absence of cold stress in contrast with wild-type plants (Figure 3A). CBF1-ox plants exhibited similar phenotypic alterations as previously described (Gilmour et al., 2004); the plants were shorter, flowered later, and their leaves were smaller and darker than those of wild-type plants. First, we investigated the responsiveness of CBF1-ox plants to exogenous GA. We found that GA treatment both rescued the normal growth and abolished the late-flowering phenotype of CBF1-ox plants (Figures 3B to 3D). Thus, as suggested above, CBF1 represses plant growth via a GA-dependent mechanism. The developmental effects of GA on plant growth are mediated by the destruction of DELLAs (Harberd, 2003; McGinnis et al., 2003; Sasaki et al., 2003; Dill et al., 2004; Fu et al., 2004). Because GA overcomes CBF1-ox–conferred dwarfism and late flowering, we next tested the hypothesis that CBF1 restrains growth via a DELLA-dependent mechanism. We found that lack of two DELLAs, GAI and RGA, in a CBF1-ox gai-t6 rga-24 genetic background suppressed the growth inhibition and late flowering phenotype (Figures 3B to 3D). Thus, CBF1 does indeed restrain growth via a DELLA-dependent mechanism.
Figure 3.
CBF1 Represses Growth and Delays Floral Transition through the GA-Signaling Pathway.
(A) Expression of CBF1 gene in wild-type, CBF1-ox, and CBF1-ox gai-t6 rga-24 plants. Each lane was loaded with 15 μg of total RNA prepared from 2-week-old wild-type and transgenic plants. The RNA gel blot was hybridized with a specific probe for CBF1.
(B) Representative 4-week-old wild-type, CBF1-ox, and CBF1-ox gai-t6 rga-24 plants treated with GA (+GA) or control (–GA).
(C) and (D) Mean (±se) plant height (cm) (C), and mean (±se) rosette leaf number (D) of 4-week-old wild-type, CBF1-ox, and CBF1-ox gai-t6 rga-24 plants GA-treated (violet) or control (blue).
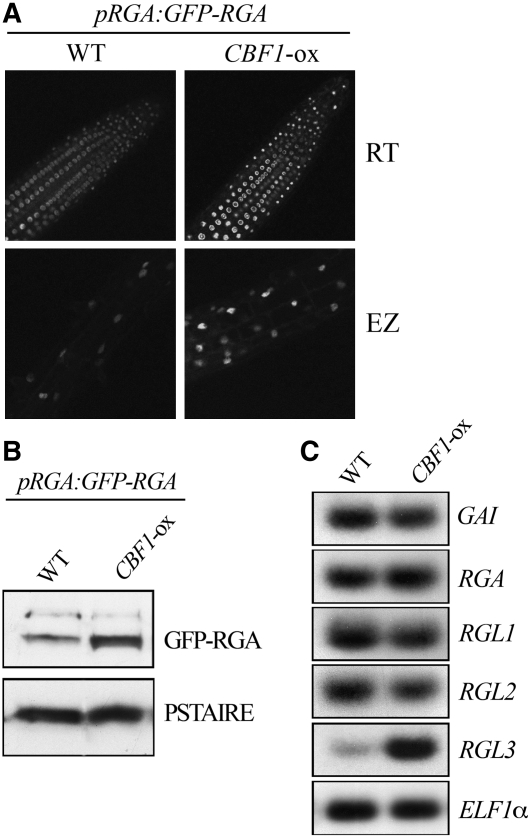
In a subsequent experiment, we determined the effect of CBF1 on DELLA protein accumulation. For this study, we crossed CBF1-ox with the pRGA:GFP-RGA line expressing a GFP-RGA fusion protein that is detectable in root cell nuclei (Silverstone et al., 2001). We found that the nuclear fluorescence attributable to GFP-RGA was brighter in cells of CBF1-ox root seedlings, particularly in the elongating zone of the root (Figure 4A). Consistently, the increase in GFP-RGA fluorescence observed in CBF1-ox plants was associated with an increase in GFP-RGA protein level (Figure 4B). Thus, CBF1 enhances DELLA protein accumulation and consequently restrains plant growth.
Figure 4.
CBF1 Represses Plant Growth by Enhancing DELLA Accumulation.
(A) GFP fluorescence (viewed by fluorescence confocal microscopy) in nuclear cells of the root tip (RT) and elongating zone (EZ) of 7-d-old seedlings of the pRGA:GFP-RGA (wild-type) and CBF1-ox pRGA:GFP-RGA lines. Images were taken with identical parameters to allow for comparison of fluorescence intensities.
(B) Immunodetection of GFP-RGA (with an antibody to GFP) in 7-d-old seedlings of pRGA:GFP-RGA (wild-type) and CBF1-ox pRGA:GFP-RGA lines. PSTAIRE serves as sample loading control.
(C) Levels of GAI, RGA, RGL1, RGL2, and RGL3 gene transcripts determined by quantitative RT-PCR experiment in 7-d-old wild-type and CBF1-ox seedlings. ELF1α transcripts provide loading control.
Finally, we investigated if CBF1-induced DELLA accumulation was attributable to an increase in DELLA transcript levels or to a posttranslational effect. Thus, we compared the levels of DELLA gene expression in wild-type and CBF1-ox plants. We found that expression of the CBF1-ox transgene, like cold treatment, had no detectable effect on either DELLA transcript levels, except for RGL3, which accumulated (Figure 4C). These results suggest that CBF1 enhances RGA accumulation (and most likely other DELLA proteins) through a posttranslational mechanism but also specifically enhances RGL3 transcript accumulation.
GA 2-Oxidase Gene Transcripts Are Upregulated upon Exposure to Cold and in CBF1-ox Plants
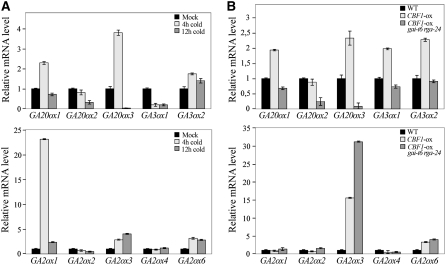
Reduced bioactive GA levels cause an increase in DELLA protein accumulation and consequently growth inhibition (Silverstone et al., 2001; Fu et al., 2004). Moreover, GA metabolism is regulated by both developmental and environmental stimuli (Hedden and Phillips, 2000). We next investigated the possibility that cold/CBF1-induced accumulation of DELLAs might be the result of CBF1-mediated transcriptional regulation of genes involved in GA metabolism. GA metabolism is tightly regulated through the modulation of the expression of members of small gene families encoding GA 20-oxidases (GA20ox) and GA 3-oxidases (GA3ox), which catalyze successive steps in the synthesis of bioactive GAs (Phillips et al., 1995, Chiang et al., 1995), but also GA 2-oxidases (GA2ox), which deactivate bioactive GAs (Thomas et al., 1999). We speculated that cold-mediated CBF1 induction either reduces GA20ox and/or GA3ox transcript levels and/or enhances GA2ox transcript levels. To test this hypothesis, we first determined the effect of cold and CBF1-ox on the accumulation of selected GA20ox and GA3ox gene transcripts. Surprisingly, we found that 2-week-old wild-type plants, exposed to cold for 4 h, and CBF1-ox plants accumulate substantially higher levels of GA20ox1, GA20ox3, and GA3ox2 transcripts than control plants (non-cold-treated wild-type plants) (Figures 5A and 5B). Interestingly, these genes were only transiently upregulated by cold, as their expression levels dropped to normal after 12 h of cold exposure. By contrast, the level of GA20ox2 transcripts was unchanged and the level of GA3ox1 transcripts was reduced upon cold treatment but increased in CBF1-ox plants. Thus, the cold/CBF1-induced accumulation of DELLAs is not due to a transcriptional repression of GA20ox or GA3ox genes. It is noteworthy that previous work has shown that increased DELLA activity (as seen in GA-deficient and GA-insensitive mutants) results in increased levels of GA20ox1 and GA3ox1 transcripts and in decreased levels of GA2ox transcripts due to a feedback mechanism (Hedden and Kamiya, 1997; Cowling et al., 1998; Xu et al., 1999; Thomas et al., 1999). Thus, it is likely that the increase in GA20ox and GA3ox transcript levels observed in 4 h cold-treated wild-type plants and in CBF1-ox plants is the result of such a mechanism (Figure 5A). Accordingly, CBF1-ox gai-t6 rga-24 plants exhibited reduced levels of GA20ox and GA3ox transcripts in comparison to CBF1-ox and wild-type plants.
Figure 5.
Cold and CBF1-ox Upregulate GA 2-oxidase Transcript Levels.
(A) Relative levels of GA biosynthesis GA20ox1, GA20ox2, GA20ox3, GA3ox1, and GA3ox2 and GA deactivation of GA2ox1, GA2ox2, GA2ox3, GA2ox4, and GA2ox6 gene transcripts (determined by real-time RT-PCR) in 2-week-old wild-type seedlings that had been subjected to cold for 4 h (light gray) and 12 h (dark gray) and control non-cold-treated (Mock, black). Data are means ± se.
(B) Relative levels of GA metabolism gene transcripts determined by real-time RT-PCR (as in [A]) in 2-week-old wild-type (black), CBF1-ox (light gray), and CBF1-ox gai-t6 rga-24 (dark gray) plants.
Next, we determined the level of GA2ox gene transcripts in cold-treated wild-type plants and CBF1-ox plants (Figures 5A and 5B). We found that 2-week-old 4 h cold-treated wild-type plants contain relatively higher amounts of GA2ox1, GA2ox3, and GA2ox6 transcripts in comparison to non-cold-treated plants. By contrast, CBF1-ox plants exhibited increased levels of GA2ox3 and GA2ox6 transcripts but not of GA2ox1 transcripts. Thus, it is likely that cold enhances GA2ox1 transcript levels in a CBF1-independent manner. Moreover, the increase in GA2ox3 and GA2ox6 transcripts is not related to DELLA status (to the feedback mechanism), as CBF1-ox gai-t6 rga-24 plants also exhibited higher GA2ox transcript levels. Thus, in contrast with GA20ox and GA3ox, the increase in GA2ox transcripts observed in cold-treated wild-type plants and in CBF1-ox plants is not due to a feedback mechanism.
The upregulation of GA2ox expression in cold-treated and CBF1-ox plants may lead to a decrease in bioactive GAs. To determine if this was the case, we compared in separate experiments the endogenous GA contents in cold-treated and untreated wild-type plants and in wild-type and CBF1-ox plants. As shown in Table 1, the level of the major biologically active GA, GA4, was significantly reduced in CBF1-ox plants compared with wild-type plants, while the content of its inactive 2β-hydroxylated product, GA34, was increased. GA4, as well as GA1, contents were also reduced after cold treatment by ∼50 and 30%, respectively. While the content of GA8, the 2β-hydroxylated product of GA1, increased after cold treatment, there was a slight reduction in GA34 content after this treatment. Some reduction in GA content in the cold-treated plants might be anticipated as a result of lower metabolic rates, but GA4 content was reduced much more than that of GA34, consistent with a higher rate of 2β-hydroxylation under these conditions. Taken together, the above results indicate that cold-mediated CBF1 induction enhances DELLA protein accumulation through a posttranslational mechanism mediated by a reduction of bioactive GA (via upregulation of GA2ox gene transcript levels). In turn, accumulation of DELLAs enhances GA20ox and GA3ox gene transcript levels by a feedback mechanism.
Table 1.
Cold and CBF1-ox Reduce Bioactive GA Levels via Upregulation of GA 2-Oxidase Activity
| ng/g Dry Weight | GA4 | GA1 | GA34 | GA8 |
|---|---|---|---|---|
| Wild type | 11.37 ± 0.26 | 0.68 ± 0.03 | 19.41 ± 0.13 | 1.82 ± 0.02 |
| Wild type cold 4 h | 5.97 ± 0.01 | 0.46 ± 0.01 | 16.95 ± 0.24 | 2.28 ± 0.04 |
| Wild type cold 12 h | 6.02 ± 0.02 | 0.44 ± 0.01 | 17.21 ± 0.14 | 2.30 ± 0.08 |
| ng/g Dry Weight
|
GA4
|
GA34
|
||
| Wild type | 4.59 ± 0.07 | 44.94 ± 0.45 | ||
| CBF1-ox | 2.74 ± 0.03 | 66.58 ± 0.16 | ||
Concentrations of GAs (ng g dry weight−1 ±sd) in wild-type plants subjected to cold for 4 and 12 h (and controls, wild-type) and in separate experiments in wild-type and CBF1-ox plants. The values are means of three and two biological replicates, respectively.
DELLAs Contribute to CBF1-Induced Freezing Tolerance and Cold Acclimation
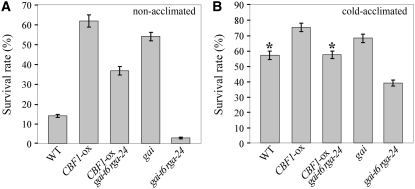
It has recently been established that DELLAs promote salt tolerance (Achard et al., 2006, 2008). However, the contribution of DELLAs in promoting tolerance to other abiotic stresses, such as freezing, is unknown. To determine the role of DELLAs in freezing tolerance and cold acclimation, we examined the freezing tolerance of 2-week-old GA-signaling mutants before and after cold acclimation (4°C, 3 d). Plants were exposed for 24 h to −10°C, and survival was scored after 4 d of recovery under controlled growth conditions (see Supplemental Figure 3 online). Strikingly, we found that the GA response mutant gai (Koornneef et al., 1985; Peng et al., 1997) exhibited increased tolerance toward freezing, whereas gai-t6 rga-24 mutant plants were less tolerant than wild-type plants (Figure 6). Thus, DELLA function is not restricted to salt stress and promotes tolerance to other environmental stresses. Furthermore, gai-t6 rga-24 mutant plants were significantly less freezing tolerant than wild-type plants after cold acclimation, despite the fact that gai-t6 rga-24 plants were still responding to cold acclimation. Overall, these observations suggest that DELLAs contribute significantly in a cold adaptive response.
Figure 6.
DELLAs Contribute to CBF1-Induced Freezing Tolerance and Cold Acclimation.
Freezing tolerance (expressed as rate of survival [%]; three replicates; n = 24) of 2-week-old wild-type, CBF1-ox, CBF1-ox gai-t6 rga-24, gai, and gai-t6 rga-24 plants nonacclimated (A) or cold-acclimated (4°C, 3 d) (B). Plants were exposed for 24 h at −10°C, and survival was scored after 4 d of recovery under controlled growth conditions. The asterisks indicate no significant difference (P < 0.05) between wild-type and CBF1-ox gai-t6 rga-24 cold-acclimated plants.
Next, we determined whether CBF1 promoted freezing tolerance in a DELLA-dependent fashion. Thus, we compared the freezing tolerance of CBF1-ox and CBF1-ox gai-t6 rga-24 plants. Remarkably, we found that CBF1-ox gai-t6 rga-24 plants exhibited lower levels of freezing tolerance than CBF1-ox plants, suggesting that DELLAs contribute at least in part to CBF1-induced freezing tolerance. The fact that the survival rates of nonacclimated CBF1-ox gai-t6 rga-24 mutant plants were higher (36.7%) than gai-t6 rga-24 plants (2.8%) shows either that additional DELLAs (these plants retain RGL1, RGL2, and RGL3) mediate the effect of CBF1 on freezing tolerance or more likely that, in addition to the DELLA-dependent mechanism, CBF1 also promotes freezing tolerance via a DELLA-independent mechanism.
CBF1 Activates the CBF Regulon via a DELLA-Independent Mechanism
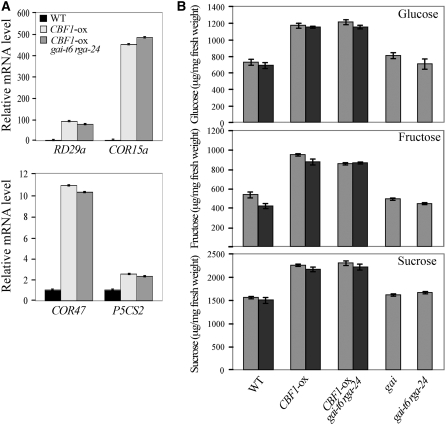
As a next step, we investigated whether DELLAs contribute to CBF1-induced freezing tolerance via the well-characterized CBF regulon. When plants are exposed to low temperature, CBF target genes are rapidly induced and act to increase freezing tolerance (see Supplemental Figure 4A online; Jaglo-Ottosen et al., 1998; Gilmour et al., 2004). As previously shown, we found that transcript levels of RD29a (COR78), COR15a, and COR47, three known CBF1-inducible COR genes (Gilmour et al., 2004), were upregulated in CBF1-ox plants in compared with wild-type plants (Figure 7A). Interestingly, there was no significant difference in transcript levels for these genes between the wild type and gai or gai-t6 rga-24 (see Supplemental Figure 4A online) or between CBF1-ox and CBF1-ox gai-t6 rga-24 plants (Figure 7A). The levels of Pro, an important protective osmolyte in higher plants, are also increased during cold acclimation or by CBF1 overexpression in nonacclimated plants (Gilmour et al., 2004). It was previously shown that expression of the gene encoding Δ1-pyrroline-5-carboxylate synthase 2 (P5CS2), an enzyme involved in Pro biosynthesis, was upregulated in CBF-ox plants. Accordingly, we found that P5CS2 transcript levels were elevated in CBF1-ox plants compared with wild-type plants (Figure 7A). However, lack of GAI and RGA in CBF1-ox gai-t6 rga-24 plants did not alter P5CS2 transcript accumulation, which remains at the level of CBF1-ox plants. Thus, DELLAs do not modulate the expression levels of the COR and Pro biosynthesis genes tested here, suggesting that CBF1-induced DELLA accumulation enhances stress tolerance via a different mechanism.
Figure 7.
CBF1 Enhances Both CBF Regulon and Sugar Levels in a DELLA-Independent Fashion.
(A) Relative levels of RD29a, COR15a, COR47, and P5CS2 gene transcripts (determined by real-time RT-PCR) in 2-week-old wild-type, CBF1-ox, and CBF1-ox gai-t6 rga-24 plants. Data are means ± se.
(B) Mean (±se) total soluble sugars (glucose, fructose, and sucrose) were determined from leaf tissue of 2-week-old wild-type, CBF1-ox, CBF1-ox gai-t6 rga-24, gai, and gai-t6 rga-24 plants GA-treated (dark gray) or not (light gray). The levels of individual sugars were determined by enzymatic assays.
The accumulation of simple sugars is another commonly observed biochemical change that occurs in cold-acclimated plants or in CBF-overexpressing plants (Gilmour et al., 2000, 2004). Soluble sugars, such as sucrose, glucose, or fructose, have cryoprotective properties (Gilmour et al., 2000; Rolland et al., 2002). Thus, it was of interest to determine whether GA/DELLA contributes to CBF1-mediated stress tolerance by modulating sugar levels. We found that cold-acclimated wild-type plants and CBF1-ox plants accumulated soluble sugars (glucose, fructose, and sucrose) at a higher level (∼1.5-fold higher) than those found in control plants (non-cold-acclimated and wild-type plants, respectively) (see Supplemental Figure 4B online; Figure 7B). However, there was no difference between the wild type and gai or gai-t6 rga-24 or between CBF1-ox and CBF1-ox gai-t6 rga-24 plants. Thus, DELLAs do not contribute to CBF1-mediated increase in sugars levels.
Overall, these results strongly suggest that DELLAs contribute to CBF1-induced freezing tolerance by a mechanism that is distinct from the CBF regulon.
DISCUSSION
The process of cold acclimation involves numerous physiological and biochemical changes. The most notable changes include reduction or cessation of growth and accumulation of cryoprotective molecules, such as Pro and soluble sugars. The GA-regulated growth-repressing DELLA proteins integrate endogenous and environmental cues in the regulation of plant growth (Alvey and Harberd, 2005; Achard et al., 2006, 2007a, 2007b). It was proposed that DELLAs permit flexible and appropriate modulation of plant growth in response to changes in natural environments (Alvey and Harberd, 2005; Achard et al., 2006). The experiments described here investigate the relationship between the GA pathway and the CBF1-dependent cold signaling pathway.
Low temperature is one of the most important factors affecting plant performance and thus plant growth. All biochemical processes run more slowly during long exposure to cold (Stitt and Hurry, 2002). Low temperatures slow down enzyme-catalyzed reactions and modify the conformation of lipids and other macromolecules, with consequences for most biological processes. To minimize the impact of such processes, we initially looked at the effect of low temperatures in fast-growing tissue, such as seedling roots. We found that low temperature promoted the accumulation of a GFP-RGA fusion protein and that growth of gai-t6 rga-24 and quadruple-DELLA mutant roots was less affected by a short exposure to cold than was growth of wild-type roots. The difference in root growth between these lines decreases progressively with increase in cold period length, prolonged exposure to cold mainly reducing growth by slowing metabolism. Accordingly, cold treatment retarded at the same level the growth of wild-type and quadruple-DELLA mutant adult plants (see Supplemental Figure 5 online). Thus, low temperature represses growth in part via a DELLA-dependent pathway, the growth repression being more severe than that due simply to a slowing of metabolism at low temperature. This mechanism could have its importance particularly when plants are exposed to alternating periods of high and low temperatures.
The CBF cold response pathway plays a central role in cold acclimation (Cook et al., 2004). CBF1, CBF2, and CBF3 genes have previously been shown to respond rapidly to low temperature and to induce the expression of various proteins (CBF regulon) that protect plants from freezing temperatures (Agarwal et al., 2006; Nakashima and Yamaguchi-Shinozaki, 2006). Accordingly, overexpression of CBF genes in transgenic Arabidopsis plants conferred tolerance to freezing but also resulted in severe growth retardation under normal growth conditions (Jaglo-Ottosen et al., 1998; Liu et al., 1998; Gilmour et al., 2000, 2004). Moreover, transgenic tomato plants overexpressing Arabidopsis CBF1 exhibited dwarfism, which could be reversed by GA treatment (Hsieh et al., 2002). Dwarfism was also observed in transgenic Arabidopsis plants overexpressing CBF4 and DDF1 genes, close homologs of the CBF/DREB1 gene family members, and it was reported that DDF1 is involved in the regulation of GA metabolism (Haake et al., 2002; Magome et al., 2004, 2008). Here, we have shown that DELLA activity is substantially responsible for the dwarfism of Arabidopsis CBF1-ox plants, as lack of GAI and RGA (in CBF1-ox gai-t6 rga-24) suppressed this phenotype. Furthermore, we showed that a GFP-RGA protein accumulated to higher levels in CBF1-ox plants. Thus, we conclude that CBF1 enhances DELLA accumulation and thereby inhibits plant growth. Because DDF1 also modulates plant growth through a GA-dependent mechanism (Magome et al., 2004, 2008), it is likely that a large number of DREB factors regulate growth by interfering with the GA pathway.
Bioactive GA relieves DELLA restraint by promoting the degradation of DELLAs by the proteasome (Silverstone et al., 2001; Dill et al., 2004; Fu et al., 2004). We next determined the levels of gene transcripts encoding GA metabolic enzymes. This analysis revealed that cold and CBF1 induction increase the level of GA2ox3 and GA2ox6 transcripts encoding an enzyme that deactivates bioactive GAs. Accordingly, we found that cold-treated wild-type plants and CBF1-ox plants contained reduced levels of bioactive GAs. We also found that exogenous GA rescued the growth of CBF1-ox plants. Thus, we conclude that cold-mediated CBF1 induction modulates GA metabolism via upregulation of GA2ox3 and GA2ox6 gene transcript levels, contributing to the accumulation of DELLAs. Analyses of GA2ox gene promoter sequences have not revealed the presence of CRT/DRE-like cis-elements, suggesting that CBF1 upregulates GA2ox gene expression indirectly. Interestingly and in contrast with CBF1-ox plants, we found that cold-treated wild-type plants also exhibited a transient increase in GA2ox1 transcript levels, suggesting that cold enhances GA2ox levels via both CBF1-dependent and -independent manners.
It was recently suggested that DELLA-dependent growth restraint is advantageous in adverse environments (Achard et al., 2006). Thus, we speculated that DELLAs may contribute to the CBF1-mediated freezing tolerance. Indeed, we showed that lack of GAI and RGA (in CBF1-ox gai-t6 rga-24) substantially suppressed the freezing tolerance exhibited by CBF1-ox plants. Furthermore, cold-acclimated wild-type plants were more resistant to freezing than gai-t6 rga-24 plants. Thus, cold-induced CBF1 promotes freezing tolerance and cold acclimation at least in part by enhancing DELLA accumulation. However, because CBF1-ox gai-t6 rga-24 plants were still more tolerant than gai-t6 rga-24 plants, it is likely that CBF1 also promotes freezing tolerance through DELLA-independent pathway(s).
Cold-induced or constitutive expression of CBF1 results in upregulation of COR and Pro biosynthesis genes and the accumulation of soluble sugars, including sucrose, glucose, and fructose via modulation of the CBF regulon (Gilmour et al., 2004). There is evidence that COR polypeptides, Pro, and sugars contribute to freezing tolerance (Thomashow, 1998; Rolland et al., 2002; Gilmour et al., 2004). Our results suggest that CBF1 modulates the CBF regulon via a DELLA-independent pathway. Taken together, these findings lead us to propose that CBF1 activates multiple mechanisms, DELLA-dependent and -independent, that work in concert to enhance freezing tolerance (Figure 8). The precise way by which DELLAs enhance freezing tolerance remains unclear. We have recently shown that DELLAs promote survival of adversity by reducing the accumulation of reactive oxygen species (ROS) levels, and it was reported that low temperature could lead to the production of ROS (Huner et al., 1998; Fowler and Thomashow, 2002; Achard et al., 2008). Perhaps DELLAs contribute to freezing tolerance through the regulation in ROS levels.
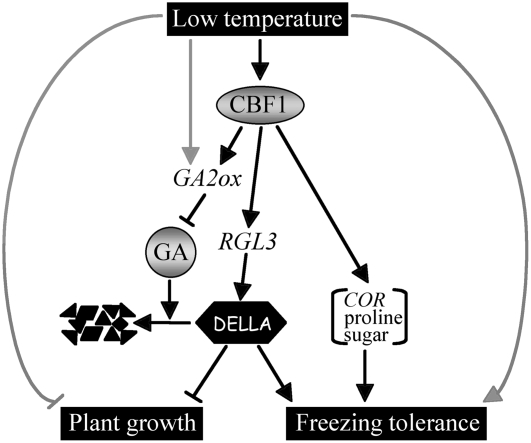
Figure 8.
Scheme Showing Contribution of DELLAs to CBF1-Mediated Growth Regulation and Freezing Tolerance.
Cold-induced or constitutive expression of CBF1 reduces bioactive GA levels (by increasing of GA2ox3 and GA2ox6 transcript abundance), thus promoting the accumulation in DELLAs by change in their protein stability. In addition, cold/CBF1 specifically increases RGL3 transcript levels. Low temperature also enhances GA2ox1 expression levels via a CBF1-independent mechanism. DELLA accumulation in turn slows plant growth. CBF1 also enhances freezing tolerance through the synergistic DELLA-dependent and COR-dependent pathways. Cold also restrains growth and promotes freezing tolerance by a DELLA-independent and CBF-independent cold response pathway.
We propose a model in which the induction of CBF1 expression by low temperature affects GA metabolism via upregulation of GA2ox gene transcripts (Figure 8). An increase in GA2ox enzyme levels results in a reduction in bioactive GAs, thus causing a higher accumulation of DELLAs (by increasing their stability). Our results also indicate that CBF1 specifically regulates RGL3 gene expression. As a consequence, increased DELLA accumulation restrains plant growth. Moreover, the CBF1-induced DELLA accumulation also contributes in a synergistic manner with the CBF1-induced COR pathway to promote cold adaptation. Finally, it is important to note that genetic studies suggest that the CBF cold response pathway is not the only system that contributes to cold acclimation. For example, eskimo1 mutant plants are more compact in stature and are constitutively freezing tolerant, yet by a mechanism that is distinct from the CBF cold response pathway (Xin and Browse, 1998).
METHODS
Plant Materials and Growth Conditions
All experiments used the Landsberg erecta laboratory strain of Arabidopsis thaliana as genetic background. The gai, gai-t6 rga-24, quadruple-DELLA mutant (gai-t6 rga-t2 rgl1-1 rgl2-1), and pRGA:GFP-RGA were as previously described (Achard et al., 2006). Plants were grown on soil or GM agar medium plates (Achard et al., 2007b) under long days (16 h light/8 h dark) at 22°C. Flowering time was expressed as the number of vegetative leaves produced before flowering. For root growth experiments, wild-type, gai-t6 rga-24, and quadruple-DELLA mutant seeds were surface sterilized and placed on GM plates at 4°C for 4 d to synchronize germination. Subsequently, plates were placed vertically in a controlled environment chamber (22°C; continuous light) for 3 d before transfer periodically (for 8 h or 16 h per day) or continuously to cold (4°C; continuous light). After 5 d, plates were scanned and root length was measured using ImageJ software (public domain; http://rsb.info.nih.gov/ij). For each experiment, roots from at least 30 seedlings were measured.
Vector Construction, Transformation, and Crossing
The CBF1 cDNA was amplified by RT-PCR from mRNA extracted from 2-week-old plants that had been cold treated (4°C) for 2 h and was cloned into the pGEM-T easy vector system (Promega). A BamHI-EcoRI fragment containing the CBF1 cDNA was inserted into pGreen0229 containing a 35S CaMV cassette (derived from the p35S-2 cassette) and Basta resistance marker (http://www.pgreen.ac.uk/). The plasmid was introduced into Agrobacterium tumefaciens GV3101 by electroporation. Arabidopsis plants were transformed by floral dip. The CBF1-ox pRGA:GFP-RGA and CBF1-ox gai-t6 rga-24 lines were isolated from F3 progeny of the appropriate crosses. Genomic PCR and kanamycin resistance were used to confirm the gai-t6 rga-24 genotype and the presence of pRGA:GFP-RGA transgene, respectively (see also Achard et al., 2006).
Transcript Level Analyses
For expression analysis, total RNA was extracted using Trizol reagent (Molecular Research Center) from 7-d-old seedlings or 2-week-old rosette leaves as indicated. For RNA gel blot analysis of cold-treated seedlings, plants were placed at 4°C for 2, 4, 8, and 24 h. RNA gel blot transfers onto Hybond-N membrane (Amersham Biosciences) were performed in 10× SSC and hybridized at 42°C in SSPE containing 50% formamide with specific 32P-random labeled probe (Promega). Blots were stripped at 80°C in 0.1× SSC/0.1% SDS. For quantitative real-time RT-PCR analysis, 2 μg of total RNA was treated first with 2 units of DNase I (Promega) and then reverse transcribed in a total volume of 40 μL with 2 μM oligo(dT)20, 0.5 mM deoxynucleotide triphosphate, 5 mM DTT, and 200 units of Superscript III reverse transcriptase (Invitrogen). RT-PCR was performed using gene-specific primers in a total volume of 15 μL SYBR Green Master mix (Roche) on a Lightcycler LC480 apparatus (Roche) according to the manufacturer's instructions. The GAPDH and At4g26410 (unknown function) genes were used as internal controls. The relative expression level of each gene in transgenic plants or in cold-treated plants was compared with that in non-cold-treated wild-type plants using GenEx Pro 4.3.5. software (MultiD Analyses) after normalization using the GAPDH cDNA level and averaging over three replicates. For quantitative RT-PCR experiments, after low-cycle PCR amplification (15 cycles) using specific primers, reaction products were blotted onto Hybond-N membrane (Amersham Biosciences) and probed at 65°C in 0.25 M sodium phosphate, pH 7.2/7% SDS with the equivalent PCR amplified 32P-random–labeled fragment (Promega). Blots were exposed to film (within the linear range of detection). The images were scanned and band intensities were quantified using ImageJ software. Two replicates were performed for each experiment. Primers for PCR amplification/probe preparation are listed in Supplemental Methods online.
Observation of GFP Fluorescence
Confocal microscopy images were obtained with a Zeiss LSM510 inverted confocal laser microscope with ×40 objectives. The excitation wavelength for GFP detection was 488 nm. All images were obtained with the same modifications and intensity parameters. GFP-RGA fluorescence was determined on 7-d-old pRGA:GFP-RGA and CBF1-ox pRGA:GFP-RGA root tip and elongating zone of the primary root.
Immunoblot Analyses
Tissues for protein analyses were ground in 2× SDS-PAGE sample buffer followed by boiling for 5 min. After centrifugation, the protein extracts were fractionated on a 10% SDS-PAGE gel and blotted onto membrane. Immunoblots were performed using a 2500-fold dilution of anti-GFP monoclonal antibodies from mouse (BD Biosciences) and a 5000-fold dilution of peroxidase-conjugated goat anti-mouse IgG (Molecular Probes). Signals were detected by film (within the linear range of detection) using the enhanced chemiluminescence protein gel blot analysis system (Amersham Biosciences). The blot was subsequently stripped with 0.2 n glycine, pH 2.5, and reprobed with anti-cdc2 (PSTAIRE) antibody (Santa Cruz Biotechnology) for loading control. Signals of GFP-RGA (Figure 2A) were scanned and then quantified using ImageJ software and normalized with the respective signals of PSTAIRE. Each immunoblot assay was repeated at least twice.
GA Determinations
GA determinations were performed on soil-grown 2-week-old vegetative rosettes. When indicated, the plants were subjected to cold for 4 and 12 h. Biological replicates (0.1 to 0.4 g dry weight) were analyzed by gas chromatography–mass spectrometry selected reaction monitoring with [2H2]GA internal standards as described elsewhere (Rieu et al., 2008).
Sugar Analysis
Glucose, fructose, and sucrose were measured with a sequential enzymatic assay (production of NADH) using glucose (G2020), fructose (F2668), and sucrose assay (S1299) kits (Sigma-Aldrich). Production of NADH was monitored at 340 nm with a spectrophotometer ELx808 (Bio-Tek Instruments). The experiment was repeated three times (n = 10).
Freezing Tolerance Assays
Two-week-old plants grown in a growth room (22°C; 16 h photoperiod) were placed at −10°C for 24 h. Plants were then placed at 4°C for 6 h for recovery before being returned to the original growth room. Plant survival was scored after 4 d of recovery under normal growth conditions. Cold acclimation was performed at 4°C for 3 d. The experiment was repeated three times (n = 24).
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: CBF1/DREB1b, AT4G25490; GAI, AT1G14920; RGA, AT2G01570; RGL1, AT1G66350; RGL2, AT3G03450; RGL3, AT5G17490; GA20ox1, AT4G25420; GA20ox2, AT5G51810; GA20ox3, AT5G07200; GA3ox1, AT1G15550; GA3ox2, AT1G80340; GA2ox1, AT1G78440; GA2ox2, AT1G30040; GA2ox3, AT2G34555; GA2ox4, AT1G47990; GA2ox6, AT1G02400; RD29a/COR78, AT5G52310; COR15a, AT2G42540; COR47, AT1G20440; and P5CS2, AT3G55610.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. The pRGA:GFP-RGA Transgenic Line Expresses a GFP-RGA Fusion Protein That Is Detectable by Protein Gel Blot Using an Anti-GFP Antibody.
Supplemental Figure 2. Expression of CBF1 in T3 Transgenic CBF1-ox Lines.
Supplemental Figure 3. Arabidopsis DELLAs Contribute Significantly to Freezing Tolerance.
Supplemental Figure 4. Cold Enhances Both CBF Regulon and Sugar Levels via a DELLA-Independent Mechanism.
Supplemental Figure 5. Cold Treatment Slows at the Same Level the Vegetative Growth of Wild-Type and Quadruple-DELLA Mutant Plants.
Supplemental Methods. Primer List.
Supplementary Material
Acknowledgments
We thank Nick Harberd for the gai, gai-t6 rga-24, and quadruple DELLA mutant lines, Tai-ping Sun for the pRGA:GFP-RGA line, Karim Essabri for assistance with the quantitative real-time PCR analyses, and Léon Otten and Bernadette Clement for their help with sugar analyses. This work was supported by the Centre National de la Recherche Scientifique, the European Molecular Biology Organization (Grant ALTF-414-2005), and Agence Nationale de la Recherche Grant 07-JCJC-0118. Rothamsted Research receives grant-aided support from the Biotechnology and Biological Sciences Research Council of the UK.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Pascal Genschik (pascal.genschik@ibmp-ulp.u-strasbg.fr).
Online version contains Web-only data.
References
- Achard, P., Cheng, H., De Grauwe, L., Decat, J., Schoutteten, H., Moritz, T., Van Der Straeten, D., Peng, J., and Harberd, N.P. (2006). Integration of plant responses to environmentally activated phytohormonal signals. Science 331 91–94. [DOI] [PubMed] [Google Scholar]
- Achard, P., Baghour, M., Chapple, A., Hedden, P., Van Der Straeten, D., Genschik, P., Moritz, T., and Harberd, N.P. (2007. b). The plant stress hormone ethylene controls floral transition via DELLA-dependent regulation of floral meristem-identity genes. Proc. Natl. Acad. Sci. USA 104 6484–6489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Achard, P., Liao, L., Jiang, C., Desnos, T., Bartlett, J., Fu, X., and Harberd, N.P. (2007. a). DELLAs contribute to plant photomorphogenesis. Plant Physiol. 143 1163–1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Achard, P., Renou, J.-P., Berthomé, R., Harberd, N.P., and Genschik, P. (2008). Plant DELLAs restrain growth and promote survival of adversity by reducing the levels of reactive oxygen species. Curr. Biol. 18 656–660. [DOI] [PubMed] [Google Scholar]
- Agarwal, P.K., Agarwal, P., Reddy, M.K., and Sopory, S.K. (2006). Role of DREB transcription factors in abiotic and biotic stress tolerance in plants. Plant Cell Rep. 25 1263–1274. [DOI] [PubMed] [Google Scholar]
- Alvey, L., and Harberd, N.P. (2005). DELLA proteins: Integrators of multiple plant growth regulatory inputs? Plant Physiol. 123 153–160. [Google Scholar]
- Bolle, C. (2004). The role of GRAS proteins in plant signal transduction and development. Planta 218 683–692. [DOI] [PubMed] [Google Scholar]
- Cao, D., Hussain, A., Cheng, H., and Peng, J. (2005). Loss of function of four DELLA genes leads to light- and gibberellin-independent seed germination in Arabidopsis. Planta 223 105–113. [DOI] [PubMed] [Google Scholar]
- Chiang, H.H., Hwang, I., and Goodman, H.M. (1995). Isolation of the Arabidopsis GA4 locus. Plant Cell 7 195–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng, H., Qin, L., Lee, S., Fu, X., Richards, D.E., Cao, D., Luo, D., Harberd, N.P., and Peng, J. (2004). Gibberellin regulates Arabidopsis floral development via suppression of DELLA protein function. Development 131 1055–1064. [DOI] [PubMed] [Google Scholar]
- Cook, D., Fowler, S., Fiehn, O., and Thomashow, M. (2004). A prominent role for the CBF cold response pathway in configuring the low-temperature metabolome of Arabidopsis. Proc. Natl. Acad. Sci. USA 101 15243–15248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowling, R.J., Kamiya, Y., Seto, H., and Harberd, N.P. (1998). Gibberellin dose–response regulation of GA4 gene transcript levels in Arabidopsis. Plant Physiol. 117 1195–1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Lucas, M., Davière, J.M., Rodrigues-Falcon, M., Iglesias-Pedraz, J.M., Lorrain, S., Fankhauser, C., Blazquez, M.A., Titarenko, E., and Prat, S. (2008). A molecular framework for light and gibberellin control of cell elongation. Nature 451 480–484. [DOI] [PubMed] [Google Scholar]
- Dill, A., and Sun, T.P. (2001). Synergistic derepression of gibberellin signaling by removing RGA and GAI function in Arabidopsis thaliana. Genetics 159 777–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dill, A., Thomas, S.G., Hu, J., Steber, C.M., and Sun, T.P. (2004). The Arabidopsis F-box protein SLEEPY1 targets gibberellin signaling repressors for gibberellin-induced degradation. Plant Cell 16 1392–1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng, S., et al. (2008). Coordinated regulation of Arabidopsis thaliana development by light and gibberellins. Nature 451 475–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu, X., Richards, D.E., Fleck, B., Xie, D., Burton, N., and Harberd, N.P. (2004). The Arabidopsis mutant sleepy1gar2–1 protein promotes plant growth by increasing the affinity of the SCFSLY1 E3 ubiquitin ligase for DELLA protein substrates. Plant Cell 16 1406–1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowler, S., and Thomashow, M.F. (2002). Arabidopsis transcriptome profiling indicates that multiple regulatory pathways are activated during cold acclimation in addition to the CBF cold response pathway. Plant Cell 14 1675–1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmour, S.J., Fowler, S.G., and Thomashow, M.F. (2004). Arabidopsis transcriptional activators CBF1, CBF2, and CBF3 have matching functional activities. Plant Mol. Biol. 54 767–781. [DOI] [PubMed] [Google Scholar]
- Gilmour, S.J., Sebolt, A.M., Salazar, M.P., Everard, J.D., and Thomashow, M.F. (2000). Overexpression of the Arabidopsis CBF3 transcriptional activator mimics multiple biochemical changes associated with cold acclimation. Plant Physiol. 124 1854–1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmour, S.J., Zarka, D.G., Stockinger, E.J., Salazar, M.P., Houghton, J.M., and Thomashow, M.F. (1998). Low temperature regulation of the Arabidopsis CBF family of AP2 transcriptional activators as an early step in cold-induced COR gene expression. Plant J. 16 433–442. [DOI] [PubMed] [Google Scholar]
- Griffiths, J., Murase, K., Rieu, I., Zentella, R., Zhang, Z.L., Powers, S.J., Gong, F., Phillips, A.L., Hedden, P., Sun, T.P., and Thomas, S.G. (2006). Genetic characterization and functional analysis of the GID1 gibberellin receptors in Arabidopsis. Plant Cell 18 3399–3414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guy, C.L. (1990). Cold acclimation and freezing stress tolerance: Role of protein metabolism. Annu. Rev. Plant Physiol. Plant Mol. Biol. 41 187–223. [Google Scholar]
- Haake, V., Cook, D., Riechmann, J.L., Pineda, O., Thomashow, M.F., and Zhang, J.Z. (2002). Transcription factor CBF4 is a regulator of drought adaptation in Arabidopsis. Plant Physiol. 130 639–648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harberd, N.P., King, K.E., Carol, P., Cowling, R.J., Peng, J., and Richards, D.E. (1998). Gibberellin: Inhibitor of an inhibitor of…? Bioessays 20 1001–1008. [DOI] [PubMed] [Google Scholar]
- Harberd, N.P. (2003). Relieving DELLA restraint. Science 299 1853–1854. [DOI] [PubMed] [Google Scholar]
- Hedden, P., and Kamiya, Y. (1997). Gibberelin biosynthesis: Enzymes, genes and their regulation. Annu. Rev. Plant Physiol. Plant Mol. Biol. 48 431–460. [DOI] [PubMed] [Google Scholar]
- Hedden, P., and Phillips, A.L. (2000). Gibberellin metabolism: New insights revealed by the genes. Trends Plant Sci. 5 523–530. [DOI] [PubMed] [Google Scholar]
- Hsieh, T.H., Lee, J.T., Yang, P.T., Chiu, L.H., Charng, Y.Y., Wang, Y.C., and Chan, M.T. (2002). Heterology expression of the Arabidopsis C-repeat/dehydration response element binding factor 1 gene confers elevated tolerance to chilling and oxidative stresses in transgenic tomato. Plant Physiol. 129 1086–1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huner, P.A., Oquist, G., and Sarhan, F. (1998). Energy balance and acclimation to light and cold. Trends Plant Sci. 3 224–230. [Google Scholar]
- Jaglo-Ottosen, K.R., Gilmour, S.J., Zarka, D.G., Schabenberger, O., and Thomashow, M.F. (1998). Arabidopsis CBF1 overexpression induces COR genes and enhances freezing tolerance. Science 280 104–106. [DOI] [PubMed] [Google Scholar]
- Kasuga, M., Liu, Q., Miura, S., Yamaguchi-Shinozaki, K., and Shinozaki, K. (1999). Improving plant drought, salt, and freezing tolerance by gene transfer of a single stress-inducible transcription factor. Nat. Biotechnol. 17 287–291. [DOI] [PubMed] [Google Scholar]
- King, K., Moritz, T., and Harberd, N.P. (2001). Gibberellins are not required for normal stem growth in Arabidopsis thaliana in the absence of GAI and RGA. Genetics 159 767–776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koornneef, M., Elgersma, A., Hanhart, C.J., van Loenen-Martinet, E.P., van Rijn, L., and Zeevaart, J.A.D. (1985). A gibberellin insensitive mutant of Arabidopsis thaliana. Physiol. Plant. 65 33–39. [Google Scholar]
- Lee, B.H., Henderson, D.A., and Zhu, J.K. (2005). The Arabidopsis cold-responsive transcriptome and its regulation by ICE1. Plant Cell 17 3155–3175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, S., Cheng, H., King, K.E., Wang, W., Husssain, A., Lo, J., Harberd, N.P., and Peng, J. (2002). Gibberellin regulates Arabidopsis seed germination via RGL2, a GAI/RGA-like gene whose expression is up-regulated following imbibition. Genes Dev. 16 646–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Q., Kasuga, M., Sakuma, Y., Abe, H., Miura, S., Yamaguchi-Shinozaki, K., and Shinozaki, K. (1998). Two transcription factors, DREB1 and DREB2, with an EREBP/AP2 DNA binding domain separate two cellular signal transduction pathways in drought- and low-temperature-responsive gene expression, respectively, in Arabidopsis. Plant Cell 10 1391–1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magome, H., Yamaguchi, S., Hanada, A., Kamiya, Y., and Oda, K. (2004). dwarf and delayed-flowering 1, a novel Arabidopsis mutant deficient in gibberellin biosynthesis because of overexpression of a putative AP2 transcription factor. Plant J. 37 720–729. [DOI] [PubMed] [Google Scholar]
- Magome, H., Yamaguchi, S., Hanada, A., Kamiya, Y., and Oda, K. (2008). DDF1 transcriptional activator upregulates expression of a gibberellin deactivating gene, GA2ox7, under high-salinity stress in Arabidopsis. Plant J., in press. [DOI] [PubMed]
- McGinnis, K.M., Thomas, S.G., Soule, J.D., Strader, L.C., Zale, J.M., Sun, T.P., and Steber, C.M. (2003). The Arabidopsis SLEEPY1 gene encodes a putative F-box subunit of an SCF E3 ubiquitin ligase. Plant Cell 15 1120–1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakajima, M., et al. (2006). Identification and characterization of Arabidopsis gibberellin receptors. Plant J. 46 880–889. [DOI] [PubMed] [Google Scholar]
- Nakashima, K., and Yamaguchi-Shinozaki, K. (2006). Regulons involved in osmotic stress-responsive and cold stress-responsive gene expression in plants. Physiol. Plant. 126 62–71. [Google Scholar]
- Oh, E., Yamaguchi, S., Hu, J., Yusuke, J., Jung, B., Paik, I., Lee, H.S., Sun, T.P., Kamiya, Y., and Choi, G. (2007). PIL5, a phytochrome-interacting bHLH protein, regulates gibberellin responsiveness by binding directly to the GAI and RGA promoters in Arabidopsis seeds. Plant Cell 19 1192–1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penfield, S., Gilday, A.D., Halliday, K.J., and Graham, I.A. (2006). DELLA-mediated cotyledon expansion breaks coat-imposed seed dormancy. Curr. Biol. 16 2366–2370. [DOI] [PubMed] [Google Scholar]
- Peng, J., Carol, P., Richards, D.E., King, K.E., Cowling, R.J., Murphy, G.P., and Harberd, N.P. (1997). The Arabidopsis GAI gene defines a signaling pathway that negatively regulates gibberellin responses. Genes Dev. 11 3194–3205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips, A.L., Ward, D.A., Uknes, S., Appleford, N.E., Lange, T., Huttly, A.K., Gaskin, P., Graebe, J.E., and Hedden, P. (1995). Isolation and expression of three gibberellin 20-oxidase cDNA clones from Arabidopsis. Plant Physiol. 108 1049–1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabbani, M.A., Maruyama, K., Abe, H., Khan, M.A., Katsura, K., Ito, Y., Yoshiwara, K., Seki, M., Shinozaki, K., and Yamaguchi-Shinozaki, K. (2003). Monitoring expression profiles of rice genes under cold, drought, and high-salinity stresses and abscisic acid application using cDNA microarray and RNA gel-blot analyses. Plant Physiol. 133 1755–1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards, D.E., King, K.E., Ait-ali, T., and Harberd, N.P. (2001). How gibberellin regulates plant growth and development: A molecular genetic analysis of gibberellin signaling. Annu. Rev. Plant Physiol. Plant Mol. Biol. 52 67–88. [DOI] [PubMed] [Google Scholar]
- Riechmann, J.L., and Meyerowitz, E.M. (1998). The AP2/EREBP family of plant transcription factors. Biol. Chem. 379 633–646. [DOI] [PubMed] [Google Scholar]
- Rieu, I., Ruiz-Rivero, O., Fernandez-Garcia, N., Griffiths, J., Powers, S.J., Gong, F., Linhartova, T., Eriksson, S., Nilsson, O., Thomas, S.G., Phillips, A.L., and Hedden, P. (2008). The gibberellin biosynthetic genes AtGA20ox1 and AtGA20ox2 act, partially redundantly, to promote growth and development throughout the Arabidopsis life cycle. Plant J. 53 488–504. [DOI] [PubMed] [Google Scholar]
- Rolland, F., Moore, B., and Sheen, J. (2002). Sugar sensing and signaling in plants. Plant Cell 14 S185–S205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki, A., Itoh, H., Gomi, K., Ueguchi-Tanaka, M., Ishiyama, K., Kobayashi, M., Jeong, D.H., An, G., Kitano, H., Ashikari, M., and Matsuoka, M. (2003). Accumulation of phosphorylated repressor for gibberellin signaling in an F-box mutant. Science 299 1896–1898. [DOI] [PubMed] [Google Scholar]
- Shinozaki, K., and Yamaguchi-Shinozaki, K. (2000). Molecular responses to dehydration and low temperature: Differences and cross-talk between two stress signaling pathways. Curr. Opin. Plant Biol. 3 217–223. [PubMed] [Google Scholar]
- Silverstone, A.L., Ciampaglio, C.N., and Sun, T.P. (1998). The Arabidopsis RGA gene encodes a transcriptional regulator repressing the gibberellin signal transduction pathway. Plant Cell 10 155–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverstone, A.L., Jung, H.S., Dill, A., Kawaide, H., Kamiya, Y., and Sun, T.P. (2001). Repressing a repressor: gibberellin-induced rapid reduction of the RGA protein in Arabidopsis. Plant Cell 13 1555–1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stitt, M., and Hurry, V. (2002). A plant for all seasons: Alterations in photosynthetic carbon metabolism during cold acclimation in Arabidopsis. Curr. Opin. Plant Biol. 5 199–206. [DOI] [PubMed] [Google Scholar]
- Stockinger, E.J., Gilmour, S.J., and Thomashow, M.F. (1997). Arabidopsis thaliana CBF1 encodes an AP2 domain-containing transcriptional activator that binds to the C-repeat/DRE, a cis-acting DNA regulatory element that stimulates transcription in response to low temperature and water deficit. Proc. Natl. Acad. Sci. USA 94 1035–1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas, S.G., Phillips, A.L., and Hedden, P. (1999). Molecular cloning and functional expression of gibberellin 2-oxidases, multifunctional enzymes involved in gibberellin deactivation. Proc. Natl. Acad. Sci. USA 96 4638–4703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomashow, M.F. (1998). Role of cold-responsive genes in plant freezing tolerance. Plant Physiol. 118 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomashow, M.F. (1999). Plant cold acclimation: Freezing tolerance genes and regulatory mechanisms. Annu. Rev. Plant Physiol. Plant Mol. Biol. 50 571–599. [DOI] [PubMed] [Google Scholar]
- Tyler, L., Thomas, S.G., Hu, J., Dill, A., Alonso, J.M., Ecker, J.R., and Sun, T.P. (2004). DELLA proteins and gibberellin-regulated seed germination and floral development in Arabidopsis. Plant Physiol. 135 1008–1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueguchi-Tanaka, M., Ashikari, M., Nakajima, M., Itoh, H., Katoh, E., Kobayashi, M., Chow, T.Y., Hsing, Y.I., Kitano, H., Yamaguchi, I., and Matsuoka, M. (2005). GIBBERELLIN INSENSITIVE DWARF1 encodes a soluble receptor for gibberellin. Nature 437 693–698. [DOI] [PubMed] [Google Scholar]
- Ueguchi-Tanaka, M., Nakajima, M., Katoh, E., Ohmiya, H., Asano, K., Saji, S., Hongyu, X., Ashikari, M., Kitano, H., Yamaguchi, I., and Matsuoka, M. (2007). Molecular interactions of a soluble gibberellin receptor, GID1, with a rice DELLA protein, SLR1, and gibberellin. Plant Cell 19 2140–2155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen, C.K., and Chang, C. (2002). Arabidopsis RGL1 encodes a negative regulator of gibberellin responses. Plant Cell 14 87–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willige, B.C., Ghosh, S., Nill, C., Zourelidou, M., Dohmann, E.M., Maier, A., and Schwechheimer, C. (2007). The DELLA domain of GA INSENSITIVE mediates the interaction with the GA INSENSITIVE DWARF1A gibberellin receptor of Arabidopsis. Plant Cell 19 1209–1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin, Z., and Browse, J. (1998). eskimo1 mutants of Arabidopsis are constitutively freezing-tolerant. Proc. Natl. Acad. Sci. USA 95 7799–7804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, Y.L., Li, L., Gage, D.A., and Zeevaart, J.A. (1999). Feedback regulation of GA5 expression and metabolic engineering of gibberellin levels in Arabidopsis. Plant Cell 11 927–936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zentella, R., Zhang, Z.L., Park, M., Thomas, S.G., Endo, A., Murase, K., Fleet, C.M., Jikumaru, Y., Nambara, E., Kamiya, Y., and Sun, T.P. (2007). Global analysis of della direct targets in early gibberellin signalling in Arabidopsis. Plant Cell 19 3037–3057. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.