Abstract
To complete the metabolic map for an entire class of compounds, it is essential to identify gene–metabolite correlations of a metabolic pathway. We used liquid chromatography–mass spectrometry (LC-MS) to identify the flavonoids produced by Arabidopsis thaliana wild-type and flavonoid biosynthetic mutant lines. The structures of 15 newly identified and eight known flavonols were deduced by LC-MS profiling of these mutants. Candidate genes presumably involved in the flavonoid pathway were delimited by transcriptome coexpression network analysis using public databases, leading to the detailed analysis of two flavonoid pathway genes, UGT78D3 (At5g17030) and RHM1 (At1g78570). The levels of flavonol 3-O-arabinosides were reduced in ugt78d3 knockdown mutants, suggesting that UGT78D3 is a flavonol arabinosyltransferase. Recombinant UGT78D3 protein could convert quercetin to quercetin 3-O-arabinoside. The strict substrate specificity of UGT78D3 for flavonol aglycones and UDP-arabinose indicate that UGT78D3 is a flavonol arabinosyltransferase. A comparison of flavonol profile in RHM knockout mutants indicated that RHM1 plays a major role in supplying UDP-rhamnose for flavonol modification. The rate of flavonol 3-O-glycosylation is more affected than those of 7-O-glycosylation by the supply of UDP-rhamnose. The precise identification of flavonoids in conjunction with transcriptomics thus led to the identification of a gene function and a more complete understanding of a plant metabolic network.
INTRODUCTION
Plants employ many diverse metabolic pathways to produce >200,000 compounds (Dixon and Strack, 2003). Completion of the Arabidopsis thaliana and rice (Oryza sativa) genome sequencing projects has allowed (1) the annotation of genes involved in metabolic pathways based on homology and (2) genome-wide identification of whole metabolic pathways. Assigning functions to each of the genes that correlate with metabolites in a pathway is a prerequisite for developing a comprehensive understanding of complicated metabolic pathways. Because of its small genome and the development of extensive genetic, molecular, and physiological tools, Arabidopsis may be the only plant in which the complete network of secondary metabolism can currently be characterized within a single plant species.
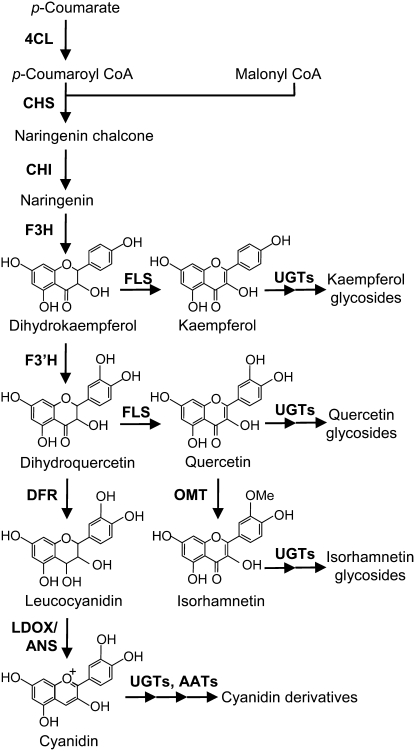
The flavonoids comprise one of the major secondary metabolite groups, with >7000 known compounds (Harborne et al., 1999; Andersen and Markham, 2006). Formation of the basic flavonoid skeleton has been well studied in terms of molecular biology and natural products chemistry. Flavonol and anthocyanidin regulatory and biosynthetic pathways have been characterized, and the corresponding genes have been isolated from various plants (Figure 1; Andersen and Markham, 2006; Tanaka and Filippa, 2006). However, the pathways for sequential modification, such as glycosylation, acylation, and methylation, are still relatively unexplored even though modification produces a huge chemical diversity and is essential for the stable accumulation of flavonoids. A wide variety of flavonoids have been isolated and identified from specific organs, such as flowers or fruits (Andersen and Markham, 2006), but there has been no comprehensive description of flavonoid metabolism in the organs of a single plant species. Although the flavonoids are important as flower pigments, UV-B protectants, phytoalexins, signaling molecules, and regulators of auxin transport (Dooner et al., 1991; Dixon and Paiva, 1995), the precise relationship between structure and function is also largely unclear.
Figure 1.
The Arabidopsis Flavonoid Biosynthetic Pathway.
4CL, 4-coumarate:CoA ligase; CHS, chalcone synthase; CHI, chalcone isomerase; F3H, flavanone 3-hydroxylase; F3′H, flavonoid 3′-hydroxylase; FLS, flavonol synthase; OMT1, O-methyltransferase; UGT, UDP-dependent glycosyltransferase; DFR, dihydroflavonol 4-reductase; LDOX/ANS, leucoanthocyanidin dioxygenase/anthocyanidin synthase; AAT, anthocyanin acyltransferase.
In Arabidopsis, at least 13 structures of flavonoids have been identified by nuclear magnetic resonance (NMR) and/or liquid chromatography–mass spectrometry (LC-MS ) with standards, and more flavonoid compounds were reported to be present (Graham, 1998; Veit and Pauli, 1999; Bloor and Abrahams, 2002; Kerhoas et al., 2006). Thirty-five flavonoid biosynthetic genes, including genes encoding 12 transcription factors, 10 structural enzymes, and 10 modification enzymes, have been identified (Tohge et al., 2005, 2007; Lepiniec et al., 2006; Fraser et al., 2007; Luo et al., 2007; Stracke et al., 2007; Yonekura-Sakakibara et al., 2007), but even in this highly studied species, the full details of the flavonoid pathway, including modification, are still unknown. The major flavonoid structures in Arabidopsis suggest that there are as yet unidentified modification enzyme genes and pathways in flavonoid metabolism. Generally, modification enzymes are encoded by multigene families. For example, there are 107 UDP-dependent glycosyltransferase (UGT) genes in Arabidopsis (D'Auria and Gershenzon, 2005). The presence of multiple genes hampers efforts to determine precise gene functions by sequence homology and to draw a complete pathway, even in Arabidopsis. The presence of unidentified minor flavonoids also suggests additional missing genes and pathways (Tohge et al., 2007).
Because of the rich research resources available for Arabidopsis, it is the best candidate for cataloging the entire flavonoid pathway. The integration of metabolome analysis with transcriptome analysis has been successfully used in Arabidopsis to decipher gene functions in biological processes (Hirai et al., 2005, 2007; Tohge et al., 2005). Transcriptome coexpression analysis is also a powerful tool for the efficient targeting of genes within a multigene family (Yonekura-Sakakibara et al., 2007; Saito et al., 2008). Use of these novel tools in combination with reverse genetics and a biochemical approach using recombinant proteins can provide robust evidence from which a whole pathway can be constructed.
In this study, we attempted to complete the identification of flavonols in each of the organs and flavonol annotation using flavonoid mutant lines. Taken together with previous reports, a total of 32 flavonol end or intermediate products were listed. Thirty of 32 flavonols were detected, and the structures of 19 compounds were annotated. Transcriptome coexpression analysis using known flavonoid genes as queries suggested 139 genes that may be involved in flavonoid metabolism. Finally, integration of comprehensive flavonol identification/annotation with transcriptome coexpression analysis identified a gene encoding flavonol 3-O-arabinosyltransferase and revealed the physiological role of a gene coding for UDP-rhamnose synthase in flavonoid biosynthesis.
RESULTS
Comprehensive Flavonol Profiling in Wild-Type and tt4 Mutants
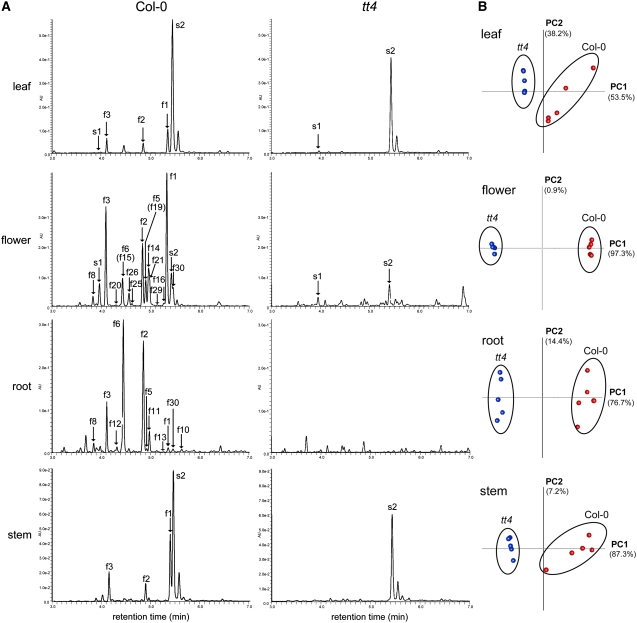
To catalog all of the flavonol derivatives in Arabidopsis, flavonol profiling in the flowers, leaves, stems, and roots of the wild type and tt4 mutants were performed using ultraperformance liquid chromatography (UPLC)–photodiode array (PDA)–electrospray ionization (ESI)/quadrupole time-of-flight (Q-TOF)/MS (Figure 2A). The metabolites detected only in the wild type should be flavonoid derivatives because TT4 encodes chalcone synthase, the first committed enzyme in flavonoid biosynthesis, and no flavonoid derivatives were detected in the tt4 mutant (Shirley et al., 1995; von Roepenack-Lahaye et al., 2004).
Figure 2.
UPLC-PDA-MS Analyses of the Extracts from Leaves, Flowers, Roots, and Stems of Wild-Type (Col-0) and tt4 Plants.
(A) UPLC-PDA and mass chromatograms of aqueous methanol extracts of the Arabidopsis wild type and the tt4 knockout mutant. Absorbance at 320 nm was used for the detection of flavonols. Labels correspond to compounds shown in Figure 3.
(B) Principal component analyses of the data obtained by LC-MS. Proportions of the first and second components are in parentheses. Wild-type results are in red, and those from the tt4 mutant are in blue.
The flavonols were differentially distributed in Arabidopsis organs (Figure 3). Kaempferol derivatives (f1 to f3 in Figure 3) were the major flavonols in leaves, stems, and flowers. In roots, there were high levels of quercetin 3-O-glucoside-7-O-rhamnoside (f6) in addition to kaempferol glycosides (f2 and f3), but there was very little kaempferol 3-O-rhamnoside-7-O-rhamnoside (f1). As for other phenolic compounds, a sinapoyl derivative s2 was detected predominantly in the leaves and stems of both the wild type and tt4.
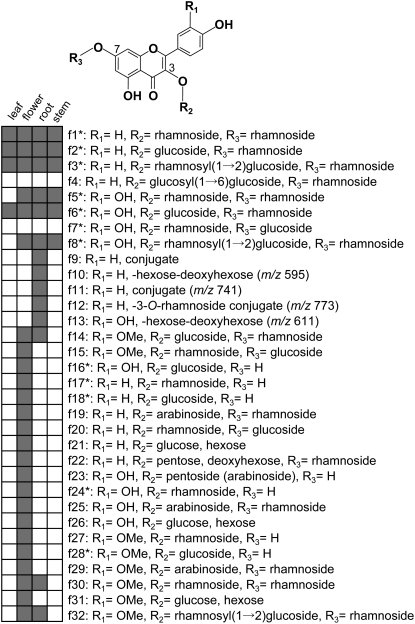
Figure 3.
Flavonol Glycosides in Arabidopsis.
Asterisks indicate that the compounds were identified based on a comparison of retention times and UV/mass spectra of the standards used in this study. R1=H, kaempferol; R1=OH, quercetin; R1=OMe, isorhamnetin. The presence of a compound in the indicated tissue is denoted by a gray box.
The MS-detectable peaks in the wild type and tt4 were compared to extract all flavonoid-related peaks (see Supplemental Figure 1 and Supplemental Data Set 1 online). Principal component analysis was conducted with 1804 MS peaks in leaves, 1864 in flowers, 1576 in roots, and 2138 in stems, indicating the presence of clear clusters according to the wild-type and tt4 genotypes (Figure 2B). The wild-type–specific peaks (53 peaks in leaves, 322 in flowers, 152 in roots, and 75 in stems) were picked up using a high PC1 loading value, and most of the peaks are due to the ions of flavonols (see Supplemental Figure 1 online). For example, 42 of 53 peaks are three major kaempferol glycosides in leaves (f1 to f3). A total of 30 flavonol derivatives were detected in Arabidopsis (four in leaves, 25 in flowers, 14 in roots, and four in stems). Flowers, in which most flavonol derivatives were detected, were used for further analysis.
Peak Annotation by Comparison with Flavonoid Biosynthetic Gene Knockout Mutants
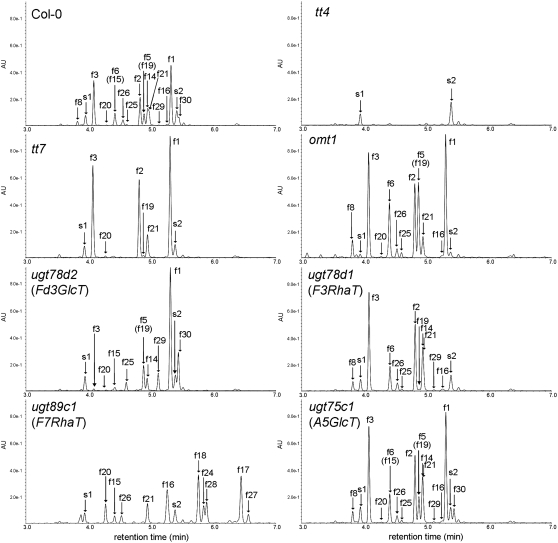
Twelve flavonols (f1 to f8, f17, f18, f20, and f24) in Arabidopsis have been identified by UV spectroscopy, MS, 1H-NMR, and 13C-NMR spectroscopy or annotated using UPLC-PDA-ESI/Q-TOF/MS (Veit and Pauli, 1999; Jones et al., 2003; Tohge et al., 2005, 2007; Kerhoas et al., 2006). We identified two additional flavonols (f16 and f28) by comparing retention times and mass-to-charge ratio (m/z) values with the standard compounds (Table 1; see Supplemental Data Set 2 online). To annotate the remaining 18 uncharacterized flavonols, mutants lacking CHS, F3′H, flavonol 3′-O-methyltransferase (OMT1), flavonoid 3-O-glucosyltransferase (UGT78D2), flavonol 3-O-rhamnosyltransferase (UGT78D1), flavonol 7-O-rhamnosyltransferase (UGT89C1), or anthocyanin 5-O-glucosyltransferase (UGT75C1) were used for flavonol analysis. Of the 14 identified (f1 to f8, f16 to f18, f20, f24, and f28), flavonol derivatives missing from each mutant were well correlated with the loss of gene function (Figure 4, Table 1). No quercetin and isorhamnetin conjugates (f5, f6, f8, f16, f24, and f28) accumulated in tt7, which lacks F3′H. No isorhamnetin conjugates (f28) were detected in omt1 mutants. The mutants ugt78d1 and ugt89c1 had no conjugates of flavonol 3-O-rhamnoside (f1, f5, f17, f20, and f24) and flavonol 7-O-rhamnoside (f1 to f3, f5, f6, and f8), respectively. Interestingly, in the ugt78d2 mutant, accumulation of the kaempferol/quercetin 3-O-glucosides (f2, f3, f6, and f8) was drastically reduced but detectable. Other kaempferol/quercetin 3-O-glucosides (f16 and f18) also decreased. This result is in agreement with the finding that cyanidin derivatives were detected at low levels in the ugt78d2 mutant (Tohge et al., 2005). On the other hand, accumulation of the isorhamnetin 3-O-glucosides (f28) was similar or slightly increased. These data indicate that another enzyme is involved in 3-O-glucosylation of isorhamnetin. Flavonol profiles in the flowers of the wild type and ugt75c1 were essentially identical, demonstrating that UGT75C1 is not involved in flavonol glycosylation at any detectable level.
Table 1.
The Flavonol Profiles in Acidic Methanol-Water Flower Extracts of Wild-Type Plants and Flavonoid Mutant Lines
| Peak No. | Rt (min) | ESI-MS (m/z) | Fragment (m/z)a | Col-0 | tt4 | tt7 | omt1 | ugt78d2 (Fd3GlcT) | ugt78d1 (F3RhaT) | ugt89c1 (F7RhaT) | ugt75c1 (A5GlcT) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| f1b | 5.38 | 579 | 433, 287 | 5.14 | ND | 6.05 | 5.71 | 5.61 | ND | ND | 4.63 |
| f2b | 4.88 | 595 | 433, 287 | 3.33 | ND | 4.87 | 3.96 | 0.19 | 4.25 | ND | 2.94 |
| f3b | 4.14 | 741 | 595, 449, 287 | 6.47 | ND | 7.75 | 7.49 | 0.29 | 7.33 | ND | 5.93 |
| f5b | 4.94 | 595 | 449, 303 | 2.09 | ND | ND | 4.14 | 2.04 | ND | ND | 1.86 |
| f6b | 4.47 | 611 | 449, 303 | 2.16 | ND | ND | 3.51 | 0.12 | 2.54 | ND | 1.97 |
| f8b | 3.88 | 757 | 611, 449, 303 | 1.11 | ND | ND | 2.37 | 0.04 | 1.58 | ND | 1.09 |
| f14 | 4.99 | 625 | 463, 317 | 2.71 | ND | ND | ND | 2.08 | 3.47 | ND | 2.51 |
| f15 | 4.49 | 625 | 479, 317 | 0.05 | ND | ND | ND | 0.41 | ND | 1.12 | 0.05 |
| f16b | 5.33 | 465 | 303 | 0.07 | ND | ND | 0.12 | ND | 0.10 | 2.36 | 0.08 |
| f17b | 6.52 | 433 | 287 | 0.03 | ND | 0.13 | 0.04 | 0.02 | ND | 3.54 | 0.02 |
| f18b | 5.84 | 449 | 287 | 0.08 | ND | 0.19 | 0.06 | ND | 0.10 | 3.18 | 0.05 |
| f19 | 4.95 | 565 | 433, 287 | 0.07 | ND | 0.28 | 0.11 | 0.66 | 0.13 | ND | 0.08 |
| f20 | 4.34 | 595 | 449, 287 | 0.10 | ND | 0.32 | 0.18 | 0.14 | ND | 1.96 | 0.09 |
| f21 | 5.02 | 611 | 449, 287 | 2.37 | ND | 3.27 | 3.10 | ND | 2.07 | 3.00 | 1.97 |
| f22 | 4.83 | 711 | 565, 433, 287 | 0.02 | ND | 0.03 | 0.03 | 0.18 | 0.02 | ND | 0.02 |
| f23 | 5.72 | 435 | 303 | 0.01 | ND | ND | 0.02 | 0.02 | 0.01 | 0.50 | 0.01 |
| f24b | 5.92 | 449 | 303 | 0.05 | ND | ND | 0.14 | 0.07 | ND | 2.09 | 0.06 |
| f25 | 4.66 | 581 | 449, 303 | 0.36 | ND | ND | 0.72 | 1.16 | 0.40 | ND | 0.33 |
| f26 | 4.60 | 627 | 465, 303 | 1.66 | ND | ND | 1.86 | 0.06 | 1.74 | 1.77 | 1.35 |
| f27 | 6.64 | 463 | 317 | 0.05 | ND | ND | ND | 0.07 | ND | 1.89 | 0.04 |
| f28b | 5.97 | 479 | 317 | 0.14 | ND | ND | ND | 0.15 | 0.16 | 2.99 | 0.15 |
| f29 | 5.17 | 595 | 463, 317 | 0.25 | ND | ND | ND | 2.46 | 0.34 | ND | 0.24 |
| f30 | 5.50 | 609 | 463, 317 | 2.09 | ND | ND | ND | 4.38 | ND | ND | 1.83 |
| f31 | 5.07 | 641 | 479, 317 | Trace | ND | ND | Trace | 0.11 | 0.01 | 0.01 | 0.01 |
| f32 | 4.27 | 771 | 625, 463, 317 | 0.03 | ND | ND | ND | 0.03 | 0.05 | ND | 0.05 |
| s1b | 4.00 | 387 | 207 | 0.06 | 0.12 | 0.07 | 0.02 | 0.07 | 0.05 | 0.07 | 0.05 |
| s2b | 5.46 | 341 | 207 | 0.08 | 0.21 | 0.08 | 0.03 | 0.10 | 0.10 | 0.12 | 0.07 |
Flavonols were quantified by measuring mass peak area using a response value of the peak area of an internal standard (naringenin 7-O-glucoside). ND, not detected; trace, <0.01; s1 and s2, sinapoyl derivatives.
Detected in mass and/or tandem mass data.
Identified based on a comparison of retention times and UV/mass spectra of the standards.
Figure 4.
UPLC-PDA-MS Analyses of the Extracts from Flowers of Wild-Type (Col-0) and Flavonoid-Defective Mutants.
Absorbance at 320 nm was used for detection. A5GlcT, anthocyanin 5-O-glucosyltransferase; F3RhaT, flavonol 3-O-rhamnosyltransferase; F7RhaT, flavonol 7-O-rhamnosyltransferase; Fd3GlcT, flavonoid 3-O-glucosyltransferase. Labels correspond to compounds shown in Figure 3. The labels in parentheses indicate compounds at <5% in the peak.
The basic flavonol structures and glycosylation patterns were revealed by superimposing the flavonol profiles of mutants over the wild type (Table 1). The structures of additional nine flavonols, including pentosides (f14, f15, f19, f23, f25, f27, f29, f30, and f32), were identified in Arabidopsis. A total of 23 flavonol structures and their distribution in the mutants show that at least four flavonol glycosyltransferases, an additional flavonol 3-O-glucosyltransferase, a flavonol 3-O-pentosyltransferase, a flavonol 3-O-glycoside:2′′-O-rhamnosyltransferase, and a flavonol 3-O-glycoside:6′′-O-glucosyltransferase remain to be identified.
New Flavonoid Biosynthetic Genes Deduced by Transcriptome Coexpression Analysis
Transcriptome coexpression analysis (Saito et al., 2008) was used to identify all of the flavonoid-related genes and was executed with the 24 genes encoding flavonoid biosynthetic enzymes or transcription factors listed in Table 2 as query. The lowest correlation coefficient (r) between query genes, except for the flavonol 7-O-glucosyltransferase gene (UGT73C6) and OMT1 is 0.527. UGT73C6 and OMT1 were excluded because UGT73C6 may cross-hybridize with UGT73C5, a brassinosteroid-related UGT, and the OMT1 coexpression profile suggests that it is under the regulation of lignin biosynthesis but not of flavonoid biosynthesis. The coexpression relationships of all Arabidopsis genes exhibiting correlation coefficients > 0.525 (r > 0.525, all data sets version 3) with the query genes are shown in Supplemental Figure 2 online and Table 3. The cutoff value was set according to Aoki et al. (2007). The clusters of coexpressed flavonoid genes were classified into two major groups. One cluster contains the general flavonoid/flavonol subgroup; another cluster represents the anthocyanin subgroup. UGT73C6 and OMT1 did not belong to either of these clusters. The 139 genes were highly correlated with the general flavonoid-related or anthocyanin-related genes (see Supplemental Figure 2 online). Twenty-four genes had more than two correlations with flavonoid-related genes. One-third of the genes listed in Table 3 were coregulated with flavonoid-related genes by flavonoid-specific MYBs (PAP1, MYB11, MYB12, and MYB111; Tohge et al., 2005, Stracke et al., 2007). The genes encoding embryo-specific protein (At5g62210), 3-keto-acyl-CoA thiolase (At5g48880), cryptochrome 3 (At5g24850), UGT84A1 (At4g15480), cinnamoyl-CoA reductase-like protein (At2g23910), early light-induced protein (At3g22840), TT8 (At4g09820), and glutathione S-transferase (GST; At1g10370) were regulated in a manner similar to the flavonoid biosynthetic genes by high irradiance and blue light in a CRYPTOCHROME1 (CRY1) photoreceptor and/or LONG HYPOCOTYL5 (HY5) transcription factor-dependent manner (Kleine et al., 2007). Other genes were also regulated by HY5 under UV-B radiation, much like the flavonoid biosynthetic genes (Table 3; Brown et al., 2005). For further analysis, we focused on UGT78D3 as a putative flavonol glycosyltransferase and RHM1, which was selected from the genes highly correlated with the flavonol biosynthetic genes.
Table 2.
Query Genes Used for Transcriptome Coexpression Analysis
| Name | AGI No. | Gene Annotation |
|---|---|---|
| 4CL3 | At1g65060 | 4-Coumarate:CoA ligase |
| CHS, TT4 | At5g13930 | Chalcone synthase |
| CHI, TT5 | At3g55120 | Chalcone isomerase |
| F3H, TT6 | At3g51240 | Flavanone 3-hydroxylase |
| F3′H, TT7 | At5g07990 | Flavonoid 3′-hydroxylase |
| FLS1 | At5g08640 | Flavonol synthase |
| DFR, TT3 | At5g42800 | Dihydroflavonol reductase |
| ANS, TT18 | At4g22880 | Anthocyanidin synthase |
| GST, TT19 | At5g17220 | GST |
| UGT73C6 | At2g36790 | Flavonol 7-O-glucosyltransferase |
| UGT75C1 | At4g14090 | Anthocyanin 5-O-glucosyltransferase |
| UGT78D1 | At1g30530 | Flavonol 3-O-rhamnosyltransferase |
| UGT78D2 | At5g17050 | Flavonoid 3-O-glucosyltransferase |
| UGT89C1 | At1g06000 | Flavonol 7-O-rhamnosyltransferase |
| A5G6′′′MaT | At3g29590 | Anthocyanin 5-O-glucoside:6′′′-O-malonyltransferase |
| A3G6′′p-CouT | At1g03940 | Anthocyanin 3-O-glucoside:6′′-O-p-coumaroyltransferase |
| A3G6′′p-CouT | At1g03495 | Anthocyanin 3-O-glucoside:6′′-O-p-coumaroyltransferase |
| SCPL10 | At2g23000 | Sinapoylglucose:anthocyanin acyltransferase |
| OMT1 | At5g54160 | Caffeic acid/5-hydroxyferulic acid O-methyltransferase, flavonol 3′-O-methyltransferase |
| MYB11 | At3g62610 | R2R3 MYB protein |
| MYB12 | At2g47460 | R2R3 MYB protein |
| MYB111 | At5g49330 | R2R3 MYB protein |
| PAP1, MYB75 | At1g56650 | R2R3 MYB protein |
| PAP2, MYB90 | At1g66390 | R2R3 MYB protein |
Table 3.
The Genes Coexpressed with Those in Flavonoid Metabolism
| Correlated Gene No. | AGI No. | PAP1 | MYB11 MYB12 MYB111 | CRY1 HY5 | |
|---|---|---|---|---|---|
| Flavonoid/phenylpropanoid biosynthesis | |||||
| 9 | At5g05270 | Chalcone isomerase family | + | + | + |
| 7 | At5g54060 | UDP-glucosyltransferase, UGT79B1 | + | ||
| 5 | At4g15480 | UDP-glucosyltransferase, UGT84A1 | + | + | |
| 3 | At2g23910 | Cinnamoyl-CoA reductase-like, CCRL9 | + | + | |
| Transporter | |||||
| 6 | At5g17010 | Sugar transporter family | |||
| 2 | At1g10370 | Glutathione S-transferase, GST30 | + | ||
| 2 | At5g02270 | ABC transporter, AtNAP9 | + | ||
| 2 | At5g44110 | ABC transporter, AtNAP2 | + | ||
| Protein kinase | |||||
| 2 | At1g10850 | Ser/Thr kinase family | |||
| 2 | At3g56370 | Leu-rich repeat transmembrane protein kinase | |||
| Transcription factor | |||||
| 2 | At2g37260 | WRKY transcription factor, TTG2 | + | ||
| 2 | At4g09820 | Basic helix-loop-helix family, TT8 | + | + | |
| 2 | At5g60140 | Transcriptional factor B3 family | |||
| Others | |||||
| 8 | At5g62210 | Embryo-specific protein | + | + | |
| 7 | At1g65560 | Allyl alcohol dehydrogenase | |||
| 7 | At5g48880 | 3-Keto-acyl-CoA thiolase, KAT5 | + | + | |
| 6 | At1g78570 | Rhamnose synthase, RHM1 | |||
| 5 | At5g24850 | Cryptochrome 3, CRY3 | + | ||
| 4 | At1g06550 | Enoyl-CoA hydratase/isomerase family | |||
| 3 | At5g20070 | Nudix hydrolase homolog, ATNUDT19 | |||
| 2 | At1g36160 | Acetyl-CoA carboxylase | |||
| 2 | At1g72970 | HOTHEAD | |||
| 2 | At3g22840 | Early light-induced protein1, ELIP1 | + | ||
| 2 | At5g39220 | Hydrolase family | |||
Only the genes that correlated with at least two genes are shown. The plus sign means the similar regulatory behavior with flavonoid biosynthetic genes in knockout mutants (CRY1/HY5, HY5 or MYB11/MYB12/MYB111) or an overexpressed plant (PAP1).
UGT78D3 Encodes Flavonol 3-O-Arabinosyltransferase
UGT78D3 is correlated with MYB111 (r = 0.572) but not with the other query genes. However, UGT78D3 belongs to the UGT78D subfamily, which also contains flavonoid 3-O-glucosyltransferase (UGT78D2) and flavonol 3-O-rhamnosyltransferase (UGT78D1), suggesting that UGT78D3 encodes a flavonoid 3-O-glycosyltransferase. UGT78D3 has high sequence identity with UGT78D2 (75.7% at the amino acid level) and UGT78D1 (68.3%). UGT78D2 has an amino acid sequence identity with UGT78D1 of 71.7%.
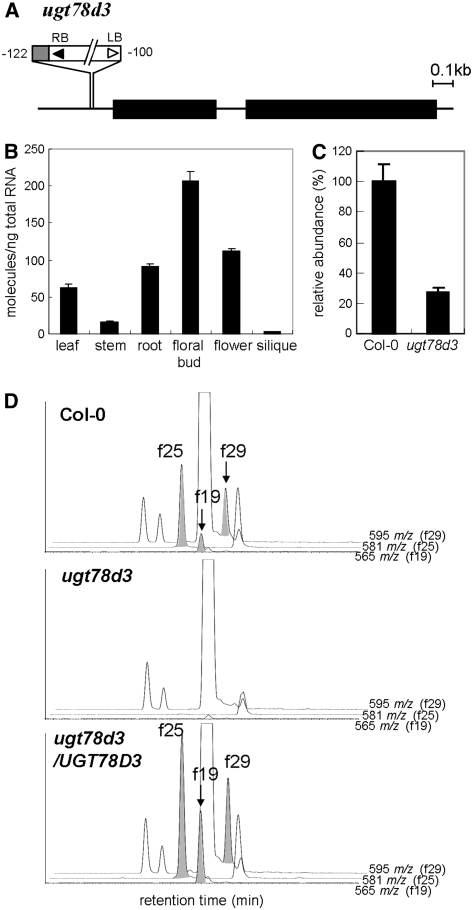
To test our hypothesis, T-DNA insertion mutant SALK_114099, designated as ugt78d3, was used for reverse genetics analysis. T-DNA was inserted between −122 and −100 bp upstream from the start codon of UGT78D3 (Figure 5A). UGT78D3 transcripts were abundant in floral buds but present at low levels in siliques and stems (Figure 5B). Real-time PCR showed that the accumulation level of UGT78D3 transcripts in flowers of the homozygous ugt78d3 mutant decreased to 30% of those in the wild type (Figure 5C). Flavonol target analysis revealed that three flavonol 3-O-pentoside conjugates (f19, f25, and f29) were under the detection limit in flowers of the ugt78d3 mutant (Figure 5D). This flavonol 3-O-pentoside conjugate-deficient phenotype was complemented by the constitutive expression of UGT78D3 cDNA under the control of the cauliflower mosaic virus 35S promoter (Figure 5D), indicating that UGT78D3 encodes a flavonol 3-O-pentosyltransferase.
Figure 5.
A T-DNA Insertion Mutant of UGT78D3.
(A) Schematic representation of UGT78D3 with a T-DNA insertion mutant used in this work. The thick black line indicates coding sequence. Numbers indicate the position of the T-DNA insertion. The gray box adjacent to T-DNA indicates the region containing the UGT78D3 promoter from −77 bp to +4 bp. LB, left border; RB, right border.
(B) Real-time PCR analysis of UGT78D3 transcripts in organs of the Arabidopsis wild type (Col-0). Error bars represent sd of three technical replicates per sample.
(C) Real-time PCR analysis of UGT78D3 transcripts in wild-type (Col-0) and ugt78d3 mutant flowers. Error bars represent sd of three technical replicates per sample.
(D) Extracted fragment mass chromatograms of aqueous methanol extracts from flowers of the wild type (Col-0), the ugt78d3 mutant, and ugt78d3 complemented with p35S:UGT78D3. The extracted fragment mass ions m/z 565, m/z 581, and m/z 595 correspond to compounds f19, f25, and f29 in Figure 3, respectively.
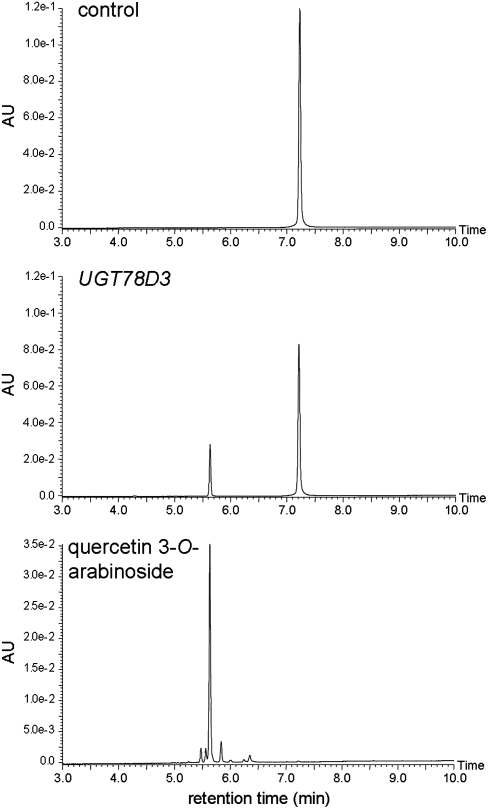
Flavonoid analysis of ugt78d3 mutants provided no additional information on the structure of flavonol 3-pentoside conjugates (f19, f25, and f29). Therefore, a biochemical approach was used to identify the exact structure of the pentose at the C-3 position. UGT78D3 recombinant protein was produced in Escherichia coli as a GST fusion and purified. The sugar donor specificity of UGT78D3 was examined with UDP-arabinose and UDP-xylose as UDP-pentoses. UDP-galactose, UDP-glucose, and UDP-rhamnose were also tested. The GST-UGT78D3 fusion protein catalyzed the conversion of quercetin to a quercetin 3-O-arabinoside, as confirmed by UPLC retention time, UV absorbance, and MS (Figure 6). No UGT activity was observed with UDP-xylose, UDP-galactose, UDP-glucose, or UDP-rhamnose as substrates, indicating that UGT78D3 is specific for UDP-arabinose as a sugar donor (Table 4). The sugar acceptor specificity of UGT78D3 was examined with kaempferol, quercetin, myricetin, isorhamnetin, cyanidin, and their glycosides as substrates. UGT78D3 activity was specific for flavonol aglycones (kaempferol, quercetin, myricetin, and isorhamnetin) and kaempferol 7-O-rhamnoside but had no activity on cyanidin or the flavonoid 3-O-glycosides. These results indicate that UGT78D3 arabinosylates flavonols at the C-3 position.
Figure 6.
UPLC Analyses of the Reaction Products of UGT78D3 Recombinant Protein.
Elution profiles of reaction products of GST protein (control) or GST-fused UGT78D3 protein (UGT78D3) and the standards (quercetin 3-O-arabinoside) are shown.
Table 4.
Substrate Specificity of Arabidopsis UGT78D3
| Relative Activity (%) | |
|---|---|
| Sugar donora | |
| UDP-arabinose | 100.0 ± 10.0 |
| UDP-glucose | ND |
| UDP-rhamnose | ND |
| UDP-xylose | ND |
| UDP-galactose | ND |
| Sugar acceptorb | |
| Kaempferol (Kae) | 100.0 ± 8.5 |
| Kae 3-O-glucoside | ND |
| Kae 7-O-rhamnoside | 140.4 ± 13.9 |
| Quercetin (Que) | 282.2 ± 9.6 |
| Que 3-O-glucoside | ND |
| Que 3-O-rhamnoside | ND |
| Myricetin | 148.2 ± 2.0 |
| Isorhamnetin | 164.9 ± 34.4 |
| Cyanidin (Cya) | ND |
| Cya 3-O-glucoside | ND |
| Cya 3-O-rhamnoside | ND |
| Cya 3,5-O-diglucoside | ND |
ND, not detected.
The reactions were performed with kaempferol as the sugar acceptor.
The reactions were performed with UDP-arabinose as the sugar donor.
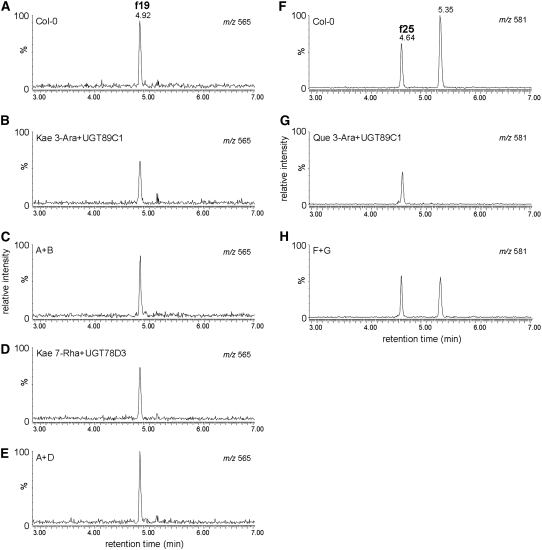
To confirm UGT78D3 function in planta, the structures of f19 and f25 were confirmed by direct comparison with the enzymatically synthesized standard compounds. Kaempferol 3-O-arabinoside-7-O-rhamnoside was prepared by the reaction catalyzed by UGT89C1 with kaempferol 3-O-α-l-arabinoside and UDP-β-l-rhamnose as substrates. An enzymatic product (1.8 mg) was isolated by multistep chromatography, and its structure was determined by 1H-NMR and rotating frame overhauser enhancement (ROE). The ROE correlation of protons at C-6 (δ 6.43) and C-8 (δ 6.73) by the irradiation of anomeric proton of rhamnose (rhamnose H-1, δ 5.52) indicated that rhamnose was attached at the C-7 position of kaempferol (Mulinacci et al., 1995). The chromatographic behavior and MS data of kaempferol 3-O-l-arabinoside-7-O-l-rhamnoside were identical with those of f19 and enzymatically synthesized products by UGT78D3 with kaempferol 7-O-α-l-rhamnoside and UDP-β-l-arabinose (Figure 7). These results indicated that f19 is kaempferol 3-O-l-arabinoside-7-O-l-rhamnoside. The component of f25 was also identified to be quercetin 3-O-l-arabinoside-7-O-l-rhamnoside by the similar procedure.
Figure 7.
UPLC-MS Analyses of the Reaction Products of UGT78D3 or UGT89C1 Recombinant Protein.
Extracted fragment mass chromatograms of aqueous methanol extracts from flowers of wild type (Col-0) ([A] and [F]), the reaction products of UGT89C1 with kaempferol 3-O-arabinoside (B), (A) coeluted with (B) (C), the reaction products of UGT78D3 with kaempferol 7-O-rhamnoside (D), (A) coeluted with (D) (E), the reaction products of UGT89C1 with quercetin 3-O-arabinoside (G), and (F) coeluted with (G) (H). The extracted fragment mass ions m/z 565 ([A] to [E]) and m/z 581 ([F] to [H]) correspond to compounds f19 and f25 in Figure 3, respectively. Kae 3-Ara, kaempferol 3-O-arabinoside; Kae 7-Rha, kaempferol 7-O-rhamnoside; Que 3-Ara, quercetin 3-O-arabinoside.
The UDP-Rhamnose Synthase 1 Gene Plays a Role in Rhamnosylation of Flavonoids
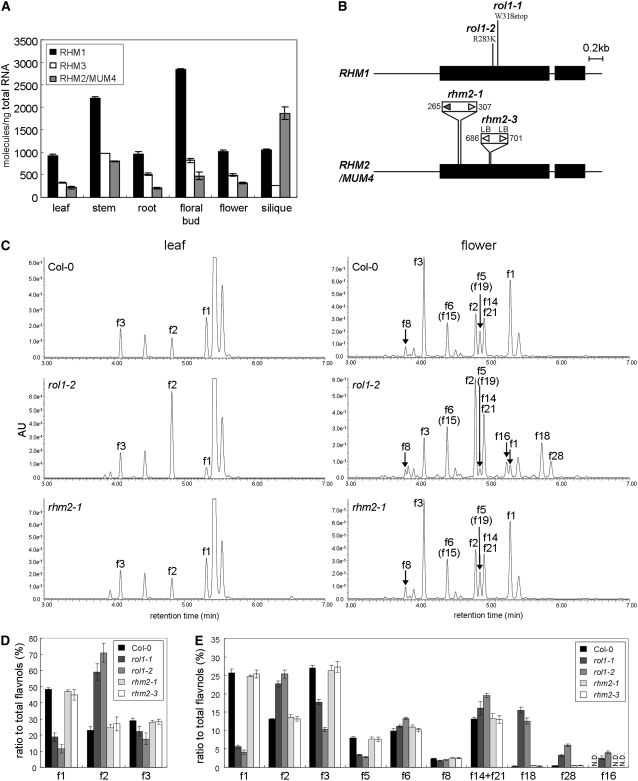
In Arabidopsis, there are three UDP-rhamnose synthase genes (RHM1, RHM2, and RHM3) (Usadel et al., 2004; Western et al., 2004; Oka et al., 2007). Expression of the UDP-Rhamnose Synthase 1 (RHM1) gene was highly correlated with the general flavonoid pathway genes, for example, 4CL3 (r = 0.736), FLS1 (r = 0.696), UGT89C1 (r = 0.669), CHI (r = 0.647), F3H (r = 0.611), and CHS (r = 0.603), based on data from all experiments. RHM2/MUM4 and RHM3 expression profiles differ from the flavonoid biosynthetic genes (r < 0.4). The accumulation of RHM1-RHM3 transcripts in the target organs was analyzed by real-time PCR (Figure 8A). Major flavonol glycosides were predominantly accumulated in floral buds and flowers (Figure 2; Yonekura-Sakakibara et al., 2007), which is consistent with accumulation pattern of RHM1 transcripts. RHM1 transcripts also made up the bulk of RHM transcript accumulation in all of the tested organs except for siliques. In siliques, the transcripts of RHM2/MUM4, which are involved in seed mucilage pectin synthesis (Usadel et al., 2004; Western et al., 2004), were predominantly accumulated.
Figure 8.
T-DNA Insertion Mutants of RHM1 and RHM2, and UPLC-MS Analyses of the rhm Mutant Lines.
(A) Real-time PCR analysis of RHMs transcripts in Arabidopsis organs. Error bars represent sd of three technical replicates per sample.
(B) Schematic representation of RHM1 and RHM2 with the mutations used in this work. rol1-1, a nonsense mutation at position 318; rol1-2, a missense mutation changing an Arg at position 283 to a Lys; rhm2-1 and rhm2-3, T-DNA insertion mutants. LB/RB, left/right borders. Numbers indicate the position of T-DNA insertion.
(C) UPLC-PDA-MS analyses of the extracts from leaves and flowers in the wild type (Col-0) and the rhm mutants. Absorbance at 320 nm was used for detection. Labels correspond to compounds shown in Figure 3. The labels in parentheses indicate compounds at <5% in the peak.
(D) The ratio of flavonol derivatives to total flavonol in leaves of the wild type (Col-0) and the mutants. Error bars represent sd for five independent experiments.
(E) The ratio of flavonol derivatives to total flavonol in flowers of the wild type (Col-0) and the mutants. Error bars represent sd for five independent experiments. N.D., not determined.
To verify the physiological relationship between RHM1 and the flavonoid biosynthetic genes, the flavonol profiles of the knockout mutants of RHM1 (rol1-1 and rol1-2) and RHM2 (rhm2-1 and rhm2-3) were analyzed (Figure 8). Flavonol profiles in leaves and flowers of the rhm2 mutants showed no significant alteration. However, in leaves of rol-1 and rol1-2, accumulation levels of kaempferol 3-O-rhamnoside-7-O-rhmanoside (f1) were considerably reduced and those of kaempferol 3-O-glucoside-7-O-rhmanoside (f2) were elevated. In flowers of the rol1 mutants, levels of kaempferol/quercetin 3-O-rhamnoside-7-O-rhamnosides (f1 and f5) and kaempferol 3-O-rhamnosyl(1→2)-glucoside-7-O-rhamnoside (f3) were reduced and those of flavonol 3-O-glucosides (f16, f18, and f28) were elevated considerably. These results imply that UDP-rhamnose produced by RHM1 is used for flavonol rhamnosylation and RHM1 plays a major role in supplying UDP-rhamnose for modification of flavonols.
DISCUSSION
The Power of Detailed Targeted Flavonol Analysis and Transcriptome Coexpression Analysis for Functional Genomics
Here, we used a novel strategy for functional genomics in Arabidopsis using flavonol as a case study (Figure 9). Coupling the detailed analysis of secondary metabolites with coexpression can be used as a more general method for identifying the biosynthetic pathways of plant compounds associated with specific organs and developmental stages as well as their genetic components. The use of T-DNA mutations in genes encoding flavonoid biosynthetic enzymes to provide comprehensive annotation of the flavonols in Arabidopsis is effective and can provide an efficient method for elucidating flavonol structures. The integration of metabolic profiling and transcriptome coexpression could efficiently narrow the pool of candidate genes. Using these methods, a flavonol 3-O-arabinosyltransferase gene was found, and the relationship between flavonols and UDP-rhamnose synthesis has emerged. After listing the candidates potentially involved in the reactions or regulation of a metabolic pathway, the specific identification of a gene function could be accomplished using a traditional reverse genetics approach with knockout mutants and/or by biochemical characterization with the recombinant proteins, as has been exemplified in this study. The above strategy will be much improved as large metabolome data sets follow transcriptome data sets into to the public domain.
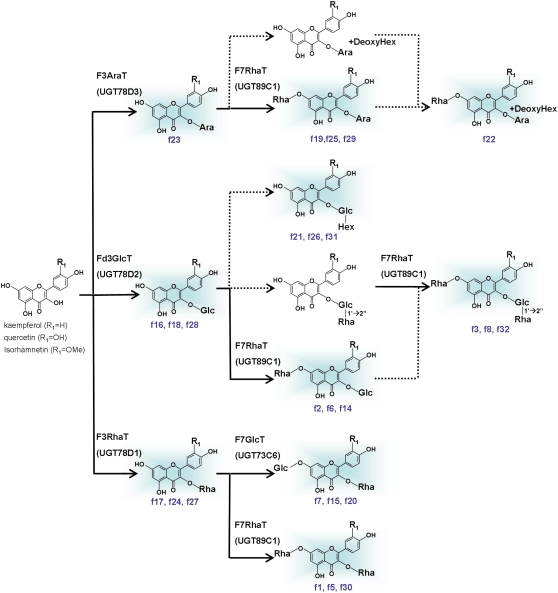
Figure 9.
Proposed Comprehensive Flavonol Pathway in Arabidopsis.
Each number in blue corresponds to the compounds shown in Figure 3. The compounds detected in Arabidopsis are shown in a blue background. Dotted lines indicate proposed but unidentified pathways. F3AraT, flavonol 3-O-arabinosyltransferase; Fd3GlcT, flavonoid 3-O-glucosyltransferase; F3RhaT, flavonol 3-O-rhamnosyltransferase; F7GlcT, flavonol 7-O-glucosyltransferase; F7RhaT, flavonol 7-O-rhamnosyltransferase; Ara, arabinose; DeoxyHex, deoxyhexose; Glc, glucose; Hex, hexose; Rha, rhamnose.
This combined strategy can also be useful for the detailed characterization of relatively minor flavonoid pathways that are evident only in mutants. The detailed flavonoid analyses of ugt78d2 mutants suggest the existence of another flavonoid 3-O-glucosyltransferase because a small amount of flavonoid 3-O-glucoside is present in the knockout mutant of UGT78D2. The ugt89c1 mutants accumulate flavonol 3-O-glucosides but not 3-O-rhamnosyl(1→2)glucosides, suggesting that 7-O-rhamnosylation likely occurs prior to 2′′-O-rhamnosylation. Accumulation of f19, f25, and f29 may also suggest that 7-O-rhamnosylation of kaempferol 3-O-arabinoside likely occurs prior to further glycosylation with deoxyhexose. The presence of f21, f26, and f31 suggests that a novel flavonol 3-O-glucoside:hexosyltransferase using glucose, galactose, or mannose may be involved in flavonol modification because those sugars have been so far identified as a sugar moiety of flavonol glycosides and have the molecular weight corresponding to unidentified hexose of f21, f26, and f30 (Andersen and Markham, 2006).
Transcriptome Coexpression Analysis Delimits Genes Related to Flavonoid Metabolism
Transcriptome coexpression analyses suggest other candidate genes that might be related to flavonoid metabolism. Eight of 24 candidate genes were regulated in PAP1-overexpressing or in MYB11/MYB12/MYB111 triple knockout plants. CHIput (At5g05270) may play a supplementary role to the primary CHI (At3g55120). The coordinated expression of CHIput with CHI tends to support this hypothesis. UGT79B1 belongs to a cluster containing the anthocyanin biosynthetic group. Phylogenetic trees of flavonoid UGTs suggest that UGT79B1 encodes anthocyanin 3-O-glucoside 2′′-xylosyltransferase (Tohge et al., 2005). These data suggest that CHIput and UGT79B1 are involved in the flavonoid pathway. UGT84A1 (At4g15480) has 1-O-glucosyltransferase activity for phenylpropanoids such as p-coumaric acid and sinapic acid (Lim et al., 2001). Sinapoylmalate converted from 1-O-sinapoylglucose by malate sinapoyltransferase (SNG1), is a UV-B protective compound much as the flavonoids are (Ruegger and Chapple, 2001). Under UV stress conditions, accumulation of sinapoylmalate was higher in tt4 mutants than in the wild type (Kusano et al., 2007), suggesting that sinapoylmalate biosynthesis acts as a backup system for flavonoid metabolism under stress conditions. The positive correlation between MYB12 and UGT84A1 (r = 0.781) and the negative correlation of MYB12/UGT84A1 and SNG1 (r = −0.357/−0.323) tends to support this hypothesis. 1-O-Sinapoylglucose is also a substrate for anthocyanin sinapoyltransferase (Fraser et al., 2007), and anthocyanins accumulate with UV exposure and high sucrose stress. Arabidopsis may thus coregulate 1-O-sinapoylglucose with the flavonoids for prompt responses to stress. Cinnamoyl-CoA reductase–related protein (At2g23910) also may be involved in phenylpropanoid biosynthesis. Although coexpression does not necessarily indicate a functional relationship (Stuart et al., 2003), we believe that the significance of transcriptome coexpression analysis for functional gene identification has been established, at least for secondary metabolism.
A Flavonol Glycosyltransferase:Flavonol 3-O-Arabinosyltransferase
We demonstrated that UGT78D3 encodes a flavonol 3-O-arabinosyltransferase. The UGT78D3 function in planta was established by the identification of f19 and f25 as kaempferol/quercetin 3-O-arabinoside-7-O-rhamnoside, respectively. The conformation of substrates (kaempferol/quercetin 3-O-α-l-arabinosides and UDP-β-l-rhamnose) and previous data are consistent with the hypothesis that f19 and f25 are kaempferol/quercetin 3-O-α-l-arabinoside-7-O-α-l-rhamnosides, respectively (Mulinacci et al., 1995; Sakar et al., 2005). Compared with UGT78D1 and UGT78D2, UGT78D3 is weakly correlated with genes in the flavonoid metabolic pathway, which is consistent with the anatomical distribution of flavonol glycosides. The flavonol 3-O-arabinosides make up only a minor fraction of flavonols or are under the detection limit in organs except for flowers, whereas the flavonol 3-O-glucosides and 3-O-rhamnosides are distributed in all tested organs as major flavonols. Among the 6850 flavonoids registered in the Flavonoid Viewer database (http://www.metabolome.jp/software/FlavonoidViewer/viewer), 2.9% are arabinosylated flavonoids (M. Arita, personal communication). To find genes involved in such minor metabolite biosynthesis, an in-depth analysis of gene-to-metabolite correlation is essential.
Functional identification of three Arabidopsis flavonol 3-O-glycosyltransferases (3GlyTs) in the UGT78D subfamily with distinct UDP-sugar specificity allowed us to deduce the amino acid residues involved in substrate recognition. The three-dimensional structure of grape (Vitis vinifera) flavonoid 3-O-glucosyltransferase (3GlcT) was determined, and a structural analysis suggests the presence of several key residues that interact with UDP-sugar and the flavonoid backbone (Offen et al., 2006). The amino acids Gln-375, Asp-374, and Thr-141 have been proposed to interact with hydroxyl groups at the C-2 and C-3, C-3, and C-4, and C-6 positions of the glucose moiety of UDP-glucose, respectively, but Gln-375 and Thr-141 are not conserved in the Arabidopsis 3GlyTs (see Supplemental Figure 3 online). His residues corresponding to Gln-375 are conserved among the galactosyltransferases (Kubo et al., 2004). A His residue was also found as the corresponding residue in UGT78D3. The configurations of hydroxyl groups at C-2, C-3, and C-4 positions are the same in arabinose and galactose. However, UGT78D3 could not use UDP-galactose. Asp-374 in grape flavonoid 3GlcT is conserved in the three Arabidopsis 3GlyTs, although the configurations of hydroxyl groups at the C-3 and C-4 positions are different (see Supplemental Figure 3 online). Given this disparity, the residues involved in sugar donor specificity cannot be ascribed to a single amino acid residue.
RHM1 Is Involved in Flavonol Biosynthesis
Rhamnosylation is a major glycosylation of flavonols in Arabidopsis. Twenty-one of 32 detected flavonol derivatives in wild-type Arabidopsis are rhamnosides (Figure 3). A comparison of flavonol profile indicates that RHM1 plays a major role in supplying UDP-rhamnose for flavonol biosynthesis. RHM1 transcripts are accumulated as the major RHM species in most of the tested Arabidopsis organs. The expression pattern of RHM1 was well coordinated with the known flavonoid biosynthetic genes. The RHM1 correlation coefficients for flavonol rhamnosyltransferase (UGT89C1, r = 0.67; UGT78D1, r = 0.51) are higher than those for flavonol glucosyltransferase (UGT78D2, r = 0.45; UGT78D3, r = 0.32). In leaves, glycosylation at the C-3 position was strongly affected by the restriction of UDP-rhamnose supply because subsequent glycosylations at the C-7 or C-2′′ positions for major flavonols are rhamnosylation only. In flowers of rol1s, additional flavonol 3-O-glucosides were detected, suggesting that more UDP-rhamnose may be necessary in flowers and glycosylation at the C-7 position is also affected in the severe limitation of UDP-rhamnose. In fact, major six flavonol contents in flowers are ∼10- to 70-fold higher than in leaves (Yonekura-Sakakibara et al., 2007). The role of RHM1 for UDP-rhamnose supply to flavonols was also supported by a recent report with young shoots of the rol1 mutants (Ringli et al., 2008) and our data with RHM1 knockdown mutants (SALK_027926 and SALK_143589; see Supplemental Figure 4 online).
The extent of the RMH3 contributions is unclear, and it may be committed to other pathways. RHM2/MUM4 is involved in the synthesis of pectinaceous rhamnogalacturonan I in seed mucilage (Usadel et al., 2004; Western et al., 2004). Interestingly, RHM3 has a low correlation coefficient with RHM1 (r = 0.46, all data sets version 3). The molecular evolution of RHMs may explain this correlation. The Arabidopsis genome contains a number of large duplicated chromosomal segments. Blanc et al. (2003) proposed that the Arabidopsis lineage underwent at least two distinct episodes (old and recent) of duplication. According to Blanc et al. on their website (http://wolfe.gen.tcd.ie/athal/dup) for exploring paralogous blocks identified within the Arabidopsis genome, RHM1 is the ancestral gene form. RHM3 was duplicated and diverged from RHM1 during the old duplication, and RHM2/MUM4 emerged from RHM3 by some recent duplication (see Supplemental Figure 5 online). Comprehensive analysis of rhamnose-containing metabolites using RHM knockout mutants will be helpful to clarify the additional functions of RHM1, RHM2/MUM4, and RHM3.
METHODS
Plant Materials
Arabidopsis thaliana (accession Columbia-0; Lehle Seeds) was used as the wild type in this study. The mutants ugt75c1, ugt78d1, ugt78d2, ugt89c1, rol1-1, rol1-2, and rhm2-1 were described previously (Jones et al., 2003; Usadel et al., 2004; Tohge et al., 2005; Diet et al., 2006; Yonekura-Sakakibara et al., 2007). The T-DNA–inserted knockout mutants of omt1 (CS25167) and tt7 (CS6509) were obtained from the ABRC. The tt4 (C1) mutant was obtained from tt mutant lines induced by ion beam irradiation of Arabidopsis (Shikazono et al., 2003). The T-DNA–inserted mutant of Arabidopsis, lines SALK_114099 for UGT78D3 and SALK_085051 (rhm2-3, a homozygous knockout line) for RHM2, were obtained from the Salk Institute. T-DNA insertion lines were screened by PCR using specific primers for UGT78D3, RHM2 ,and T-DNA: UGT78D3f, UGT78D3r, RHM2f, RHM2r, LBa1, and RBa1 (see Supplemental Table 1 online). PCR products were sequenced to determine the exact insertion points.
Plants were cultured on Murashige and Skoog–agar medium containing 1% sucrose (Valvekens et al., 1988) in a growth chamber at 22°C with 16 h/ 8 h light and dark cycles for 18 d or in a greenhouse at 22°C with 16 h/ 8 h light/dark for 4 weeks. Plants were harvested, immediately frozen with liquid nitrogen, and stored at −30°C until use. Five biological replicates were used for flavonoid profiling.
Chemicals
Flavonoid standards, including quercetin 3-O-arabinoside, were purchased from Extrasynthese and Analyticon. Kaempferol 3-O-α-l-rhamnoside-7-O-α-l-rhamnoside, kaempferol 3-O-α-l-rhamnosyl(1→2)-β-d-glucoside-7-O-α-l-rhamnoside, quercetin 3-O-α-l-rhamnoside-7-O-α-l-rhamnoside, and quercetin 3-O-α-l-rhamnosyl(1→2)-β-d-glucoside-7-O-α-l-rhamnoside were from our laboratory collection. UDP-β-l-arabinose and UDP-α-d-xylose were purchased from CarboSource Services (supported in part by National Science Foundation–Plant Cell Wall Biosynthesis Research Network Grant 0090281).
Flavonol Profiling by UPLC-PDA-ESI/Q-TOF/MS
Flavonol analyses were performed in quintuplicate. Frozen leaves were homogenized in 5 μL of extraction solvent (methanol: CH3COOH:H2O = 9:1:10, 0.02 mM naringenin-7-O-glucoside) per milligram of fresh weight of tissue in a mixer mill (MM300; Retsch) for 5 min at 30 Hz. After centrifugation at 12,000g, the supernatants were immediately used for flavonoid analysis.
For flavonol profiling, a Waters Acquity UPLC system (Waters) fitted with a Q-TOF Premier mass spectrometer (Micromass MS Technologies) was used. UPLC was performed on a UPLC phenyl C18 column (Φ2.1 mm × 100 mm; Waters) at a flow rate of 0.5 mL/min at 35°C. Compounds were separated with a linear elution gradient with solvent A (0.1% formic acid in water) and solvent B (0.1% formic acid in acetonitrile) from 0 min, 100% A to 10 min, 40% B. PDA was used for the detection of UV-visible absorption in the range of 210 to 500 nm. The TOF mass analyzer was used for the detection of flavonol glycosides [M + H]+ and the peak of fragment ions in a positive ion scanning mode with the following setting: desolvation temperature, 400°C with a nitrogen gas flow of 600 L/h; capillary spray, 3.0 kV; source temperature, 150°C; and cone voltage of 10 V for flavonoid glycosides [M + H]+ or 30 V for fragment ions. For comprehensive flavonol profiling, nonprocessed MS data were converted to NetCDF format by MassLynx software (Micromass MS Technologies). Data analyses including principal component analysis were performed by the Phenomenome Profiler (Phenomenome Discoveries). Flavonol glycoside standards were used for the identification of the peaks in the plant extracts based on retention times, UV-visible absorption spectra, and mass fragmentation by tandem MS analysis. Other flavonol peaks were annotated by comparing their UV-visible absorption spectra, elution times, m/z values, and MS2 fragmentation patterns with 85 reference flavonoid compounds and the reported data (Tohge et al., 2005, 2007; Routaboul et al., 2006; Yonekura-Sakakibara et al., 2007). The mass spectrum data of standard compounds (see Supplemental Data Set 2 online) were recorded in the MASSBANK Database (http://www.massbank.jp/index-e.html).
Transcriptome Coexpression Analysis
Coexpression analyses were performed using a Coexpression Gene Search algorithm on the RIKEN PRIMe website (http://prime.psc.riken.jp/; Akiyama et al., 2008) as described previously (Yonekura-Sakakibara et al., 2007; Saito et al., 2008). Twenty-four genes (listed in Table 2) were used as core query genes. The genes exhibiting positive correlations (r > 0.525) against the flavonoid biosynthetic genes were extracted using ATTED-II (Obayashi et al., 2007) based on all data sets version 3 in AtGenExpress. The coexpression graph was depicted by the Pajek program.
Evaluation of T-DNA Insertion Mutants
For complementation tests, full-length UGT78D3 cDNA was amplified by PCR using Arabidopsis flower cDNA as a template with primers UGT78D3-GWf and UGT78D3-GWr (see Supplemental Table 1 online). The Gateway system was used for construction of the binary vectors for Arabidopsis transformation. cDNA was cloned into the pCR8/GW/TOPO vector (Invitrogen) as an entry vector and sequenced to confirm the absence of PCR errors. pGWB2 was used as a destination vector. Transformation into Agrobacterium tumefaciens and Arabidopsis and the selection of transformants were performed as described previously (Yonekura-Sakakibara et al., 2007).
Arabidopsis plants were grown at 22°C under 16 h/8 h light and dark cycles in a plant growth room for ∼5 weeks. The leaves and flowers of plants were harvested, immediately frozen with liquid nitrogen, and stored at −30°C until use for metabolite profiles.
Quantitative Real-Time PCR
RNA Extraction and cDNA synthesis were performed as described previously (Yonekura-Sakakibara et al., 2004). The developmental stage of each organ used for analysis was as described previously (Yonekura-Sakakibara et al., 2007). Accumulation levels of UGT78D3, RHM1, RHM2, and RHM3 transcripts were analyzed by real-time PCR with an ABI PRISM 7500 real-time PCR system (Applied Biosystems), monitoring amplification with SYBR-Green I dye (Applied Biosystems). The primers UGT78D3-RTf, UGT78D3-RTr, RHM1-RTf, RHM1-RTr, RHM2-RTf, RHM2-RTr, RHM3-RTf, and RHM3-RTr (see Supplemental Table 1 online) were designed using Primer Express software (Applied Biosystems) and checked for specific product formation by a dissociation program. In each case, plasmid DNA containing the corresponding gene was used as a template to generate a calibration curve. Real-time PCR was performed in triplicate on a single biological sample for each genotype.
Production of Recombinant UGT78D3 Protein and Assay of Glycosyltransferase
Full-length UGT78D3 cDNA was amplified and cloned into pCR8/GW/TOPO (Invitrogen) as described above. To construct the protein expression vector, the resultant plasmid was amplified by PCR with the primers UGT78D3-pET41bf and UGT78D3-pET41br (see Supplemental Table 1 online), the PCR product was digested with StuI and XhoI, and ligated with StuI-XhoI-digested pET-41b(+) vector (Novagen). The sequence of the resultant plasmid, pKYS342, was confirmed. Escherichia coli strain BL21star(DE3) was used as a host for expression. Transformed cells were cultivated at 37°C until A600 reached 0.5. After the addition of isopropyl-β-d-thiogalactopyranoside to a final concentration of 1 mM, cells were cultured at 25°C for 4 h. The cells were collected, and the protein was purified as a GST fusion according to the manufacturer's instructions (Qiagen).
The standard enzyme assay reaction mixture (final volume of 50 μL) consisted of 50 mM HEPES-KOH, pH 7.5, 150 μM flavonoid substrate, and 500 μM UDP-sugar. The mixture was preincubated at 30°C for 2 min, and the reaction was started by the addition of enzyme. Reactions were stopped after 0, 4, 8, 12, or 30 min of incubation at 30°C by the addition of 50 μL ice-cold 0.5% (v/v) trifluoroacetic acid/methanol for flavonols or 50 μL ice-cold 1.0% (v/v) HCl/methanol for anthocyanidins and anthocyanins, and the supernatant was recovered by centrifugation at 12,000g for 3 min. Flavonoids in the resultant solution were analyzed as described above.
Preparation and NMR Analysis of Kaempferol 3-O-Arabinoside-7-O-Rhamnoside and Quercetin 3-O-Arabinoside-7-O-Rhamnoside
A reaction mixture of UGT89C1 with kaempferol 3-O-α-l-arabinoside and UDP-β-l-rhamnose as substrates (total 300 mL) was concentrated and chromatographed on COSMOSIL 75C18-OPN (Nacalai Tesque) and stepwise eluted with water, 10% aqueous CH3CN, 30% aqueous CH3CN, 50% aqueous CH3CN, 70% aqueous CH3CN, and 100% methanol. Kaempferol glycosides in the fractions of 30% aqueous CH3CN, 50% aqueous CH3CN, 70% aqueous CH3CN, and 100% methanol were further separated by preparative HPLC using a C18 reversed-phase column (10 mm ×150 mm I.D., particle size 5 μm, Inertsil ODS-EP; GL Science) using 30% aqueous CH3CN with flow rates of 2.0 mL/min at 30°C. Kaempferol 3-O-l-arabinoside-7-O-l-rhamnoside (1.8 mg) was obtained and the structure was supported by 1H-NMR (600 MHz, methanol-d4): δ 5.16 (1H, d, J = 6.6 Hz, arabinose H-1), δ 5.52 (1H, br. s, rhamnose H-1), δ 6.43 (1H, d, J = 1.8 Hz, H-6), δ 6.73 (1H, d, J = 1.2 Hz, H-8), δ 6.86 (2H, d, J = 9.0 Hz, H-3′and H-5′), δ 8.06 (2H, d, J = 8.4 Hz, H-2′ and H-6′), ROE correlation between the anomeric proton of rhamnose (rhamnose H-1, δ 5.52), and the protons at C-6 (H-6, δ 6.43) and C-8 position (H-8, δ 6.73), ESI-TOF-MS (positive ion mode): m/z 565.1554 [M+H]+, calculated for C26H29O14, 565.1557, ESI-TOF-MS/MS (positive ion mode, collision energy 30 eV): m/z (relative intensity) 433.1145 [M+H-arabinose]+ (39), 287.0605 [M+H-arabinose-rhamnose]+ (100), and UV λmax nm: 345 (on flow).
A large-scale enzymatic product of UGT89C1 with quercetin 3-O-α-l-arabinoside and UDP-β-l-rhamnose was isolated by a similar procedure except for the mobile phase (20% aqueous CH3CN) of preparative HPLC step. Quercetin 3-O-l-arabinoside-7-O-l-rhamnoside (2.1 mg) was obtained and the structure was supported by 1H-NMR (600 MHz, methanol-d4): δ 5.22 (1H, d, J = 6.0 Hz, arabinose H-1), δ 5.56 (1H, br. s, rhamnose H-1), δ 6.46 (1H, br. s, H-6), δ 6.75 (1H, br. s, H-8), δ 6.88 (1H, d, J = 7.8 Hz, H-5′), δ 7.61 (1H, br. d, J = 8.4 Hz, H-6′), δ 7.76 (1H, br. s, H-2′). ROE (irradiation δ 5.56 [rhamnose H-1]): H-6 (δ 6.46) and H-8 (δ 6.75), ROE correlation between the anomeric proton of rhamnose (rhamnose H-1, δ 5.52) and the protons at C-6 (H-6, δ 6.43) and C-8 position (H-8, δ 6.73), ESI-TOF-MS (positive ion mode): m/z 581.1496 [M+H]+, calculated for C26H29O15, 581.1506. ESI-TOF-MS/MS (positive ion mode, collision energy 30 eV): m/z (relative intensity) 449.1086 [M+H-arabinose]+ (41), 303.0554 [M+H-arabinose-rhamnose]+ (100), and UV λmax nm:340 (on flow). Monitoring was accomplished by PDA. 1H-NMR spectra were recorded on a JNM ECP-600 spectrometer (JEOL).
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative database under accession numbers TT4 (At5g13930), TT7 (At5g07990), UGT75C1 (At4g14090), UGT78D1 (At1g30530), UGT78D2 (At5g17050), UGT78D3 (At5g17030), UGT89C1 (At1g06000), OMT1 (At5g54160), RHM1 (At1g78570), RHM2 (At1g53500), and RHM3 (At3g14790).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Venn Diagrams of MS Peaks Detected in Various Organs of Col-0 and tt4.
Supplemental Figure 2. Coexpression Relationships of Genes Involved in the Flavonoid Pathway in Arabidopsis.
Supplemental Figure 3. Multiple Alignment of the Deduced Amino Acid Sequences of Flavonoid 3-O-Glucosyltransferases from Arabidopsis and Grape (Vitis vinifera).
Supplemental Figure 4. T-DNA Insertion Mutants of RHM1 and UPLC/MS Analyses of the rhm1 Mutant Lines.
Supplemental Figure 5. Schematic Representation of Duplicated Regions Containing RHMs in the Arabidopsis Genome.
Supplemental Table 1. Primers Used in This Study.
Supplemental Data Set 1. MS-Detectable Peaks in the Wild Type and tt4.
Supplemental Data Set 2. Characteristics of Standard Compounds.
Supplementary Material
Acknowledgments
We thank C. Ringli (University of Zurich) for the rol1-1 and rol1-2 mutants, M. Pauly (Michigan State University) for the rhm2-1 mutant, S. Kitamura (Japan Atomic Energy Research Institute) for the tt4 mutant, T. Nakagawa (Shimane University) for the pGWB2 vector, Y. Sekiyama (RIKEN) for helpful comments on NMR analysis, T. Obayashi (University of Tokyo), H. Suzuki, and N. Sakurai (Kazusa DNA Research Institute) for advice on coexpression analysis, and K. Akiyama (RIKEN) for excellent technical support for using the RIKEN-PRIMe website. This study was supported in part by grants-in-aid from the Ministry of Education, Culture, Sports, Science, and Technology of Japan and the Uehara Memorial Foundation, Japan.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Kazuki Saito (ksaito@faculty.chiba-u.jp).
Online version contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Akiyama, K., Chikayama, E., Yuasa, H., Shimada, Y., Tohge, T., Shinozaki, K., Hirai, M.Y., Sakurai, T., Kikuchi, J., and Saito, K. (2008). PRIMe: A Web site that assembles tools for metabolomics and transcriptomics. In Silico Biol. 8 0027. [PubMed] [Google Scholar]
- Andersen, Ø.M., and Markham, K.R. (2006). Flavonoids: Chemistry, Biochemistry, and Applications. (Boca Raton, FL: CRC Taylor & Francis).
- Aoki, K., Ogata, Y., and Shibata, D. (2007). Approaches for extracting practical information from gene co-expression networks in plant biology. Plant Cell Physiol. 48 381–390. [DOI] [PubMed] [Google Scholar]
- Blanc, G., Hokamp, K., and Wolfe, K.H. (2003). A recent polyploidy superimposed on older large-scale duplications in the Arabidopsis genome. Genome Res. 13 137–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloor, S.J., and Abrahams, S. (2002). The structure of the major anthocyanin in Arabidopsis thaliana. Phytochemistry 59 343–346. [DOI] [PubMed] [Google Scholar]
- Brown, B.A., Cloix, C., Jiang, G.H., Kaiserli, E., Herzyk, P., Kliebenstein, D.J., and Jenkins, G.I. (2005). A UV-B-specific signaling component orchestrates plant UV protection. Proc. Natl. Acad. Sci. USA 102 18225–18230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Auria, J.C., and Gershenzon, J. (2005). The secondary metabolism of Arabidopsis thaliana: Growing like a weed. Curr. Opin. Plant Biol. 8 308–316. [DOI] [PubMed] [Google Scholar]
- Diet, A., Link, B., Seifert, G.J., Schellenberg, B., Wagner, U., Pauly, M., Reiter, W.D., and Ringli, C. (2006). The Arabidopsis root hair cell wall formation mutant lrx1 is suppressed by mutations in the RHM1 gene encoding a UDP-L-rhamnose synthase. Plant Cell 18 1630–1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon, R.A., and Paiva, N.L. (1995). Stress-induced phenylpropanoid metabolism. Plant Cell 7 1085–1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon, R.A., and Strack, D. (2003). Phytochemistry meets genome analysis, and beyond. Phytochemistry 62 815–816. [DOI] [PubMed] [Google Scholar]
- Dooner, H.K., Robbins, T.P., and Jorgensen, R.A. (1991). Genetic and developmental control of anthocyanin biosynthesis. Annu. Rev. Genet. 25 173–199. [DOI] [PubMed] [Google Scholar]
- Fraser, C.M., Thompson, M.G., Shirley, A.M., Ralph, J., Schoenherr, J.A., Sinlapadech, T., Hall, M.C., and Chapple, C. (2007). Related Arabidopsis serine carboxypeptidase-like sinapoylglucose acyltransferases display distinct but overlapping substrate specificities. Plant Physiol. 144 1986–1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham, T.L. (1998). Flavonoid and flavonol glycoside metabolism in Arabidopsis. Plant Physiol. Biochem. 36 135–144. [Google Scholar]
- Harborne, J.B., Baxter, H., and Moss, G.P. (1999). Phytochemical Dictionary: A Handbook of Bioactive Compounds from Plants, 2nd ed. (Londong: Taylor & Francis).
- Hirai, M.Y., et al. (2005). Elucidation of gene-to-gene and metabolite-to-gene networks in Arabidopsis by integration of metabolomics and transcriptomics. J. Biol. Chem. 280 25590–25595. [DOI] [PubMed] [Google Scholar]
- Hirai, M.Y., et al. (2007). Omics-based identification of Arabidopsis Myb transcription factors regulating aliphatic glucosinolate biosynthesis. Proc. Natl. Acad. Sci. USA 104 6478–6483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, P., Messner, B., Nakajima, J., Schaffner, A.R., and Saito, K. (2003). UGT73C6 and UGT78D1, glycosyltransferases involved in flavonol glycoside biosynthesis in Arabidopsis thaliana. J. Biol. Chem. 278 43910–43918. [DOI] [PubMed] [Google Scholar]
- Kerhoas, L., Aouak, D., Cingoz, A., Routaboul, J.M., Lepiniec, L., Einhorn, J., and Birlirakis, N. (2006). Structural characterization of the major flavonoid glycosides from Arabidopsis thaliana seeds. J. Agric. Food Chem. 54 6603–6612. [DOI] [PubMed] [Google Scholar]
- Kleine, T., Kindgren, P., Benedict, C., Hendrickson, L., and Strand, A. (2007). Genome-wide gene expression analysis reveals a critical role for CRYPTOCHROME1 in the response of Arabidopsis to high irradiance. Plant Physiol. 144 1391–1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubo, A., Arai, Y., Nagashima, S., and Yoshikawa, T. (2004). Alteration of sugar donor specificities of plant glycosyltransferases by a single point mutation. Arch. Biochem. Biophys. 429 198–203. [DOI] [PubMed] [Google Scholar]
- Kusano, M., Fukushima, A., Arita, M., Jonsson, P., Moritz, T., Kobayashi, M., Hayashi, N., Tohge, T., and Saito, K. (2007). Unbiased characterization of genotype-depedent metabolic regulations by metabolomic approach in Arabidopsis thaliana. BMC Syst Biol. 1 53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lepiniec, L., Debeaujon, I., Routaboul, J.M., Baudry, A., Pourcel, L., Nesi, N., and Caboche, M. (2006). Genetics and biochemistry of seed flavonoids. Annu. Rev. Plant Biol. 57 405–430. [DOI] [PubMed] [Google Scholar]
- Lim, E.K., Li, Y., Parr, A., Jackson, R., Ashford, D.A., and Bowles, D.J. (2001). Identification of glucosyltransferase genes involved in sinapate metabolism and lignin synthesis in Arabidopsis. J. Biol. Chem. 276 4344–4349. [DOI] [PubMed] [Google Scholar]
- Luo, J., et al. (2007). Convergent evolution in the BAHD family of acyl transferases: Identification and characterization of anthocyanin acyl transferases from Arabidopsis thaliana. Plant J. 50 678–695. [DOI] [PubMed] [Google Scholar]
- Mulinacci, N., Vincieri, F.F., Baldi, A., Bambagiotti-alberti, M., Sendl, A., and Wagner, H. (1995). Flavonol glycosides from Sedum telephium subspecies maximum leaves. Phytochemistry 38 531–533. [Google Scholar]
- Obayashi, T., Kinoshita, K., Nakai, K., Shibaoka, M., Hayashi, S., Saeki, M., Shibata, D., Saito, K., and Ohta, H. (2007). ATTED-II: A database of co-expressed genes and cis elements for identifying co-regulated gene groups in Arabidopsis. Nucleic Acids Res. 35 D863–D869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Offen, W., Martinez-Fleites, C., Yang, M., Kiat-Lim, E., Davis, B.G., Tarling, C.A., Ford, C.M., Bowles, D.J., and Davies, G.J. (2006). Structure of a flavonoid glucosyltransferase reveals the basis for plant natural product modification. EMBO J. 25 1396–1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oka, T., Nemoto, T., and Jigami, Y. (2007). Functional analysis of Arabidopsis thaliana RHM2/MUM4, a multidomain protein involved in UDP-D-glucose to UDP-L-rhamnose conversion. J. Biol. Chem. 282 5389–5403. [DOI] [PubMed] [Google Scholar]
- Ringli, C., Bigler, L., Kuhn, B.M., Leiber, R.M., Diet, A., Santelia, D., Frey, B., Pollmann, S., and Klein, M. (2008). The modified flavonol glycosylation profile in the Arabidopsis rol1 mutants results in alterations in plant growth and cell shape formation. Plant Cell 20. [DOI] [PMC free article] [PubMed]
- Routaboul, J.M., Kerhoas, L., Debeaujon, I., Pourcel, L., Caboche, M., Einhorn, J., and Lepiniec, L. (2006). Flavonoid diversity and biosynthesis in seed of Arabidopsis thaliana. Planta 224 96–107. [DOI] [PubMed] [Google Scholar]
- Ruegger, M., and Chapple, C. (2001). Mutations that reduce sinapoylmalate accumulation in Arabidopsis thaliana define loci with diverse roles in phenylpropanoid metabolism. Genetics 159 1741–1749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito, K., Hirai, M.Y., and Yonekura-Sakakibara, K. (2008). Decoding genes with coexpression networks and metabolomics – ‘Majority report by precogs.’ Trends Plant Sci. 13 36–43. [DOI] [PubMed] [Google Scholar]
- Sakar, M.K., Şöhretoğlu, D., Özalp, M., Ekizoğlu, M., Piacente, S., and Pizza, C. (2005). Polyphenolic compounds and antimicrobial activity of Quercus aucheri leaves. Turk. J. Chem. 29 555–559. [Google Scholar]
- Shikazono, N., Yokota, Y., Kitamura, S., Suzuki, C., Watanabe, H., Tano, S., and Tanaka, A. (2003). Mutation rate and novel tt mutants of Arabidopsis thaliana induced by carbon ions. Genetics 163 1449–1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirley, B.W., Kubasek, W.L., Storz, G., Bruggemann, E., Koornneef, M., Ausubel, F.M., and Goodman, H.M. (1995). Analysis of Arabidopsis mutants deficient in flavonoid biosynthesis. Plant J. 8 659–671. [DOI] [PubMed] [Google Scholar]
- Stracke, R., Ishihara, H., Huep, G., Barsch, A., Mehrtens, F., Niehaus, K., and Weisshaar, B. (2007). Differential regulation of closely related R2R3-MYB transcription factors controls flavonol accumulation in different parts of the Arabidopsis thaliana seedling. Plant J. 50 660–677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart, J.M., Segal, E., Koller, D., and Kim, S.K. (2003). A gene-coexpression network for global discovery of conserved genetic modules. Science 302 249–255. [DOI] [PubMed] [Google Scholar]
- Tanaka, Y., and Filippa, B. (2006). Flower color. In Flowering and Its Manipulation, Vol, 20, C. Ainsworth, ed (London: Blackwell Publishers), pp. 201–239.
- Tohge, T., et al. (2005). Functional genomics by integrated analysis of metabolome and transcriptome of Arabidopsis plants over-expressing an MYB transcription factor. Plant J. 42 218–235. [DOI] [PubMed] [Google Scholar]
- Tohge, T., Yonekura-Sakakibara, K., Niida, R., Watanabe-Takahashi, A., and Saito, K. (2007). Phytochemical genomics in Arabidopsis thaliana: A case study for functional identification of flavonoid biosynthsis genes. Pure Appl. Chem. 79 811–823. [Google Scholar]
- Usadel, B., Kuschinsky, A.M., Rosso, M.G., Eckermann, N., and Pauly, M. (2004). RHM2 is involved in mucilage pectin synthesis and is required for the development of the seed coat in Arabidopsis. Plant Physiol. 134 286–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valvekens, D., Van Montagu, M., and Van Lijsebettens, M. (1988). Agrobacterium tumefaciens-mediated transformation of Arabidopsis thaliana root explants by using kanamycin selection. Proc. Natl. Acad. Sci. USA 85 5536–5540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veit, M., and Pauli, G.F. (1999). Major flavonoids from Arabidopsis thaliana leaves. J. Nat. Prod. 62 1301–1303. [DOI] [PubMed] [Google Scholar]
- von Roepenack-Lahaye, E., Degenkolb, T., Zerjeski, M., Franz, M., Roth, U., Wessjohann, L., Schmidt, J., Scheel, D., and Clemens, S. (2004). Profiling of Arabidopsis secondary metabolites by capillary liquid chromatography coupled to electrospray ionization quadrupole time-of-flight mass spectrometry. Plant Physiol. 134 548–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Western, T.L., Young, D.S., Dean, G.H., Tan, W.L., Samuels, A.L., and Haughn, G.W. (2004). MUCILAGE-MODIFIED4 encodes a putative pectin biosynthetic enzyme developmentally regulated by APETALA2, TRANSPARENT TESTA GLABRA1, and GLABRA2 in the Arabidopsis seed coat. Plant Physiol. 134 296–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yonekura-Sakakibara, K., Kojima, M., Yamaya, T., and Sakakibara, H. (2004). Molecular characterization of cytokinin-responsive histidine kinases in maize. Differential ligand preferences and response to cis-zeatin. Plant Physiol. 134 1654–1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yonekura-Sakakibara, K., Tohge, T., Niida, R., and Saito, K. (2007). Identification of a flavonol 7-O-rhamnosyltransferase gene determining flavonoid pattern in Arabidopsis by transcriptome coexpression analysis and reverse genetics. J. Biol. Chem. 282 14932–14941. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.