Abstract
Bacterial wilt, a disease impacting cultivated crops worldwide, is caused by the pathogenic bacterium Ralstonia solanacearum. PopP2 (for Pseudomonas outer protein P2) is an R. solanacearum type III effector that belongs to the YopJ/AvrRxv protein family and interacts with the Arabidopsis thaliana RESISTANT TO RALSTONIA SOLANACEARUM 1-R (RRS1-R) resistance protein. RRS1-R contains the Toll/Interleukin1 receptor–nucleotide binding site–Leu-rich repeat domains found in several cytoplasmic R proteins and a C-terminal WRKY DNA binding domain. In this study, we identified the Arabidopsis Cys protease RESPONSIVE TO DEHYDRATION19 (RD19) as being a PopP2-interacting protein whose expression is induced during infection by R. solanacearum. An Arabidopsis rd19 mutant in an RRS1-R genetic background is compromised in resistance to the bacterium, indicating that RD19 is required for RRS1-R–mediated resistance. RD19 normally localizes in mobile vacuole-associated compartments and, upon coexpression with PopP2, is specifically relocalized to the plant nucleus, where the two proteins physically interact. No direct physical interaction between RRS1-R and RD19 in the presence of PopP2 was detected in the nucleus as determined by Förster resonance energy transfer. We propose that RD19 associates with PopP2 to form a nuclear complex that is required for activation of the RRS1-R–mediated resistance response.
INTRODUCTION
Plants have evolved two lines of defense in response to pathogen attack (Jones and Dangl, 2006; De Wit, 2007). Primary or basal defense constitutes the first barrier to all potential pathogens and is triggered by pathogen-associated molecular patterns (PAMPs), conserved molecules present in a wide range of pathogens. The best known PAMPs are bacterial flagellin and elongation factor Tu (Kunze et al., 2004; Zipfel et al., 2004, 2006; Chinchilla et al., 2006) and fungal β-glucans and chitin (Fliegmann et al., 2004; Kaku et al., 2006; Miya et al., 2007). PAMPs are perceived by transmembrane pattern recognition receptors that mediate basal defense responses (Jones and Dangl, 2006; De Wit, 2007), thereby providing immunity of plants to most potentially pathogenic microbes. This primary line of defense can be overcome by microbes that have evolved various effectors with different virulence functions to avoid or suppress PAMP-triggered immune responses in the host, enabling microbes to promote disease (Mudgett, 2005; Grant et al., 2006; da Cunha et al., 2007). The second line of defense, targeted either against these effectors (now called avirulence factors [Avr]) or against effector-induced perturbations, is mediated by plant resistance (R) proteins. This results in effector-triggered immunity (ETI), which leads to strong disease resistance responses that are often associated with a hypersensitive response (HR) (Jones and Dangl, 2006; De Wit, 2007). In the absence of an R protein, the pathogen avoids or suppresses basal defense, colonizes the host plant, and causes disease. The biochemical implication of the gene-for-gene hypothesis (also called ETI) initially proposed by Flor (1971) implies that an R protein is the primary receptor of an Avr protein (Keen, 1990; De Wit, 1992). Direct interaction between an R and an Avr protein has been confirmed in a few pathosystems, including Magnaporthe grisea–rice (Oryza sativa; Jia et al., 2000), Ralstonia solanacearum–Arabidopsis thaliana (Deslandes et al., 2003), Melampsora lini–flax (Linum usitatissimum; Dodds et al., 2006), and tobacco mosaic virus (TMV)–tobacco (Nicotiana tabacum; Burch-Smith et al., 2007), but this situation is the exception rather than the rule. In most cases, an R protein does not directly interact with an effector but guards and senses perturbation of a virulence target by an effector and subsequently mediates ETI. This indirect interaction, also known as the guard hypothesis (Van der Biezen and Jones, 1998), has been demonstrated to occur in several R-Avr pairs from various host–pathogen interactions (Ren et al., 2000; Krüger et al., 2002; Mackey et al., 2002, 2003; Axtell and Staskawicz, 2003; Shao et al., 2003; Xia et al., 2004; Rooney et al., 2005).
Most gram-negative bacterial plant pathogens use the type-three secretion system (TTSS) to deliver, directly into plant cells, effectors that can either elicit disease symptoms in susceptible plants or ETI in resistant plants containing a cognate R protein (Cornelis and Van Gijsegem, 2000; Jones and Dangl, 2006). TTSS is an essential pathogenicity determinant since mutants defective in HR and pathogenicity (hrp) genes encoding the building blocks for this secretion machinery are no longer virulent. Despite the identification of a large number of bacterial effectors (Grant et al., 2006), their precise roles in colonization of the host plant, pathogenesis, and activation of plant defense responses are still largely unknown.
Perturbation of host protein activities involved in basal defense is a common strategy used by pathogens to avoid or suppress host defense responses and thereby to cause disease. Effectors can circumvent host defense responses by inducing proteolysis of plant proteins. Several bacterial effectors sharing similarities with Cys proteases have been identified (Hotson and Mudgett, 2004; van der Hoorn, 2008) and shown to be involved in crucial regulatory steps controlling defense responses and resistance. The Cys proteases AvrRpt2 and HopAR1 from Pseudomonas syringae cleave essential components of the R-mediated resistance signaling pathway. AvrRpt2 targets RPM1-interacting protein 4 (RIN4), a negative regulator of the basal defense response, whose degradation leads to activation of Resistance to Pseudomonas syringae 2 (RPS2), a nucleotide binding site–Leu-rich repeat (NBS-LRR) containing R protein (Axtell and Staskawicz, 2003). HopAR1 (previously called AvrPphB) specifically cleaves PBS1, a Ser-Thr kinase required for activation of the cognate R protein, RPS5 (Shao et al., 2003).
Host plant proteases can also play key roles in pathogen recognition and in disease resistance signaling (van der Hoorn and Jones, 2004; Mosolov and Valueva, 2006). Maturation of the effectors Avr4 and Avr9 of the fungal tomato pathogen Cladosporium fulvum is at least partially achieved in planta by yet unknown plant proteases during infection (Van den Ackerveken et al., 1993; Joosten et al., 1997), whereas perception of the Avr2 effector of this pathogen by the cognate tomato R protein Cf-2 (Dixon et al., 1996, 2000; Luderer et al., 2002) requires Rcr3 (for Required for Cladosporium fulvum resistance 3), a secreted tomato (Solanum lycopersicum) Cys protease that is inhibited through its interaction with Avr2 (Rooney et al., 2005). Recently it was shown that C. fulvum Avr2 targets several extracellular Cys proteases of tomato that are required for basal host defense (Shabab et al., 2008; van Esse et al., 2008). The diverse functions of plant Cys proteases are also illustrated by their involvement in the HR, commonly associated with plant disease resistance (D'Silva et al., 1998; Solomon et al., 1999; Mosolov et al., 2001; Chichkova et al., 2004; Rojo et al., 2004; Gilroy et al., 2007; Mur et al., 2007; van der Hoorn, 2008). Thus, manipulation of plant proteases may contribute to distinct aspects of plant responses to pathogens.
R. solanacearum is a soil-borne β-proteobacterium causing bacterial wilt disease in >200 species, including agronomically important crop plants of the Solanaceous family, such as tobacco, tomato, and potato (Solanum tuberosum; Hayward, 1991, 2000). Bacterial infection predominantly occurs at both the root elongation zone and the sites of lateral root emergence. After invasion of the intercellular spaces, bacteria cross the endoderm and reach the xylem vessels where they proliferate. Subsequently, bacteria spread into the aerial parts of the infected plants (Vasse et al., 1995). The massive production of exopolysaccharides leads to the obstruction of the vascular system that causes wilting symptoms resulting from strong reduction of water flow (Schell et al., 1994). R. solanacearum encodes up to 80 putative TTSS effectors (Salanoubat et al., 2002; Cunnac et al., 2004; Occhialini et al., 2005). Among them, Pseudomonas outer protein P2 (PopP2), an effector found in most R. solanacearum strains, elicits a specific disease resistance response in Arabidopsis mediated by RESISTANT TO R. SOLANACEARUM 1-R (RRS1-R), its cognate R protein (Deslandes et al., 2003). RRS1-R is an atypical R protein that contains not only the Toll/Interleukin1 receptor (TIR)–NBS-LRR domains found in several other R proteins but also a C-terminal WRKY domain, a signature of the zinc-finger class of WKRY transcription factors (Deslandes et al., 2002; Eulgem and Somssich, 2007). Coexpression of both PopP2 and RRS1-R in Arabidopsis revealed that RRS1-R is specifically targeted to the plant nucleus in the presence of PopP2 and that these two proteins colocalize to the nucleus (Deslandes et al., 2003).
PopP2 belongs to the YopJ/AvrRxv effector protein family whose members share structural similarities with the C55 peptidase family of the CE clan of Cys proteases (Barrett and Rawlings, 2001). Apart from RRS1-R (Deslandes et al., 2003), no other plant targets of PopP2 have yet been described. In this study, search for such targets was performed by a yeast two-hybrid screen using PopP2 as a bait, which led to the identification of RESPONSIVE TO DEHYDRATION19 (RD19), a Cys protease. We showed that PopP2 specifically relocalizes RD19 from vacuole-associated compartments to the nucleus. A fluorescence lifetime imaging (FLIM) approach allowed us to demonstrate a physical interaction between these two proteins in living cells. Since we also found that an rd19 knockout mutant is impaired in resistance to R. solanacearum, we conclude that RD19 is a crucial host factor for PopP2-triggered RRS1-R–mediated resistance.
RESULTS
Identification of RD19
To identify components of the RRS1-R/PopP2-mediated disease resistance signaling, PopP2 was used as bait in the screening of a yeast two-hybrid Arabidopsis cDNA library generated from mRNAs isolated from a mixture of root tissues of 10-d-old seedlings (from both RRS1-S and RRS1-R genetic backgrounds), challenged with the R. solanacearum GMI1000 strain expressing PopP2. After several rounds of screening, different prey cDNA clones were identified. Among those, we focused on a partial cDNA clone encoding the last 124 amino acid residues of the RD19 Cys protease. According to the MEROPS peptidase database, RD19 is a predicted papain-like Cys protease (PLCP; subfamily C1A, http://merops.sanger.ac.uk/).
The rd19 Mutant Is Compromised in RRS1-R–Mediated Resistance Signaling
To investigate the function of RD19 in the establishment of the plant response to R. solanacearum, we searched for an Arabidopsis rd19 null mutant in the SALK collection (http://signal.salk.edu/). Among various candidate lines, we were able to identify a homozygous rd19 knockout line containing a T-DNA insertion at position +55 of the predicted open reading frame (SALK_031088 line). To determine whether inactivation of RD19 could compromise RRS1-R–mediated resistance, the rd19 mutation originally identified in a Columbia (Col-0) accession (susceptible, RRS1-S) was introduced into an Nd-1 accession (resistant, RRS1-R). Interestingly, two independent F3 lines (F3-1 and F3-3; see Supplemental Figures 1A and 1B online), selected as being homozygous for both the rd19 mutation and the RRS1-R gene, developed wilting symptoms in response to the GMI1000 strain. These data suggest that RD19 is involved in RRS1-R–mediated resistance signaling. Segregation of the loss of resistance phenotype with the rd19 mutation in an RRS1-R background was confirmed by expanding this analysis on eight different segregating F3 families. Susceptibility to the GMI1000 strain correlated with the presence of the rd19 mutation in two homozygous rd19 RRS1-R lines challenged with the GMI1000 strain (F3-1 and F3-3 lines; see Supplemental Figures 1A and 1B online). For all subsequent experiments, a representative rd19 RRS1-R F3 line (F3-1) was used.
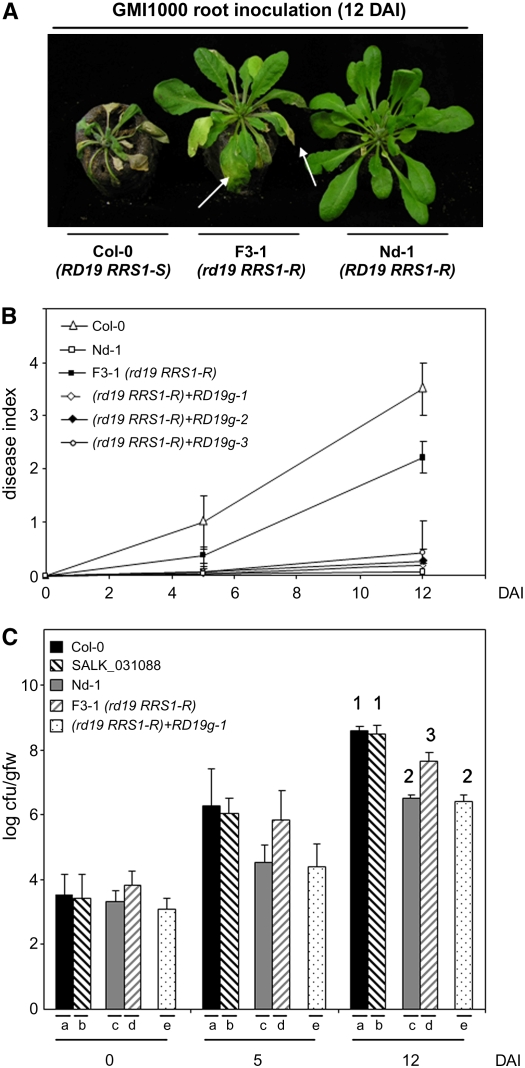
The development of disease symptoms in this rd19 RRS1-R F3 line after inoculation with the GMI1000 strain was studied in more detail. Five days after inoculation (DAI), Col-0 plants showed some wilting symptoms, whereas the rd19 RRS1-R line and wild-type Nd-1 plants remained without symptoms. Seven days later (12 DAI), 25% of the scored leaves of rd19 RRS1-R plants were wilted (disease index 1 [D1]), whereas Nd-1 plants showed no detectable symptoms and Col-0 plants were completely wilted (Figures 1A and 1B). Disease symptoms correlated with an increase of bacterial growth in rd19 RRS1-R plants. Twelve days after inoculation, the bacterial growth in the rd19 RRS1-R plants reached intermediate levels between those found in Col-0 and Nd-1 plants (Figure 1C). Taken together, these data suggest that RD19 is involved in the establishment of PopP2-triggered RRS1-R–mediated resistance. Bacterial multiplication of the GMI1000 strain monitored in the SALK_031088 line (Col-0 background) was similar to that of Col-0, indicating that rd19 loss of function did not affect the susceptibility to Ralstonia in an RRS1-S genetic background (Figure 1C).
Figure 1.
RD19 Is Required for RRS1-R–Mediated Disease Resistance Signaling.
(A) Phenotypic responses of susceptible Col-0 (RD19 RRS1-S) and resistant Nd-1 (RD19 RRS1-R) wild-type Arabidopsis plants and of the rd19 RRS1-R (F3-1) line to the GMI1000 strain of R. solanacearum 12 DAI. Arrows point to wilted leaves of the mutant line.
(B) Disease symptom development curves. Each plant was scored at 0, 5, and 12 DAI of roots using a scale between 0 and 4: 0 = no wilting, 1 = 25%, 2 = 50%, 3 = 75%, and 4 =100% of leaves were wilted. Means and sd were calculated from scores of a total of 60 plants per genotype (from three independent experiments); triangle, Col-0 (RD19 RRS1-S); open square, Nd-1(RD19 RRS1-R); closed square, rd19 mutant (F3-1 line [rd19 RRS1-R]); open diamonds, closed diamond, and open circle, three independent complemented rd19 lines (RD19g1 to -3 in an rd19 RRS1-R background).
(C) Bacterial growth inside the plant was estimated as described before (Deslandes et al., 1998). Means of colony-forming units per gram of fresh weight (cfu/gfw) and sd were calculated from triplicates of three plants for each genotype (from three independent experiments). (a) Col-0 (RD19 RRS1-S), (b) SALK_031088 line (rd19 RRS1-S), (c) Nd-1(RD19 RRS1-R), (d) rd19 RRS1-R F3-1 line, (e) complemented rd19 line (RD19g1 in an rd19 RRS1-R background). Analysis of variance and multiple between-group comparisons (Bonferroni test) were used to analyze differences between ecotypes. Groups 1 (a and b), 2 (d), and 3 (c and e) are statistically different (P value of 0.05).
Complementation of the rd19 Mutant Restores PopP2-Triggered RRS1-R–Mediated Resistance
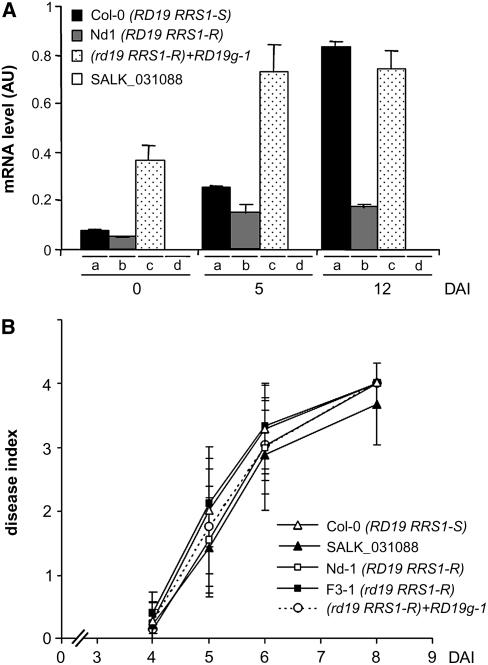
To demonstrate that the wilting symptoms observed in the rd19 RRS1-R mutant were causally related to the loss of function of RD19, genetic complementation of this mutant was performed using a 3527-bp genomic fragment from Col-0 containing the complete RD19 open reading frame driven by 1429 bp of the native 5′ regulatory sequence and 596 bp of the 3′ terminator sequence (named RD19g). Three independent homozygous RD19g-complemented T2 lines (RD19g-1 to -3) containing a single insertion of the wild-type RD19 gene were selected and root inoculated with R. solanacearum GMI1000. As shown in Figure 1B, rd19 RRS1-R plants containing the RD19g construct regained a resistance phenotype similar to that of Nd-1 plants upon inoculation. Internal bacterial multiplication in the RD19g-1 line, which we selected as a representative transgenic line, was also restored to wild-type levels (Figure 1C). These results confirm that loss of RD19 function leads to development of enhanced wilting symptoms and increased bacterial multiplication in response to R. solanacearum. We compared the level of detectable RD19 transcript, as determined by quantitative RT-PCR (Q-RT-PCR), with the gain-of-function phenotype. In unchallenged plants, RD19 expression patterns were similar in both Col-0 and Nd-1 plants. By contrast, RD19g-1 plants showed a three- to fourfold higher RD19 expression level than that of wild-type plants (Figure 2A; t = 0). Similar results were obtained for the two other independent RD19g complemented lines, RD19g-2 and -3 (see Supplemental Figure 2 online). This increased expression level of RD19 might be caused by a position effect of the T-DNA insertion.
Figure 2.
Enhanced Expression of RD19 Does Not Affect the Plant Response to the ΔPopP2 Strain.
(A) Expression analysis of the RD19 gene in three different genotypes after inoculation with the GMI1000 strain. RD19 transcript levels were determined by Q-RT-PCR from cDNAs generated from the aerial parts of three plants per genotype at 0, 5, and 12 DAI. The expression values of RD19 were normalized using the expression level of two housekeeping genes considered as internal standards. Mean expression and sd values were calculated from the results of two independent experiments (triplicate samples of three plants were taken at each time point). (a) Col-0 (RD19 RRS1-S), (b) Nd-1(RD19 RRS1-R), (c) RD19g-1 line (rd19 RRS1-R mutant complemented with RD19 genomic clone), (d) rd19 RRS1-S mutant (SALK_031088). AU, arbitrary units.
(B) Disease symptom developments were scored after root inoculation with the ΔPopP2 strain. Each plant was scored at 4, 5, 6, and 8 DAI of roots using a scale between 0 and 4: 0 = no wilting, 1 = 25%, 2 = 50%, 3 = 75%, and 4 =100% of leaves were wilted. Means and sd were calculated from scores of 30 plants per accession and mutant line. This experiment was repeated three times, and reproducible results were obtained; open triangle, Col-0 (RD19 RRS1-S); open square, Nd-1 (RD19 RRS1-R); closed triangle, SALK_031088 line (rd19 RRS1-S); closed square, F3-1 line (rd19 RRS1-R); open circle, RD19g-1 line (rd19 RRS1-R mutant complemented with RD19 genomic clone).
To rule out the possibility that the resistance phenotype of the RD19g-1 line was solely due to the higher expression of the transgene, which could confer nonspecific resistance, we challenged these plants with ΔPopP2, a GMI1000 strain that lacks PopP2 (Lavie et al., 2002). This strain is virulent on both Nd-1 and Col-0 plants (Deslandes et al., 2003). RD19g-1 plants developed wilting symptoms similar to those observed on Nd-1 plants (Figure 2B). These data demonstrate that the resistance response of RD19g-1 plants challenged with the GMI1000 strain is specific and is not due to the increased expression level of the transgene.
RD19 Expression Is Upregulated during the Wilt Disease Development
The expression of RD19 in Col-0, Nd-1, and the rd19 RRS1-R+RD19g line was analyzed upon challenge with the GMI1000 strain using Q-RT-PCR. Whereas no RD19 transcript could be detected in the rd19 mutant (SALK_031088 line), the expression of RD19 increased after inoculation with R. solanacearum in susceptible Col-0 plants (eightfold between 0 and 12 DAI; Figure 2A). The increased expression of RD19 in wilting Col-0 plants is in agreement with previous reports showing that RD19 is induced during drought conditions (Koizumi et al., 1993). By contrast, both resistant Nd-1 and RD19g-1 transgenic plants showed a twofold increase in RD19 transcripts at 5 DAI after which the levels remained constant until 12 DAI.
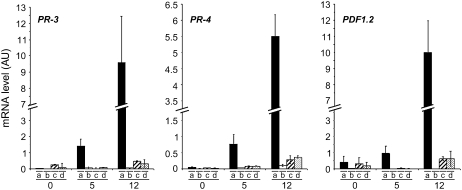
We then checked whether the appearance of wilt symptoms in the rd19 RRS1-R mutant line in response to R. solanacearum was accompanied by the induction of the ethylene/jasmonic acid signaling pathways. Previously, ethylene and jasmonic acid marker genes have been shown to be upregulated during wilt disease development in susceptible Col-0 plants challenged with the GMI1000 strain (Hirsch et al., 2002). The expression profiles of the pathogenesis-related (PR) genes PR-3, PR-4, and PDF1.2 were monitored in GMI1000-inoculated rd19 RRS1-R plants (Figure 3). Transcript levels of these PR genes increased in wilting-susceptible Col-0 plants (at 5 and 12 DAI), whereas no induction of those genes could be detected either in the partially susceptible rd19 RRS1-R mutant line and in symptomless Nd-1 or in the complemented mutant RD19g-1 line. These data indicate that partial loss of RRS1-R–mediated resistance in the rd19 mutant is not associated with the activation of the ethylene/jasmonic acid defense signaling pathways.
Figure 3.
Expression Analysis of PR Genes PR-3, PR-4, and PDF1.2.
Transcript levels of PR-3, PR-4, and PDF1.2 genes were determined by Q-RT-PCR from cDNAs generated from aerial parts of three plants per genotype at 0, 5, and 12 DAI. The expression values of the genes were normalized using the expression level of two housekeeping genes considered as internal standards. Mean expression and sd values were calculated from the results of two independent experiments (triplicate samples of three plants were taken at each time point). (a) Col-0 (RD19 RRS1-S), (b) Nd-1(RD19 RRS1-R), (c) rd19 mutant (F3-1 line, rd19 RRS1-R), (d) RD19g-1 line (rd19 RRS1-R mutant complemented with RD19).
RD19 Colocalizes with Aleurain, a Vacuole-Targeted Cys Protease
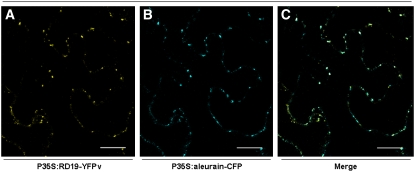
To determine the subcellular localization of the RD19 protein in plant cells, RD19 was tagged on its C terminus with the yellow fluorescent protein venus (YFPv) and transiently expressed by means of particle bombardment under the control of the constitutive 35S promoter in Arabidopsis epidermal cells. Using confocal laser scanning microscopy, the RD19-YFPv fusion protein was found to label small mobile compartments (see Supplemental Figure 3 online). However, due to the extremely low transformation efficiency, transient expression of RD19-YFPv was subsequently performed via Agrobacterium tumefaciens in Nicotiana benthamiana leaves, where a similar localization was observed in a very high number of transformed cells (Figure 4A). This localization pattern is similar to that of vacuolar proteases that are synthesized as preproteins in the rough endoplasmic reticulum and transiently transported to the vacuole through the endomembrane secretion system (Mo et al., 2006). To determine the subcellular localization of RD19 more precisely, markers of various vesicular compartments were used. A perfect colocalization was found with aleurain fused to cyan fluorescent protein (CFP). Aleurain is a PLCP described as being targeted to the lytic vacuole where it becomes active after proteolytic processing (Paris et al., 1996). Forty eight hours after coexpression in N. benthamiana leaves, overlay of YFP and CFP images demonstrated that both fluorescent protein fusions labeled the same mobile vacuole-associated compartments that may correspond to prevacuolar vesicles (Figures 4A to 4C). No fluorescence could be detected in the vacuole for both RD19 and aleurain, probably due to the light-dependent degradation of YFP under the acidic vacuolar conditions (Fluckiger et al., 2003; Tamura et al., 2003). These observations suggest that RD19 is a Cys protease targeted to the lytic vacuole through a similar secretory pathway as aleurain.
Figure 4.
Subcellular Localization of RD19-YFPv in N. benthamiana.
Confocal images of N. benthamiana epidermal cells 48 h after coexpression of P35S:RD19-YFPv and P35S:aleurain-CFP via A. tumefaciens infiltration. RD19-YFPv and aleurain-CFP colocalize in mobile vesicles ([A] and [B], respectively), moving along the endoplasmic reticulum. Colocalization of RD19-YFPv and aleurain-CFP is shown in the merged image in (C). Bars = 20 μm.
RD19 Is Relocalized to the Plant Nucleus in the Presence of PopP2
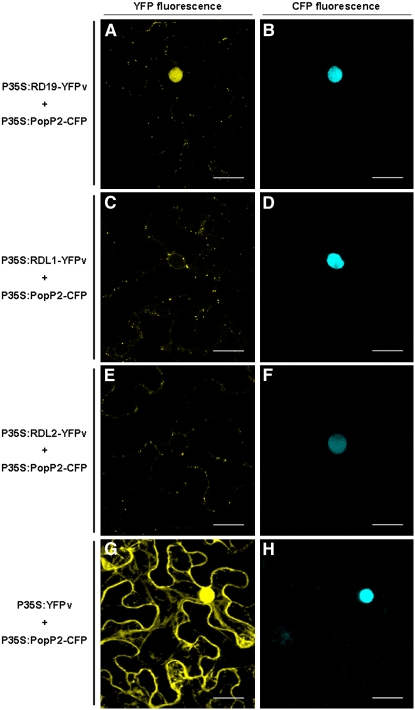
Based on the yeast two-hybrid results, we predicted a physical interaction between these two proteins in planta. When expressed individually, PopP2 is exclusively detected in the nucleus (Deslandes et al., 2003) and RD19 in mobile vacuole-associated compartments (Figure 4A). Coexpression of PopP2 fused to CFP (PopP2-CFP) and RD19 fused to YFPv (RD19-YFPv) in N. benthamiana led to YFP fluorescence detection not only in these mobile vesicles but also in the plant nucleus. This observation demonstrates partial relocalization of RD19-YFPv to the nucleus due to the presence of PopP2 (Figures 5A and 5B). By contrast, RD19 expression had no effect on the nuclear localization of PopP2-CFP. In a similar way, relocalization of RD19-YFPv was observed in epidermal cells of Arabidopsis upon delivery of the corresponding constructs by particle gun bombardment (see Supplemental Figure 3 online). However, due to the extremely low transformation efficiency of Arabidopsis cells, we decided to use the N. benthamiana expression system. This decision was also justified by the PopP2-dependent nuclear targeting of the RRS1-R protein in Arabidopsis cells reported before (Deslandes et al., 2003), which occurs also in N. benthamiana.
Figure 5.
RD19 Relocalizes to the Plant Nucleus in the Presence of PopP2.
Coexpression of RD19-YFPv with PopP2-CFP in N. benthamiana epidermal cells resulted in the relocalization of RD19 into the nucleus ([A] and [B]). YFP fluorescence was not detected in the nucleus when PopP2-CFP was coexpressed with RDL1-YFPv or RDL2-YFPv ([C] and [D] and [E] and [F], respectively). YFP fluorescence was detected in the whole cell when YFPv alone was expressed in the presence of PopP2-CFP ([G] and [H]). Bars = 20 μm.
To test the specificity of RD19 recruitment to the nucleus, we used two RD19-like proteins, RDL1 and RDL2 from Arabidopsis sharing 86 and 73% identity, respectively, with RD19 (see Supplemental Figure 4 online). Both RDL1-YFPv and RDL2-YFPv were found to localize in the same mobile vacuole-associated compartments as RD19 since they also colocalize with aleurain-CFP (see Supplemental Figure 5 online), suggesting that these two RD19-like proteins are also targeted to the lytic vacuole. However, after coexpression of RDL1 or RDL2 with PopP2, no YFP fluorescence could be detected within the plant nucleus (Figures 5C to 5F). To confirm that nuclear YFP fluorescence was due to the relocalization of RD19-YFPv and not the result of a passive diffusion of a YFPv truncated form, we coexpressed YFPv alone with PopP2-CFP. This led to the labeling of the whole cell with YFP fluorescence (Figures 5G and 5H), a pattern completely different from that observed with RD19-YFPv+PopP2-CFP (Figure 5A). The presence of the full-length RD19-YPFv, RDL1-YFPv, and RDL2-YFPv fusion proteins (either coexpressed with PopP2 or alone) was further confirmed by protein gel blot analysis (see Supplemental Figure 6 online). Taken together, these data demonstrate specific recruitment of RD19 to the nucleus in the presence of PopP2.
RD19 Physically Associates with PopP2 in the Nucleus
To confirm a physical interaction between PopP2 and RD19 in the nucleus, we took a quantitative noninvasive FLIM approach to monitor the Förster resonance energy transfer (FRET) between the CFP (donor) and YFPv (acceptor) molecules fused to PopP2 and RD19, respectively. If these two proteins interact, the transfer of energy from the donor to the acceptor decreases the fluorescence lifetime (average time that a molecule remains in its excited state prior to returning to its basal state) of the donor fluorophore. The relative difference of lifetime is a measure of FRET efficiency. The average CFP lifetime in nuclei expressing PopP2-CFP was 2.136 ± 0.028 ns (mean ± sd, n = 86 nuclei). A significant reduction of the average CFP lifetime to 1.945 ± 0.024 ns (n = 91) was recorded in nuclei coexpressing the PopP2-CFP and RD19-YFPv fusion proteins (Table 1). FRET efficiencies corresponding to the different combinations tested are shown in Table 1. In nuclei expressing both PopP2-CFP and free YFPv, even with the high levels of YFPv in the nucleus (Figures 5G and 5H), no physical interaction could be detected between PopP2-CFP and YFPv, as indicated by an average CFP lifetime of 2.197 ± 0.017 ns (n = 32), which is not significantly different from that of CFP or PopP2-CFP alone. Taken together, these data confirm that reduction of PopP2-CFP lifetime, in the presence of RD19-YFPv, is not due to nonspecific transfer of energy between the two fluorophores and provide strong evidence for both a specific nuclear relocalization of RD19 (Figure 5A) and a physical interaction between PopP2 and RD19 in planta.
Table 1.
FLIM Measurements Showing That PopP2 Physically Interacts with RD19 in the Nucleus of N. benthamiana Epidermal Cells
| Donor | Acceptor | Cofactor | Lifetimea | sdb | nc | Ed |
|---|---|---|---|---|---|---|
| PopP2-CFP | – | – | 2.136 | 0.024 | 86 | – |
| PopP2-CFP | YFPv | – | 2.197 | 0.017 | 32 | – |
| PopP2-CFP | RD19-YFPv | – | 1.945 | 0.024 | 91 | 8.94 |
| PopP2-CFP | RD19-YFPv | RRS1-R | 1.936 | 0.026 | 30 | 9.36 |
| RRS1-R-CFP | – | PopP2 | 1.847 | 0.024 | 30 | – |
| RRS1-R-CFP | RD19-YFPv | PopP2 | 1.836 | 0.022 | 30 | – |
Mean lifetime in nanoseconds.
sd.
Total number of nuclei measured.
FRET efficiency percentage (E = 1 − τDA/τD) was calculated by comparing the lifetime of the donor in the presence of the acceptor (τDA) with its lifetime in the absence of the acceptor (τD).
Since RRS1-R and PopP2 physically interact in yeast (Deslandes et al., 2003), we checked whether the presence of RRS1-R could modulate the physical association between PopP2 and RD19 in N. benthamiana. Thus, the FRET efficiency of the PopP2-CFP/RD19-YFPv donor/acceptor couple was monitored in the presence of RRS1-R. As shown in Table 1, RRS1-R did not significantly alter the average CFP lifetime of PopP2-CFP coexpressed with RD19-YFPv. This indicates that the physical interaction between PopP2 and RD19 is not significantly altered in the presence of RRS1-R. Finally, when RRS1-R-CFP, RD19-YFPv, and PopP2 were simultaneously coexpressed, the average CFP lifetime of RRS1-R-CFP did not significantly differ from that found in nuclei coexpressing only RRS1-R-CFP and PopP2, suggesting that there is no detectable physical interaction between RRS1-R and RD19 in the nucleus (Table 1).
DISCUSSION
We report here the identification of the Arabidopsis Cys protease RD19 as a PopP2-interacting partner that is required for RRS1-R function triggered by PopP2, as demonstrated by the loss of full resistance of an rd19 RRS1-R mutant to the GMI1000 strain of R. solanacearum (Figure 1). Confocal fluorescence microscopy studies demonstrated that RD19 colocalizes with aleurain, a papain-like Cys protease known to locate in the lytic vacuole (Figure 4). Coexpression studies revealed that in the presence of PopP2, RD19 is specifically relocalized to the nucleus (Figure 5). FLIM analysis performed in living cells showed that both proteins physically interact in this key compartment (Table 1).
RD19 encodes a drought-inducible Cys protease (Koizumi et al., 1993) whose transcript levels increase strongly after R. solanacearum infection causing wilt disease development and water deprivation. The function of this Cys protease during drought stress is not known. RD19 inactivation leads to a loss of full RRS1-R–mediated resistance that is associated with increased bacterial multiplication in infected plants (Figure 1), which is comparable to the effect of RIN4 inactivation on RPM1-mediated resistance (Mackey et al., 2002). Upon challenge with the GMI1000 strain, an rd19 mutant developed disease symptoms comparable to those observed in the corresponding wild-type parental Col-0 plants (Figure 1), suggesting that, unlike RIN4, RD19 is not an essential component of basal plant defense. This assumption is strengthened by the observation that the susceptibility of RD19-overexpressing plants (RD19g-1 complemented line) is comparable to that of Nd-1 plants, after challenge with a virulent strain of R. solanacearum lacking PopP2 (ΔPopP2) (Figure 2B). Interestingly, loss of full RRS1-R–mediated resistance due to RD19 inactivation is not associated with the induction of marker genes of the ethylene and jasmonic acid signaling pathways, whereas these genes are induced in wilted Col-0 plants. This observation indicates that wilting symptom appearance is not tightly correlated with the activation of these signaling pathways.
Subcellular localization of RD19 was studied by fluorescence microscopy. We found that RD19 colocalizes with aleurain (Figure 4), a vacuolar PLCP (Paris et al., 1996). Although RD19-YFP was only detected in mobile vacuole-associated vesicles, our colocalization data are consistent with it being destined to the lytic vacuole. The failure to detect both RD19-YFP and aleurain-CFP fusion proteins in the lytic vacuole could be due to the light-dependent degradation of both fluorescent proteins in this acidic compartment. Green fluorescent protein (GFP) is indeed unstable in the acidic vacuoles of higher plants in the presence of light (Tamura et al., 2003). In the lytic vacuole, absorption of the blue light at low pH induces a conformational change of the fluorescent protein, making it more sensitive to degradation by vacuolar proteinases. Thus, the clear visualization of RD19-YFP and aleurain-CFP may be explained by the higher stability of the fluorescent proteins in prevacuolar vesicles.
In this study, we demonstrated that PopP2 partially modifies the targeting of RD19 from mobile vacuole-associated compartments to the plant nucleus. The specificity of this bacterial effector for its target was demonstrated by showing that the localization of RDL1 and RDL2, two closely related Arabidopsis homologs of RD19, predicted to be targeted to the lytic vacuole, was not altered in the presence of PopP2 (Figure 5). Together with RRS1-R, whose nuclear targeting is dependent on the presence of PopP2 (Deslandes et al., 2003), RD19 constitutes the second example of a PopP2-interacting plant component whose subcellular localization is affected by this bacterial effector. Changes in subcellular protein localization, reorganization of the plant cytoskeleton, organelle positioning, cell trafficking, and cytoplasmic-nuclear transport and partitioning represent some of the many dynamic cellular processes accompanying plant defense (Lipka and Panstruga, 2005; Robatzek, 2007; Shen et al., 2007; Shen and Schulze-Lefert, 2007; Wiermer et al., 2007). Dynamic trafficking of many defense regulators and R proteins between the cytoplasm and the nucleus is crucial for the integration of distinct signaling pathways in innate immunity in plants (Shen and Schulze-Lefert, 2007; Wiermer et al., 2007). In Arabidopsis, essential components of the immune response, such as NONEXPRESSOR OF PR GENES1, which is also known as NO IMMUNITY1, ENHANCED DISEASE SUSCEPTIBILITY1, and some of its interacting partners, such as PHYTOALEXIN DEFICIENT4 (Dong, 2004; Feys et al., 2005; Wiermer et al., 2005), localize to both the cytoplasm and the nucleus. Intracellular distribution of predicted cytoplasmic NBS-LRR R proteins can also be modified in effector-triggered cells. For instance, the nuclear pool of MLA1, a barley (Hordeum vulgare) coiled coil–NB-LRR R protein, which recognizes the Blumeria graminis f. sp hordei effector AVRA1, is strongly increased during an incompatible interaction (Shen et al., 2007).
The nuclear relocalization of RD19, which is activated by an unknown mechanism, is surprising since, unlike RRS1-R, which carries a bipartite nuclear localization signal (NLS) sequence, no obvious NLS is present within the RD19 sequence. Nevertheless, nuclear localization of Cys proteases that possess no NLS has been reported previously (Tabaeizadeh et al., 1995; Harrak et al., 2001; Goulet et al., 2004). Among them, the tomato LeCp vacuolar protease acts as a transcription factor that activates the expression of the tomato ACC sythase2 gene upon treatment with the ETHYLENE-INDUCING XYLANASE (EIX) elicitor (Matarasso et al., 2005). Nuclear targeting of Cp occurs via a still unexplained mechanism. EIX might trigger a vacuole membrane collapse leading to the release of Cp into the cytoplasm where it becomes available for SUMOylation, thereby allowing its nuclear targeting (Rosin et al., 2005). Similarly, specific release of RD19 from mobile vacuole-associated compartments to the nucleus may be triggered by a PopP2-induced permeabilization of membranes by an unknown mechanism in Arabidopsis and N. benthamiana cells. Nuclear relocalization of RD19 might also involve a SUMOylation process. Alternatively, we cannot rule out the possibility that PopP2 may intercept RD19 on its way to the vacuole via retrograde signaling from the endomembrane system, which has some continuity with the nuclear envelope.
Physical association between PopP2 and RD19 was demonstrated using a FLIM-based approach, allowing the observation of these proteins in their natural location (Table 1). We recently detected a physical interaction between PopP2 and RRS1-R using FLIM (M. Bernoux, C. Tasset, Y. Marco, and L. Deslandes, unpublished data). Despite the requirement of RD19 for the RRS1-R–mediated disease resistance response triggered by PopP2, various attempts to show a physical interaction between RRS1-R and RD19, in the presence of PopP2, were unsuccessful. In addition, the presence of RRS1-R seems to have no significant effect on the physical interaction between PopP2 and RD19 (Table 1). However, the possibility that other unknown host factors mediate interaction between the three proteins cannot be ruled out. Some bacterial type III effectors do indeed target multiple host components (Belkhadir et al., 2004; Chisholm et al., 2006; Grant et al., 2006; Lin et al., 2006), and some additional PopP2-interacting candidate proteins from Arabidopsis currently under investigation were recently identified (M. Bernoux, C. Tasset, Y. Marco, and L. Deslandes, unpublished data). Contribution of these additional PopP2-interacting host factors to the full perception of PopP2 by RRS1-R could then explain the partial loss of resistance of the rd19 mutant in response to Ralstonia infection.
What could be the biological significance of the relocalization of a Cys protease by PopP2 to the plant nucleus? Interaction of PopP2 with both RRS1-R and RD19 could serve a dual recruitment function. PopP2 localizes both proteins to the nucleus where RD19 could, as in the case of tomato Le Cp, act as a transcription factor and compete with RRS1-R for similar or overlapping cis-elements of promoters of defense genes. RD19 and RRS1-R could act as positive and negative regulators, respectively. RRS1-R might indeed act as a negative transcription factor, as a mutation in its WRKY domain (addition of one amino acid) seems to impair its DNA binding activity leading to constitutive expression of salicylic acid–dependent defense genes (Noutoshi et al., 2005). These authors found that under low humidity, the sensitive to low humidity1 mutant of Arabidopsis, which essentially contains the RRS1-R gene with the above-mentioned mutation in the WRKY domain, accumulates callose, autofluorescent compounds, salicylic acid, and salicylic acid–induced PR proteins like PR-1, PR-2, and PR-5. Direct targeting of these putative transcriptional regulators by PopP2 would suggest a short signaling pathway leading to R-mediated ETI.
Localization of immune receptors and effector proteins and identification of associated host proteins is of crucial importance for the deciphering of plant innate immunity. Recently, the host N RECEPTOR-INTERACTING PROTEIN1 (NRIP1) that directly interacts with both the TIR domain of the N immune receptor and the 50-kD helicase (p50) domain of TMV was identified (Caplan et al., 2008). NRIP1 has an in vitro sulfurtransferase activity and is required for N-mediated resistance to TMV. NRIP1 normally localizes to the chloroplast but is recruited to the cytoplasm and the nucleus by the p50 effector. Caplan et al. (2008) proposed that this recruitment forms a mature p50-NRIP1 complex that is recognized through N's TIR domain to activate successful defense signaling. RRS1-R represents an atypical fusion of NB-LRR and WRKY domains, which provides the host with a transcription machinery-associated receptor whose transcriptional activity needs likely to be tightly regulated by association with multiple host proteins. In this context, the recruitment of RD19 within the nucleus by PopP2 could lead to the formation of an active RRS1-R/PopP2 perception complex. The modification recognized by RRS1-R could be the nuclear targeting of RD19, a Cys protease normally targeted to the lytic vacuole. On the other hand, RRS1-R could perceive a posttranslational modification provoked by PopP2 in the nucleus. This TTSS effector belongs to the YopJ/AvrRxv effector protein family whose members share structural similarities with the C55 peptidase family of the CE clan of Cys proteases (Barrett and Rawlings, 2001). YopJ, identified in Yersinia pestis, the causal agent of the bubonic plague, is a TTSS effector harboring both deubiquitinating and acetyltransferase activities, which are essential for the death of infected macrophages and for the inhibition of host proinflammatory responses (Mittal et al., 2006; Mukherjee et al., 2006; Sweet et al., 2007). Characterization of such enzymatic activities of PopP2 that may play a role in both RD19 and RRS1-R relocalization is currently under investigation.
Among the plant proteases involved in disease resistance, Rcr3, required for Cf2-mediated resistance in tomato, is inhibited by Avr2, the cognate avirulence protein of C. fulvum (Luderer et al., 2002; Rooney et al., 2005). It has been proposed that inhibition of Rcr3 by Avr2 induces a conformational change in Rcr3 that causes the Cf-2 protein to activate HR. Whether PopP2 harbors enzymatic activities similar to that of YopJ or acts as an inhibitor of RD19 remains to be determined. RD19 protease profiling in the presence of PopP2 is under investigation and should help us to answer this question. Alternatively, RD19 protease could be required for PopP2 avirulence activity. In the case of the AvrRpt2 bacterial effector, activation by a host factor such as cyclophilin is necessary for protease activity and recognition by the R protein RPS2 (Coaker et al., 2005). Detailed analysis of resistasome protein complexes will enable us to broaden our understanding of activation and execution of plant innate immunity.
METHODS
All experiments reported in this article were performed at least three times with similar results.
Yeast Two-Hybrid Screening
An Arabidopsis thaliana Gal4 Gateway yeast two-hybrid cDNA prey library (MatchMaker; Clontech) was generated from mRNA isolated from a mixture of root tissue of wild-type Col-0 seedlings (RRS1-S genetic background) and transgenic Col-0 (complemented with the RRS1-R gene) challenged or not with the Ralstonia solanacearum GMI1000 strain (PopP2) and harvested at different times up to 24 h after infection. The full-length PopP2 protein (488 amino acids) was used as a bait for screening 3.2 × 106 independent transformants exhibiting His auxotrophy on selective plates.
Plant and Bacterial Materials
Arabidopsis ecotype Col-0 (Nottingham Arabidopsis Stock Centre [NASC] accession number N1093) and Niedersenz (Nd-1; NASC accession number N1636) were used as the wild types. Seeds were germinated on Murashige and Skoog medium, and plants were grown in Jiffy pots in a growth chamber at 22°C, with a 9-h light period and a light intensity of 190 μmol·m−2·s−1. Experiments were performed on 4-week-old plants. The rd19 T-DNA insertion line was derived from the SALK collection (line N531088 in Col-0 background). The position of the T-DNA insertion was confirmed by PCR (forward RD19-specific primer, 5′-ATTATCCAAGACAACACGGCACTGCTA-3′; T-DNA left border primer, 5′-CCCTTTAGGGTTCCGATTTAGTGCT-3′) and sequencing. Individual homozygous lines for both rd19 and RRS1-R (rd19 RRS1-R line) were obtained by crossing the rd19 line with Nd-1. The selection of F2 plants homozygous for both the T-DNA insertion (forward RD19-specific primer, 5′-ATTATCCAAGACAACACGGCACTGCTA-3′; RD19-rev2, 5′-GAGAGAACTGTGAATATCTAGGA-3′) and the RRS1-R gene (RT1, 5′-GGCTATAGACGAGGGAGATCTATGGA-3′; RT3, 5′-GAACGAGTGGAGTCAGCGAGAGCCT-3′) was performed by PCR. Absence of RD19 transcripts on homozygous rd19 RRS1-R lines was verified by RT-PCR with RD19 gene-specific primers. Plant phenotypic responses toward the GMI1000 strain of R. solanacearum were determined by root inoculation of 4-week-old plants, and bacterial internal growth curves were performed as described previously (Deslandes et al., 1998).
Plasmid Constructions
Plasmids used in this study were constructed by Gateway technology (GW; Invitrogen) following the instructions of the manufacturer. PCR products flanked by the attB sites were recombined into the pDONR 207 vector (Invitrogen) via a BP reaction to create the corresponding entry clones with attL sites. Inserts cloned into the entry clones (pENTR) were subsequently recombined into the destination vectors via an LR reaction to create the expression constructs.
Col-0 genomic DNA was used as a template for the amplification of the RD19 gene (RD19g, a 3527-bp fragment extending 1429 bp before the start codon and 596 bp after the stop codon). The sense primer (AttB1-RD19g) used in the amplification was 5′-GGGGACAAGTTTGTACAAAAAAGCAGGCTCAGGTTTCATCCTTCTTGTA-3′, and the antisense primer (AttB2-RD19g) was 5′-GGGGACCACTTTGTACAAGAAAGCTGGGTCCGAGTTTGAGCCCATATAA-3′. pENTR-RD19g was recombined with the pAM-PAT-GW destination vector to generate the pAM-PAT-RD19g binary plasmid.
The full-length RD19 cDNA clone was amplified from first-strand cDNAs synthesized from 1 μg of total RNA (Col-0; 4-week-old plants) using oligo(dT) primer and SuperScript reverse transcriptase II (Invitrogen). The sense primer (AttB1-RD19) used in the amplification was 5′-GGGGACAAGTTTGTACAAAAAAGCAGGCTTAATGGATCGTCTTAAGCTTTATTTCT-3′, and the antisense primer (AttB2-RD19) was 5′-GGGGACCACTTTGTACAAGAAAGCTGGGTCATGGGCGGTGGTTGAGACGGTGGCT-3′. The AttB1-RD19-AttB2 PCR product was recombined into the pDONR 207 vector (Invitrogen) via a BP reaction to produce the pENTR-RD19 construct. The same procedure was followed for the generation of pENTR-RDL1 (the sense primer [AttB1-RDL1] was 5′-GGGGACAAGTTTGTACAAAAAAGCAGGCTTAATGGATTATCATCTTAGGGTTTTG-3′, and the antisense primer [AttB2-RDL1] was 5′-GGGGACCACTTTGTACAAGAAAGCTGGGTCAGAAGTGGTAGCAGCGACGGTGGAGAC-3′) and pENTR-RDL2 (the sense primer [AttB1-RDL2] was 5′-GGGGACAAGTTTGTACAAAAAAGCAGGCTTAATGGATCGTGTGGTCTTCTTCTTC-3′, and the antisense primer [AttB2-RDL2] was 5′-GGGGACCACTTTGTACAAGAAAGCTGGGTCCTTGGGTGAGGTATGAACAGCAGCAAC-3′). pENTR-PopP2 and pENTR-RRS1-R have been previously described (Deslandes et al., 2003).
The PopP2-3HA construct was derived from the recombination of the corresponding pENTR constructs with the pAM-PAT-P35S-GW-3HA destination vector. CFP- and YFPv-tagged proteins were generated from recombination of the corresponding pENTR constructs with the pAM-PAT-P35S-GW-CFP and pAM-PAT-P35S-GW-YFPv destination vectors, respectively (YFPv for YFPvenus, an enhanced form of the YFP [Nagai et al., 2002]). The aleurain-CFP construct has been previously described (Humair et al., 2001).
Transgenic Plants
Transgenic Arabidopsis plants (rd19 RRS1-R line) complemented with the genomic clone of RD19 (pAM-PAT-RD19g binary vector) were produced by the floral dipping method according to the protocol described by Clough and Bent (1998). Primary transformants were selected on Murashige and Skoog medium supplemented with 5 μg⋅mL−1 of dl-phosphinothricin (Duchefa).
Transient Transfection in Arabidopsis
Particle bombardments were done as previously described (Shirasu et al., 1999). Ten mature Arabidopsis leaves were transfected 4 h after detachment with 3 μg of 35S:RD19-YFPv together with 3 μg of 35S-PopP2-CFP plasmids. Bombardments were done at 900 p.s.i. under vacuum with a PDS-1000/He particule delivery system (Bio-Rad).
Agroinfiltration
Agrobacterium tumefaciens cells were grown overnight in YEB media containing appropriate antibiotic selections. Cells were pelleted at 7500 rpm and resuspended in infiltration medium (10 mM MgCl2, 10 mM MES, and 150 μM acetosyringone) and incubated for 2 h at room temperature. Resuspended cells were infiltrated into leaves of 4-week-old N. benthamiana plants at an optical density of OD600 = 0.5 with a 1-mL needleless syringe. For coinfiltration, equal volumes of A. tumefaciens were mixed prior to infiltration. The infiltrated plants were incubated for 48 h in growth chambers for a 16-h daylength at 20°C.
RNA Extraction and Q-RT-PCR Analysis
Material for RNA analysis was ground in liquid nitrogen, and total RNA was isolated using the Nucleospin RNA plant kit (Macherey-Nagel) according to the manufacturer's recommendations. Reverse transcription was performed using 1 μg of total RNA and SuperScript reverse transcriptase II (Invitrogen). Real-time PCR analysis was performed using the LightCycler FastStart DNA MasterPlus SYBR Green I kit (Roche Applied Science) according to the manufacturer's instructions. Each reaction was performed with 2 μL of a 1:20 (v/v) dilution of the first cDNA strand, with 0.5 μM of each primer in a total reaction volume of 10 μL, with the following conditions: 1 cycle of 9 min at 95°C and 45 cycles of 5 s at 95°C, 10 s at 65°C, and 20 s at 72°C. The following primer sets were used: RD19 (forward, 5′-ATTATCCAAGACAACACGGCACTGCTA-3′; reverse, 5′-CACCTTCCAAAGCTCCAGTG-3′), PR-3 (forward, 5′-CGCTTGTCCTGCTAGAGGTT-3′; reverse, 5′-GCTCGGTTCACAGTAGTCTGA-3′), PR-4 (forward, 5′-TTGCTCCACGTGGGATGCTGAT-3′; reverse, 5′-AGCTCATTGCCACAGTCGACAA-3′), and PDF1.2 (forward, 5′-TCATGGCTAAGTTTGCTTCC-3′; reverse, 5′-AATACACACGATTTAGCACC-3′). Two housekeeping genes (At1g13320 and At5g09810), whose transcript levels have been previously demonstrated not to change in susceptible and resistant genetic backgrounds challenged with the GMI1000 strain, were both used as internal standards for data normalization (Hu et al., 2008). To amplify the housekeeping genes, the following primers were used: At1g13320 (forward, 5′-GACCGGAGCCAACTAGGAC-3′; reverse, 5′-AAAACTTGGTAACTTTTCCAGCA-3′) and At5g09810 (forward, 5′-GTGGTCGTACAACCGGTATT-3′; reverse, 5′-AAGGATAGCATGAGGAAGAGCA-3′). PCR amplification specificity was verified by analysis of a dissociation curve at the end of the PCR cycles of each experiment as well as by ethidium bromide–stained agarose gel resolution and sequencing of the corresponding PCR products.
Protein Gel Blot Analysis
Forty-eight hours after infiltration, five discs (8 mm) of Nicotiana benthamiana leaves expressing proteins of interest were homogenized in loading buffer. Proteins were transferred to Protran BA85 membranes (Whatman) and visualized by Ponceau S red staining. YFP-tagged and HA-tagged proteins were detected using an anti-GFP rabbit polyclonal antibody (Invitrogen) and an anti-HA rat monoclonal antibody (clone 3F10; Roche), respectively. Goat-anti-rabbit antibody conjugated with horseradish peroxidase was used as secondary antibody (Santa Cruz).
Fluorescence Microscopy
The CFP and YFP fluorescence in N. benthamiana leaves was analyzed with a confocal laser scanning microscope (TCS SP2-SE; Leica) using a ×63 water immersion objective lens (numerical aperture 1.20; PL APO). CFP fluorescence was excited with the 458-nm ray line of the argon laser and recorded in one of the confocal channels in the 465- to 520-nm emission range. YFP fluorescence was excited with the 514-nm line ray of the argon laser and detected in the range between 520 and 575 nm. Images were acquired in the sequential mode using Leica LCS software (version 2.61).
FLIM and Data Analysis
Fluorescence lifetime of the donor was experimentally measured in the presence and absence of the acceptor. FRET efficiency (E) was calculated by comparing the lifetime of the donor in the presence (τDA) or abscence (τD) of the acceptor: E = 1 − (τDA)/(τD). We performed FLIM measurements using a multiphoton FLIM system coupled to a streak camera (Krishnan et al., 2003). The light source was a mode-locked Ti:sapphire laser (Tsunami, model 3941; Spectra-Physics), pumped by a 10-W diode laser (Millennia Pro; Spectra-Physics), delivering ultrafast femtosecond pulses with a fundamental frequency of 80 MHz. A pulsepicker (model 3980; Spectra-Physics) was used to reduce the repetition rate to 2 MHz. All the experiments reported in this work were performed at λ = 820 nm, the optimal wavelength to excite CFP in multiphoton mode while minimizing the excitation of YFP (Chen and Periasamy, 2004). The power delivered at the entrance of the FLIM optics was 14 mW. All images were acquired with a ×60 oil immersion lens (Plan Apo 1.4 numerical aperture, IR) mounted on an inverted microscope (Eclipse TE2000E; Nikon) coupled to the FLIM system. The fluorescence emission was directed back out into the detection unit through a short-pass filter (λ < 750 nm). The FLIM unit was composed of a streak camera (Streakscope C4334; Hamamatsu Photonics) coupled to a fast and high-sensitivity CCD camera (model C8800-53C; Hamamatsu) (Krishnan et al., 2003; Biener et al., 2005). For each nucleus, average fluorescence decay profiles were plotted and lifetimes were estimated by fitting data with biexponential function using a nonlinear least squares estimation procedure with Origin 7.5 software (OriginLab).
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL data libraries under the following accession numbers: At5g44420 (PDF1.2), CAD14570 (PopP2), At3g12500 (PR-3), At3g04720 (PR-4), At4g39090 (RD19), At2g21430 (RDL1), At4g16190 (RDL2), and At5g45260 (RRS1-R).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Genotyping and Phenotyping of F3 Families Segregating for RD19/rd19 and RRS1-S/RRS1-R.
Supplemental Figure 2. RD19 Expression in Wild-Type Plants and in Three Independent Transgenic RD19g Lines.
Supplemental Figure 3. RD19-YFPv Subcellular Localization in Arabidopsis.
Supplemental Figure 4. Comparison of the Deduced Amino Acid Sequences of RD19 with RDL1 and RDL2.
Supplemental Figure 5. RDL1-YFPv and RDL2-YFPv Colocalize with Aleurain-CFP.
Supplemental Figure 6. Detection of Full-Length YFPv-Tagged Proteins.
Supplementary Material
Acknowledgments
We thank the Salk Institute Genomic Analysis Laboratory for providing the Arabidopsis T-DNA insertion mutant. We thank Nadine Paris for providing plasmids used for colocalization studies. We also thank Susana Rivas for very helpful discussion and critical reading of the manuscript. This work was supported by funding from the Agence Nationale de la Recherche, Grant NT05-3_42336 (ICARE). M.B. was supported by a grant from the Centre National de la Recherche Scientifique (Bourse Docteur-Ingénieur).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Laurent Deslandes (laurent.deslandes@toulouse.inra.fr).
Online version contains Web-only data.
References
- Axtell, M.J., and Staskawicz, B.J. (2003). Initiation of RPS2-specified disease resistance in Arabidopsis is coupled to the AvrRpt2-directed elimination of RIN4. Cell 112 369–377. [DOI] [PubMed] [Google Scholar]
- Barrett, A.J., and Rawlings, N.D. (2001). Evolutionary lines of Cysteine peptidases. Biol. Chem. 382 727–733. [DOI] [PubMed] [Google Scholar]
- Belkhadir, Y., Nimchuk, Z., Hubert, D.A., Mackey, D., and Dangl, J.L. (2004). Arabidopsis RIN4 negatively regulates disease resistance mediated by RPS2 and RPM1 downstream or independent of the NDR1 signal modulator and is not required for the virulence functions of bacterial type III effectors AvrRpt2 or AvrRpm1. Plant Cell 16 2822–2835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biener, E., Charlier, M., Ramanujan, V.K., Daniel, N., Eisenberg, A., Bjorbaek, C., Herman, B., Gertler, A., and Djiane, J. (2005). Quantitative FRET imaging of leptin receptor oligomerization kinetics in single cells. Biol. Cell 97 905–919. [DOI] [PubMed] [Google Scholar]
- Burch-Smith, T.M., Schiff, M., Caplan, J.L., Tsao, J., Czymmek, K., and Dinesh-Kumar, S.P. (2007). A novel role for the TIR domain in association with pathogen-derived elicitors. PLoS Biol. 5 e68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caplan, J.L., Mamillapalli, P., Burch-Smith, T.M., Czymmek, K., and Dinesh-Kumar, S.P. (2008). Chloroplastic protein NRIP1 mediates innate immune receptor recognition of a viral effector. Cell 132 449–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, Y., and Periasamy, A. (2004). Characterization of two-photon excitation fluorescence lifetime imaging microscopy for protein localization. Microsc. Res. Tech. 63 72–80. [DOI] [PubMed] [Google Scholar]
- Chichkova, N.V., Kim, S.H., Titova, E.S., Kalkum, M., Morozov, V.S., Rubtsov, Y.P., Kalinina, N.O., Taliansky, M.E., and Vartapetian, A.B. (2004). A plant caspase-like protease activated during the hypersensitive response. Plant Cell 16 157–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinchilla, D., Bauer, Z., Regenass, M., Boller, T., and Felix, G. (2006). The Arabidopsis receptor kinase FLS2 binds flg22 and determines the specificity of flagellin perception. Plant Cell 18 465–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chisholm, S.T., Coaker, G., Day, B., and Staskawicz, B.J. (2006). Host-microbe interactions: Shaping the evolution of the plant immune response. Cell 124 803–814. [DOI] [PubMed] [Google Scholar]
- Clough, S.J., and Bent, A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16 735–743. [DOI] [PubMed] [Google Scholar]
- Coaker, G., Arnold Falick, A., and Staskawicz, B.J. (2005). Activation of a phytopathogenic bacterial effector protein by a eukaryotic cyclophilin. Science 308 548–550. [DOI] [PubMed] [Google Scholar]
- Cornelis, G.R., and Van Gijsegem, F. (2000). Assembly und function of type III secretory systems. Annu. Rev. Microbiol. 54 735–774. [DOI] [PubMed] [Google Scholar]
- Cunnac, S., Occhialini, A., Barberis, P., Boucher, C., and Genin, S. (2004). Inventory and functional analysis of the large Hrp regulon in Ralstonia solanacearum: Identification of novel effector proteins translocated to plant host cells through the type III secretion system. Mol. Microbiol. 53 115–128. [DOI] [PubMed] [Google Scholar]
- da Cunha, L., Sreerekha, M.V., and Mackey, D. (2007). Defence suppression by virulence effectors of bacterial phytopathogens. Curr. Opin. Plant Biol. 10 349–357. [DOI] [PubMed] [Google Scholar]
- Deslandes, L., Olivier, J., Peeters, N., Feng, D.X., Khounlotham, M., Boucher, C., Somssich, I., Genin, S., and Marco, Y. (2003). Physical interaction between RRS1-R, a protein conferring resistance to bacterial wilt, and PopP2, a type III effector targeted to the plant nucleus. Proc. Natl. Acad. Sci. USA 100 8024–8029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deslandes, L., Olivier, J., Theulières, F., Hirsch, J., Feng, D.X., Bittner-Eddy, P., Beynon, J., and Marco, Y. (2002). Resistance to Ralstonia solanacearum is conferred by the recessive RRS1-R gene, a member of a novel family of resistance genes. Proc. Natl. Acad. Sci. USA 99 2404–2409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deslandes, L., Pileur, F., Liaubet, L., Camut, S., Can, C., Williams, K., Holub, E., Beynon, J., Arlat, M., and Marco, Y. (1998). Genetic characterization of RRS1, a recessive locus in Arabidopsis thaliana that confers resistance to the bacterial soilborne pathogen Ralstonia solanacearum. Mol. Plant Microbe Interact. 11 659–667. [DOI] [PubMed] [Google Scholar]
- De Wit, P.J.G.M. (1992). Molecular characterization of gene-for-gene systems in plant-fungus interactions and the application of avirulence genes in control of plant pathogens. Annu. Rev. Phytopathol. 30 391–418. [DOI] [PubMed] [Google Scholar]
- De Wit, P.J.G.M. (2007). How plants recognize pathogens and defend themselves. Cell. Mol. Life Sci. 64 2726–2732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon, M.S., Golstein, C., Thomas, C.M., van Der Biezen, E.A., and Jones, J.D. (2000). Genetic complexity of pathogen perception by plants: the example of Rcr3, a tomato gene required specifically by Cf-2. Proc. Natl. Acad. Sci. USA 97 8807–8817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon, M.S., Jones, D.A., Keddie, J.S., Thomas, C.M., Harrison, K., and Jones, J.D. (1996). The tomato Cf-2 disease resistance locus comprises two functional genes encoding leucine-rich repeat proteins. Cell 84 451–459. [DOI] [PubMed] [Google Scholar]
- Dodds, P.N., Lawrence, G.J., Catanzariti, A.M., Teh, T., Wang, C.I., Ayliffe, M.A., Kobe, B., and Ellis, J.G. (2006). Direct protein interaction underlies gene-for-gene specificity and coevolution of the flax resistance genes and flax rust avirulence genes. Proc. Natl. Acad. Sci. USA 103 8888–8893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong, X. (2004). NPR1, all things considered. Curr. Opin. Plant Biol. 7 547–552. [DOI] [PubMed] [Google Scholar]
- D'Silva, I., Poirier, G.G., and Heath, M.C. (1998). Activation of Cysteine proteases in cowpea plants during the hypersensitive response-a form of programmed cell death. Exp. Cell Res. 245 389–399. [DOI] [PubMed] [Google Scholar]
- Eulgem, T., and Somssich, I.E. (2007). Networks of WRKY transcription factors in defence signalling. Curr. Opin. Plant Biol. 10 366–371. [DOI] [PubMed] [Google Scholar]
- Feys, B.J., Wiermer, M., Bhat, R.A., Moisan, L.J., Medina-Escobar, N., Neu, C., Cabral, A., and Parker, J.E. (2005). Arabidopsis SENESCENCE-ASSOCIATED GENE101 stabilizes and signals within an ENHANCED DISEASE SUSCEPTIBILITY1 complex in plant innate immunity. Plant Cell 17 2601–2613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fliegmann, J., Mithofer, A., Wanner, G., and Ebel, J. (2004). An ancient enzyme domain hidden in the putative beta-glucan elicitor receptor of soybean may play an active part in the perception of pathogen-associated molecular patterns during broad host resistance. J. Biol. Chem. 279 1132–1140. [DOI] [PubMed] [Google Scholar]
- Fluckiger, R., De Caroli, M., Piro, G., Dalessandro, G., Neuhaus, J.M., and Di Sansebastiano, G.P. (2003). Vacuolar system distribution in Arabidopsis tissues, visualized using GFP fusion proteins. J. Exp. Bot. 54 1577–1584. [DOI] [PubMed] [Google Scholar]
- Flor, H.H. (1971). Current status of the gene-for-gene concept. Annu. Rev. Phytopathol. 9 275–296. [Google Scholar]
- Gilroy, E.M., et al. (2007). Involvement of cathepsin B in the plant disease resistance hypersensitive response. Plant J. 52 1–13. [DOI] [PubMed] [Google Scholar]
- Goulet, B., Baruch, A., Moon, N.S., Poirier, M., Sansregret, L.L., Erickson, A., Bogyo, M., and Nepveu, A. (2004). A cathepsin L isoform that is devoid of a signal peptide localizes to the nucleus in S phase and processes the CDP/Cux transcription factor. Mol. Cell 14 207–219. [DOI] [PubMed] [Google Scholar]
- Grant, S.R., Fisher, E.J., Chang, J.H., Mole, B.M., and Dangl, J.L. (2006). Subterfuge and manipulation: Type III effector proteins of phytopathogenic bacteria. Annu. Rev. Microbiol. 60 425–449. [DOI] [PubMed] [Google Scholar]
- Harrak, H., Azelmat, S., Baker, E.N., and Tabaeizadeh, Z. (2001). Isolation and characterization of a gene encoding a drought-induced Cysteine protease in tomato (Lycopersicon esculentum). Genome 44 368–374. [DOI] [PubMed] [Google Scholar]
- Hayward, A.C. (1991). Biology and epidemiology of bacterial wilt caused by Pseudomonas solanacearum. Annu. Rev. Phytopathol. 29 65–87. [DOI] [PubMed] [Google Scholar]
- Hayward, A.C. (2000). Ralstonia solanacearum. In Encyclopedia of Microbiology, J. Lederberg, ed (San Diego, CA: Academic Press), pp. 32–42.
- Hirsch, J., Deslandes, L., Feng, D.X., Balagué, C., and Marco, Y. (2002). Delayed symptom development in ein2-1, an Arabidopsis ethylene-insensitive mutant, in response to bacterial wilt caused by Ralstonia solanacearum. Phytopathology 92 1142–1148. [DOI] [PubMed] [Google Scholar]
- Hotson, A., and Mudgett, M.B. (2004). Cysteine proteases in phytopathogenic bacteria: Identification of plant targets and activation of innate immunity. Curr. Opin. Plant Biol. 7 384–390. [DOI] [PubMed] [Google Scholar]
- Hu, J., Barlet, X., Deslandes, L., Hirsch, J., Feng, D.X., Somssich, I., and Marco, Y. (2008). Transcriptional responses of Arabidopsis thaliana during wilt disease caused by the soil-borne phytopathogenic bacterium, Ralstonia solanacearum. PLoS ONE 3 e2589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humair, D., Hernández Felipe, D., Neuhaus, J.-M., and Paris, N. (2001). Demonstration in yeast of the function of BP-80, a putative plant vacuolar sorting receptor. Plant Cell 13 781–792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia, Y., McAdams, S.A., Bryan, G.T., Hershey, H.P., and Valent, B. (2000). Direct interaction of resistance gene and avirulence gene products confers rice blast resistance. EMBO J. 19 4004–4014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, J.D.G., and Dangl, J.L. (2006). The plant immune system. Nature 444 323–329. [DOI] [PubMed] [Google Scholar]
- Joosten, M.H.A.J., Vogelsang, R., Cozijnsen, T.J., Verberne, M.C., and De Wit, P.J.G.M. (1997). The biotrophic fungus Cladosporium fulvum circumvents Cf-4-mediated resistance by producing unstable AVR4 elicitors. Plant Cell 9 367–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaku, H., Nishizawa, Y., Ishii-Minami, N., Akimoto-Tomiyama, C., Dohmae, N., Takio, K., Minami, E., and Shibuya, N. (2006). Plant cells recognize chitin fragments for defence signaling through a plasma membrane receptor. Proc. Natl. Acad. Sci. USA 103 11086–11091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keen, N.T. (1990). Gene-for-gene complementary in plant-pathogen interactions. Annu. Rev. Genet. 24 447–463. [DOI] [PubMed] [Google Scholar]
- Koizumi, M., Yamaguchi-Shinozaki, K., Tsuji, H., and Shinozaki, K. (1993). Structure and expression of two genes that encode distinct drought-inducible Cysteine proteinases in Arabidopsis thaliana. Gene 129 175–182. [DOI] [PubMed] [Google Scholar]
- Krishnan, R.V., Masuda, A., Centonze, V.E., and Herman, B. (2003). Quantitative imaging of protein-protein interactions by multiphoton fluorescence lifetime imaging microscopy using a streak camera. J. Biomed. Opt. 8 362–367. [DOI] [PubMed] [Google Scholar]
- Krüger, J., Thomas, C.M., Golstein, C., Dixon, M.S., Smoker, M., Tang, S., Mulder, L., and Jones, J.D.G. (2002). A tomato Cysteine protease required for Cf-2-dependent disease resistance and suppression of autonecrosis. Science 296 744–747. [DOI] [PubMed] [Google Scholar]
- Kunze, G., Zipfel, C., Robatzek, S., Niehaus, K., Boller, T., and Felix, G. (2004). The N terminus of bacterial elongation factor Tu elicits innate immunity in Arabidopsis plants. Plant Cell 16 3496–3507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavie, M., Shillington, E., Eguiluz, C., Grimsley, N., and Boucher, C. (2002). PopP1, a new member of the YopJ/AvrRxv family of type III effector proteins, acts as a host-specificity factor and modulates aggressiveness of Ralstonia solanacearum. Mol. Plant Microbe Interact. 15 1058–1068. [DOI] [PubMed] [Google Scholar]
- Lin, N.C., Abramovitch, R.B., Kim, Y.J., and Martin, G.B. (2006). Diverse AvrPtoB homologs from several Pseudomonas syringae pathovars elicit Pto-dependent resistance and have similar virulence activities. Appl. Environ. Microbiol. 72 702–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipka, V., and Panstruga, R. (2005). Dynamic cellular responses in plant-microbe interactions. Curr. Opin. Plant Biol. 8 625–631. [DOI] [PubMed] [Google Scholar]
- Luderer, R., Takken, F.L.W., De Wit, P.J.G.M., and Joosten, M.H.A.J. (2002). Cladosporium fulvum overcomes Cf-2-mediated resistance by producing truncated AVR2 elicitor proteins. Mol. Microbiol. 45 875–884. [DOI] [PubMed] [Google Scholar]
- Mackey, D., Belkhadir, Y., Alonso, J.M., Ecker, J.R., and Dangl, J.L. (2003). Arabidopsis RIN4 is a target of the type III virulence effector AvrRpt2 and modulates RPS2-mediated resistance. Cell 112 379–389. [DOI] [PubMed] [Google Scholar]
- Mackey, D., Holt III, B.F., Wiig, A., and Dangl, J.L. (2002). RIN4 interacts with Pseudomonas syringae type III effector molecules and is required for RPM1-mediated resistance in Arabidopsis. Cell 108 743–754. [DOI] [PubMed] [Google Scholar]
- Matarasso, N., Schuster, S., and Avni, A. (2005). A novel plant Cysteine protease has a dual function as a regulator of 1-aminocyclopropane-1-carboxylic acid synthase gene expression. Plant Cell 17 1205–1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittal, R., Peak-Chew, S.Y., and McMahon, H.T. (2006). Acetylation of MEK2 and I kappa B kinase (IKK) activation loop residues by YopJ inhibits signaling. Proc. Natl. Acad. Sci. USA 103 18574–18579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miya, A., Albert, P., Shinya, T., Desaki, Y., Ichimura, K., Shirasu, K., Narusaka, Y., Kawakami, N., Kaku, H., and Shibuya, N. (2007). Cerk1, a lysM receptor kinase, is essential for chitin elicitor signaling in Arabidopsis. Proc. Natl. Acad. Sci. USA 104 19613–19618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mo, B., Tse, Y.C., and Jiang, L. (2006). Plant prevacuolar/endosomal compartments. Int. Rev. Cytol. 253 95–129. [DOI] [PubMed] [Google Scholar]
- Mosolov, V.V., Grigor'eva, L.I., and Valueva, T.A. (2001). The role of proteolytic enzymes and their inhibitors in plant protection (review). Prikl. Biokhim. Mikrobiol. 37 131–140. [PubMed] [Google Scholar]
- Mosolov, V.V., and Valueva, T.A. (2006). Participation of proteolytic enzymes in the interaction of plants with phytopathogenic microorganisms. Biochemistry (Mosc.) 71 838–845. [DOI] [PubMed] [Google Scholar]
- Mudgett, M.B. (2005). New insights to the function of phytopathogenic bacterial type III effectors in plants. Annu. Rev. Plant Biol. 56 509–531. [DOI] [PubMed] [Google Scholar]
- Mukherjee, S., Keitany, G., Li, Y., Wang, Y., Ball, H.L., Goldsmith, E.J., and Orth, K. (2006). Yersinia YopJ acetylates and inhibits kinase activation by blocking phosphorylation. Science 312 1211–1214. [DOI] [PubMed] [Google Scholar]
- Mur, L.A., Kenton, P., Lloyd, A.J., Ougham, H., and Prats, E.J. (2007). The hypersensitive response; the centenary is upon us but how much do we know? J. Exp. Bot. 59 501–520. [DOI] [PubMed] [Google Scholar]
- Nagai, T., Ibata, K., Park, E.S., Kubota, M., Mikoshiba, K., and Miyawaki, A. (2002). A variant of yellow fluorescent protein with fast and efficient maturation for cell-biological applications. Nat. Biotechnol. 20 87–90. [DOI] [PubMed] [Google Scholar]
- Noutoshi, Y., Ito, T., Seki, M., Nakashita, H., Yoshida, S., Marco, Y., Shirasu, K., and Shinozaki, K. (2005). A single amino acid insertion in the WRKY domain of the Arabidopsis TIR-NBS-LRR-WRKY-type disease resistance protein SLH1 (sensitive to low humidity 1) causes activation of defence responses and hypersensitive cell death. Plant J. 43 873–888. [DOI] [PubMed] [Google Scholar]
- Occhialini, A., Cunnac, S., Reymond, N., Genin, S., and Boucher, C. (2005). Genome-wide analysis of gene expression in Ralstonia solanacearum reveals that the hrpB gene acts as a regulatory switch controlling multiple virulence pathways. Mol. Plant Microbe Interact. 18 938–949. [DOI] [PubMed] [Google Scholar]
- Paris, N., Stanley, C.M., Jones, R.L., and Rogers, J.C. (1996). Plant cells contain two functionally distinct vacuolar compartments. Cell 85 563–572. [DOI] [PubMed] [Google Scholar]
- Ren, T., Qu, F., and Morris, T.J. (2000). HRT gene function requires interaction between a NAC protein and viral capsid protein to confer resistance to turnip crinkle virus. Plant Cell 12 1917–1926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robatzek, S. (2007). Vesicle trafficking in plant immune responses. Cell. Microbiol. 9 1–8. [DOI] [PubMed] [Google Scholar]
- Rojo, E., Martin, R., Carter, C., Zouhar, J., Pan, S., Plotnikova, J., Jin, H., Paneque, M., Sanchez-Serrano, J.J., Baker, B., Ausubel, F.M., and Raikhel, N.V. (2004). VPEgamma exhibits a caspase-like activity that contributes to defence against pathogens. Curr. Biol. 14 1897–1906. [DOI] [PubMed] [Google Scholar]
- Rosin, F.M., Watanabe, N., and Lam, E. (2005). Moonlighting vacuolar proteases: multiple jobs for a busy protein. Trends Plant Sci. 11 516–518. [DOI] [PubMed] [Google Scholar]
- Rooney, H.C., Van't Klooster, J.W., van der Hoorn, R.A.L., Joosten, M.H.A.J., Jones, J.D.G., and de Wit, P.J.G.M. (2005). Cladosporium Avr2 inhibits tomato Rcr3 protease required for Cf-2-dependent disease resistance. Science 308 1783–1786. [DOI] [PubMed] [Google Scholar]
- Salanoubat, M., et al. (2002). Genome sequence of the plant pathogen Ralstonia solanacearum. Nature 415 497–502. [DOI] [PubMed] [Google Scholar]
- Schell, M.A., Denny, T.P., and Huang, J. (1994). Extracellular virulence factors of Pseudomonas solanacearum: Role in disease and their regulation. Molecular Mechanisms of Bacterial Virulence, C.I. Kado and J.H. Crosa, eds (Dordrecht, The Netherlands: Kluwer Academic Publishers), pp. 311–324.
- Shabab, M., Shindo, T., Gu, C., Kaschani, F., Pansuriya, T., Chintha, R., Harzen, A., Colby, T., Kamoun, S., and van der Hoorn, R.A.L. (2008). Fungal effector protein AVR2 targets diversifying defense-related Cys proteases of tomato. Plant Cell 20 1169–1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao, F., Golstein, C., Ade, J., Stoutemyer, M., Dixon, J.E., and Innes, R.W. (2003). Cleavage of Arabidopsis PBS1 by a bacterial type III effector. Science 301 1230–1233. [DOI] [PubMed] [Google Scholar]
- Shen, Q.H., Saijo, Y., Mauch, S., Biskup, C., Bieri, S., Keller, B., Seki, H., Ulker, B., Somssich, I.E., and Schulze-Lefert, P. (2007). Nuclear activity of MLA immune receptors links isolate-specific and basal disease-resistance responses. Science 315 1098–1103. [DOI] [PubMed] [Google Scholar]
- Shen, Q.H., and Schulze-Lefert, P. (2007). Rumble in the nuclear jungle: Compartmentalization, trafficking, and nuclear action of plant immune receptors. EMBO J. 26 4293–4301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirasu, K., Nielsen, K., Piffanelli, P., Oliver, R., and Schulze-Lefert, P. (1999). Cell-autonomous complementation of mlo resistance using a biolistic transient expression system. Plant J. 17 293–300. [Google Scholar]
- Solomon, M., Belenghi, B., Delledonne, M., Menachem, E., and Levine, A. (1999). The involvement of Cysteine proteases and protease inhibitor genes in the regulation of programmed cell death in plants. Plant Cell 11 431–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweet, C.R., Conlon, J., Golenbock, D.T., Goguen, J., and Silverman, N. (2007). YopJ targets TRAF proteins to inhibit TLR-mediated NF-kappaB, MAPK and IRF3 signal transduction. Cell. Microbiol. 9 2700–2715. [DOI] [PubMed] [Google Scholar]
- Tabaeizadeh, Z., Chen, R.D., Yu, L.X., and Lafontaine, J.G. (1995). Identification and immunolocalization of a 65 kDa drought induced protein in cultivated tomato Lycopersicon esculentum. Protoplasma 186 208–219. [Google Scholar]
- Tamura, K., Shimada, T., Ono, E., Tanaka, Y., Nagatani, A., Higashi, S.I., Watanabe, M., Nishimura, M., and Hara-Nishimura, I. (2003). Why green fluorescent fusion proteins have not been observed in the vacuoles of higher plants. Plant J. 35 545–555. [DOI] [PubMed] [Google Scholar]
- Van den Ackerveken, A.F.J.M., Vossen, J.P.M.J., and De Wit, P.J.G.M. (1993). The AVR9 race-specific elicitor of Cladosporium fulvum is processed by endogenous and plant proteases. Plant Physiol. 103 91–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Biezen, E.A., and Jones, J.D.G. (1998). Plant disease-resistance proteins and the gene-for-gene concept. Trends Biochem. Sci. 23 454–456. [DOI] [PubMed] [Google Scholar]
- van der Hoorn, R.A.L. (2008). Plant proteases: From phenotypes to molecular mechanisms. Annu. Rev. Plant Biol. 59 191–223. [DOI] [PubMed] [Google Scholar]
- van der Hoorn, R.A.L., and Jones, J.D.G. (2004). The plant proteolytic machinery and its role in defence. Curr. Opin. Plant Biol. 7 400–407. [DOI] [PubMed] [Google Scholar]
- van Esse, H.P., van't Klooster, J.W., Bolton, M.D., Yadeta, K.A., van Baarlen, P., Boeren, S., Vervoort, J., de Wit, P.J.G.M., and Thomma, B.P.H.J. (2008). The Cladosporium fulvum virulence protein Avr2 inhibits host proteases required for basal defense. Plant Cell 20 1948–1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasse, J., Frey, P., and Trigalet, A. (1995). Microscopic studies of intercellular infection and protoxylem invasion of tomato roots by Pseudomonas solanacearum. Mol. Plant Microbe Interact. 8 259–267. [Google Scholar]
- Wiermer, M., Feys, B.J., and Parker, J.E. (2005). Plant immunity: The EDS1 regulatory node. Curr. Opin. Plant Biol. 8 383–389. [DOI] [PubMed] [Google Scholar]
- Wiermer, M., Palma, K., Zhang, Y., and Li, X. (2007). Should I stay or should I go? Nucleocytoplasmic trafficking in plant innate immunity. Cell. Microbiol. 9 1880–1890. [DOI] [PubMed] [Google Scholar]
- Xia, Y., Suzuki, H., Borevitz, J., Blount, J., Guo, Z., Patel, K., Dixon, R.A., and Lamb, C. (2004). An extracellular aspartic protease functions in Arabidopsis disease resistance signaling. EMBO J. 23 980–988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zipfel, C., Kunze, G., Chinchilla, D., Caniard, A., Jones, J.D.G., Boller, T., and Felix, G. (2006). Perception of the bacterial PAMP EF-Tu by the receptor EFR restricts Agrobacterium-mediated transformation. Cell 125 749–760. [DOI] [PubMed] [Google Scholar]
- Zipfel, C., Robatzek, S., Navarro, L., Oakeley, E.J., Jones, J.D.G., Felix, G., and Boller, T. (2004). Bacterial disease resistance in Arabidopsis through flagellin perception. Nature 428 764–767. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.