Abstract
MADS box genes in plants consist of MIKC-type and type I genes. While MIKC-type genes have been studied extensively, the functions of type I genes are still poorly understood. Evidence suggests that type I MADS box genes are involved in embryo sac and seed development. We investigated two independent T-DNA insertion alleles of the Arabidopsis thaliana type I MADS box gene AGAMOUS-LIKE61 (AGL61) and showed that in agl61 mutant ovules, the polar nuclei do not fuse and central cell morphology is aberrant. Furthermore, the central cell begins to degenerate before fertilization takes place. Although pollen tubes are attracted and perceived by the mutant ovules, neither endosperm development nor zygote formation occurs. AGL61 is expressed in the central cell during the final stages of embryo sac development. An AGL61:green fluorescent protein–β-glucoronidase fusion protein localizes exclusively to the polar nuclei and the secondary nucleus of the central cell. Yeast two-hybrid analysis showed that AGL61 can form a heterodimer with AGL80 and that the nuclear localization of AGL61 is lost in the agl80 mutant. Thus, AGL61 and AGL80 appear to function together to differentiate the central cell in Arabidopsis. We renamed AGL61 DIANA, after the virginal Roman goddess of the hunt.
INTRODUCTION
The MADS box family of transcription factors in plants is well known for its role in developmental processes. Members of this family fulfill important functions in vegetative development, regulation of flowering time, control of meristem identity, development of the floral organs, and fruit and seed development (Becker and Theissen, 2003; Ferrario et al., 2004). All of these well-characterized MADS box genes belong to the MIKC type of MADS box genes, which share, in addition to the MADS box, the I (Intervening), K (Keratin-like), and C (C-terminal) regions. Remarkably, analysis of the Arabidopsis thaliana genome revealed the existence of a second type of MADS box genes in plants (Alvarez-Buylla et al., 2000; Arabidopsis Genome Initiative, 2000) that had not appeared in forward genetic studies. This group of genes, named type I MADS box genes (Alvarez-Buylla et al., 2000) or M-type MADS box genes (Kofuji et al., 2003), have no domains in common except for the MADS box.
The type I MADS box genes from Arabidopsis and rice (Oryza sativa) can be further subdivided into Mα, Mβ, and Mγ subclasses, which are probably not monophyletic (De Bodt et al., 2003b; Kofuji et al., 2003; Pařenicová et al., 2003; Nam et al., 2004). Despite the presumed polyphyletic origin of the type I MADS box genes, several characteristics are shared by the three subclasses. In contrast with the MIKC-type genes, the majority of the type I genes contain no introns (De Bodt et al., 2003a), are weakly expressed (Kofuji et al., 2003; Pařenicová et al., 2003), and were duplicated after the divergence of monocots and dicots (De Bodt et al., 2003a).
Although the type I genes outnumber the MIKC type genes in the Arabidopsis genome (61 versus 46; Pařenicová et al., 2003), little is known about the function of these genes. Recently, the functional characterization of four Arabidopsis type I genes contributed considerably to the understanding of the type I subfamily. The first type I gene to be characterized, PHERES1 (PHE1; also known as AGAMOUS-LIKE37 [AGL37], an Mγ-type gene), was found to be regulated by the Polycomb group gene MEDEA (MEA) and is expressed in the embryo and endosperm shortly after fertilization (Köhler et al., 2003b). Although phe1 mutants show a wild-type phenotype, reduced expression levels of PHE1 in mea seed development mutants partially restored the mutant phenotype, indicating a role of PHE1 repression in seed development (Köhler et al., 2003b).
More evidence for the functionality of Mγ-type genes was provided by the agl80 mutant (Portereiko et al., 2006). agl80 mutant megagametophytes show a defect in the maturation of the central cell and fail to develop endosperm after fertilization. In accordance with the mutant phenotype, AGL80 is expressed in the central cell and in the endosperm before cellularization. The Mα-type gene AGL62, which interacts with AGL80 (De Folter et al., 2005), was found to be expressed during the syncytial phase of endosperm development and is suppressed by the FERTILIZATION-INDEPENDENT SEED (FIS) Polycomb group complex just before cellularization of the endosperm occurs (Kang et al., 2008). In agl62 mutants, the endosperm undergoes precocious cellularization, indicating that AGL62 plays an important role in endosperm development. Finally, the Mα-type gene AGL23 was found to be involved in both female gametophyte and embryo development. The agl23 mutant is arrested after megasporogenesis, although this defect is not completely penetrant. In addition, the gene was found to be essential for the development of chloroplasts during embryogenesis (Colombo et al., 2008).
The functional studies of PHE1, AGL80, AGL62, and AGL23 suggest a role for type I MADS box genes in female gametophyte development or early seed development, processes that are molecularly not very well understood. In the last decade, progress has been achieved in the genetic characterization of megagametogenesis, predominantly by forward genetic screens for Arabidopsis mutants that exhibit a distorted segregation (Moore et al., 1997; Howden et al., 1998). Many of the mutants identified affect cell cycle progression in early megagametogenesis, when the megaspore mother cell undergoes three rounds of mitosis to produce the eight-nucleus embryo sac (Moore et al., 1997; Christensen et al., 1998; Springer et al., 2000; Kwee and Sundaresan, 2003; Pagnussat et al., 2005). In the final stages of Arabidopsis megagametogenesis, the eight nuclei of the embryo sac migrate and acquire a specific fate, followed by cellularization of the syncytium. The resulting seven-celled female gametophyte consists of three antipodal cells at the chalazal pole, the egg cell and two synergid cells at the micropylar pole, and the binucleate central cell in the center. Upon maturation, the nuclei of the central cell fuse to form the diploid secondary nucleus and the antipodal cells degenerate, resulting in a four-celled mature female gametophyte (Schneitz et al., 1995; Christensen et al., 1997).
The molecular processes that regulate the migration of the nuclei and determine cell fate and function are not very well understood, although recent studies have revealed several mutants that exhibit defects in the final stages of female gametophyte development. The mutants myb domain protein98 (Kasahara et al., 2005) and feronia (which is allelic to sirene) (Huck et al., 2003; Rotman et al., 2003) are affected in pollen tube guidance and reception, probably due to a defect in synergid cell functioning. Several other mutants were found to be defective in central cell functioning. These include the gametophytic factor2 (gfa2), gfa3, and gfa7 mutants, in which fusion of the polar nuclei does not occur (Christensen et al., 1998), agl80, in which central cell functioning is impaired, and fertilization-independent endosperm (fie), fis2, mea, and multicopy suppressor of ira1 (msi1), which develop endosperm in the absence of fertilization (Ohad et al., 1996; Chaudhury et al., 1997; Grossniklaus et al., 1998; Köhler et al., 2003a). Mutants specifically associated with egg cell fate or functioning have not been described, but in a screen for regulators of egg cell fate, Gross-Hardt et al. (2007) identified the lachesis mutant, in which egg cell fate is extended to the accessory cells of the embryo sac. Moreover, misexpression of the BEL1-LIKE HOMEODOMAIN gene in the eostre mutant leads to the conversion of synergid into egg cell fate (Pagnussat et al., 2007). Despite the identification of several mutants, the molecular processes underlying cell fate adoption in the female gametophyte and specifically the determination of gametic cell (egg cell and central cell) fate and functioning remain elusive.
Here, we report the characterization of the type I MADS box gene AGL61. AGL61 is an Mα-type MADS box gene that is specifically expressed in the central cell of the female gametophyte, and the protein is exclusively targeted to the polar nuclei and the secondary nucleus of the central cell. We studied two T-DNA insertion alleles of AGL61 and demonstrate that both lines fail to transmit the mutant allele via the female gametophyte. A thorough investigation of the mutant phenotype showed that central cell formation is impaired in agl61 female gametophytes. Although the identities of the synergids, egg cell, and antipodal cells are not affected, agl61 embryo sacs start to degenerate before fertilization and fail to initiate embryo and endosperm development after pollen tube penetration. We performed yeast two-hybrid interaction studies and found that AGL61 can form a heterodimer with AGL80. Moreover, we showed that the nuclear localization of AGL61 is lost in the agl80 mutant, indicating that transport of AGL61 to the nucleus is dependent on dimerization with AGL80. These data suggest that AGL61 and AGL80 act together in the differentiation of the central cell. We renamed AGL61 DIANA (DIA), after the virginal Roman goddess of the hunt, also the goddess of nature and childbirth.
RESULTS
DIA (AGL61) Gene Structure
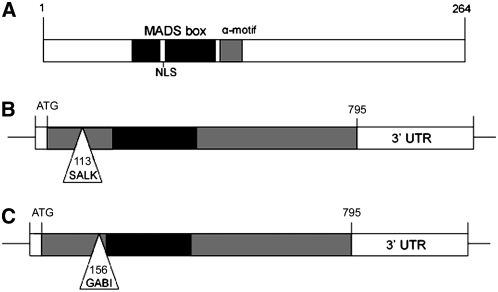
The type I MADS box gene DIA, formerly known as AGL61, is a member of the Mα subclass that contains 25 genes in the Arabidopsis genome (Pařenicová et al., 2003). Like the majority of the type I genes, DIA is a relatively small protein of 264 amino acids that is encoded by a single exon, as is depicted schematically in Figure 1A. The first ATG in the open reading frame is present 177 bp upstream of the conserved MADS box, suggesting that DIA contains a distinct N-terminal region, whereas in most Mα-type proteins the MADS domain is located close to the N terminus. Analysis of the protein sequence in PSORT (http://psort.nibb.ac.jp/form.html) revealed a nuclear localization signal within the MADS domain, in agreement with a putative role as a transcription factor. Except for the MADS domain, no conserved domains are present in the type I proteins, but several motifs can be recognized that are specific for the different subclasses (De Bodt et al., 2003a). DIA contains a distinct Mα-type motif of 11 residues just C-terminal from the MADS box (Figure 1A).
Figure 1.
Structure of the DIA Protein and Positions of the T-DNA Insertions in the DIA Gene.
(A) DIA protein structure. The MADS box is indicated in black (amino acids 62 to 110), and the α-motif is indicated in gray (amino acids 116 to 126). A predicted nuclear localization signal (NLS) is present in the MADS box (amino acids 83 to 86).
(B) DIA gene structure, demonstrating the position of the T-DNA insert in line SALK_009008. Only the first ATG codon in the open reading frame is indicated. The T-DNA is inserted 113 nucleotides downstream of the ATG start codon. UTR, untranslated region.
(C) DIA gene structure, demonstrating the position of the T-DNA insert in line GK_642H10. The T-DNA is inserted 156 nucleotides downstream of the ATG start codon.
DIA Is Expressed in the Final Stages of Embryo Sac Development
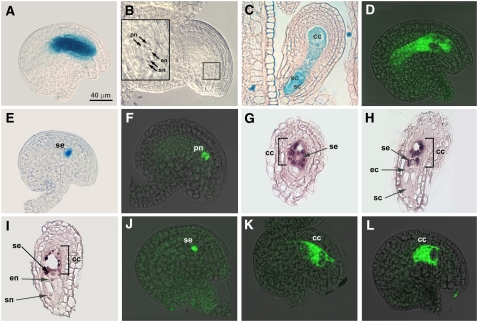
To determine the spatial and temporal expression patterns of DIA, we analyzed Arabidopsis plants transformed with the pDIA:GFP-GUS construct. This construct contains a 1.9-kb fragment upstream of the DIA start codon transcriptionally fused to the green fluorescent protein (GFP) and β-glucuronidase (GUS) reporter genes. All plant tissues from six GUS-expressing transformants were tested for GUS staining, but staining was only observed in a small region where the stamen filament is attached to the anther (see Supplemental Figure 1A online) and in the female gametophyte. For detailed analysis of DIA expression, pistils were harvested from various developmental stages and stained for GUS activity. Staining was only observed in the final stages of embryo sac development, from stage FG5 onward (stages after Christensen et al., 1997) (Figures 2A and 2C). At stage FG5, the female gametophyte consists of eight nuclei that migrate to specific positions within the embryo sac (Christensen et al., 1997) (Figure 2B). At late stage FG5, the female gametophyte cellularizes to give rise to the four distinct cell types of the female gametophyte: the synergid cells, the egg cell, the central cell, and the antipodal cells. At stage FG6, the polar nuclei of the central cell fuse to form the secondary nucleus and the three antipodal cells start to degenerate, eventually resulting in a four-celled embryo sac (stage FG7). Ovules expressing the pDIA:GFP-GUS construct exhibited a high GUS signal throughout the embryo sac already after a couple hours of staining (Figure 2A). Sectioning of the GUS-stained ovules showed signal in the central cell, synergids (Figure 2C), and egg cell (see Supplemental Figure 1B online). GFP analysis of the transformed ovules revealed the same expression pattern (Figure 2D).
Figure 2.
DIA Expression Analysis in the Female Gametophyte.
(A) GUS expression under the control of the DIA promoter in a stage FG6 ovule after 4 h of staining. GUS signal is visible throughout the embryo sac.
(B) Cleared nonstained ovule at stage FG5. The micropylar region is enlarged in the inset.
(C) A 7.0-μm Technovit section of a GUS-stained ovule at stage FG7. The synergid cells (sc) and the central cell (cc) are visible in this section.
(D) GFP expression under the control of the DIA promoter in a stage FG7 ovule.
(E) GUS signal in the secondary nucleus (se) of a pDIA:DIA-GFP-GUS ovule at stage FG7.
(F) GFP signal in the polar nuclei of a pDIA:DIA-GFP-GUS ovule at late stage FG5.
(G) to (I) In situ hybridization of sectioned stage FG7 ovules with a DIA digoxigenin-labeled mRNA probe. The hybridization signal is only visible in the central cell.
(J) GFP signal in the secondary nucleus of a stage FG7 AGL80 ovule containing pDIA:DIA-GFP-GUS.
(K) to (L) GFP signal in the cytoplasm of the central cell in stage FG7 agl80 ovules containing pDIA:DIA-GFP-GUS.
The GFP images are overlays of a CSLM image and a bright-field image. ec, egg cell; en, egg cell nucleus; pn, polar nuclei; se, secondary nucleus; sn, synergid nucleus. The scale bar in (A) is identical for all panels.
To obtain more detailed information about the embryo sac cell types expressing DIA and to investigate the subcellular localization of the DIA protein, we transformed Arabidopsis plants with the pDIA:DIA-GFP-GUS construct and determined the expression of the fusion protein. In contrast with the apparent embryo sac–wide expression of the transcriptional reporter construct, the chimeric protein was expressed exclusively in the central cell in the final stages (i.e., FG5 to FG7) of megagametogenesis (Figures 2E and 2F). We did not observe any expression in the egg cell or synergid cells, even after prolonged staining (up to 4 d). Consistent with a putative role as a transcription factor, the DIA-GFP-GUS fusion protein was found to be located in the secondary nucleus (Figure 2E). Expression was also detected in polar nuclei prior to fusion (stage FG5; Figure 2F), consistent with the earliest expression of DIA determined by the pDIA:GFP-GUS reporter construct.
To investigate the discrepancy between the observed expression patterns of the promoter–reporter construct and that of the fusion protein further, we performed in situ hybridization experiments with embryo sacs from stages FG4 to FG7 using a DIA-specific antisense probe that contained a fragment of ∼400 nucleotides downstream of the MADS box. In accordance with the other experiments, no signal was detected in female gametophytes before stage FG5. Only embryo sacs in stages FG5 to FG7 showed a hybridization signal, and this signal was specifically located in the central cell (Figures 2G to 2I). Female gametophytes that were hybridized with a sense probe showed no signal at all. These data reveal that both the DIA mRNA and the DIA protein are specifically located in the central cell and that the observed GFP-GUS expression throughout the embryo sac in pDIA:GFP-GUS plants is most likely due to diffusion of the reporter gene product. Alternatively, the coding region of DIA may contain elements that regulate differential expression of DIA in the different embryo sac cell types.
To determine whether DIA is also expressed after fertilization, we analyzed developing seeds of plants containing either pDIA:GFP-GUS or pDIA:DIA-GFP-GUS at several time points after pollination. GFP/GUS signal was visible until 48 h after pollination near the micropylar end of developing seeds containing pDIA:GFP-GUS and could occasionally be observed in the first two endosperm nuclei in ovules that contained pDIA:DIA-GFP-GUS (see Supplemental Figures 1C and 1D online, respectively). However, expression of the reporter constructs in developing seeds was not detected when the construct was introduced via the male parent, indicating that the observed signal remained from the megagametophyte.
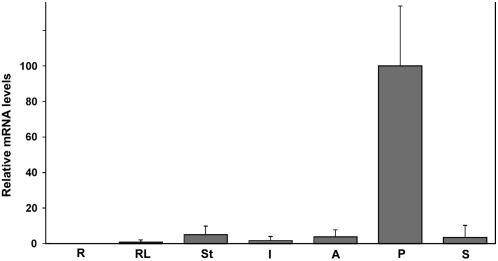
DIA expression appears to be almost completely restricted to the female gametophyte. To confirm the exclusive expression of the gene, we performed real-time RT-PCR analysis with RNA from different tissues (Figure 3). Expression of DIA was detected in rosette leaves, stems, inflorescences, stamens, and siliques at 5 d after pollination, but the relative expression in these tissues was <5% of the expression measured in pistils, demonstrating that DIA is indeed predominantly expressed in the female gametophyte. This expression pattern confirmed the results obtained with the promoter–reporter construct.
Figure 3.
Real-Time RT-PCR Analysis of DIA Expression.
Quantitative RT-PCR of DIA transcript using UBIQUITIN-CONJUGATING ENZYME21 as a reference gene. R, roots; RL, rosette leaves; St, stem; I, inflorescences from flowers at stages 1 to 11 (according to Smyth et al., 1990); A, anthers and filaments from flowers at stages 11 to 14; P, pistils from flowers at stages 12 to 14; S, siliques at 5 d after pollination. Error bars indicate the sd of two biological replicates.
DIA Interacts with Several Mγ-Type Proteins
The interactions between all MADS box transcription factors in Arabidopsis have been investigated by De Folter et al. (2005) in a matrix-type yeast two-hybrid screen. The ability of AGL61 (DIA) to interact with other MADS domain proteins was also investigated in their study, but no interactions were identified. To examine these results, we sequenced the DIA clone used in the study by De Folter et al. (2005) and identified a frameshift mutation, which probably explained the lack of interactions. Therefore, we performed a similar screen using newly constructed bait and prey vectors with the DIA open reading frame. Both vectors were transformed to yeast and mated with the set of MADS box prey and bait vectors used by De Folter et al. (2005).
Analysis of the interactions revealed that DIA interacts both as prey and as bait with the Mγ-type proteins AGL80, PHE1, AGL38 (PHE2), and AGL86, while homodimerization was not observed (see Supplemental Figure 2 online). Of the four identified interaction partners, AGL80 is the only gene reported to be expressed in the female gametophyte (Portereiko et al., 2006). In the context of the embryo sac, AGL80 is expressed exclusively in the polar nuclei and secondary nucleus of the central cell (Portereiko et al., 2006), and the AGL80 protein is therefore a likely candidate to interact in vivo with DIA in the central cell.
DIA and AGL80 Interact in Vivo in the Central Cell
The efficiency with which MADS box proteins are transported into the nucleus appears to be dependent on their ability to form homodimers or heterodimers (Immink et al., 2002). Since the yeast two-hybrid analysis revealed that DIA does not form homodimers, the protein probably needs to form a heterodimer for efficient transport to the central cell's secondary nucleus. To investigate whether the candidate interaction partner AGL80 is able to facilitate the transport of DIA to the nucleus of the central cell, we analyzed the localization of the DIA fusion protein in the agl80/AGL80 mutant. Homozygous pDIA:DIA-GFP-GUS plants were pollinated with pollen derived from a heterozygous agl80/AGL80 plant, and the progeny were used for the analysis. Both in agl80 and in AGL80 megagametophytes, a clear GFP signal was detected, indicating that DIA expression is not controlled by AGL80. However, while the GFP signal in AGL80 embryo sacs was localized to the nucleus (Figure 2J), we observed a cytoplasmic localization of the fusion protein in agl80 ovules (Figures 2K to 2L). These data demonstrate that DIA is dependent on the presence of AGL80 to be efficiently transported to the nucleus.
The dia Mutation Affects the Female Gametophyte
To investigate the function of DIA, we identified T-DNA insertion lines in the SALK Institute Genomic Analysis Laboratory collection (Alonso et al., 2003) and in the GABI-Kat insertion collection (Rosso et al., 2003). Figures 1B and 1C show the positions of the T-DNA insertions in DIA. The T-DNA in line SALK_009008 (dia-1) is inserted 113 bp downstream of the putative start codon and 46 bp upstream of the start of the MADS box region. Line GK_642H10 (dia-2) contains the T-DNA insertion 156 bp downstream of the putative start codon and only 3 bp upstream of the start of the MADS box.
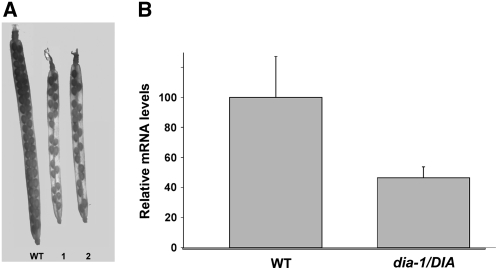
For both alleles, seeds from a segregating T3 generation were obtained. Genotyping this generation by PCR analysis using gene-specific and T-DNA–specific primer sets revealed that no homozygous mutant was present for dia-1 (n = 19) and dia-2 (n = 36). Furthermore, plants scored as heterozygous mutants had siliques in which only ∼50% of the seeds developed (Figure 4A), suggesting that the dia mutation causes gametophytic lethality. Since no difference was observed between the alleles, we continued a thorough analysis with dia-1 only.
Figure 4.
Seed Set and DIA Expression in dia/DIA Plants.
(A) Cleared siliques from wild-type, dia-1/DIA (1), and dia-2/DIA (2) plants at 7 d after pollination. Siliques of the heterozygous mutants are smaller than the wild-type siliques, and seed set is reduced to ∼50%.
(B) Quantitative RT-PCR analysis of DIA expression in wild-type and dia-1/DIA mature pistils. The bars indicate relative expression of DIA. Error bars indicate sd of three biological replicates.
To determine if the dia mutation affects the male or the female gametophyte, reciprocal crosses were performed with dia-1/DIA plants. Heterozygous mutants were selfed, used as the female parent for pollination with wild-type pollen, or used as the male donor to pollinate wild-type females, and the progeny were scored (Table 1). In all progeny plants, occurrence of the silique phenotype was linked to the presence of the dia-1 mutant allele, as determined by PCR. The progeny of the self-pollinated dia-1/DIA plants did not exhibit a Mendelian segregation (P = 9.6 × 10−40) but approximated a 1:1 segregation of DIA/DIA to dia-1/DIA plants (P = 0.48), and no homozygous mutants were observed (Table 1). When dia-1/DIA was used as the male parent, 54% of the progeny were heterozygous for the mutation, demonstrating no significant difference between the transmission of the dia-1 allele and the wild-type allele via the male gametophyte (P = 0.79). However, the use of dia-1/DIA as the female parent resulted only in wild-type progeny, indicating that the mutation affects the transmission of the mutant allele via the female gametophyte (P = 4.3 × 10−12). The absence of any homozygous mutant in the progeny indicates that the dia-1 mutation is fully penetrant.
Table 1.
Segregation of the dia-1 Mutation in Selfed and Reciprocally Backcrossed Offspring Populations
| Parental Genotypes
|
Progeny Genotypes
|
|||
|---|---|---|---|---|
| Female | Male | DIA/DIA | dia-1/DIA | dia-1/dia-1 |
| dia-1/DIA | dia-1/DIA | 128 | 116 | 0 |
| dia-1/DIA | DIA/DIA | 48 | 0 | 0 |
| DIA/DIA | dia-1/DIA | 64 | 67 | 0 |
The location of the T-DNA insertion in dia-1 downstream of the first ATG codon but upstream of the MADS box region suggests a complete disturbance of DIA function. To obtain information about the reduction of gene expression, DIA expression was analyzed by real-time RT-PCR in wild-type and dia-1/DIA mature pistils. In the heterozygous pistils, we observed a reduction in expression of ∼50% (Figure 4B) compared with the wild-type. This is in agreement with the expected loss of expression in the mutant gametophytes, which are harbored by half of the ovule population.
To verify that the mutant phenotype was indeed caused by the loss of DIA function, we transformed dia-1/DIA plants with pDIA:DIA-GFP-GUS to complement the mutant phenotype. No homozygous dia-1 mutants were obtained in the primary transformants, indicating that the transformation event occurred after the aberrations appeared or that expression of the complementation construct was insufficient. However, seven primary transformants hemizygous for both the rescue construct and for the dia-1 mutation exhibited an increased seed set to ∼75%. After selfing, the progeny of these seven primary transformants were further investigated to obtain dia-1/dia-1 plants. Indeed, genotypic analysis revealed homozygous dia-1 plants among the progeny of five of the primary lines. Fifty-five plants of the offspring (a mix from different offspring populations) were genotyped, and 15 appeared to be homozygous dia-1 plants. The dia-1/dia-1 plants that also contained the complementation construct in a homozygous state showed a wild-type seed set, consistent with a full complementation. These data demonstrate that the loss of DIA is responsible for the female gametophytic phenotype.
dia Embryo Sacs Have Aberrant Central Cell and Egg Cell Morphology
To investigate the dia phenotype in more detail, we analyzed >100 embryo sacs from wild-type and dia-1/DIA mutant plants. Approximately 50% of the ovules in dia-1/DIA ovaries contained the DIA allele and showed a wild-type phenotype, whereas the other 50% represented a mutant population in which an aberrant phenotype may be expected. To search for morphological abnormalities in this population, ovules were fixed in glutaraldehyde (Christensen et al., 1997) and observed by confocal scanning laser microscopy (CSLM). Ovules were analyzed from stage FG1 (one-nucleus embryo sac) to stage FG7 (mature, four-cell embryo sac with degenerated antipodal cells). Analysis of the early stages of female gametophyte development revealed that there is no difference between the development of wild-type and dia-1 gametophytes up to and including the four-nucleus stage (n = 18; Figure 5A). However, from stage FG5 onward, we observed an increasing number of aberrant embryo sacs in the dia-1/DIA pistils. The effect of the dia-1 mutation was most apparent at the final stage of megagametogenesis (stage FG7). In dia-1/DIA pistils harvested at 24 to 48 h after emasculation of the flower, we clearly observed a wild-type population (53%) and a mutant population (47%).
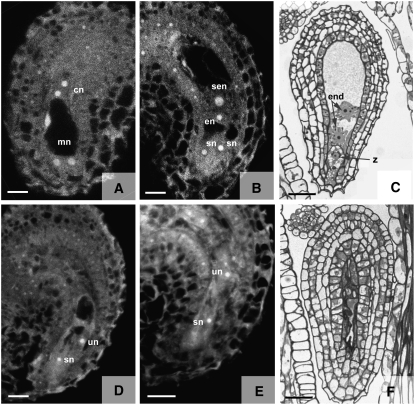
Figure 5.
Microscopic Analysis of the dia-1 Phenotype.
(A) and (D) to (F) dia-1 ovules.
(B) and (C) Wild-type ovules.
(A) Stage FG4 ovule showing a wild-type phenotype. This image is a CSLM projection of three 1.0-μm optical sections.
(B) Stage FG7 wild-type ovule. This image is a CSLM projection of two 1.0-μm sections.
(C) Wild-type ovule at 16 h after pollination. Light microscopy image of a 1.0-μm section.
(D) and (E) dia-1 ovule at stage FG7. Both images are single 1.0-μm CSLM sections.
(F) Degenerated dia-1 ovule at 12 h after pollination. Light microscopy image of a 1.0-μm section.
cn, chalazal nuclei; en, egg cell nucleus; end, endosperm nuclei; mn, micropylar nuclei; sen, secondary nucleus; sn, synergid cell nucleus; un, undefined nucleus; z, zygote. Bars in (A), (B), (D), and (E) = 20 μm; bars in (C) and (F) = 30 μm.
Wild-type embryo sacs at stage FG7 consist of three cell types (Figure 5B). The two synergid cells are localized at the micropylar end and function in the attraction and guidance of the pollen tube, while the haploid egg cell and diploid central cell represent the female gametes that will develop into the zygote and the endosperm, respectively, after fertilization. We could identify all three cell types in >90% of the wild-type ovules at stage FG7 (n = 36) but only in 53% of the dia-1/DIA ovules (n = 109). In the mutant population (51 ovules), the secondary nucleus of the central cell was never observed, whereas the appearance of the other two cell types was variable (Figures 5D and 5E). In one-third of these ovules, no cell types could be recognized because the embryo sac was filled with a fluorescent substance, indicating that the female gametophyte was completely degenerated. In the remaining mutant population (34 ovules), synergid nuclei were observed in more than two-thirds of the embryo sacs, whereas a putative egg cell nucleus could only be identified in 26% of the assumed dia-1 ovules. However, enhanced fluorescence in the embryo sacs of these ovules suggests that degeneration is initiated as well (Figures 5D and 5E).
In the majority of the mutant embryo sacs, synergid cells were present but a distinct egg cell was lacking. These embryo sacs often contained one or more undefined nuclei, whose spatial arrangement was aberrant (Figures 5D and 5E). Whether these are nuclei of egg cells is difficult to determine from these photographs. To study the functionality of the egg cell, we fixed mutant ovules shortly after fertilization and investigated if a zygote was formed. However, neither endosperm nor embryo development was initiated in dia-1 ovules after pollination, and a fast degeneration was observed (Figures 5C and 5F), suggesting that a functional egg cell is lacking in dia-1 embryo sacs.
dia-1 Embryo Sacs Fail to Establish Central Cell Identity
Microscopic analysis of the dia-1 phenotype revealed that a secondary nucleus is absent in the mutant megagametophytes. In addition, the overall embryo sac morphology is aberrant and undefined nuclei are often observed (Figures 5D and 5E). To investigate the fate of the different cell types in dia-1 embryo sacs further, we crossed dia-1/DIA plants with marker lines expressing GUS in the different cell types of the female gametophyte (Gross-Hardt et al., 2007). Seeds harvested from each cross were sown and embryo sacs of the F1 generation, hemizygous for both dia-1 and the reporter construct, were analyzed. Because the reporter construct was present in a hemizygous state, GUS signal could only be observed in 50% of the ovules from DIA/DIA plants. For each cross, the expression of the reporter line in DIA/DIA plants was compared with the expression in dia-1/DIA plants and the number of expressing ovules was scored at 40 h after emasculation or, in case of the antipodal marker, in stage FG5 to FG6 (Figures 6A to 6L).
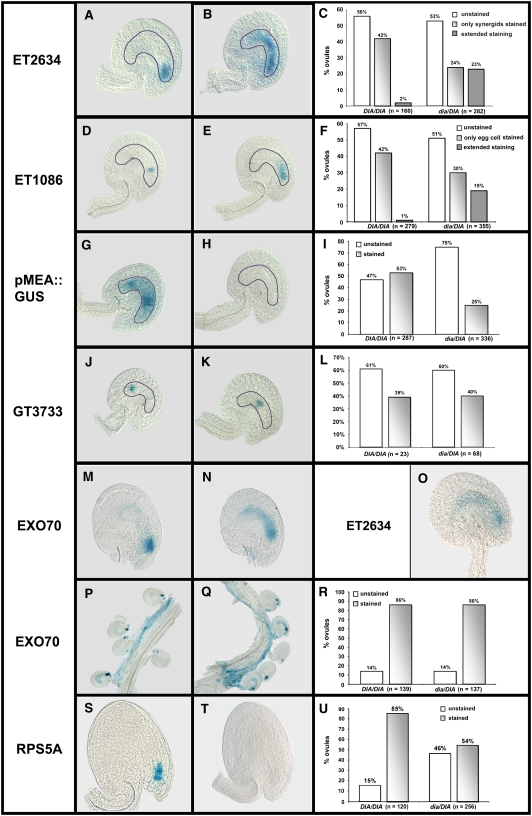
Figure 6.
Characterization of the dia Mutant Ovules Using Marker Lines.
Expression analysis of cell-specific marker genes in DIA/DIA and dia-1/DIA pistils. The marker lines are present in a hemizygous state. The pistils were harvested at 2 d after emasculation, unless indicated otherwise.
(A) to (C) Expression of the synergid cell marker ET2634 in DIA (A) and dia-1 (B) stage FG7 ovules. In (C), the bars show the percentage of GUS-stained ovules in DIA/DIA and dia-1/DIA pistils (n = 442).
(D) to (F) Expression of the egg cell marker ET1086 in DIA (D) and dia-1 (E) stage FG7 ovules. In (F), the bars show the percentage of GUS-stained ovules in DIA/DIA and dia-1/DIA pistils (n = 634).
(G) to (I) Expression of the central cell marker pMEA:GUS in DIA (G) and dia-1 (H) stage FG7 ovules. In (I), the bars show the percentage of GUS-stained ovules in DIA/DIA and dia-1/DIA pistils (n = 623).
(J) to (L) Expression of the antipodal marker GT3733 in DIA (J) and dia-1 (K) ovules between stages FG5 and FG6. In (L), the bars show the percentage of GUS-stained ovules in DIA/DIA and dia-1/DIA pistils (n = 88).
(M) and (N) GUS staining in DIA (M) and dia-1 (N) ovules at 8 h after pollination with pollen expressing the EXO70 marker.
(O) GUS staining of the synergid cell marker ET2634 at 8 h after pollination of DIA (M) ovules.
(P) to (R) GUS staining in DIA/DIA (P) and dia-1/DIA (R) pistils at 16 h after pollination with pollen expressing the EXO70 pollen tube marker. In (R), the bars show the percentage of GUS-stained ovules in DIA/DIA and dia-1/DIA ovules (n = 276).
(S) to (U) GUS staining in DIA (S) and dia-1 (T) developing seeds at 20 h after pollination with pollen from the RPS5A reporter line. In (U), the bars show the percentage of GUS-stained ovules in DIA/DIA and dia-1/DIA ovules (n = 376).
Consistent with the microscopic analysis, we were unable to detect expression of the central cell marker pMEA:GUS in dia embryo sacs (Figures 6G to 6I). Although dia female gametophytes also show an aberrant morphology of the micropylar cell types, we observed similar levels of expression of both the synergid marker ET2634 and the egg cell marker ET1086 in DIA/DIA and dia/DIA pistils (Figures 6A to 6F). This suggests that synergid cell fate and egg cell fate are not compromised in dia embryo sacs. However, in dia megagametophytes, the GUS signal of both the synergid and the egg cell markers appeared to be extended to adjacent cells and sometimes filled the embryo sac completely (Figures 6B and 6E, respectively). This extension of the signal may be explained by an increased diffusion of GUS, caused by the initiation of degeneration in the mutant embryo sacs. The analysis of the dia phenotype by confocal microscopy revealed that degeneration had occurred in approximately one-third of the FG7 embryo sacs. Moreover, a fluorescent substance was often visible in the remaining population (Figures 5D and 5E), indicating that these embryo sacs were also subjected to degeneration. To investigate if the observed signal diffusion could indeed be a result of degeneration, we analyzed expression of the ET2634 marker in DIA/DIA ovules at 8 h after pollination. At this time point, one of the synergids has undergone cell death in response to the perception of the pollen tube (Sandaklie-Nikolova et al., 2007). In many ovules, we observed diffusion of the GUS signal that resembled the staining observed in dia embryo sacs (Figure 6O), supporting the idea that the extension of ET2634 and ET1086 signals in dia ovules is caused by the degeneration of embryo sac cells and subsequent diffusion of GUS.
To provide additional evidence that the broad GUS signal from markers ET1086 and ET2634 in dia embryo sacs was a result of degeneration, we analyzed the expression of the markers at an earlier stage. Flowers were harvested from stages 12c and 13 (according to Smyth et al., 1990) and the pistils were stained for GUS activity. Flowers in these stages contain embryo sacs that range from the four-nucleus stage (FG4) to the four-cell stage (FG7) and do not yet all express the cell identity markers. The egg cell marker ET1086 and the synergid cell marker ET2634 are both expressed from approximately late stage FG5 onward. We found a distinct correlation between the number of ovules expressing the markers and the number of ovules with extended staining. If only a few ovules were expressing the marker (i.e., the ovules were in a relatively early stage), the GUS signal was always restricted to synergids or the egg cell (comparable with Figures 6A and 6D, respectively). However, as more ovules reached the final developmental stage, the number of embryo sacs that showed extension of the GUS signal increased. In the analyzed embryo sacs that expressed GUS (stages FG5 to FG7), marker ET1086 was restricted to the egg cell in 81% of the analyzed dia/DIA ovules (n = 53), while diffusion was observed in the remaining 19%. The number of embryo sacs that showed extension of the ET2634 marker was reduced to 16% in the tested stages (FG5 to FG7; n = 165), indicating that the egg cell and synergid cell identity are normally established in dia megagametophytes. At maturity (FG7), the degeneration of the embryo sacs is most likely responsible for the extension of the GUS signal (Figures 6B and 6E).
To study the fate of the antipodal cells in dia embryo sacs, we analyzed dia-1/DIA plants crossed with the antipodal marker GT3733. We did not find a difference between the number of FG5 and FG6 female gametophytes expressing the marker in dia/DIA and DIA/DIA plants (Figures 6J to 6L), indicating that the differentiation of the antipodals is not affected in dia mutants. Also, diffusion of the signal was not observed in dia embryo sacs, suggesting that degeneration had not yet initiated and that cellularization occurred normally in stage FG5 mutant embryo sacs. However, we did observe a difference in the location of the GUS signal in ovules from dia/DIA and DIA/DIA plants. In a number of ovules from dia-1/DIA plants, the expression of the antipodal marker appeared to be shifted toward the micropylar pole (cf. Figure 6J with Figure 6K), probably because of the absence of a large central cell. Yet, due to the weak and transient expression of the GT3733 marker, we were not able to quantify this observation.
Taken together, these data suggest that DIA is important for the differentiation of the central cell but does not determine the fate of the synergids, the egg cell, and the antipodal cells. However, our data indicate that the absence of the large central cell triggers degeneration of the embryo sac at stage FG7 and thus indirectly affects the synergid cells and egg cell.
dia Embryo Sacs Perceive Pollen Tubes but Do Not Form a Zygote
To investigate if the initiation of degeneration in stage FG7 affects the ability of the synergids and egg cell to fulfill their functions, we performed additional experiments. Several female gametophytic mutants, exhibiting a defect in fertilization, have been reported to be affected in pollen tube guidance or reception due to malfunctioning of the synergid cells (Huck et al., 2003; Rotman et al., 2003; Kasahara et al., 2005). To address the question of whether dia female gametophytes are able to attract a pollen tube and to determine if the pollen tube penetrates dia ovules, we pollinated wild-type and dia-1/DIA ovaries with pollen tubes expressing the pollen tube marker EXO70. This marker line carries a 1.4-kb promoter fragment of the EXO70 gene fused to the GUS reporter gene and shows a strong GUS signal in the pollen tube that is released in one of the synergid cells when the pollen tube penetrates the embryo sac. Ovules were harvested at 8 and 16 h after pollination and stained for GUS activity. In both experiments, the number of stained ovules from dia-1/DIA siliques (188 of 207) was similar to the number of stained ovules in the wild type (187 of 206), indicating that pollen tube guidance by dia mutant embryo sacs is normal. Also, we did not observe uncontrolled growth of the pollen tube inside the female gametophyte, as was reported for the feronia and sirene mutants (Huck et al., 2003; Rotman et al., 2003). However, approximately half of the ovules from dia-1/DIA ovaries exhibited diffusion of the GUS signal at 8 h after pollination (29 of 67), whereas we observed extension of the signal only in a small portion of the wild-type ovules (9 of 70) (Figures 6M and 6N). Nonetheless, this difference had disappeared at 16 h after pollination, when diffusion of the signal had also occurred in wild-type ovules, due to the degeneration of one of the synergids upon pollination (Figures 6P and 6Q). Our data indicate that the penetration of the synergid cell by the pollen tube is normal in dia embryo sacs and that the synergid function is not impaired.
Microscopic analysis revealed that embryo initiation does not occur in dia ovules. To confirm this observation, we analyzed zygote formation by pollinating wild-type and dia-1/DIA pistils with pollen from a pRPS5A:GUS marker line (Weijers et al., 2001b). The RPS5A gene is strongly expressed in dividing cells, and activity of the paternal allele can be detected from the two-cell stage onward (Weijers et al., 2001a). Wild-type and dia-1/DIA pistils were pollinated by the homozygous marker line, harvested after 20 h, and stained for GUS activity. A clear GUS signal was present in 85% of the ovules from wild-type plants but in only 53% of the ovules from dia/DIA plants (Figures 6S to 6U). These data indicate that the majority of the dia ovules remained unfertilized, although we cannot exclude the possibility that some dia ovules manage to activate embryogenesis.
DISCUSSION
DIA Is Required for Central Cell Development
DIA is an Mα-type MADS box gene that is almost exclusively expressed in the final stages of female gametophyte development. We investigated two independent T-DNA insertion alleles that disrupt DIA gene functioning and found a distorted Mendelian segregation of 1:1 instead of 3:1. Reciprocal crosses revealed that the mutant allele is fully transmitted through the male gametophyte but fails to transmit through the female gametophyte.
We investigated the dia phenotype in megagametophytes both by microscopy and by marker line analysis and found that the dia mutation predominantly affects the differentiation of the central cell. Up to the four-nucleus stage, no differences were observed between embryo sacs from wild-type and dia/DIA ovaries, but from the eight-nucleus stage onward (stage FG5), aberrant embryo sacs were present in the dia/DIA pistils. However, the embryo sacs within one pistil do not develop completely synchronously, which made it difficult to precisely determine the stage in which abnormalities were observed first. The results obtained from the cross with the cell-specific markers suggest that cellularization occurs normally in dia embryo sacs. The first distinct defect in dia embryo sacs was observed in stage FG6 ovules, in which the secondary nucleus is not formed. The percentage of ovules that succeed to form a secondary nucleus in dia/DIA ovaries is 53%, which is much lower than the 92% observed in wild-type ovaries; however, it cannot be excluded that a few dia embryo sacs do manage to develop a secondary nucleus.
In the final stages of megagametogenesis, dia embryo sacs contain a variable number of unidentified nuclei with an aberrant position. Marker line analysis revealed that both the egg cell and the synergid cell fates are established in the mutant embryo sacs, whereas the central cell marker pMEA:GUS is not expressed. Both synergid and egg cell nuclei thus appear to be present and the observed aberrant morphology of the egg cell may be caused by the absence of a normal central cell. The nature of the additional nuclei that were observed in several dia embryo sacs was not completely elucidated by the marker line analysis. Probably, these nuclei represent remaining polar nuclei. The expression of the egg cell and synergid cell markers in dia ovules was indistinguishable from the expression in wild-type ovules at the seven-cell embryo sac stage, but the GUS signal extended to the other cells in mature ovules. Therefore, we presume that the extended GUS signal is a result of diffusion caused by degeneration of the dia embryo sacs in response to the absence of a normal central cell. Although the dia mutation primarily affects central cell development, the initiation of degeneration prior to fertilization has an effect on the micropylar cell types as well. Synergid cells are able to attract and receive pollen tubes, but the failure of zygote formation in the majority of the dia embryo sacs reveals that egg cell function is impaired, which is consistent with the absence of an egg cell in the majority of the mature dia embryo sacs.
DIA Protein Exclusively Accumulates in the Central Cell
Analysis of plants transformed with a pDIA:DIA-GFP-GUS construct revealed that the fusion protein is present exclusively in the polar nuclei and the secondary nucleus of the central cell, consistent with the function of DIA in central cell differentiation. Although the pDIA:GFP-GUS reporter construct appeared to be expressed in the egg cell and synergids as well, in situ hybridization revealed that the DIA mRNA is only present in the central cell. Therefore, the broad GFP/GUS signal we observed in pDIA:GFP-GUS plants is most likely due to diffusion of the GFP-GUS mRNA or protein to the other cells of the embryo sac. However, it cannot be excluded that DIA transcription is differentially regulated via elements that are present in the coding region of the gene.
DIA Interacts with Mγ-Type MADS Domain Proteins
We used a yeast two-hybrid assay to identify the interaction partners of DIA in order to better understand the gene regulatory networks involved in megagametogenesis. The assay revealed that DIA strongly interacts with the Mγ-type MADS box proteins AGL80, PHE1, PHE2 (AGL38), and AGL86. This finding supports the observation of De Folter et al. (2005) that Mα-type MADS box proteins preferentially dimerize with Mβ- or Mγ-type proteins. Interestingly, among the interaction partners of DIA are AGL80 and PHE1, the only two Mγ-type proteins that have been characterized to date. PHE1 and PHE2 have been reported to be expressed in the developing seed after fertilization and are thus probably not coexpressed with DIA (Köhler et al., 2005). However, AGL80 is known to be expressed in the central cell and therefore is a likely candidate to interact with DIA in planta (Portereiko et al., 2006).
To learn more about the AGL80-DIA complex, we introduced the pDIA:DIA-GFP-GUS construct in agl80 mutant ovules (Portereiko et al., 2006). In the agl80 embryo sacs, the DIA fusion protein appeared to be localized in the cytoplasm instead of the nucleus, which revealed that DIA is dependent on AGL80 for efficient transport to the nucleus. These data also demonstrate that DIA and AGL80 interact in planta and may act together to form the central cell.
In Arabidopsis, AGL80 functions in central cell and endosperm development and is first expressed in the central cell just prior to the fusion of the polar nuclei (Portereiko et al., 2006). Maternal expression of AGL80 can be detected in the endosperm nuclei until 3 d after pollination (Portereiko et al., 2006). In the central cell, AGL80 and DIA are coexpressed and form a complex that probably plays an important role in central cell differentiation. If both genes act together, the phenotypes of the dia and agl80 mutants in the embryo sac are expected to be similar. Comparison of phenotypes, however, reveals that the agl80 phenotype appears less severe than the dia phenotype, although both mutants are impaired in central cell function (Portereiko et al., 2006). In contrast with dia mutants, a secondary nucleus is formed in agl80 embryo sacs, although the size of the nucleolus and that of the vacuole are smaller than in the wild type. Moreover, a zygote-like structure is present in agl80 ovules after fertilization, indicating that fertilization and egg cell function are not affected (Portereiko et al., 2006). However, instead of endosperm nuclei, the central cell cavity of agl80 ovules contains highly fluorescent material after fertilization, probably marking the degeneration of the central cell. Fluorescent substances were not observed in mature agl80 embryo sacs before fertilization (Portereiko et al., 2006). The absence of a secondary nucleus and the premature initiation of degeneration in dia ovules may suggest that DIA has an additional function early in central cell formation that is independent of AGL80. Unlike AGL80, which is not expressed until the polar nuclei are immediately adjacent to one another (Portereiko et al., 2006), we also observed DIA expression in separated polar nuclei, indicating that DIA is expressed slightly earlier than AGL80. Possibly, DIA interacts with other factors at this earlier stage, such as AGL86 or non-MADS box proteins. Analysis of the AGL86 expression pattern in future experiments will explore this possibility.
The AGL80-DIA complex probably functions in central cell development by regulating a set of genes. Unfortunately, the severe central cell phenotype of dia does not allow for a reliable identification of downstream genes, since most genes expressed in the central cell are likely affected by the malformation of the central cell. Therefore, we can only speculate about the downstream genes that are regulated by the AGL80-DIA dimer. However, the absence of DEMETER (DME) and DD46 expression in agl80 mutants (Portereiko et al., 2006) suggests that the AGL80-DIA complex is directly or indirectly involved in the regulation of many genes. DME positively regulates MEA (Gehring et al., 2006), while MEA is involved in a complex with FIE, FIS2, and MSI1 to repress the expression of genes required for endosperm development, including PHE1 and PHE2. To obtain more information about the role of AGL80-DIA in the central cell network, additional studies involving target gene identification need to be performed.
DIA Is Specifically Involved in Female Gametophyte Development
A large proportion of the mutants with a defect in the formation or functioning of the female gametophyte are disrupted in housekeeping genes or genes that encode for cell cycle proteins (Dresselhaus, 2006). These genes usually function throughout the plant but exhibit female sterility in a hemizygous state and are thus identified as female gametophyte mutants. This category comprises cytokynin-independent1 (Pischke et al., 2002), nomega (Kwee and Sundaresan, 2003), lachesis (Gross-Hardt et al., 2007), and many of the mutants identified by Pagnussat et al. (2005). In contrast with the genes disrupted in these mutants, DIA is specifically and almost exclusively expressed in the female gametophyte (Figures 2 and 3) and is involved in megagametogenesis. The identification of transcription factors like DIA that play a specific role in female gametophyte development is essential for unraveling the gene regulatory networks involved in megagametogenesis. To date, knowledge about these networks is largely missing and only fragmentary information is available. Expression studies performed by Yu et al. (2005), Johnston et al. (2007), and Steffen et al. (2007) provide a source for the identification of cell type–specific transcription factors in Arabidopsis and can be used as a starting point for the analysis of gene regulatory networks underlying female gametogenesis.
Type I MADS Box Genes Are Involved in Megagametogenesis and Early Seed Development
Functional analysis of MIKC-type MADS box genes revealed that they are predominantly involved in specifying the identities of meristems, tissues, and specific cells throughout the life cycle of a plant. By contrast, the type I genes seem to be more confined to a particular developmental process, and clear homeotic functions have not been identified (Becker and Theissen, 2003; De Bodt et al., 2003b). Of the 61 type I genes in Arabidopsis, functional information has only been reported for the Mα-type genes AGL23 and AGL62 and the Mγ-type genes PHE1 (Kofuji et al., 2003) and AGL80 (Portereiko et al., 2006), which all play a role in megagametogenesis or seed development.
The functional importance of type I MADS box genes has been under debate since their discovery in 2000. This study provides evidence, together with the functional information on AGL23, AGL62, AGL80, and PHE1, that both Mα- and Mγ-type genes can play important roles in plants. Moreover, it suggests a role for type I genes in reproduction and, more specifically, in female gametophyte development. Expression studies of the type I MADS box genes in Arabidopsis revealed that the majority of the type I genes are very weakly expressed, although transcripts were detected in the inflorescences and siliques for most of the genes by RT-PCR (Köhler et al., 2003b; Pařenicová et al., 2003). In the study of Pařenicová et al. (2003), DIA expression was not detected, probably due to the very specific spatial and temporal expression of the gene. Similarly, other type I MADS box genes, the expression of which was reported to be weak, may exhibit a very specific expression in the female gametophyte as well.
A combinatorial role for both Mα- and Mγ-type MADS box genes in female gametophyte development would be consistent with the interaction data reported by De Folter et al. (2005). The interaction map of the Arabidopsis MADS box proteins revealed that Mα-type MADS box proteins interact predominantly with Mγ-type proteins, whereas the heterodimerization of proteins belonging to the same class is rare, suggesting that both types of genes are required for the formation of functional regulatory complexes.
In conclusion, based on expression studies, mutant analysis, and protein–protein interaction studies, we hypothesize that the type I MADS box genes play important roles in female gametophyte and early seed development. Further studies are needed to confirm this hypothesis.
METHODS
Growth Conditions
Arabidopsis thaliana plants were grown in the greenhouse with a 16-h-light/8-h-dark cycle at 22°C. Seeds resulting from floral dip transformation were sterilized for 1 min in 100% ethanol and for 5 min in 1% bleach, washed three times in sterile water, and germinated on half-strength Murashige and Skoog selective plates (2.2 g of Murashige and Skoog salts including Gamborg B5 vitamins, 0.5 g of MES, and 40 mg/L kanamycin). After 10 d of incubation in a growth chamber (16 h of light/8 h of dark, 22°C), resistant plants were transferred to soil.
Segregation Analysis
SALK_009008 plants were genotyped by PCR using the following primers: Lba1 (5′-TGGTTCACGTAGTGGGCCATCG-3′), 61for (5′-AAGGCAAGCCGAGTAATTACAA-3′), and 61rev (5′-CGGCTCTGCGTTTGGAGAATGT-3′). GABI_642H10 plants were genotyped using primers pAC161for (5′-GATGAAATGGGTATCTGGGAATGG-3′), GABI-61for (5′-AAGGCAAGCCGAGTAATTACA-3′), and GABI-61rev (5′-CTTGGATGTCCGAATGAGAAAGG-3′). Segregation of the phenotype was tested by clearing the siliques overnight in 70% ethanol and counting the number of developed seeds.
For reciprocal cross analysis, dia-1/DIA (SALK_009008.51.65.x) plants were self-pollinated or crossed with the wild type as outlined in Table 1. The genotype of the progeny was tested with PCR; the phenotype was tested by ethanol clearing of the siliques, as described above. The outcome of these crosses was statistically analyzed using the χ2 test.
Expression Analysis
The pDIA:GFP-GUS and pDIA:DIA-GFP-GUS constructs were generated using the Gateway system (Invitrogen). For pDIA:GFP-GUS, a 1.9-kb promoter fragment was amplified from genomic DNA using primers pAGL61for (5′-CACCAACCGATTTGACAAATGCCCGAAACCGA-3′) and pAGL61rev (5′-TTTTTGTATGGAGGGTTTTAGTTGCTTTTCT-3′). A genomic fragment (2.5 kb) containing the DIA open reading frame and upstream sequences was amplified with the primers pAGL61for and fusAGL61rev (5′-TGAAACAACCATTTCCATTGGCAAAATT-3′). Both fragments were cloned into the pENTR/D-TOPO vector, and an LR reaction was performed to recombine the fragments in the binary vectors pKGWFS7 (pDIA) and pBGWFS7 (pDIA:DIA) (Karimi et al., 2002). The resulting vectors were transformed into Agrobacterium tumefaciens using freeze-thaw transformation (Chen et al., 1994). Transformation of Arabidopsis wild-type Columbia plants or dia-1/DIA plants was performed using the floral dip method as described by Clough and Bent (1998).
GUS activity was analyzed by staining various tissues overnight at 37°C in staining solution (0.1% Triton X-100, 2 mM Fe2+CN, 2 mM Fe3+CN, and 1 mM 5-bromo-4-chloro-3-indolyl-β-glucuronic acid in 50 mM phosphate buffer, pH 7.0). For ovule staining, pistils were incised on both sides and incubated in staining solution for 2 to 16 h. Ovules were cleared in 20% lactic acid/20% glycerol and observed on a Zeiss Axiovert 135 microscope using differential interference contrast optics.
For sectioning, pistils were fixed in 90% ice-cold acetone, incubated on ice for 20 min, and washed two times in 50 mM phosphate buffer. Pistils were incised on both sides, stained in staining solution for 4 h, and fixed in FAA fixative (50 mL ethanol, 5 mL of acetic acid, 10 mL of 37% formaldehyde, and 35 mL of water) for 2 h. The tissue was dehydrated in an ethanol series (50, 70, 90, 100, and 100%) for 10 min each and slowly infiltrated with preparation solution A (100 mL of Technovit 7100, one package of Hardener I [Kulzer], and 2.5 mL of polyethylene glycol 400) as follows: 1 h at 1:3 solution A:100% ethanol; 1 h at 1:1 solution A:100% ethanol; 1 h at 3:1 solution A:100% ethanol; and then solution A overnight. The pistils were embedded in a mix of 15 mL of solution A and 1 mL of Hardener II (Kulzer). Sections (7 μm) were cut, stained with 0.2% safranine, and observed with light microscopy.
For analysis of GFP expression, pistils were dissected on a microscope slide in 50 mM phosphate buffer (pH 7.0) and observed with CSLM using a Leica TCS-SP5 microscope. GFP was excited with an argon laser (488 nm), and emission was detected between 500 and 530 nm.
For in situ hybridization, pistils from flower stages 12c and 13 were embedded in BMM (40 mL of butyl methacrylate, 10 mL of methyl methacrylate, and 0.5% [w/v] benzoinethylether). The tissue was fixed in 10% paraformaldehyde and 0.25% glutaraldehyde in 10 mM phosphate buffer/0.1 M NaCl, washed in phosphate buffer, and dehydrated in an ethanol series. Infiltration with BMM was performed as follows: 2 h at 3:1 ethanol:BMM; 2 h at 1:1 ethanol:BMM; 2 h at 1:3 ethanol:BMM; and then 100% BMM overnight, all at 4°C. After incubation in fresh BMM (2 h), the material was poured in gelatin capsules, overfilled with BMM, and closed with a cap. Polymerization was induced for 24 h at −20°C in UV light. Sections (3 μm) were processed and hybridized with a DIA antisense or sense RNA probe. The 443-nucleotide probe fragment was amplified from genomic DNA using primers QAGL61for (5′-TGAATCTGTATTGGATCGCTACG-3′) and TOPO61rev (5′-AAAGCATTATTATGAATCAGAAACA-3′) and cloned into pGEM-T Easy (Promega). The resulting construct was either digested with NcoI (antisense) or with SpeI (sense), and the runoff transcript was produced using the SP6 (antisense) or T7 (sense) RNA polymerases and digoxigenin-labeled UTP (Roche Applied Science).
Hybridization was performed as follows: the BMM was washed off with acetone, and the sections were rehydrated in an ethanol series before proteinase K treatment (30 min, 37°C, 1 mg/μL). After dehydration in an ethanol series, hybridization mix was added to the sections (10 mM Tris, pH 7.5, 300 mM NaCl, 1 mM EDTA, 10% dextran sulfate, 250 ng/mL tRNA, 50% formamide, 1× Denhardt's solution [1× Denhardt's solution is 0.02% Ficoll, 0.02% polyvinylpyrrolidone, and 0.02% BSA], 10 mM DTT, and 50 ng of probe fragment per slide) and incubated overnight at 50°C. The slides were washed in 2× SSC (1× SSC is 150 mM NaCl and 15 mM sodium citrate) (10 min, 50°C), 1× SSC (10 min, 50°C), and 0.5× SSC (20 min, room temperature). After blocking with 1% BSA, a 1:500 dilution of anti-digoxigenin/AP in 1% BSA was pipetted on each slide, which was incubated for 2 h at 37°C. After washing with BSA (once for 20 min) and Tris-buffered saline (10× Tris-buffered saline is 1.5 M NaCl and 1 M Tris, pH 7.5; twice for 15 min), the color–substrate solution (5 μL of 5-bromo-4-chloro-3-indolyl-phosphate, 7.5 μL of nitroblue tetrazolium [Roche Applied Science] in 1.5 mL of 0.1 M Tris, pH 9.0, 0.1 M NaCl, and 0.05 M MgCl2) was applied to the sections, which were incubated in the dark at room temperature until signal appeared.
For quantitative RT-PCR analysis of DIA expression, different tissues were harvested from Columbia wild-type Arabidopsis plants. RNA was extracted using the Qiagen RNeasy Plant mini kit, and cDNA was synthesized with the iScript cDNA synthesis kit (Bio-Rad). Real-time RT-PCR was performed with the iQ SYBR Green Supermix from Bio-Rad using primers QAGL61for (5′-TGAATCTGTATTGGATCGCTACG-3′) and QAGL61rev (5′-CCCTTCTTCTTCTCTTCTTCTACC-3′) for AGL61 and Ath UBCfor (5′-ATGCTTGGAGTCCTGCTTGG-3′) and Ath UBCrev (5′-TGCCATTGAATTGAACCCTCTC-3′) for the reference gene UBIQUITIN-CONJUGATING ENZYME21 (Czechowski et al., 2005). The following PCR program was used: 1 min at 95°C; 40 cycles of 10 s at 95°C and 45 s at 57°C; 1 min at 95°C; and 1 min at 57°C. Two biological and two technical replicates were performed.
To test the reduction in DIA expression in the dia-1 allele, pistils of wild-type and dia-1/DIA plants were emasculated and harvested 48 h later. RNA extraction and quantitative RT-PCR analysis were performed with the same protocol and primers as the DIA expression analysis.
Yeast Two-Hybrid Analysis
The yeast two-hybrid screening was performed as described by De Folter et al. (2005). The original AGL61 (DIA) clone used by De Folter et al. (2005) lacks the first 113 bp of the open reading frame, causing a frameshift at that position. A new full-length clone was used for the construction of the yeast two-hybrid vectors. Bait (pBDGAL4) vectors transformed in yeast strain PJ69-4A and prey (pADGAL4) vectors transformed in strain PJ69-4α were mated on minimal synthetic defined (SD) medium containing all essential amino acids and grown overnight at 30°C. Subsequently, the yeast was transferred to SD plates without Leu (L) and Trp (W) to select for yeast containing both plasmids. After 2 d of growth at 30°C, the yeast was transferred to three different selective media (SD – LW and adenine [A]; SD – LW and His [H], 5 mM 3-amino-1,2,4-triazole; and SD – LWH, 10 mM 3-amino-1,2,4-triazole). These plates were incubated at room temperature and scored for yeast growth after 4 d. For the identified interactions, distinct yeast growth was observed on all media for both orientations (DIA as bait or prey). The AGL61 (DIA) bait used in the analysis of De Folter et al. (2005) contained a frameshift mutation and was used as a negative control in these experiments.
Phenotypic Analysis
Wild-type and dia-1/DIA pistils and siliques were harvested at different time points and fixed for 2 h in 2% glutaraldehyde in 50 mM phosphate buffer (pH 7.2). The tissue was rinsed three times with phosphate buffer and incubated for 60 min in 1% OsO4. The tissue was dehydrated in an ethanol series (10% steps, 30 min each) until 100% ethanol was reached. Subsequently, the tissue was incubated in 100% propylene oxide (30 min, two times), 25% Spurr's resin in propylene oxide (4 h), and 50% Spurr's resin in propylene oxide (overnight). The vials were left opened in the fume hood until the propylene oxide was evaporated and incubated in fresh 100% Spurr's resin for 4 to 8 h. The tissue was embedded in flat embedding molds and polymerized overnight at 60°C. Sections (1 μm) were cut, stained with toluidine blue, and observed with light microscopy.
CSLM analysis of wild-type and dia-1/DIA pistils and siliques was performed as described by Christensen et al. (1997).
Marker Line Analysis
The gametophytic markers used in this study are enhancer detector (ET) and gene trap (GT) lines that were generated using the system of Sundaresan and colleagues (1995). All insertions are in the Landsberg erecta accession and were described by Gross-Hardt and colleagues (2007) with the exception of ET1086, which stains the egg cell and the pollen tube. The different reporter lines were pollinated with dia-1/DIA pollen, and the harvested seeds were sown on soil. DIA/DIA plants were distinguished from dia-1/DIA plants by PCR using primers Lba1, 61for, and 61rev. Pistils were dissected and incubated for 2 to 3 d in GUS staining buffer containing 10 mM EDTA, 0.1% Triton X-100, 0.5 mM Fe2+CN, 0.5 mMFe3+CN, 2 mg/mL 5-bromo-4-chloro-3-indolyl-β-glucuronic acid, and 0.1 mg/mL chloramphenicol in 50 mM phosphate buffer, pH 7.0. Ovules were cleared in 20% lactic acid/20% glycerol and observed on a Zeiss Axiovert 135 microscope using differential interference contrast optics.
The pEXO70:GUS marker line contained a 1.4-kb promoter fragment of the EXO70 gene fused to the GUS reporter gene. The pRPS5A:GUS marker line has been described by Weijers et al. (2001b). For analysis of the EXO70 and RPS5A reporter lines, dia-1/DIA pistils were pollinated with the respective lines and the pistil was harvested at 8 h, 16 h (EXO70), or 20 h (RPS5A) after pollination. Staining was performed from 4 h (in the case of EXO70) to overnight (for RPS5A).
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: DIA (AGL61), At2g24840 (Arabidopsis Genome Initiative) or EU836691 (GenBank); AGL80, At5g48670 or DQ406752; AGL62, At5g60440 or EU493093; PHE1 (AGL37), At1g65330 or AF528580; PHE2 (AGL38), At1g65300 or AY141245; AGL86, At1g31630; EXO70, At2g28640.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. DIA Expression Analysis Using pDIA:GFP-GUS and pDIA:DIA-GFP-GUS Reporter Lines.
Supplemental Figure 2. Yeast Two-Hybrid Interaction Assay.
Supplementary Material
Acknowledgments
We thank the Salk Institute Genomic Analysis Laboratory and the Max Planck Institute for Breeding Research in Cologne for providing the sequence-indexed Arabidopsis T-DNA insertion mutants. We thank Richard Immink for his help with the yeast two-hybrid experiments and Gary Drews, Dolf Weijers, and Shipeng Li for providing the agl80 mutant, the RPS5A marker line, and the EXO70 marker line, respectively. We thank Liesbeth Pierson, John Franken, and Marco Busscher for assistance with the microscopic analysis and Olga Kirioukhova for suggestions regarding GUS staining in female gametophytes.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Gerco C. Angenent (gerco.angenent@wur.nl).
Online version contains Web-only data.
References
- Alonso, J.M., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301 653–657. [DOI] [PubMed] [Google Scholar]
- Alvarez-Buylla, E.R., Pelaz, S., Liljegren, S.J., Gold, S.E., Burgeff, C., Ditta, G.S., de Pouplana, L.R., Martinez-Castilla, L., and Yanofsky, M.F. (2000). An ancestral MADS-box gene duplication occurred before the divergence of plants and animals. Evolution Int. J. Org. Evolution 97 5328–5333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arabidopsis Genome Initiative (2000). Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408 796–815. [DOI] [PubMed] [Google Scholar]
- Becker, A., and Theissen, G. (2003). The major clades of MADS-box genes and their role in the development and evolution of flowering plants. Mol. Phylogenet. Evol. 29 464–489. [DOI] [PubMed] [Google Scholar]
- Chaudhury, A.M., Ming, L., Miller, C., Craig, S., Dennis, E.S., and Peacock, W.J. (1997). Fertilization-independent seed development in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 94 4223–4228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, H., Nelson, R.S., and Sherwood, J.L. (1994). Enhanced recovery of transformants of Agrobacterium tumefaciens after freeze-thaw transformation and drug selection. Biotechniques 16 664–670. [PubMed] [Google Scholar]
- Christensen, C.A., King, E.J., Jordan, J.R., and Drews, G.N. (1997). Megagametogenesis in Arabidopsis wild type and the Gf mutant. Sex. Plant Reprod. 10 49–64. [Google Scholar]
- Christensen, C.A., Subramanian, S., and Drews, G.N. (1998). Identification of gametophytic mutations affecting female gametophyte development in Arabidopsis. Dev. Biol. 202 136–151. [DOI] [PubMed] [Google Scholar]
- Clough, S.J., and Bent, A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16 735–743. [DOI] [PubMed] [Google Scholar]
- Colombo, M., Masiero, S., Vanzulli, S., Lardelli, P., Kater, M.M., and Colombo, L. (2008). AGL23, a type I MADS-box gene that controls female gametophyte and embryo development in Arabidopsis. Plant J. 54 1037–1048. [DOI] [PubMed] [Google Scholar]
- Czechowski, T., Stitt, M., Altmann, T., Udvardi, M.K., and Scheible, W.-R. (2005). Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol. 139 5–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Bodt, S., Raes, J., Florquin, K., Rombauts, S., Rouzé, P., Theissen, G., and Van de Peer, Y. (2003. a). Genomewide structural annotation and evolutionary analysis of the type I MADS-box genes in plants. J. Mol. Evol. 56 573–586. [DOI] [PubMed] [Google Scholar]
- De Bodt, S., Raes, J., Van de Peer, Y., and Theissen, G. (2003. b). And then there were many: MADS goes genomic. Trends Plant Sci. 8 475–483. [DOI] [PubMed] [Google Scholar]
- De Folter, S., Immink, R.G.H., Kiefer, M., Parenicova, L., Henz, S.R., Weigel, D., Busscher, M., Kooiker, M., Colombo, L., Kater, M.M., Davies, B., and Angenent, G.C. (2005). Comprehensive interaction map of the Arabidopsis MADS box transcription factors. Plant Cell 17 1424–1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dresselhaus, T. (2006). Cell-cell communication during double fertilization. Curr. Opin. Plant Biol. 9 41–47. [DOI] [PubMed] [Google Scholar]
- Ferrario, S., Immink, R.G.H., and Angenent, G.C. (2004). Conservation and diversity in flower land. Curr. Opin. Plant Biol. 7 84–91. [DOI] [PubMed] [Google Scholar]
- Gehring, M., Huh, J.H., Hsieh, T.F., Penterman, J., Choi, Y., Harada, J.J., Goldberg, R.B., and Fischer, R.L. (2006). DEMETER DNA glycosylase establishes MEDEA polycomb gene self-imprinting by allele-specific demethylation. Cell 124 495–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross-Hardt, R., Kägi, C., Baumann, N., Moore, J.M., Baskar, R., Gagliano, W.B., Jürgens, G., and Grossniklaus, U. (2007). LACHESIS restricts gametic cell fate in the female gametophyte of Arabidopsis. PLoS Biol. 5 e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossniklaus, U., Vielle-Calzada, J.P., Hoeppner, M.A., and Gagliano, W.B. (1998). Maternal control of embryogenesis by MEDEA, a Polycomb group gene in Arabidopsis. Science 280 446–450. [DOI] [PubMed] [Google Scholar]
- Howden, R., Park, S.K., Moore, J.M., Orme, J., Grossniklaus, U., and Twell, D. (1998). Selection of T-DNA-tagged male and female gametophytic mutants by segregation distortion in Arabidopsis. Genetics 149 621–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huck, N., Moore, J., Federer, M., and Grossniklaus, U. (2003). The Arabidopsis mutant feronia disrupts the female gametophytic control of pollen tube reception. Development 130 2149–2159. [DOI] [PubMed] [Google Scholar]
- Immink, R.G.H., Gadella, T.W.J., Ferrario, S., Buscher, M., and Angenent, G.C. (2002). Analysis of MADS box protein-protein interactions in living plant cells. Proc. Natl. Acad. Sci. USA 99 2416–2421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston, A.J., Meier, P., Gheyselinck, J., Wuest, S.E., Federer, M., Schlagenhauf, E., Becker, J.D., and Grossniklaus, U. (2007). Genetic subtraction profiling identifies genes essential for Arabidopsis reproduction and reveals interaction between the female gametophyte and the maternal sporophyte. Genome Biol. 8 R204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang, I.-H., Steffen, J.G., Portereiko, M.F., Lloyd, A., and Drews, G.N. (2008). The AGL62 MADS domain protein regulates cellularization during endosperm development in Arabidopsis. Plant Cell 20 635–647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi, M., Inzé, D., and Depicker, A. (2002). Gateway vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci. 7 193–195. [DOI] [PubMed] [Google Scholar]
- Kasahara, R.D., Portereiko, M.F., Sandaklie-Nikolova, L., Rabiger, D.S., and Drews, G.N. (2005). MYB98 is required for pollen tube guidance and synergid cell differentiation in Arabidopsis. Plant Cell 17 2981–2992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kofuji, R., Sumikawa, N., Yamasaki, M., Kondo, K., Ueda, K., Ito, M., and Hasebe, M. (2003). Evolution and divergence of MADS-box gene family based on genome-wide expression analysis. Mol. Biol. Evol. 20: 1963– 1977. [DOI] [PubMed] [Google Scholar]
- Köhler, C., Hennig, L., Bouveret, R., Gheyselinck, J., Grossniklaus, U., and Gruissem, W. (2003. a). Arabidopsis MSI1 is a component of the MEA/FIE Polycomb group complex and required for seed development. EMBO J. 22 4804–4814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köhler, C., Hennig, L., Spillane, C., Pien, S., Gruissem, W., and Grossniklaus, U. (2003. b). The Polycomb-group protein MEDEA regulates seed development by controlling expression of the MADS-box gene PHERES1. Genes Dev. 17 1540–1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köhler, C., Page, D., Gagliardini, V., and Grossniklaus, U. (2005). The Arabidopsis thaliana MEDEA Polycomb group protein controls expression of PHERES1 by parental imprinting. Nat. Genet. 37 28–30. [DOI] [PubMed] [Google Scholar]
- Kwee, H., and Sundaresan, V. (2003). The NOMEGA gene required for female gametophyte development encodes the putative APC6/CDC16 component of the anaphase promoting complex in Arabidopsis. Plant J. 36 853–866. [DOI] [PubMed] [Google Scholar]
- Moore, J.M., Calzada, J.P., Gagliano, W.B., and Grossniklaus, U. (1997). Genetic characterization of hadad, a mutant disrupting female gametogenesis in Arabidopsis thaliana. Cold Spring Harb. Symp. Quant. Biol. 62 35–47. [PubMed] [Google Scholar]
- Nam, J., Kim, J., Lee, S., An, G., Ma, H., and Nei, M. (2004). Type I MADS-box genes have experienced faster birth-and-death evolution than type II MADS-box genes in angiosperms. Proc. Natl. Acad. Sci. USA 101 1910–1915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohad, N., Yadegari, R., Margossian, L., Hannon, M., Michaeli, D., Harada, J.J., Goldberg, R.B., and Fischer, R.L. (1996). Mutations in FIE, a WD Polycomb group gene, allow endosperm development without fertilization. Plant Cell 11 407–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagnussat, G., Yu, H., and Sundaresan, V. (2007). Cell-fate switch of synergid to egg cell in Arabidopsis eostre mutant embryo sacs arises from misexpression of the BEL1-like homeodomain gene BLH1. Plant Cell 19 3578–3592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagnussat, G.C., Yu, H.-J., Ngo, Q.A., Rajani, S., Mayalagu, S., Johnson, C.S., Capron, A., Xie, L.-F., Ye, D., and Sundaresan, V. (2005). Genetic and molecular identification of genes required for female gametophyte development and function in Arabidopsis. Development 132 603–614. [DOI] [PubMed] [Google Scholar]
- Pařenicová, L., de Folter, S., Kieffer, M., Horner, D.S., Favalli, C., Busscher, J., Cook, H.E., Ingram, R.M., Kater, M.M., Davies, B., Angenent, G.C., and Colombo, L. (2003). Molecular and phylogenetic analyses of the complete MADS-box transcription factor family in Arabidopsis: New openings to the MADS world. Plant Cell 15 1538–1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pischke, M.S., Jones, L.G., Otsuga, D., Fernandez, D.E., Drews, G.N., and Sussman, M.R. (2002). An Arabidopsis histidine kinase is essential for megagametogenesis. Proc. Natl. Acad. Sci. USA 99 15800–15805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portereiko, M.F., Lloyd, A., Steffen, J.G., Punwani, J.A., Otsuga, D., and Drews, G.N. (2006). AGL80 is required for central cell and endosperm development in Arabidopsis. Plant Cell 18 1862–1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosso, M.G., Li, Y., Strizhov, N., Reiss, B., Dekker, K., and Weisshaar, B. (2003). An Arabidopsis thaliana T-DNA mutagenized population (GABI-Kat) for flanking sequence tag-based reverse genetics. Plant Mol. Biol. 53 247–259. [DOI] [PubMed] [Google Scholar]
- Rotman, N., Rozier, F., Boavida, L., Dumas, C., Berger, F., and Faure, J.E. (2003). Female control of male gamete delivery during fertilization in Arabidopsis thaliana. Curr. Biol. 13 432–436. [DOI] [PubMed] [Google Scholar]
- Sandaklie-Nikolova, L., Palanivelu, R., King, E.J., Copenhaver, G.P., and Drews, G.N. (2007). Synergid cell death in Arabidopsis is triggered following direct interaction with the pollen tube. Plant Physiol. 144 1753–1762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneitz, K., Hulskamp, M., and Pruitt, R.E. (1995). Wild-type ovule development in Arabidopsis thaliana: A light microscope study of cleared whole-mount tissue. Plant J. 7 731–749. [Google Scholar]
- Smyth, D.R., Bowman, J.L., and Meyerowitz, E.M. (1990). Early flower development in Arabidopsis. Plant Cell 2 755–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Springer, P.S., Holding, D.R., Groover, A., Yordan, C., and Martienssen, R.A. (2000). The essential Mcm7 protein PROLIFERA is localized to the nucleus of dividing cells during the G(1) phase and is required maternally for early Arabidopsis development. Development 127 1815–1822. [DOI] [PubMed] [Google Scholar]
- Steffen, J.G., Kang, I.-H., Macfarlane, J., and Drews, G.N. (2007). Identification of genes expressed in the Arabidopsis female gametophyte. Plant J. 51 281–292. [DOI] [PubMed] [Google Scholar]
- Sundaresan, V., Springer, P., Volpe, T., Haward, S., Jones, J.D.G., Dean, C., Ma, H., and Martienssen, R. (1995). Patterns of gene action in plant development revealed by enhancer trap and gene trap transposable elements. Genes Dev. 9 1797–1810. [DOI] [PubMed] [Google Scholar]
- Weijers, D., Franke-van Dijk, M., Vencken, R.-J., Quint, A., Hooykaas, P., and Offringa, R. (2001. b). An Arabidopsis Minute-like phenotype caused by a semi-dominant mutation in a RIBOBOSOMAL PROTEIN S5 gene. Development 128 4289–4299. [DOI] [PubMed] [Google Scholar]
- Weijers, D., Geldner, N., Offringa, R., and Jürgens, G. (2001. a). Seed development: Early paternal gene activity in Arabidopsis. Nature 414 709–710. [DOI] [PubMed] [Google Scholar]
- Yu, H., Hogan, P., and Sundaresan, V. (2005). Analysis of the female gametophyte transcriptome of Arabidopsis by comparative expression profiling. Plant Physiol. 139 1853–1869. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.