Abstract
Apical meristems play a central role in plant development. Self-renewing cells in the central region of the shoot meristem replenish the cell population in the peripheral region, where organ primordia emerge in a predictable pattern, and in the underlying rib meristem, where new stem tissue is formed. While much is known about how organ primordia are initiated and their lateral boundaries established, development at the interface between the stem and the meristem or the lateral organs is poorly understood. Here, we show that the BELL-type ARABIDOPSIS THALIANA HOMEOBOX GENE1 (ATH1) is required for proper development of the boundary between the stem and both vegetative and reproductive organs and that this role partially overlaps with that of CUP-SHAPED COTYLEDON genes. During the vegetative phase, ATH1 also functions redundantly with light-activated genes to inhibit growth of the region below the shoot meristem. Consistent with a role in inhibiting stem growth, ATH1 is downregulated at the start of inflorescence development and ectopic ATH1 expression prevents growth of the inflorescence stem by reducing cell proliferation. Thus, ATH1 modulates growth at the interface between the stem, meristem, and organ primordia and contributes to the compressed vegetative habit of Arabidopsis thaliana.
INTRODUCTION
Much of plant architecture is established in the apical meristems. In the shoot meristem, stem cells in the central region replenish the cell population in the peripheral region, where organ primordia emerge reiteratively (Fletcher, 2002). Below the central and peripheral regions, the rib meristem generates the pith of the stem and petioles (Vaughan, 1955). The pattern of organ initiation in the peripheral region is controlled by auxin transport (Fleming, 2005), and subsequent internode growth completes the process of establishing the arrangement of organs around the stem (Peaucelle et al., 2007). During the last several decades, cellular and genetic analysis of meristem function has focused on the central and peripheral regions, on organ initiation, and on the establishment of the lateral boundaries of primordia (Tooke and Battey, 2003). By contrast, events at the interface between the stem and the meristem or primordia have remained largely uncharacterized.
In Arabidopsis thaliana, the homeodomain protein SHOOTMERISTEMLESS (STM) is required for multiple functions of the shoot meristem. In combination with WUSCHEL, STM is required to maintain the stem cell population in the central zone (Gallois et al., 2002; Lenhard et al., 2002). In the periphery of the meristem, STM delays differentiation and antagonizes primordium development. STM also functions together with CUP-SHAPED COTYLEDON (CUC) genes, which repress growth locally to establish organ boundaries (Aida et al., 1999). BREVIPEDICELLUS (BP) encodes a close homolog of STM that has partially redundant function in meristem maintenance in addition to having a more specialized role in stem development (Byrne et al., 2002; Douglas et al., 2002; Venglat et al., 2002). Exactly how KNOTTED-LIKE HOMEOBOX (KNOX) proteins such as STM and BP control the behavior of meristem cells is still unclear, but one of the mechanisms is the localized control of phytohormone levels. STM activates the biosynthesis of cytokinin, which maintains cell division in the meristem (Jasinski et al., 2005; Yanai et al., 2005). In addition, STM represses the biosynthesis and activates the catabolism of gibberellin, which would otherwise antagonize meristem functions (Hay et al., 2002; Jasinski et al., 2005).
The multiple roles of KNOX proteins in the meristem raise the question of whether these proteins function together with localized factors to control specific aspects of meristem function. Candidate cofactors are homeodomain proteins of the BELL family, which form heterodimers with STM/BP and have been proposed to control distinct aspects of meristem function (Bellaoui et al., 2001; Muller et al., 2001; Smith et al., 2002; Byrne et al., 2003; Smith and Hake, 2003; Bhatt et al., 2004; Cole et al., 2006; Kanrar et al., 2006). The only example that has been functionally characterized, however, is the BELL-type protein most often referred to as BELLRINGER (BLR) (Byrne et al., 2003) but also known as PENNYWISE (Smith and Hake, 2003), REPLUMLESS (Roeder et al., 2003), VAAMANA (Bhatt et al., 2004), LARSON (Bao et al., 2004), and BLH9 (Cole et al., 2006). BLR is required for correct phyllotaxis and interacts with BP to promote inflorescence stem development. The roles of other KNOX-interacting BELL proteins in meristem development remain unknown.
ARABIDOPSIS THALIANA HOMEOBOX GENE1 (ATH1) encodes a BELL-type homeodomain protein that was initially described as a light-regulated transcription factor (Quaedvlieg et al., 1995) and more recently as an activator of the flowering repressor FLOWERING LOCUS C (FLC) (Proveniers et al., 2007). The ATH1 protein has been reported to form heterodimers with STM and BP and to interact synergistically with ectopically expressed STM, suggesting that ATH1 functions as a partner of STM and BP in the meristem (Cole et al., 2006). Here, we investigate the roles of ATH1 in the shoot apex. We show that ATH1 is required for development of the boundaries between shoot organs and the stem and that this function partially overlaps with that of CUC genes. In addition, ATH1 functions redundantly with light-activated genes to prevent stem growth during the vegetative phase. At the transition to reproductive development, when stem growth is activated, ATH1 is downregulated; conversely, constitutive ATH1 inhibits growth of the inflorescence stem by inhibiting cell proliferation. We conclude that ATH1 represses growth at the interface between the meristem, the stem, and lateral organs to establish the basal boundaries of shoot organs and the compressed rosette habit of Arabidopsis.
RESULTS
ATH1 Is Required for Development of the Basal Region of All Shoot Organs
To reveal possible roles of ATH1 in the shoot meristem, we analyzed in detail the development of homozygous ath1 mutant plants. Both ath1-1 and ath1-3 have been described as likely null alleles (Proveniers et al., 2007). While our analysis was focused on ath1-3, the same defects were seen with ath1-1 (see Supplemental Figure 1 online). To confirm that all phenotypes described below were caused by ath1-3, we complemented the mutant by transformation with a wild-type genomic DNA fragment (see Supplemental Figure 2 online).
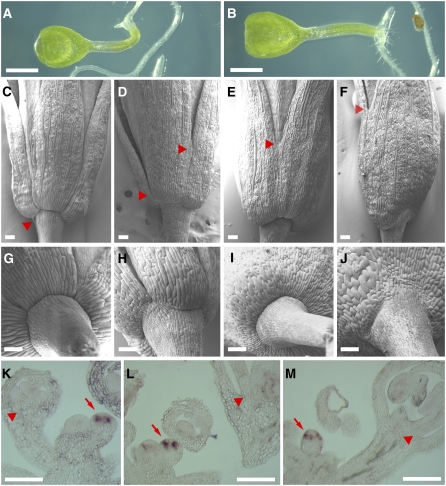
The most readily visible defect of ath1-3 plants was that the stamens were not shed after fertilization (Figures 1A and 1B). In accordance with this phenotype, sections through the base of mature stamens showed that the small cells that characterize the abscission zone were absent in ath1-3 (Figures 1C and 1D); sections through developing buds revealed that this defect became visible relatively late in development, at floral stages 10 and 11 (see Supplemental Figure 3 online). Closer examination, however, revealed that the defective abscission zone in ath1-3 stamens was only one aspect of a general defect in the basal region of the flowers. The mutant showed partial fusion at the base of stamens (Figures 1E and 1F), which was also visible at stages 10 and 11 (Figures 1G and 1H). Scanning electron microscopy revealed that the sepals were similarly fused at the base and that development of their basal boundary was affected: from stages 7 to 11, the constriction that separates sepals from the floral pedicel in the wild type was much less pronounced inthe mutant (Figures 1I and 1J). In sections, defects in this boundary region were first visible at stages 8 and 9 (see Supplemental Figures 3A and 3B online). As the flowers matured, however, the basal constriction and a functional abscission zone eventually developed, and sepals were shed after fertilization (Figures 2D and 2H; see Supplemental Figures 2A and 2B online).
Figure 1.
ATH1 Is Required to Form the Basal Boundaries of Shoot Organs.
(A) and (B) Inflorescences of wild-type (A) and ath1-3 (B) plants, showing that stamens remain attached to the developing fruits of ath1-3 (arrows).
(C) and (D) Sections through the base of flowers, with arrows indicating the dehiscence zone at the base of a wild-type stamen (C) and the corresponding region in ath1-3 (D).
(E) and (F) Closeups of the base of mature stamens in the wild type (E) and in ath1-3, which has partially fused stamens in this region (F).
(G) and (H) Scanning electron micrographs of stage 11 wild-type (G) and ath1-3 (H) flowers; the arrow shows the partial fusion at the base of stamens in ath1-3.
(I) and (J) Scanning electron micrographs of wild-type (I) and ath1-3 (J) floral buds, with arrowheads indicating the boundary between sepals and pedicel.
(K) and (L) Whole-mount staining of BP:GUS in wild-type (K) and ath1-3 (L) backgrounds.
(M) and (N) Scanning electron micrographs of the base of cauline leaves of the wild type (M) and ath1-3 (N); arrows indicate the boundary between the leaf and inflorescence stem.
(O) and (P) Base of the rosette of 2-week-old wild-type (O) and ath1-3 (P) plants; h and c mark the hypocotyls and the petioles of cotyledons, respectively, and the arrowheads show the boundary at the base of leaf petioles.
Bars = 1 cm in (A) and (B), 1 mm in (K), (L), (O), and (P), and 100 μm in (C) to (J), (M), and (N).
Figure 2.
Interaction between ath1-3 and cuc Genes.
(A) and (B) One-week-old homozygous cuc1 cuc2 (A) and ath1-3 cuc1 cuc2 (B) seedlings.
(C) to (F) Scanning electron micrographs of mature (stage 13) flowers of the wild type (C), ath1-3 (D), cuc2/cuc2 cuc1/+ (E), and ath1-3/ath1-3 cuc2/cuc2 cuc1/+ (F); the arrowheads show the point where sepals become separate.
(G) to (J) Scanning electron micrographs of the base of flowers comparable to (C) to (F), respectively; note that the partial loss of the basal boundaries in (H) is enhanced in (J).
(K) to (M) RNA in situ hybridization showing expression of CUC1 at the developing organ boundaries of a stage 3 bud (arrows) but absent from the basal region of organs at later stages (arrowheads) in the wild type (K), ath1-3 (L), and 35S:ATH1 (M).
Bars = 1 mm in (A) and (B) and 100 μm in (C) to (M).
To determine whether the morphological changes described above reflected changes in cell identity, we used BP:GUS (for β-glucuronidase) as a marker gene that is strongly expressed in the basal region of developing buds (Ori et al., 2000) (Figure 1K). In ath1-3, expression of BP:GUS was reduced at the base of early buds and absent from mature flowers, although it was comparable to the wild type in pedicels and in the inflorescence stem (Figure 1L). Sections through the GUS-stained inflorescences showed that the ath1-3 mutation caused a decrease in BP:GUS expression in the inflorescence meristem, without diminishing expression in the subtending stem (see Supplemental Figures 4A and 4B online). At the base of developing buds, BP:GUS expression was visibly decreased relative to the wild type at stages 6 and 7 (see Supplemental Figures 4C and 4D online) and abolished at later stages (see Supplemental Figures 4E and 4F online). Thus, changes in BP:GUS expression preceded the morphological changes at the base of ath1-3 floral organs, revealing that ATH1 functions at the base of floral organs earlier than suggested by the histological analysis. However, it must be noted that loss of BP alone cannot be the cause of the morphological defects of ath1-3, because these defects are not seen in bp mutants (Douglas et al., 2002; Venglat et al., 2002).
Considering that ath1-3 had defects at the base of sepals that were not obvious macroscopically, we next searched more thoroughly for defects at the base of other shoot organs. At the boundary between cauline leaves and the inflorescence stem, the wild type develops a groove with small cells that has been interpreted as a vestigial abscission zone (Stenvik et al., 2006). This was absent in the mutant, in which epidermal cells formed continuous files across the leaf–stem junction (Figures 1M and 1N). The constricted boundary at the base of rosette leaves was also less pronounced in the mutant than in the wild type (Figures 1O and 1P). Therefore, ATH1 has a general role in the development of the basal region of shoot organs, both in the vegetative and reproductive phases.
Since the partial fusion at the base of floral organs raised the possibility that ATH1 might function in the organ boundary pathway controlled by CUC genes (Aida and Tasaka, 2006), we tested for genetic interaction between ath1-3 and cuc mutants. Combined loss of CUC1 and CUC2 function was epistatic over ath1-3: the triple mutant seedling looked similar to the cuc1-5 cuc2-1 double mutant, with fused cotyledons and loss of the shoot meristem (Figures 2A and 2B). In a mutant background with partial loss of cuc function, however, we saw that the ath1-3 and cuc mutations enhanced each other's phenotype. As described before, the sepals of cuc2/cuc2 cuc1/+ flowers were partially fused (Aida et al., 1997) but had a normal basal boundary (Figures 2E and 2I). In this background, loss of ATH1 function (ath1-3/ath1-3 cuc2-1/cuc2-1 cuc1-5/+) caused a more severe sepal fusion and a basal boundary defect that was more pronounced than in the single ath1-3 mutant (cf. Figures 2F and 2J with Figures 2D and 2H). Control crosses for the mixed Columbia/Landsberg erecta (Col/Ler) background did not show enhancement of cuc1, cuc2, or ath1-3 phenotypes (this control was necessary because the cuc2-1 mutation was originally in a Ler background, whereas cuc1-5 and ath1-3 were in Col). Thus, both ATH1 and CUC genes contribute to the establishment of lateral and basal organ boundaries, even though the single mutant showed that ATH1 function is more important for the development of the basal than the lateral boundaries.
Because loss of ATH1 function enhanced the floral phenotype of cuc2/cuc2 cuc1-5/+, we tested whether ATH1 might be required for normal CUC1 expression. As reported (Takada et al., 2001), in situ hybridization showed clear CUC1 expression at the organ boundaries of early floral buds in the wild type (Figure 2K). This expression pattern was not changed in ath1-3 or in plants overexpressing ATH1 (35S:ATH1; described below) (Figures 2L and 2M). Furthermore, although strong expression was detected at the organ boundaries in early buds (up to stage 5), it was not detectable above background at the basal boundaries of organs at later stages, when the development of ath1-3 diverged from the wild type. We conclude that ATH1 converges on the regulation of basal organ boundaries downstream of CUC expression.
The Expression of ATH1 Is Consistent with the Mutant Phenotypes
We next tested whether the expression pattern of ATH1 is consistent with a role in the development of the basal region of shoot organs. Previous studies of the developmental regulation of ATH1 have shown expression in the shoot apical meristem and young leaves (Proveniers et al., 2007), in the inflorescence and floral meristems (Cole et al., 2006), and in developing stamens (Gomez-Mena et al., 2005). These partial analyses of ATH1 expression were not sufficient to verify correlation with the phenotypes described above, so we examined ATH1 expression in more detail by in situ hybridization. In addition, to account for the possibility that ATH1 accumulation might be regulated at the transcriptional and posttranscriptional levels, we used a translational fusion between ATH1 and the GUS reporter (ATH1:ATH1-GUS; containing the genomic ATH1 sequence starting 1.2 kb upstream of the start codon, with GUS fused in-frame at the end of the ATH1 coding sequence).
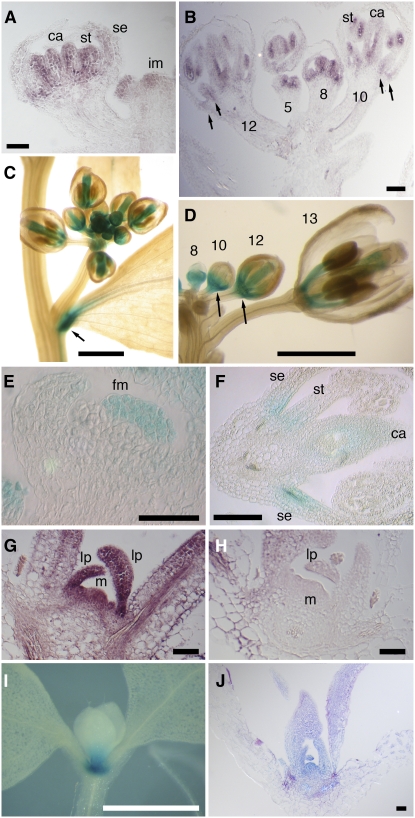
In situ hybridization confirmed that ATH1 was weakly expressed in the inflorescence and floral meristems and more strongly in the developing stamens and carpels of buds at stages 5 to 8 (Smyth et al., 1990) (Figures 3A and 3B). From stages 8 to 12, ATH1 continued to be expressed in developing stamens and carpels and in addition was expressed in sepals and petals, initially throughout the organs, but progressively restricted to the basal region (Figures 3A and 3B). A comparable expression pattern was seen in ATH1:ATH1-GUS plants: whole-mount staining (Figure 3C) showed expression in young buds, in developing stamens and carpels, and at the base of cauline leaves (matching the phenotype shown in Figure 1N). Closer examination of cleared buds revealed expression at the base of developing flowers (Figure 3D), while sections through the GUS-stained inflorescences confirmed expression in the floral meristem in early buds (Figure 3E) and at the bases of sepals and stamens at later stages, although the GUS reporter did not show the strong expression in developing pollen seen by in situ hybridization (cf. Figure 3F with Figure 3B). In the vegetative phase, ATH1 was expressed throughout the shoot apex, including the meristem, leaf primordia, and the base of developing leaves (Figures 3G and 3H). Whole-mount staining of ATH1:ATH1-GUS plants confirmed expression in the shoot apex (Figure 3I), consistent with publicly available expression data (Genevestigator; https://www.genevestigator.ethz.ch/). Within the shoot apex, the ATH1:ATH1-GUS expression pattern was the same as that seen by in situ hybridization, with strong expression throughout the meristem and leaf primordia and weaker expression at the base of young leaves (cf. Figure 3J with Figure 3G).
Figure 3.
ATH1 Expression Pattern.
(A) and (B) In situ hybridization showing the expression pattern of ATH1 in the inflorescence tip. (A) shows a section through a stage 6 floral bud, showing ATH1 expression in developing stamens (st), carpels (ca), and the base of sepals (se); im indicates the inflorescence meristem. (B) shows a section through buds at different stages (numbers marked on the floral pedicels); arrows indicate expression at the base of sepals and stamens at stages 10 to 12.
(C) and (D) Whole-mount staining of ATH1:ATH1-GUS inflorescences. (C) shows a lateral inflorescence showing expression in young buds, in the carpels and stamens of older buds, and at the base of a cauline leaf (arrow). (D) shows an inflorescence with most buds removed, showing expression at different stages (numbered); arrows indicate expression at the base of stage 10 to 12 buds.
(E) and (F) Sections through ATH1:ATH1-GUS at an early stage ([E]; stage 3), showing expression in the floral meristem (fm), and at a late stage ([F]; stage 10 to 11), showing expression in carpels (ca) and at the bases of sepals (se) and stamens (st).
(G) and (H) Sections through seedling apices, hybridized with ATH1 antisense probe (G) and sense control (H); m marks the shoot apical meristem, and lp indicates leaf primordia.
(I) and (J) Staining of ATH1:ATH1-GUS seedlings, showing expression in the shoot apex and at the base of developing leaves. (I) shows whole-mount staining. (J) shows a section through a seedling comparable to that in (I), showing GUS expression throughout the meristem and leaf primordia.
Bars = 50 μm in (A), (E), (G), (H), and (J), 100 μm in (B) and (F), or 1 mm in (C), (D), and (I).
In summary, the expression pattern of ATH1 was consistent with the ath1-3 phenotype, although it was not restricted to the basal organ boundaries.
ATH1 Functions Redundantly with Light-Activated Genes to Restrict Growth of the Region below the Shoot Meristem
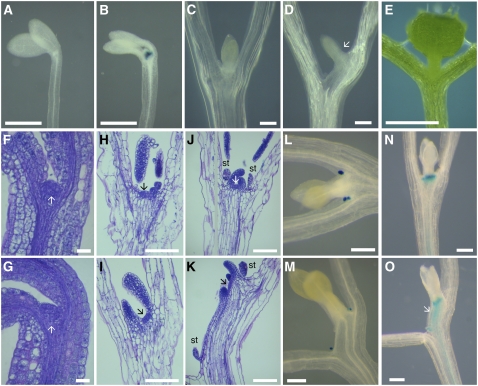
In addition to the developmental regulation described above, ATH1 has been reported to be light-regulated (Genevestigator; https://www.genevestigator.ethz.ch/) (Quaedvlieg et al., 1995). For this reason, we asked whether ATH1 might mediate the input of light signals into meristem development. The shoot meristem does not function in dark-grown plants, but the light requirement can be bypassed if the seedlings are grown in medium containing sugar (Roldan et al., 1999). In agreement with reports that ATH1 is not expressed in the dark (Quaedvlieg et al., 1995), we saw that ATH1:ATH1-GUS expression was absent in seedlings grown in the dark without sucrose (Figure 4A). However, ATH1:ATH1-GUS was strongly expressed in dark-grown seedlings when meristem development was activated by sucrose (Figure 4B). Thus, the apparent activation of ATH1 by light may be an indirect consequence of the fact that meristem development is normally light-dependent.
Figure 4.
ATH1 Functions Redundantly with Light-Activated Genes to Inhibit Growth of the Subapical Region of the Shoot.
(A) and (B) Expression of ATH1:ATH1-GUS in the shoot apex of seedlings grown in the dark for 1 week without sucrose (A) or with 1% sucrose (B).
(C) and (D) Shoot apices of wild-type (C) and ath1-3 (D) seedlings after 2 weeks of growth in the dark with 1% sucrose; the arrow in (D) indicates the displaced meristem and leaf primordia in ath1-3.
(E) A 2-week-old ath1-3 seedling grown in standard light conditions; note that the leaf primordia emerged from the normal position at the junction of the cotyledon petioles.
(F) to (K) Sections through the shoot apex of wild-type ([F], [H], and [J]) or ath1-3 ([G], [I], and [K]) seedlings grown in the dark at different times after germination on medium with 1% sucrose ([F] and [G], 0 weeks; [H] and [I], 1 week; [J] and [K], 2 weeks); arrows indicate the shoot meristem, and st indicates stipules.
(L) and (M) Expression of the stipule marker PFCA:GUS (Laurie, 2003) in wild-type (L) and ath1-3 (M) seedlings after 2 weeks of growth in the dark with 1% sucrose.
(N) and (O) BP:GUS expression in wild-type (N) and ath1-3 (O) seedlings grown for 2 weeks in the dark on medium with 1% sucrose; the arrow in (O) indicates the extended BP:GUS-expressing region below the meristem of ath1-3.
Bars = 0.5 mm in (A), (B), and (E), 20 μm in (F) and (G), and 100 μm in (C), (D), and (H) to (O).
Consistent with the expression of ATH1:ATH1-GUS in dark-grown, sucrose-treated seedlings, ATH1 was required for proper development of the shoot apex under these conditions. While 98% (n = 55) of wild-type seedlings had the meristem and primordia located in the normal position at the junction between the cotyledon petioles (Figure 4C), 88% (n = 81) of ath1-3 seedlings had the meristem and leaf primordia displaced toward one of the cotyledon petioles (Figure 4D). This displacement of the shoot apex was not seen in light-grown ath1-3 seedlings (Figure 4E), revealing an aspect of ATH1 function that can be covered by other, light-activated genes.
The normal position of the shoot apex in the light-grown ath1-3 seedlings also suggested that the meristem had been positioned normally during embryogenesis and that the displacement of the apex in the dark must have resulted from abnormal growth after germination. This was confirmed by sections through seedlings at different stages of growth in the dark: during germination, the meristem was in the same position in ath1-3 and in the wild type (Figures 4F and 4G), but displacement of the shoot apex was visible 1 week later (Figures 4H and 4I). In 2-week-old mutant seedlings, the extended region below the meristem was composed of two distinct parts: files of large cells that were continuous with the cotyledon petiole, and files of smaller cells converging on the meristem and leaf primordia (Figure 4K). The files of small cells leading to the leaf primordium were present between a displaced stipule and the leaf primordium of dark-grown ath1-3 (Figures 4K and 4M); by contrast, the wild-type seedlings always had stipules in the vicinity of leaf primordia (Figures 4J and 4L). This suggested that additional cell divisions had occurred in the dark-grown ath1-3 mutant between each stipule and its adjacent leaf primordium, in the region that normally gives rise to the leaf petiole (Bell, 1998). Other cell files underlying the displaced apex in ath1-3 clearly converged at the base of the meristem, as expected for the inner stem tissues (Figure 4K). Unlike the cells of cotyledon petioles, the region below the displaced apex expressed BP:GUS, which as described above is a marker for the base of the meristem and the stem (Figures 4N and 4O) (it was also noticeable that, as in flowers, BP:GUS expression was lower in the seedling apex). We conclude that in ath1-3 seedlings grown in the dark, the shoot meristem was displaced by growth of leaf petiole and stem tissues that remained fused with one of the cotyledon petioles.
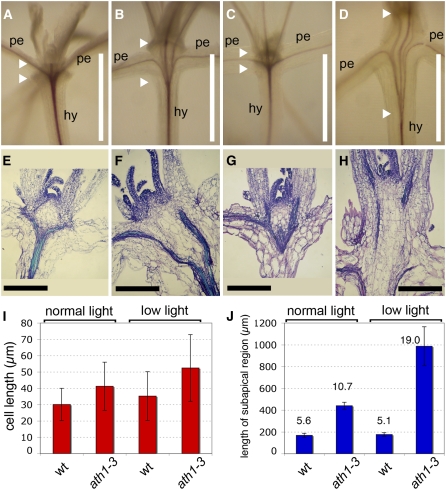
The results above showed that ATH1 and light-dependent genes can function redundantly to regulate growth of the region below the shoot meristem. To confirm this under conditions more similar to natural growth, we compared mutant and wild-type plants grown under low light intensity (15 μmol·m−2·s−1 white light, instead of the standard 100 μmol·m−2·s−1). We found that in low light, the subapical region of the shoot (defined as the region between the base of the meristem and the point where vascular strands converge at the top of the hypocotyl) was enlarged in ath1-3 seedlings (Figures 5C and 5D). Although the enlargement of the subapical region was much more obvious in low light, it was also detectable in the mutant grown in standard light (Figures 5A and 5B). Sections through the shoot apex showed that this enlargement was caused primarily by an increase in cell number, with only a minor contribution of cell expansion (Figures 5E to 5J). Based on the vascular pattern (Figures 5A to 5D), at least part of the enlarged subapical region in ath1-3 plants grown in low light likely corresponded to fused leaf petioles. However, some cell files in the enlarged subapical region converged at the rib zone of the meristem (Figure 5H), as expected for stem tissue (in fact, simple fusion of petioles without additional growth below the meristem would be expected to result in a meristem surrounded by fused petiole tissues, which is not the case in ath1-3).
Figure 5.
ATH1 Inhibits Cell Proliferation in the Subapical Region of the Shoot.
(A) to (D) Whole-mount staining of vascular strands of wild-type ([A] and [C]) or ath1-3 ([B] and [D]) seedlings grown in standard light ([A] and [B]) or in low light ([C] and [D]); the arrowheads mark the distance between the base of the shoot meristem and the point where the vascular strands converge at the top of the hypocotyls. hy, hypocotyl; pe, petiole.
(E) to (H) Sections through the shoot apices of seedlings comparable to those shown in (A) to (D), respectively.
(I) Cell length in the region indicated between arrowheads in (A) to (D), measured in sections comparable to those shown in (E) to (H). Bars represent averages ± sd (n = 16 to 18 for each treatment).
(J) Total length of the region between arrowheads in (A) to (D), measured in sections equivalent to those in (E) to (H). Bars represent averages ± sd (n = 4 for each treatment). Numbers over each bar are ratios between total length and average cell length.
Bars = 1 mm in (A) to (D) and 200 μm in (E) to (H).
In summary, ATH1 limits growth of the subapical region of the shoot by preventing the inclusion of petiole cells into this region and by restricting growth of the region below the meristem. This function of ATH1 is not light-dependent but is partially redundant with a light-dependent pathway.
ATH1 Is Downregulated at the Start of Inflorescence Development, and Constitutive ATH1 Inhibits Cell Proliferation in the Inflorescence Stem
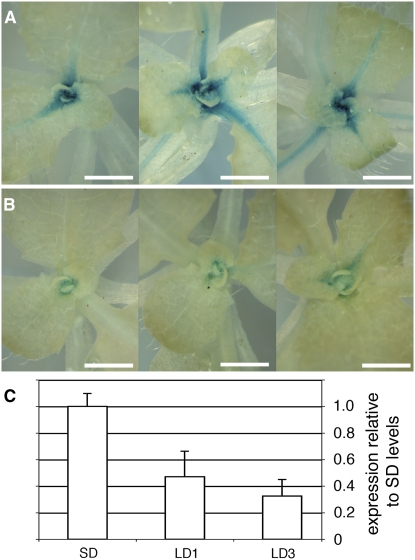
During the transition to reproductive development, subapical growth is activated to produce the inflorescence stem. Given the evidence that ATH1 represses stem growth during the vegetative stage, its activity would be expected to change at the transition to flowering. ATH1 has been reported to be downregulated during floral induction, although the actual data were not shown (Proveniers et al., 2007). We found that ATH1:ATH1-GUS was downregulated in the shoot apex when flowering was induced by a shift from short days to long days (Figure 6).
Figure 6.
ATH1 Is Downregulated at the Transition to Bolting.
(A) and (B) Expression of ATH1:ATH1-GUS in the shoot apex of three different plants grown for 33 d in short days (A) or for 30 d in short days followed by 3 d in long days to induce flowering (B). Bars = 1 mm.
(C) Expression of ATH1 measured by quantitative RT-PCR in plants grown for 33 d in short days (SD), 32 d in short days followed by 1 long day (LD1), or 30 short days followed by 3 long days (LD3). The vertical axis shows expression levels relative to the average of the SD treatment; the bars show averages ± sd for three independent RNA extractions (each from three to four whole rosettes).
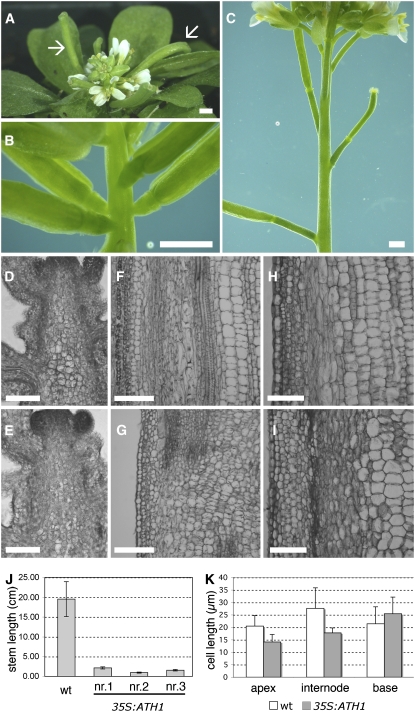
If downregulation of ATH1 has a role in promoting stem growth, then constitutive expression of ATH1 should inhibit the rapid increase in cell division that sustains stem growth during the reproductive phase (Sachs et al., 1959). In fact, expression of ATH1 using the viral 35S promoter has been reported to inhibit growth of the inflorescence stem (Cole et al., 2006). To determine whether this was due to inhibited cell proliferation, we examined 35S:ATH1 plants in more detail. Fifteen 35S:ATH1 lines were generated, which showed very similar phenotypes: vegetative growth appeared normal, flowering occurred at the same time as in the wild-type, but growth of the inflorescence stem and of the floral pedicels was severely inhibited, nearly reducing the inflorescence to a rosette of flowers and siliques (Figure 7A). The reduction in stem length was due primarily to inhibition of internode growth (Figures 7B and 7C). Sections through apical, internode, and basal regions of the inflorescence stem (Figures 7D to 7I) showed that although cell elongation was somewhat inhibited in the internodes of 35S:ATH1 (Figure 7K), this inhibition was far too little to account for the approximately 10-fold reduction in stem length (Figure 7J). Therefore, 35S:ATH1 inhibited stem growth mostly by limiting cell proliferation.
Figure 7.
Ectopic Expression of ATH1 Inhibits Cell Proliferation in the Stem.
(A) Inflorescence of a 35S:ATH1 plant. Note the maturing siliques (arrows) in spite of very little stem elongation.
(B) and (C) Closeups of the inflorescence stem of a 35S:ATH1 plant (B) and a wild-type control (C). Note that the internodes that separate developing siliques are much shorter in 35S:ATH1.
(D) to (I) Sections through the inflorescence apex ([D] and [E]), through the internode between siliques at positions 9 and 10 (Bleecker and Patterson, 1997) ([F] and [G]), or through the base of the inflorescence stem ([H] and [I]) of wild-type ([D], [F], and [H]) and 35S:ATH1 (line nr.3) ([E], [G], and [I]) plants.
(J) Length of the inflorescence stem measured at the same stage of development in the wild type and three independent 35S:ATH1 lines (nr.1, nr.2, and nr.3). The bars show averages ± sd (n = 11 for the wild type, n = 5 for each 35S:ATH1 line).
(K) Length of pith cells near the apical region (100 to 500 μm from the meristem), internode (positions 9 and 10), and basal region of the stem of wild-type or 35S:ATH1 plants. The bars represent averages ± sd (n = 13 to 17).
Bars = 1 mm in (A) to (C) and 100 μm in (D) to (I).
One way that ATH1 might inhibit stem growth would be by antagonizing gibberellin activity, because gibberellin promotes stem growth by stimulating both cell division and cell expansion (Jacobs, 1997). The levels of active gibberellin are controlled by the balance between biosynthesis and inactivation by GIBBERELLIC ACID (GA) 2-oxidases (Olszewski et al., 2002; Fleet and Sun, 2005). To determine if ATH1 regulates genes involved in gibberellin homeostasis, we tested whether the ath1-3 mutant had altered expression levels of GA4, GA5, GA2ox2, and GA2ox4. The levels of GA4 and GA5 were not significantly changed in ath1-3 (see Supplemental Figure 5 online), and external application of gibberellin did not restore growth of the inflorescence stem in 35S:ATH1 plants (see Supplemental Figure 6 online), suggesting that ATH1 does not simply repress gibberellin biosynthesis. Expression of GA2ox2 and GA2ox4 was lower in the mutant (Student's t test, P < 0.05), suggesting that ATH1 might promote gibberellin catabolism (see Supplemental Figure 5 online). If ATH1 functioned as an activator of gibberellin catabolism genes, then 35S:ATH1 would be expected to show high levels of GA2-oxidase expression. Surprisingly, however, 35S:ATH1 plants showed decreased expression of GA2ox2 and GA2ox4 (see Supplemental Figure 5 online). Thus, the expression of ATH1 did not correlate directly with that of GA2ox2 and GA2ox4, and the reduced expression of these genes may have been an indirect consequence of the developmental changes in ath1-3 and in 35S:ATH1. We conclude that ATH1 did not inhibit stem growth by inhibiting the biosynthesis of gibberellin or by activating its catabolism.
DISCUSSION
Our results revealed that ATH1 controls the development of the boundary region between shoot lateral organs and the stem. The defects seen at the boundary between leaves and the stem in ath1-3 are reminiscent of the phenotype caused by the maize (Zea mays) liguleless mutations (lg1 and lg2), which blur the boundary between the leaf blade and the sheath, at the point where leaves are attached to the main shoot axis (Moreno et al., 1997; Walsh et al., 1998). This similarity, however, is only superficial, because the boundary between leaf blade and sheath in grasses is not anatomically equivalent to the boundary between the leaf and stem in dicotyledons (Bell, 1998) and because the LG1 and LG2 proteins are unrelated to ATH1. Other Arabidopsis mutants show defects at the junction between cauline leaves and the stem or between the floral pedicel and the stem (Smith et al., 2004; Hibara et al., 2006; McKim et al., 2008), but to our knowledge, no mutations have been described that affect development of the basal boundary of lateral organs throughout shoot development.
We saw that in addition to controlling development of the basal region of shoot organs, ATH1 restricted growth of the region underlying the shoot meristem. The rapid development of a stem during the reproductive phase of rosette plants has been shown to be due initially to a large increase in cell division in the subapical region of the shoot, followed by cell expansion (Sachs et al., 1959). In Arabidopsis, the cell proliferation that sustains growth of the inflorescence stem is promoted by BP (Douglas et al., 2002; Venglat et al., 2002) and by the putative receptor ERECTA (ER) and close homologs of ER (Torii et al., 1996; Shpak et al., 2004). An opposite role for ATH1 in limiting cell proliferation in the stem is suggested by three lines of evidence. First, growth of the region below the shoot meristem was enhanced in ath1-3 seedlings grown in the dark or in low light, suggesting that ATH1 functions redundantly with light-activated genes to repress stem growth. The presence of a light-dependent pathway that inhibits stem growth has been shown by the change from rosette to caulescent growth in Arabidopsis plants with combined mutation of multiple photoreceptors (Devlin et al., 1996; Mazzella et al., 2000), although the exact genes involved downstream of the photoreceptors remain to be identified. Second, ATH1 was rapidly downregulated at the transition to flowering, when stem growth is released. Third, constitutive expression of ATH1 inhibited growth of the inflorescence stem and floral pedicels, with no obvious effect on flowering time or other aspects of inflorescence development (including the development of organ boundaries) (Cole et al., 2006; Proveniers et al., 2007).
Repression of growth may be the common theme linking the functions of ATH1 in the formation of basal organ boundaries and in repressing stem development. The CUC genes, whose function partially overlaps that of ATH1, establish organ boundaries by locally inhibiting cell proliferation (Aida and Tasaka, 2006). In the inflorescence stem, expression of a microRNA-resistant version of CUC2 also reduced stem growth by inhibiting both cell proliferation and cell expansion (Peaucelle et al., 2007). However, ATH1 alone is not sufficient to repress growth or cell proliferation, because overexpression did not affect the growth of leaves and floral organs. Either the processes targeted by ATH1 are relevant only to the basal region of organs and the stem or its function is dependent on other localized factors.
Another indication that ATH1 function depends on localized cofactors was that the ATH1 expression domain was wider than the regions affected by the ath1-3 mutation. Although ATH1 expression included the basal region of lateral organs and of the shoot meristem, other prominent aspects of the expression pattern did not correspond to an obvious mutant phenotype. The expression throughout the shoot meristem may relate to the role of ATH1 in activating FLC, which in turns controls the transition from vegetative to inflorescence meristem, although this role of ATH1 is only revealed when ath1-3 is combined with mutation of additional regulators of FLC (Proveniers et al., 2007). Expression at late stages of developing stamens and carpels is consistent with the activation of ATH1 by AGAMOUS (Gomez-Mena et al., 2005), but if this later expression of ATH1 is important for the development of reproductive organs, this role must also be obscured by functional redundancy in the single ath1-3 mutant.
STM and BP are good candidates for localized cofactors, because both interact with ATH1 in yeast two-hybrid experiments (Cole et al., 2006) and because expression of both genes overlaps with that of ATH1, including the subapical region of the shoot during the vegetative phase (Lincoln et al., 1994; Long et al., 1996). BP would appear to be a particularly good candidate partner for ATH1, because it is preferentially expressed at the base of the shoot meristem and organ primordia (Lincoln et al., 1994; Douglas et al., 2002). Our own results showed that this aspect of BP expression actually requires ATH1 (Figures 1K and 1L). If ATH1 and BP form a heterodimer to regulate target genes, then a positive feedback loop could maintain BP expression at the base of the flowers. However, if ATH1 does function as a heterodimer with BP, the latter must function redundantly, because bp mutants do not show the basal boundary defects seen in the ath1 mutants (Douglas et al., 2002; Venglat et al., 2002). It is also noteworthy that BP controls stem development in combination with BLR (Venglat et al., 2002; Smith and Hake, 2003). Although BLR is related to ATH1, its role in stem development seems to be opposite: the blr bp double mutant was very similar to 35S:ATH1 plants, with compact inflorescences and short pedicels (Byrne et al., 2003). It will be interesting to investigate whether ATH1 and BLR function as antagonistic partners of BP in the control of stem growth.
What are the downstream processes that mediate the inhibition of growth by ATH1? In the case of the stem, an obvious possibility was that ATH1 might antagonize gibberellin activity. Our data did not support a role for ATH1 in repressing the gibberellin biosynthetic genes GA4 and GA5, but expression of GA2-oxidase genes was partially dependent on ATH1, suggesting that ATH1 could stimulate gibberellin catabolism. Regulation of GA2-oxidase expression by ATH1 would be consistent with the fact that GA2ox2 and GA2ox4 are expressed specifically at the base of the meristem and in the basal boundaries of organ primordia (Jasinski et al., 2005), where ATH1 is also expressed (Figures 3B, 3D, and 3F). In addition, GA2ox2 and GA2ox4 were activated by ectopic expression of STM (Jasinski et al., 2005), which, as mentioned above, interacts physically with ATH1. However, there was not a simple correlation between growth repression by ATH1 and activation of GA2-oxidase genes, because 35S:ATH1 strongly inhibited growth of the inflorescence stem while expression of GA2ox2 and GA2ox4 actually decreased. One explanation for this could be that gibberellin catabolism genes do not mediate the growth effects of ATH1 and that expression of these genes decreases as an indirect consequence of the changes in the basal organ boundaries of ath1-3 and in the inflorescence stem of 35S:ATH1. Thus, ATH1 must repress stem growth by other means, such as antagonizing downstream responses to gibberellin or processes independent of gibberellin. Mutation of the polyamine biosynthetic gene ACAULIS5 causes a phenotype very similar to that of 35S:ATH1 (Hanzawa et al., 2000), so polyamine production or response would be plausible alternatives.
In conclusion, we have shown that ATH1 is required for the development of the basal boundaries of shoot organs. The enhanced subapical growth seen in the ath1 mutant in low light, combined with the inhibition of the inflorescence stem by ectopic expression of ATH1, indicate that ATH1 also has a role in inhibiting stem development. Our results show that, as hypothesized previously (Cole et al., 2006), a BLH protein that interacts with the master meristem regulators STM and BP controls a subset of meristem functions. These functions highlighted aspects of shoot meristem development that had previously been poorly characterized but that play a role in establishing important features of plant architecture.
METHODS
Mutant Lines and Genotyping
Arabidopsis thaliana ath1-3 (SALK_113353) and ath1-1 (GK-114A12) were ordered from the Nottingham Arabidopsis Stock Centre (lines N13353 and N303729, respectively). Plants were genotyped using the following primers: ATH1-RP (5′-GGCGGGTTTCGGATCTACATT-3′) and ATH1-LP (5′-CCAATACCGGTTTTTCAGACATGA-3′) for the wild-type fragment and LBb1 (5′-GCGTGGACCGCTTGCTGCAACT-3′) and ATH1-LP to identify the presence of the T-DNA insertion. cuc2-1 (Aida et al., 1997) and cuc1-5 (Hibara et al., 2006) were kindly provided by Mitsuhiro Aida (Nara Institute of Science and Technology) and Sylvestre Marillonet (Sainsbury Laboratory), respectively. The cuc2-1 mutant was genotyped with primers Robs172 (5′-CATTATTAACCACGCCCCTTACTCAAG-3′), RobS173 (5′-GTAACATTGAGAGTAAAGATTTCAGAAACC-3′), and RobS174 (5′-CGGGTCGGGCGTGAAAACATTG-3′), which amplify 410 bp in the wild type and 160 bp in the mutant. cuc1-5 was selected based on BASTA resistance using phosphinotricine (Duchefa) at a final concentration of 10 μg/mL on plates or by PCR using the oligonucleotides BARfor (5′-ATCAGATTTCGGTGACGGGC-3′) and BARrev (5′-GATCTACCATGAGCCCAGAAC-3′).
Plant Growth Conditions
Plants were grown on a mix of vermiculite:soil:sand at 18°C with 16-h-light/8-h-dark cycles. For in vitro growth, seeds were surface-sterilized by chlorine gas by being kept in a desiccator with a mixture of 100 mL of commercial bleach and 3 mL of concentrated hydrochloric acid for 7 h in a fume hood. Sterile seeds were plated on GM medium (Valvekens et al., 1988) containing 1% glucose, stratified for 2 d at 4°C, and grown in 16-h-light/8-h-dark cycles at 18 to 20°C. To grow seedlings in the dark, after sterilization and sowing, seeds were exposed to light for 12 h to trigger germination, and then the plates were wrapped with aluminum foil and stored vertically at 18 to 20°C. For growth in low light, the plates were wrapped in six layers of Miracloth, which reduced the total light level from 100 to 15 μmol·m−2·s−1 (measured with a Macam spectroradiometer). For experiments on induction of flowering, seeds were plated as above but germinated in an environmental test chamber (Sanyo MLR-350) under short-day conditions (8-h-light/16-h-dark cycles, light level 4). After 10 d on the plates, the seedlings were transplanted to soil as described above and returned to short days for another 20 d. Flowering was then induced by 3 long days (16-h-light/8-h-dark cycles). Induced plants and control plants that had remained for another 3 d in short days were harvested at the same time, 1 h after the start of the light cycle.
Construction of Transgenes
For ath1-3 complementation, a 6-kb DNA fragment (Ch4 nucleotides 15914300 to 15920300, including 3434 bp before the start codon and 567 bp after the stop codon) was amplified using Long Expand DNA polymerase (Roche) from Col DNA using oligonucleotides 5′-aaaaggatccCATTCGCCGTAAAAGGCTCCGTC-3′ and 5′-ttttgagctcACACGATCAGTGTGACCTTCAAG-3′ (sequences added for subcloning are in lowercase letters and those corresponding to genomic DNA are in uppercase letters). The amplified fragment was cloned as a BamHI to SacI insert in pGEM-T Easy (Promega), sequenced, then moved to pGreen 0229 (Hellens et al., 2000). Homozygous ath1-3 plants were transformed by infiltration with Agrobacterium tumefaciens GV3101 pSOUP (Hellens et al., 2000).
To make the GUS reporter fusion, a 3.2-kb fragment containing the ATH1 promoter plus the coding region was amplified using Long Expand DNA polymerase (Roche) and oligonucleotides 5′-GGGGACAAGTTTGTACAAAAAAGCAGGCTacatgtaaatagtaaaatgt-3′ and 5′-GGGGACCACTTTGTACAAGAAAGCTGGGTCtttatgcattgcttggctca-3′, then cloned in the pDONR207 vector (Invitrogen). Destination vector pGWB3 (Nakagawa et al., 2007) was used to generate the pATH1:ATH1-GUS construct. For ectopic expression, ATH1 cDNA was amplified with oligonucleotides 5′-TTTCATAGAAACCCAATGGACAACAACA-3′ and 5′-aggatccTTATTTATGCATTGCTTGGCTC-3′, cloned into pGEM-T, and sequenced. The cDNA was placed downstream of the cauliflower mosaic virus 35S promoter in the binary vector pGWB2 (Nakagawa et al., 2007) using Gateway cloning technology (Invitrogen). Transgenic plants were generated by agroinfiltration using the floral dip method (Clough and Bent, 1998) after electroporating plasmids into Agrobacterium strain GV3101 or ASE.
Quantitative Real-Time PCR
Total RNA was extracted using TRI reagent (Sigma-Aldrich) according to the manufacturer's instructions and subjected to DNase treatment. RNA (1 μg) was then reverse-transcribed using oligo(dT) (20-mer) and SuperScript II RNase H reverse transcriptase (Invitrogen) according to the manufacturer's instructions, and the reaction mixture was diluted to 200 μL. Each primer pair was designed to span introns in order to detect and eliminate amplified genomic DNA products. Each PCR contained 10 μL of SYBR Green JumpStart Taq Ready Mix (Sigma-Aldrich) containing Hot Start Taq polymerase, 0.4 μL of each primer in the pair (primers were at 10 μM; primer pairs are listed in Supplemental Table 1 online), 5 μL of the diluted cDNA solution described above, and 4.2 μL of water for a reaction volume of 20 μL. Reactions were performed in triplicate on a Bio-Rad Chromo4 system. Data were analyzed using the 2−ΔΔCT method and normalized using the expression of ADENINE PHOSPHORIBOSYLTRANSFERASE (APT) as a constitutive control (Moffatt et al., 1994). The amplification efficiencies for the APT and GA4, GA5, and GA2ox primers were found to be approximately equal.
In Situ Hybridization
RNA was hybridized in situ (Fobert et al., 1996; Gomez-Mena et al., 2005) using digoxigenin-labeled probes transcribed with T7 polymerase from linearized plasmid (pGEM-T Easy; Promega) containing a 3′ cDNA fragment of ATH1 (nucleotides 773 to 1422 of the ATH1 coding sequence) or the complete coding sequence for CUC1. Color detection was performed with 5-bromo-4-chloro-3-indolyl phosphate/nitroblue tetrazolium according to the manufacturer's instructions (Boehringer).
Histological Techniques
For histological studies, tissue was fixed, sectioned, and stained with 0.05% toluidine blue in 0.1 M phosphate buffer at pH 6.8 (Obrien et al., 1964). For vascular staining, sections were stained for 2 min in a 2% phloroglucinol solution in 95% ethanol, then photographed in 50% hydrochloric acid. For whole-mount GUS detection, tissues were fixed for 10 min in ice-cold 90% acetone, stained for GUS (Sieburth et al., 1998), and cleared in a solution of chloral hydrate:water:glycerol (8:3:1, by weight). For GUS detection in sectioned tissues, seedlings were first stained for GUS, followed by fixation and sectioning as for in situ hybridization. Digital images were processed (cropping, brightness, contrast, and color balance) with Adobe Photoshop (Adobe Systems) and analyzed quantitatively using Image J (http://rsb.info.nih.gov/ij/).
Scanning Electron Microscopy
Plants were fixed in 2.5% glutaraldehyde in PBS at 4°C overnight, dehydrated in an ethanol series, and critical-point dried in liquid CO2. Sepals were removed from flower samples, sputter-coated with gold palladium, and analyzed with a Philips XL 30 FEG scanning electron micrograph. For cryo-scanning electron microscopy, flowers were frozen in nitrogen slush at −190°C. Ice was sublimated at −90°C, and the specimen was sputter-coated and examined on a Zeiss Supra 55 VP FEG scanning electron micrograph fitted with a Gatan Alto 2500 cryo system for cryofixed samples.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative database under the following accession numbers: At4g32980 (ATH1), At3g15170 (CUC1), At4g08150 (BP), At1g15550 (GA4), At4g25420 (GA5), At1g30040 (GA2ox2), and At1g47990 (GA2ox4).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. ath1-1 Phenotypes Are Comparable to Those of ath1-3.
Supplemental Figure 2. Complementation of ath1-3.
Supplemental Figure 3. Development of the Basal Regions of Stamens and Sepals in the Wild Type and in ath1-3.
Supplemental Figure 4. Sections through GUS-Stained Wild-Type BP:GUS and ath1-3 BP:GUS Inflorescences.
Supplemental Figure 5. Expression of Gibberellin Homeostasis Genes in the Wild Type, ath1-3, and 35S:ATH1, Measured by Quantitative RT-PCR.
Supplemental Figure 6. External Gibberellin Treatment Did Not Restore Inflorescence Stem Growth in 35S:ATH1 Plants.
Supplemental Table 1. Oligonucleotides Used for Quantitative RT-PCR.
Supplementary Material
Acknowledgments
We thank the Biotechnology and Biological Sciences Research Council for financial support (Grant BBS/B/04234), Mary Byrne for BP:GUS, Claire Lister for PFCA:GUS, and Lars Østergaard and Mary Byrne for critical comments on the manuscript.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Robert Sablowski (robert.sablowski@bbsrc.ac.uk).
Online version contains Web-only data.
References
- Aida, M., Ishida, T., Fukaki, H., Fujisawa, H., and Tasaka, M. (1997). Genes involved in organ separation in Arabidopsis: An analysis of the cup-shaped cotyledon mutant. Plant Cell 9 841–857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aida, M., Ishida, T., and Tasaka, M. (1999). Shoot apical meristem and cotyledon formation during Arabidopsis embryogenesis: Interaction among the CUP-SHAPED COTYLEDON and SHOOT MERISTEMLESS genes. Development 126 1563–1570. [DOI] [PubMed] [Google Scholar]
- Aida, M., and Tasaka, M. (2006). Genetic control of shoot organ boundaries. Curr. Opin. Plant Biol. 9 72–77. [DOI] [PubMed] [Google Scholar]
- Bao, X., Franks, R.G., Levin, J.Z., and Liu, Z. (2004). Repression of AGAMOUS by BELLRINGER in floral and inflorescence meristems. Plant Cell 16 1478–1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell, A.D. (1998). Plant Form—An Illustrated Guide to Flowering Plant Morphology. (Oxford, UK: Oxford University Press).
- Bellaoui, M., Pidkowich, M.S., Samach, A., Kushalappa, K., Kohalmi, S.E., Modrusan, Z., Crosby, W.L., and Haughn, G.W. (2001). The Arabidopsis BELL1 and KNOX TALE homeodomain proteins interact through a domain conserved between plants and animals. Plant Cell 13 2455–2470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatt, A.A., Etchells, J.P., Canales, C., Lagodienko, A., and Dickinson, H. (2004). VAAMANA—A BEL1-like homeodomain protein, interacts with KNOX proteins BP and STM and regulates inflorescence stem growth in Arabidopsis. Gene 328 103–111. [DOI] [PubMed] [Google Scholar]
- Bleecker, A.B., and Patterson, S.E. (1997). Last exit: Senescence, abscission, and meristem arrest in Arabidopsis. Plant Cell 9 1169–1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrne, M.E., Groover, A.T., Fontana, J.R., and Martienssen, R.A. (2003). Phyllotactic pattern and stem cell fate are determined by the Arabidopsis homeobox gene BELLRINGER. Development 130 3941–3950. [DOI] [PubMed] [Google Scholar]
- Byrne, M.E., Simorowski, J., and Martienssen, R.A. (2002). ASYMMETRIC LEAVES1 reveals knox gene redundancy in Arabidopsis. Development 129 1957–1965. [DOI] [PubMed] [Google Scholar]
- Clough, S.J., and Bent, A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16 735–743. [DOI] [PubMed] [Google Scholar]
- Cole, M., Nolte, C., and Werr, W. (2006). Nuclear import of the transcription factor SHOOT MERISTEMLESS depends on heterodimerization with BLH proteins expressed in discrete sub-domains of the shoot apical meristem of Arabidopsis thaliana. Nucleic Acids Res. 34 1281–1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devlin, P.F., Halliday, K.J., Harberd, N.P., and Whitelam, G.C. (1996). The rosette habit of Arabidopsis thaliana is dependent upon phytochrome action: Novel phytochromes control internode elongation and flowering time. Plant J. 10 1127–1134. [DOI] [PubMed] [Google Scholar]
- Douglas, S.J., Chuck, G., Dengler, R.E., Pelecanda, L., and Riggs, C.D. (2002). KNAT1 and ERECTA regulate inflorescence architecture in Arabidopsis. Plant Cell 14 547–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleet, C.M., and Sun, T.P. (2005). A DELLAcate balance: The role of gibberellin in plant morphogenesis. Curr. Opin. Plant Biol. 8 77–85. [DOI] [PubMed] [Google Scholar]
- Fleming, A.J. (2005). Formation of primordia and phyllotaxy. Curr. Opin. Plant Biol. 8 53–58. [DOI] [PubMed] [Google Scholar]
- Fletcher, J.C. (2002). Shoot and floral meristem maintenance in Arabidopsis. Annu. Rev. Plant Biol. 53 45–66. [DOI] [PubMed] [Google Scholar]
- Fobert, P.R., Gaudin, V., Lunness, P., Coen, E.S., and Doonan, J.H. (1996). Distinct classes of cdc2-related genes are differentially expressed during the cell division cycle in plants. Plant Cell 8 1465–1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallois, J.L., Woodward, C., Reddy, G.V., and Sablowski, R. (2002). Combined SHOOT MERISTEMLESS and WUSCHEL trigger ectopic organogenesis in Arabidopsis. Development 129 3207–3217. [DOI] [PubMed] [Google Scholar]
- Gomez-Mena, C., de Folter, S., Costa, M.M.R., Angenent, G.C., and Sablowski, R. (2005). Transcriptional program controlled by the floral homeotic gene AGAMOUS during early organogenesis. Development 132 429–438. [DOI] [PubMed] [Google Scholar]
- Hanzawa, Y., Takahashi, T., Michael, A.J., Burtin, D., Long, D., Pineiro, M., Coupland, G., and Komeda, Y. (2000). ACAULIS5, an Arabidopsis gene required for stem elongation, encodes a spermine synthase. EMBO J. 19 4248–4256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay, A., Kaur, H., Phillips, A., Hedden, P., Hake, S., and Tsiantis, M. (2002). The gibberellin pathway mediates KNOTTED1-type homeobox function in plants with different body plans. Curr. Biol. 12 1557–1565. [DOI] [PubMed] [Google Scholar]
- Hellens, R., Mullineaux, P., and Klee, H. (2000). A guide to Agrobacterium binary Ti vectors. Trends Plant Sci. 5 446–451. [DOI] [PubMed] [Google Scholar]
- Hibara, K.-i., Karim, M.R., Takada, S., Taoka, K.-i., Furutani, M., Aida, M., and Tasaka, M. (2006). Arabidopsis CUP-SHAPED COTYLEDON3 regulates postembryonic shoot meristem and organ boundary formation. Plant Cell 18 2946–2957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs, T. (1997). Why do plant cells divide? Plant Cell 9 1021–1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jasinski, S., Piazza, P., Craft, J., Hay, A., Woolley, L., Rieu, L., Phillips, A., Hedden, P., and Tsiantis, M. (2005). KNOX action in Arabidopsis is mediated by coordinate regulation of cytokinin and gibberellin activities. Curr. Biol. 15 1560–1565. [DOI] [PubMed] [Google Scholar]
- Kanrar, S., Onguka, O., and Smith, H.M.S. (2006). Arabidopsis inflorescence architecture requires the activities of KNOX-BELL homeodomain heterodimers. Planta 224 1163–1173. [DOI] [PubMed] [Google Scholar]
- Laurie, R.E. (2003). Controlling the Expression of the Arabidopsis Floral Promoter FCA. (Norwich, UK: University of East Anglia).
- Lenhard, M., Jurgens, G., and Laux, T. (2002). The WUSCHEL and SHOOTMERISTEMLESS genes fulfill complementary roles in Arabidopsis shoot meristem regulation. Development 129 3195–3206. [DOI] [PubMed] [Google Scholar]
- Lincoln, C., Long, J., Yamaguchi, J., Serikawa, K., and Hake, S. (1994). A knotted1-like homeobox gene in Arabidopsis is expressed in the vegetative meristem and dramatically alters leaf morphology when overexpressed in transgenic plants. Plant Cell 6 1859–1876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long, J.A., Moan, E.I., Medford, J.I., and Barton, M.K. (1996). A member of the KNOTTED class of homeodomain proteins encoded by the STM gene of Arabidopsis. Nature 379 66–69. [DOI] [PubMed] [Google Scholar]
- Mazzella, M.A., Bertero, D., and Casal, J.J. (2000). Temperature-dependent internode elongation in vegetative plants of Arabidopsis thaliana lacking phytochrome B and cryptochrome 1. Planta 210 497–501. [DOI] [PubMed] [Google Scholar]
- McKim, S.M., Stenvik, G.-E., Butenko, M.A., Kristiansen, W., Cho, S.K., Hepworth, S.R., Aalen, R.B., and Haughn, G.W. (2008). The BLADE-ON-PETIOLE genes are essential for abscission zone formation in Arabidopsis. Development 135 1537–1546. [DOI] [PubMed] [Google Scholar]
- Moffatt, B.A., McWhinnie, E.A., Agarwal, S.K., and Schaff, D.A. (1994). The adenine phosphoribosyltransferase-encoding gene of Arabidopsis thaliana. Gene 143 211–216. [DOI] [PubMed] [Google Scholar]
- Moreno, M.A., Harper, L.C., Krueger, R.W., Dellaporta, S.L., and Freeling, M. (1997). liguleless1 encodes a nuclear-localized protein required for induction of ligules and auricles during maize leaf organogenesis. Genes Dev. 11 616–628. [DOI] [PubMed] [Google Scholar]
- Muller, J., Wang, Y., Franzen, R., Santi, L., Salamini, F., and Rohde, W. (2001). In vitro interactions between barley TALE homeodomain proteins suggest a role for protein-protein associations in the regulation of Knox gene function. Plant J. 27 13–23. [DOI] [PubMed] [Google Scholar]
- Nakagawa, T., Kurose, T., Hino, T., Tanaka, K., Kawamukai, M., Niwa, Y., Toyooka, K., Matsuoka, K., Jinbo, T., and Kimura, T. (2007). Development of series of Gateway binary vectors, pGWBs, for realizing efficient construction of fusion genes for plant transformation. J. Biosci. Bioeng. 104 34–41. [DOI] [PubMed] [Google Scholar]
- Obrien, T.P., Feder, N., and McCully, M.E. (1964). Polychromatic staining of plant cell walls by toluidine blue O. Protoplasma 59 368–373. [Google Scholar]
- Olszewski, N., Sun, T.-p., and Gubler, F. (2002). Gibberellin signaling: Biosynthesis, catabolism, and response pathways. Plant Cell 14 S61–S80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ori, N., Eshed, Y., Chuck, G., Bowman, J.L., and Hake, S. (2000). Mechanisms that control knox gene expression in the Arabidopsis shoot. Development 127 5523–5532. [DOI] [PubMed] [Google Scholar]
- Peaucelle, A., Morin, H., Traas, J., and Laufs, P. (2007). Plants expressing a miR164-resistant CUC2 gene reveal the importance of post-meristematic maintenance of phyllotaxy in Arabidopsis. Development 134 1045–1050. [DOI] [PubMed] [Google Scholar]
- Proveniers, M., Rutjens, B., Brand, M., and Smeekens, S. (2007). The Arabidopsis TALE homeobox gene ATH1 controls floral competency through positive regulation of FLC. Plant J. 52 899–913. [DOI] [PubMed] [Google Scholar]
- Quaedvlieg, N., Dockx, J., Rook, F., Weisbeek, P., and Smeekens, S. (1995). The homeobox gene Ath1 of Arabidopsis is derepressed in the photomorphogenic mutants cop1 and det1. Plant Cell 7 117–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roeder, A.H.K., Ferrandiz, C., and Yanofsky, M.F. (2003). The role of the REPLUMLESS homeodomain protein in patterning the Arabidopsis fruit. Curr. Biol. 13 1630–1635. [DOI] [PubMed] [Google Scholar]
- Roldan, M., Gomez-Mena, C., Ruiz-Garcia, L., Salinas, J., and Martinez-Zapater, J.M. (1999). Sucrose availability on the aerial part of the plant promotes morphogenesis and flowering of Arabidopsis in the dark. Plant J. 20 581–590. [DOI] [PubMed] [Google Scholar]
- Sachs, R.M., Bretz, C.F., and Lang, A. (1959). Shoot histogenesis—The early effects of gibberellin upon stem elongation in 2 rosette plants. Am. J. Bot. 46 376–384. [Google Scholar]
- Shpak, E.D., Berthiaume, C.T., Hill, E.J., and Torii, K.U. (2004). Synergistic interaction of three ERECTA-family receptor-like kinases controls Arabidopsis organ growth and flower development by promoting cell proliferation. Development 131 1491–1501. [DOI] [PubMed] [Google Scholar]
- Sieburth, L.E., Drews, G.N., and Meyerowitz, E.M. (1998). Non-autonomy of AGAMOUS function in flower development: Use of a Cre/loxP method for mosaic analysis in Arabidopsis. Development 125 4303–4312. [DOI] [PubMed] [Google Scholar]
- Smith, H.M.S., Boschke, I., and Hake, S. (2002). Selective interaction of plant homeodomain proteins mediates high DNA-binding affinity. Proc. Natl. Acad. Sci. USA 99 9579–9584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, H.M.S., Campbell, B.C., and Hake, S. (2004). Competence to respond to floral inductive signals requires the homeobox genes PENNYWISE and POUND-FOOLISH. Curr. Biol. 14 812–817. [DOI] [PubMed] [Google Scholar]
- Smith, H.M.S., and Hake, S. (2003). The interaction of two homeobox genes, BREVIPEDICELLUS and PENNYWISE, regulates internode patterning in the Arabidopsis inflorescence. Plant Cell 15 1717–1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth, D.R., Bowman, J.L., and Meyerowitz, E.M. (1990). Early flower development in Arabidopsis. Plant Cell 2 755–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenvik, G.-E., Butenko, M.A., Urbanowicz, B.R., Rose, J.K.C., and Aalen, R.B. (2006). Overexpression of INFLORESCENCE DEFICIENT IN ABSCISSION activates cell separation in vestigial abscission zones in Arabidopsis. Plant Cell 18 1467–1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takada, S., Hibara, K., Ishida, T., and Tasaka, M. (2001). The CUP-SHAPED COTYLEDON1 gene of Arabidopsis regulates shoot apical meristem formation. Development 128 1127–1135. [DOI] [PubMed] [Google Scholar]
- Tooke, F., and Battey, N. (2003). Models of shoot apical meristem function. New Phytol. 159 37–52. [DOI] [PubMed] [Google Scholar]
- Torii, K.U., Mitsukawa, N., Oosumi, T., Matsuura, Y., Yokoyama, R., Whittier, R.F., and Komeda, Y. (1996). The Arabidopsis ERECTA gene encodes a putative receptor protein kinase with extracellular leucine-rich repeats. Plant Cell 8 735–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valvekens, D., Vanmontagu, M., and Vanlijsebettens, M. (1988). Agrobacterium tumefaciens-mediated transformation of Arabidopsis thaliana root explants by using kanamycin selection. Proc. Natl. Acad. Sci. USA 85 5536–5540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaughan, J.G. (1955). The morphology and growth of the vegetative and reproductive apices of Arabidopsis thaliana (L.) Heynh., Capsella bursa-pastoris (L.) Medic. and Anagallis arvensis L. Bot. J. Linn. Soc. 55 279–301. [Google Scholar]
- Venglat, S.P., Dumonceaux, T., Rozwadowski, K., Parnell, L., Babic, V., Keller, W., Martienssen, R., Selvaraj, G., and Datla, R. (2002). The homeobox gene BREVIPEDICELLUS is a key regulator of inflorescence architecture in Arabidopsis. Proc. Natl. Acad. Sci. USA 99 4730–4735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh, J., Waters, C.A., and Freeling, M. (1998). The maize gene liguleless2 encodes a basic leucine zipper protein involved in the establishment of the leaf blade-sheath boundary. Genes Dev. 12 208–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanai, O., Shani, E., Dolezal, K., Tarkowski, P., Sablowski, R., Sandberg, G., Samach, A., and Ori, N. (2005). Arabidopsis KNOXI proteins activate cytokinin biosynthesis. Curr. Biol. 15 1566–1571. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.